Most plant lineages engage with Glomeromycotina fungi to form the ubiquitous arbuscular mycorrhizal (AM) symbiosis. Despite its wide occurrence in diverse plant–fungal species combinations, the interaction dynamics are strikingly uniform (Fig. 1). The events leading up to a successful mutualism start when plant and fungus advertise their presence in the rhizosphere by releasing diffusible chemical cues. In vascular plants this presymbiotic dialog results in physical contact whereby extraradical hyphae differentiate into hyphopodia on the surface of roots preceding fungal entry. Intraradical hyphal passage is followed by fungal accommodation in cortical cells to foster arbuscules. This is accompanied by the rapid formation of plant and fungal membranes in juxtaposition to each other, resulting in a magnified surface area. The mutualistic nature of the association manifests here as the reciprocal exchange of nutrients occurs. Subsequently formed fungal vesicles and spores are symptomatic of a sustained association. At the whole‐root level, symbiosis establishment is considered asynchronous as all AM fungal symbiotic structures can be found simultaneously. However, at the infection unit resolution, every stage depends on the preceding one reflecting that precise molecular programs dynamically coordinate the interaction.
Figure 1.
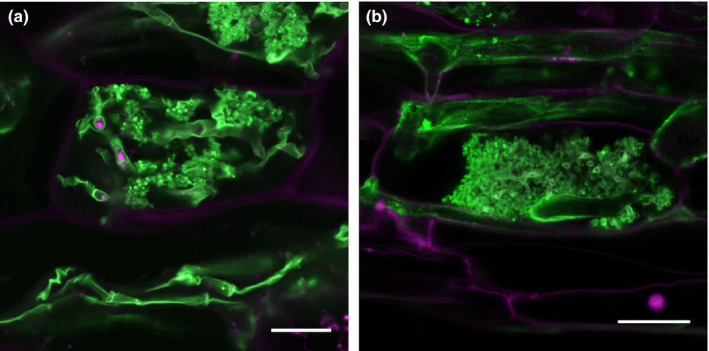
Arbuscular mycorrhizal (AM) fungal symbiotic structures are similarly devised in distantly related plant clades. (a) Marchantia palacea thalloid cell and (b) maize cortical root cell colonized with the AM fungus Rhizophagus irregularis highlighting the fundamental conservation of symbiotic traits across land plants. WGA‐Alexa Fluor 488 staining. Bars, 20 μm.
Although the knowledge of Glomeromycotina genomics has recently improved (Kamel et al., 2017), plants remain experimentally more tractable than the fungal partner leading to a more advanced understanding of the plant regulatory mechanisms. The discovery of functionally relevant genes relies on the accurate characterization of symbiotic phenotypes which often implies documenting the growth of the fungal partner resulting from alterations of plant host genes. This can be a complex task and mutant phenotypes are routinely reported in the absence of standardized procedures leading to ambiguous interpretations. Here we provide general guidelines for accurately characterizing AM phenotypes in their temporal and spatial aspects.
Importance of systemic diagnoses
AM phenotypes observed during the colonization process can be explained by gene impairment in the plant directly affecting symbiosis establishment and/or functioning, or secondarily as a side effect of abnormal plant development. The possibilities are ample. Alterations in processes related to photosynthesis, cell wall properties, root development and hormonal regulation could all potentially lead to strong symbiotic phenotypes, but the underlying disrupted plant genes causing them do not necessarily need to be dedicated to symbiosis. To distinguish pleiotropic from specific symbiotic phenotypes, plant developmental, fungal and overall symbiotic phenotypes must be comprehensively assessed. Additionally, any conclusion drawn should be contemplated based on the model organisms used. Although any plant pecies able to host AM fungi is a good model to study fundamental symbiotic processes, extrapolations should be avoided when dealing with genes involved in plant clade‐specific dynamics. For example, symbiotic LysM‐containing receptors have undergone expansion in legumes likely followed by neo‐ or sub‐functionalization adapted to the bipartite symbiotic context of this plant family (Gough et al., 2018).
Phenotyping a snowball
The encounter between the root of a young seedling and extraradical fungal hyphae results in a lifelong association in which both parallel and consecutive recolonization events are the norm. Although inherently iterative, it is currently unknown how the first colonization event compares to recolonization at the molecular and physiological level. Priming may render a fraction of the presymbiotic colonization events not needed during recolonization. By contrast, plant cell autonomous events are likely to require the same de novo‐activated set of molecular drivers in non colonized and recolonized roots. Although AM fungal colonization of a plant root is a stepwise process, the timing of the different events is plastic and dependent on environmental factors besides plant and fungal identity. Hence alterations in one step of the cyclic procedure of AM fungal colonization will necessarily affect subsequent steps, likely in a dynamic and amplified fashion (Fig. 2). From the phenotyping perspective this complexity demands a proper temporal resolution of AM fungal colonization. The inclusion of multiple time points in experimental designs should be a standard procedure when describing AM fungal colonization, aiming to capture general quantitative dynamics in the presymbiotic and symbiotic stages of the association followed by observations on the development of particular AM fungal symbiotic structures. Several techniques have been developed to stain AM fungi (Vierheilig et al., 2005) which are normally used in conjunction with standardized methods to quantify the extent of AM colonization in root tissues (Giovannetti & Mosse, 1980).
Figure 2.

Time course observations can capture the dynamic patterns of arbuscular mycorrhizal (AM) fungal colonization. (a) Success of the presymbiotic stage could affect the time of onset of AM fungal colonization. Altered AM fungal spore germination, hyphal branching and hyphopodia formation could result in accelerated or delayed AM fungal colonization, which is evident at early time points. (b) Intraradical hyphal progression including arbuscule development influences the quantity of the colonization at later time points. (c) Success of post‐arbuscule development stage includes vesicle formation and sporulation. Failure in these processes could affect maintenance of symbiosis resulting in AM fungal colonization collapse at later time points. WT, wild‐type.
To complement microscopic observations, transcript levels of plant and AM fungal marker genes known to be induced at different stages of symbiotic establishment should be monitored. While several AM fungal housekeeping genes are routinely used as a way to indirectly assess fungal biomass, it would be useful to develop a set of AM fungal marker genes induced at distinct symbiotic stages. From the host plant side, several time‐course transcriptomics studies have identified genes related to early and late stages of the association (reviewed in Choi et al., 2018; Pimprikar & Gutjahr, 2018). However, the fact that mutation in plant symbiotic genes can potentially affect the expression of marker genes highlights their use as a complement rather than an alternative to microscopic observations.
Assessing phenotypes in the presymbiotic stage
In the presymbiotic stage, plant roots and AM fungi secrete diffusible compounds in the rhizosphere allowing reciprocal recognition before physical contact. The plant host increases the secretion of signaling molecules, such as strigolactones (SLs; Akiyama et al., 2005) from root exudates, in order to metabolically activate AM fungi. Highly ramified fungal mycelia then produce carbohydrate based signaling molecules such as LipoChitoOligosaccharides (LCOs) and ChitoOligosaccharides (COs) (Maillet et al., 2011; Genre et al., 2013). Successful presymbiotic reprogramming is the prerequisite for subsequent initiation of root colonization as demonstrated by the impairment of initiation of invasion of the roots of plant mutants defective in this process (Gutjahr et al., 2015; Nadal et al., 2017).
One of the key steps monitoring the initial root colonization by AM fungi is to quantitatively and qualitatively assess hyphopodia formation defects, which are often observed in plant mutants defective in AM fungal perception. For example, mutants of the rice chitin receptor, Chitin Elicitor Receptor Kinase 1 (CERK1) and mutants of genes involved in the common symbiosis pathway showed similar aberrant swollen hyphopodia (Saito et al., 2007; Gutjahr et al., 2008; Kosuta et al., 2011; Liao et al., 2012; Miyata et al., 2014). Once defects in AM fungal hyphopodia formation are identified, subsequent AM fungal colonization assays with increasing inoculum strength are useful to test whether the phenotype can be overcome thereby demonstrating the robustness of the early defects. Conversely, some plant mutants such as those overproducing SLs (Yoshida et al., 2012; Gutjahr et al., 2015), exhibit higher AM fungal colonization compared to wild‐type plants. The use of high AM fungal inoculum strength for AM colonization assays can potentially mask these phenotypes, hence assaying increased number of replicates with reduced inoculum is advised.
Strong early phenotypes could be due to the plant's inability to either produce active compounds such as SLs to prime the fungi or to recognize fungal compounds such as LCOs or COs. To distinguish between these two scenarios, global transcriptomics of plant or AM fungi have been implemented to compare transcriptional responses to germinated spore exudates (GSEs) of the AM fungi or plant root exudates, respectively (Campos‐Soriano et al., 2011; Giovannetti et al., 2015; Gutjahr et al., 2015; Nadal et al., 2017). Alternatively, nurse plant experiments where mutants are co‐cultivated with wild‐type plants can be performed. If the presymbiosis defects are complemented in the mutant plant, its phenotype likely relates to the production of root exudates rather than being defective in perception of AM fungi. If this is the case, the next step would be to investigate the consequences on AM fungal performance, which includes spore germination, hyphal branching and directional growth towards the root. To this end, in vitro AM fungal spore germination or hyphal branching assays with root exudates collected under low phosphate conditions can be performed (Kretzschmar et al., 2012; Aroca et al., 2013; Nadal et al., 2017).
Phenotyping the arbusculated cell
The development of arbuscules is mirrored by developmental reprogramming of the hosting root cells. The reciprocal exchange of nutrients in arbusculated cells renders them a specialized cell type, which is also evidenced by their specific transcriptional activity as determined in laser‐captured microdissection studies (reviewed in Pimprikar & Gutjahr, 2018). Only a small fraction of the plant genes induced in arbusculated cells has been functionally characterized, mostly in nutrient exchange and peri‐arbuscular membrane (PAM) development (Luginbuehl & Oldroyd, 2017). Impairment in symbiotic processes from the arbusculated cell perspective can potentially fall into vast spectra of outcomes ranging from the extreme cases of complete absence of arbuscules to fully formed ones but with impaired function (Breuillin‐Sessoms et al., 2015). Decreased arbuscule numbers could be due to failures in presymbiotic processes, in which case this should be mirrored by decreased abundance of all symbiotic fungal structures. By contrast, the phenotype likely relates to the symbiotic stage if fungal entry and spread in the roots occurs but arbuscules do not form. Hence when reporting a reduced number of arbuscules it is more informative to also document hyphopodia formation and intraradical hyphal spread.
Although the interaction between plant roots and AM fungi is long‐lasting, at the cell level the commitment for symbiosis is ephemeral as arbuscules disappear in a matter of days after their inception. Arbuscule development phenotypes are, in contrast to alterations in arbuscule abundance, less likely to be linked to presymbiotic influences. Rather, they can arise from impairment of genes impacting the accommodation of initial intracellular hyphae, arbuscule size, branching or lifespan. The majority of genes that have been demonstrated to regulate arbuscule developmental processes are of plant origin but some fungal genes have been shown to participate as well (Helber et al., 2011; Tsuzuki et al., 2016; Xie et al., 2016; Sun et al., 2018). As wild‐type plants will display arbuscules at diverse developmental stages at a given time point, it is important to score the relative proportions. Quantification of the size of arbuscules in relation to that of the host cell is advised here, which is commonly performed by employing wheat germ agglutinin (WGA) fluorophore‐conjugated treated roots. However, there is a continuum in the transition from a nascent, fully branched to a degenerating arbuscule that makes the interpretation of the developmental stage in place challenging to determine. To examine arbuscule branching, efforts have focused on the implementation of live cell imaging, particularly using subcellular marker lines (Ivanov & Harrison, 2014) including those for the differential localization to PAM subdomains (Park et al., 2015). Impairments of a number of plant genes involved in lipid (Keymer & Gutjahr, 2018) and phosphate dynamics (Javot et al., 2007; Yang et al., 2012) exhibit stunted arbuscules linking nutrient homeostasis with arbuscule development. Using nurse plant systems to investigate complementation of phenotypes provides glimpses on the involvement of the underlying genes in AM fungal nutrition, hence it is advised to perform this experiment routinely.
To track arbuscule collapse, plant subcellular marker lines can be used in combination with time lapse imaging. Such markers have not been broadly implemented mainly because the study of arbuscule collapse is in its infancy. Of potential use are plant peroxisome markers as they have been shown to localize around collapsing arbuscules (Pumplin & Harrison, 2009) and the plant secretory carrier membrane protein SCAMP which forms distinctive aggregates as arbuscules collapse (Kobae & Fujiwara, 2014). The general occurrence of the patterns, however, is yet to be confirmed.
It is not over when the arbuscule is over
When an arbuscule vanishes from a root cell, new ones are born elsewhere. In parallel, fungal vesicles and spores appear. Vesicles are globular intercellular structures believed to be storage organs for lipids including phospholipids (Jabaji‐Hare et al., 1984). As phospholipids potentially contain phosphate and lipids traded between symbionts, alterations in vesicle abundance may reflect major defects in symbiotic nutrient homeostasis. Unlike arbuscules, vesicle formation is stimulated by phosphate treatment in rice (Kobae et al., 2016) suggesting them to have complex yet‐to‐be understood roles in the fungal handling of nutritional resources. Vesicles and spores are overlooked structures but they give account of the completion of fungal life cycle which is, in turn, relevant for subsequent recolonization. Post‐arbuscule defects are expected to ultimately lead to the collapse of the association which may be observable only upon prolonged symbiotic co existence. Importantly, it is at this stage that the plant and fungus are concomitantly assimilating the nutrients exchanged, conceivably leading to further modifications of plant physiological processes. This adds a layer of complexity to the phenotyping process that researchers should be aware of as more molecular players appear in this largely unexplored stage of the AM symbiosis.
Concluding remarks
Although the researcher's biological question is the ultimate driver of the method of choice for AM symbiosis phenotyping, minimum standards must be followed (Fig. 3). To achieve this, the plant systemic context should be considered alongside phenotypic assessment over plant roots colonized by AM fungi. Upon zooming into the roots, the dynamic cumulative complexity of the AM fungal colonization process cannot be captured in a snapshot. Instead, phenotyping should be performed at multiple time points to distinguish the stage during which symbiotic alterations are occurring. This, together with inspections of alterations in particular AM fungal symbiotic structures, will help unveil functional aspects of the plant genes impaired in an accurate and reproducible way.
Figure 3.
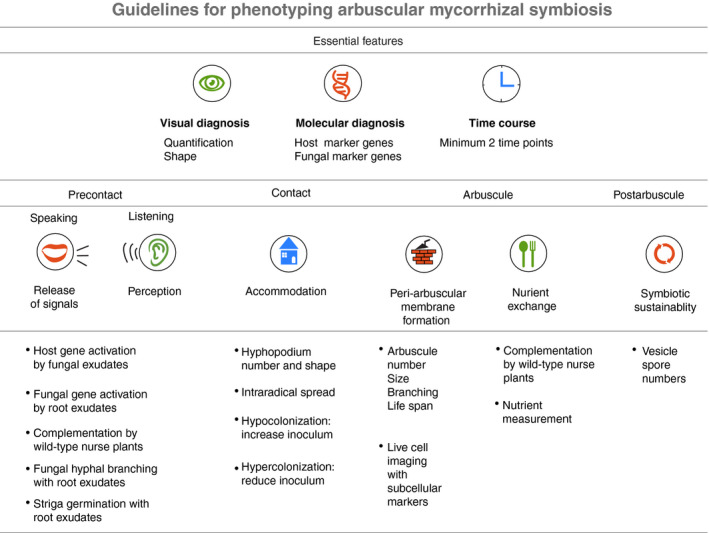
Guidelines for phenotyping arbuscular mycorrhizal (AM) symbiosis. AM phenotyping data should contain three essential features. Visual diagnosis includes AM fungal staining to quantify the colonization level and assess shapes of AM fungal structures. Stage‐specific host and fungal molecular marker gene expression should support the visual observations. Root samples should be harvested at least at two‐time points to capture the early and late stages of AM fungal colonization. This initial evaluation will reveal a specific stage when a plant host gene is likely to function. We listed assays that may help to address roles of host genes in each stage of AM fungal colonization.
Acknowledgements
HM is a CONICYT Chile‐Cambridge TRUST fellow. JC is supported by the Leverhulme Early Career Fellowship (ECF‐2016‐392), The Leverhulme Trust, UK. UP's research is supported by Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/P003419/1 and BB/N008723/1.
References
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Aroca R, Ruiz‐Lozano JM, Zamarreño ÁM, Paz JA, García‐Mina JM, Pozo MJ, López‐Ráez JA. 2013. Arbuscular mycorrhizal symbiosis influences strigolactone production under salinity and alleviates salt stress in lettuce plants. Journal of Plant Physiology 170: 47–55. [DOI] [PubMed] [Google Scholar]
- Breuillin‐Sessoms F, Floss DS, Gomez SK, Pumplin N, Ding Y, Levesque‐Tremblay V, Noar RD, Daniels DA, Bravo A, Eaglesham JB et al 2015. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter 4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. The Plant Cell 27: 1352–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos‐Soriano L, Gómez‐Ariza J, Bonfante P, San Segundo B. 2011. A rice calcium‐dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biology 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Summers W, Paszkowski U. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annual Review of Phytopathology 56: 135–160. [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech‐Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P et al 2013. Short‐chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytologist 198: 190–202. [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Mari A, Novero M, Bonfante P. 2015. Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Frontiers in Plant Science 6: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B. 1980. An evaluation of techniques for measuring vesicular–arbuscular mycorrhizal infection in roots. New Phytologist 84: 489–500. [Google Scholar]
- Gough C, Cottret L, Lefebvre B, Bono J‐J. 2018. Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Frontiers in Plant Science 9: 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi‐Anraku H, Paszkowski U. 2008. Arbuscular mycorrhiza‐specific signaling in rice transcends the common symbiosis signaling pathway. The Plant Cell 20: 2989–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang S‐Y, Nadal M et al 2015. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. 2011. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. The Plant Cell 23: 3812–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Harrison MJ. 2014. A set of fluorescent protein‐based markers expressed from constitutive and arbuscular mycorrhiza‐inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula . Plant Journal 80: 1151–1163. [DOI] [PubMed] [Google Scholar]
- Jabaji‐Hare S, Deschene A, Kendrick B. 1984. Lipid content and composition of vesicles of a vesicular‐arbuscular mycorrhizal fungus. Mycologia 76: 1024–1030. [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. 2007. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences, USA 104: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel L, Keller‐Pearson M, Roux C, Ané J‐M. 2017. Biology and evolution of arbuscular mycorrhizal symbiosis in the light of genomics. New Phytologist 213: 531–536. [DOI] [PubMed] [Google Scholar]
- Keymer A, Gutjahr C. 2018. Cross‐kingdom lipid transfer in arbuscular mycorrhiza symbiosis and beyond. Current Opinion in Plant Biology 44: 137–144. [DOI] [PubMed] [Google Scholar]
- Kobae Y, Fujiwara T. 2014. Earliest colonization events of Rhizophagus irregularis in rice roots occur preferentially in previously uncolonized cells. Plant and Cell Physiology 55: 1497–1510. [DOI] [PubMed] [Google Scholar]
- Kobae Y, Ohmori Y, Saito C, Yano K, Ohtomo R, Fujiwara T. 2016. Phosphate treatment strongly inhibits new arbuscule development but not the maintenance of arbuscule in mycorrhizal rice roots. Plant Physiology 171: 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, Morieri G, MacGillivary A, Johansen C, Antolín‐Llovera M, Parniske M, Oldroyd GED, Downie AJ et al 2011. Lotus japonicus symRK‐14 uncouples the cortical and epidermal symbiotic program. Plant Journal 67: 929–940. [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone‐dependent symbiotic signalling and branching. Nature 483: 341–344. [DOI] [PubMed] [Google Scholar]
- Liao J, Singh S, Hossain MS, Andersen SU, Ross L, Bonetta D, Zhou Y, Sato S, Tabata S, Stougaard J et al 2012. Negative regulation of CCaMK is essential for symbiotic infection. Plant Journal 72: 572–584. [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Oldroyd GED. 2017. Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Current Biology 27: R952–R963. [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech‐Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A et al 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63. [DOI] [PubMed] [Google Scholar]
- Miyata K, Kozaki T, Kouzai Y, Ozawa K, Ishii K, Asamizu E, Okabe Y, Umehara Y, Miyamoto A, Kobae Y et al 2014. The bifunctional plant receptor, OsCERK1, regulates both chitin‐triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant and Cell Physiology 55: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Nadal M, Sawers R, Naseem S, Bassin B, Kulicke C, Sharman A, An G, An K, Ahern KR, Romag A et al 2017. An N‐acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nature Plants 3: 17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H‐J, Floss DS, Levesque‐Tremblay V, Bravo A, Harrison MJ. 2015. Hyphal branching during arbuscule development requires Reduced Arbuscular Mycorrhiza1 . Plant Physiology 169: 2774–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P, Gutjahr C. 2018. Transcriptional regulation of arbuscular mycorrhiza development. Plant and Cell Physiology 59: 678–695. [DOI] [PubMed] [Google Scholar]
- Pumplin N, Harrison MJ. 2009. Live‐cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiology 151: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi‐Anraku H, Umehara Y et al 2007. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus . The Plant Cell 19: 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Song J, Xin X, Xie X, Zhao B. 2018. Arbuscular mycorrhizal fungal 14‐3‐3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Frontiers in Microbiology 9: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S, Handa Y, Takeda N, Kawaguchi M. 2016. Strigolactone‐induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis . Molecular Plant–Microbe Interactions 29: 277–286. [DOI] [PubMed] [Google Scholar]
- Vierheilig H, Schweigerb P, Brundrett M. 2005. An overview of methods for the detection and observation of arbuscular mycorrhizal fungi in roots. Physiologia Plantarum 125: 393–404. [Google Scholar]
- Xie X, Lin H, Peng X, Xu C, Sun Z, Jiang K, Huang A, Wu X, Tang N, Salvioli A et al 2016. Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Molecular Plant 9: 1583–1608. [DOI] [PubMed] [Google Scholar]
- Yang S‐Y, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N et al 2012. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. The Plant Cell 24: 4236–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kameoka H, Tempo M, Akiyama K, Umehara M, Yamaguchi S, Hayashi H, Kyozuka J, Shirasu K. 2012. The D3 F‐box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytologist 196: 1208–1216. [DOI] [PubMed] [Google Scholar]


