Abstract
The role of miRNAs in mediating insecticide resistance remains largely unknown, even for the model species Drosophila melanogaster. Building on prior research, this study used microinjection of synthetic miR-310s mimics into DDT-resistant 91-R flies and observed both a significant transcriptional repression of computationally-predicted endogenous target P450 detoxification genes, Cyp6g1 and Cyp6g2, and also a concomitant increase in DDT susceptibility. Additionally, co-transfection of D. melanogaster S2 cells with dual luciferase reporter constructs validated predictions that miR-310s bind to target binding sites in the 3ʹ untranslated regions (3ʹ-UTR) of both Cyp6g1 and Cyp6g2 in vitro. Findings in the current study provide empirical evidence for a link between reduced miRNA expression and an insecticidal resistance phenotype through reduced targeted post-transcriptional suppression of transcripts encoding proteins involved in xenobiotic detoxification. These insights are important for understanding the breadth of adaptive molecular changes that have contributed to the evolution of DDT resistance in D. melanogaster.
Subject terms: Molecular biology, Molecular evolution
Introduction
The exposure of a species to changing environmental conditions, such as variation in nutrient availability, climate, and toxic chemicals, can lead to corresponding phenotypic change(s) via adaptive directional selection. Insecticidal compounds—including synthetic chemicals, natural products, and protein toxins—represent human-imposed selection pressures upon insect populations. Specifically, insecticides have been widely used to suppress insect populations in efforts to protect human health by stabilizing the output of agricultural commodities and foodstuffs1, and reducing the incidence of insect vector-borne diseases, such as malaria and dengue fever2,3. However, frequent and widespread application of insecticides has contributed to the development of insect populations with high frequencies of phenotypic resistance to one or more classes of insecticides4. Such responses by insect populations and selection for high levels of resistance represent serious threats to many pest control programs. The evolution of insecticide resistance in insect populations involves genomic variations in the genome that, in turn, offers the scientific community an opportunity to both understand the genes directly involved in resistance and, in some cases, regulatory mechanisms associated with those genes. A model system that affords the opportunity to perform gene-by-gene analysis of traits involved in polygenic pesticide resistance is that of dichlorodiphenyltrichloroethane (DDT) resistance in Drosophila melanogaster (hereafter referred to as Drosophila).
DDT is an organochlorine insecticide that disrupts the insect nervous system by affecting the permeability of nerve cell plasma membranes and causing paralysis5. While DDT was extensively used during the post-World War II to control insect pests, deleterious side effects on non-target mammalian, bird, and insect species ultimately led to its ban by most countries by the 1980s6. Although DDT is no longer extensively used, selected laboratory colonies of Drosophila with varying levels of DDT resistance provide a model system for investigating adaptive genomic responses that lead to insecticide resistance7. The Drosophila model laboratory strain for DDT resistance, 91-R, has been under chronic exposure to DDT for over six decades and reared in parallel with the non-selected control strain 91-C; the two strains came from a common population that was split before these decades-long difference in treatment of the two populations. This high-level DDT resistance phenotype in Drosophila is polygenic and associated with multimodal resistance mechanisms8,9 including, but not limited to, involvement of phase I, II, and III detoxification enzymes. For example, the variance in protein structure and transcript expression of cytochrome P450 monooxygenases (P450s), including Cyp6g1 and ATP-binding cassette (ABC) transporters, has been reported in the DDT-resistant 91-R compared to DDT-susceptible strains 91-C and Canton-S10–13. Additionally, directional selection was predicted to contribute to selective sweeps proposed within multiple genome regions of 91-R compared to 91-C14; among these implicated genes, the role of the ABC transporter, multidrug resistant (MDR) 49, in DDT resistance was validated using a transgenic expression approach13. Moreover, the involvement of several MDR and P450 genes were implicated in DDT resistance of 91-R using transgenic knockdown lines10. Despite the implication of directional selection within multiple genome regions, the independent roles or additive/non-additive contributions to the DDT resistance phenotype in 91-R remains unknown.
Among different genetic mechanisms implicated within DDT resistant phenotypes, P450s play pivotal roles in detoxifying exogenous xenobiotics such as insecticides and plant toxins through catabolic pathways that relegate compounds into more soluble and less toxic products15. Two possible mechanisms of P450-mediated insecticide resistance have been demonstrated. First, genome-wide association studies identified DDT-associated multiple genes and signatures of adaptive selection within the genome, where the amino acid changes in Cyp6w1 were associated with DDT resistance16. Second, the increased abundance of transcripts for P450s and the likely increased (subsequent) levels of functional translated P450 enzymes have been proposed as a mechanism of resistance to several classes of insecticides17, including DDT in Drosophila18–20. For example, the potential involvement of Cyp4g1, Cyp6g1, and Cyp12d1 in DDT resistance in 91-R was demonstrated via independent RNA interface (RNAi)-mediated knockdown of transcripts using the transgenic Gal4/UAS-RNAi expression system, each resulting in increased susceptibility10. Moreover, differences in micro-RNA (miRNA) repression of P450 transcription or translation was implicated within the polygenic DDT resistance mechanism of 91-R11,21. However, relatively little is known regarding the precise mechanism(s) by which P450s are silenced by miRNAs or the downstream effects on response to xenobiotic exposure in Drosophila.
miRNAs are small endogenous non-protein coding RNAs with lengths between 19 and 23 nucleotides. They are involved in many biological processes, including regulation of cellular metabolism and organismal homeostasis22. A given miRNA can negatively regulate the translation of mRNA transcripts by reverse complementarily binding the 3′-UTR of the target mRNA, which leads to post-transcriptional degradation of the mRNA23. This mRNA decay is preceded by miRNA-mediated inhibition of translation, indicating that transcriptional and translational silencing are intertwined24. Thus, miRNAs have important roles in regulating transcript levels for genes involved in insect development, behaviour, and host–pathogen interactions23,25. To date, there are few instances in which miRNAs are known to regulate genes that mediate insecticide resistance within insect species. One study example in the cotton aphid, Aphis gossypii, demonstrated that two miRNAs, miR-276 and miR-3016, contribute to a spirotetramat resistance phenotype through the targeting of an acetyl-coenzyme A carboxylase gene26. Another study was associated with the down-regulation of the miR-2~13~71 cluster in deltamethrin-resistant adult Culex pipiens with the concomitant increase in transcript levels of putative targets Cyp325bg3 and Cyp9j35 mRNAs27. In Tetranychus cinnabarinus, tci-miR-1-3p plays a critical role in cyflumetofen resistance by targeting a GST gene, TCGSTM428.
In previous research, we identified miRNAs with significant levels of differential expression between Drosophila strains that are DDT-resistant (91-R) and -susceptible (91-C)21. The miR-310s were significantly down-regulated in the 91-R strain as compared to the 91-C strain and computational predictions identified a number of cytochrome P450 transcripts, including Cyp6g1, Cyp6g2, Cyp6w1, Cyp49a1, and Cyp12a5, as potential targets of these miR-310s21. However, empirical evidence to support these predicted regulatory roles of miR-310s in DDT resistance is currently lacking.
In the present study, we tested the hypothesis that miR-310s impact the levels of endogenous Cyp6g1 and Cyp6g2 transcripts in vivo and are associated with levels of DDT-resistance in adults for 91-R. Furthermore, reporter assay experiments allowed us to test the hypothesis that computationally-predicted miR-310s seed regions in Cyp6g1 and Cyp6g2 3ʹ -UTRs are linked to decreased levels of corresponding transcripts when co-expressed with miR-310s.
Results
Constitutive expression levels of target Cyp6g1 and Cyp6g2 genes of miR-310s
The association between the up-regulation of Cyp6g1 and Cyp6g2 expression with DDT resistance in 91-R was validated via comparison of RT-qPCR assay results with DDT–susceptible strains 91-C and Canton-S. Specifically, our results showed a significantly higher level of constitutive Cyp6g1 and Cyp6g2 transcript expression in 91-R as compared to both 91-C and Canton-S (Supplemental Fig. S1; F = 154.8, df = 2, p < 0.05 for Cyp6g1; F = 93.7, df = 2, p < 0.05 for Cyp6g2). In contrast, neither Cyp6g1 or Cyp6g2 showed any significant level of differential expression between the DDT-susceptible strains 91-C and Canton-S (p = 0.139 for Cyp6g1; p = 0.438 for Cyp6g2).
Microinjection of miR-310s impacts P450 gene regulation
miRNA mimics are small, chemically modified double-stranded RNAs that mimic endogenous miRNAs. The microinjection of mimics for the miR-310s cluster (miR-310, miR-311, miR-312, and miR-313) was used to evaluate their potential role in mediating the regulation of putatively targeted cytochrome P450s-Cyp6g1 and -Cyp6g2 transcript levels in the 91-R strain. Following the injection of miR-310s mimics into adult females of the 91-R strain, the levels of these aforementioned miRNAs increased at all time points compared to the NCsiRNA and DEPC-injected control (Fig. 1). Specifically, temporal sampling showed that the increase in cellular miR-310 levels were 8.7-, 7.4-, and 5.3-fold (F = 18.6, df = 4, p < 0.05) and cellular miR-311 levels were 80.9-, 55.7-, and 31.9-fold (F = 22.4, df = 4, p < 0.05) as compared to the controls at 24 h, 48 h, and 72 h following mimic injections, respectively (Fig. 1). However, the relative increase in cellular miR-310 and miR-311 mimic levels were not significantly different across the three time points. When miR-312 and miR-313 mimic levels were measured, their increases occurred with estimates of 139.8-, 117-, 59.7-fold (F = 37.8, df = 4, p < 0.05) for miR-312 mimic and 96.8-, 56.7-, and 35.4-fold (F = 80.2, df = 4, p < 0.05) for miR-313 mimic at 24 h, 48 h, and 72 h post-injection as compared to the controls (Fig. 1). The relative increases in cellular miR-312 and miR-313 mimic levels were the greatest at 24 h as compared to the controls (p < 0.05; Fig. 1). When measured 72 h after injection, however, the levels of miR-312 and miR-313 mimics had significantly declined as compared to 24 h post-injection interval (p < 0.05; Fig. 1).
Figure 1.
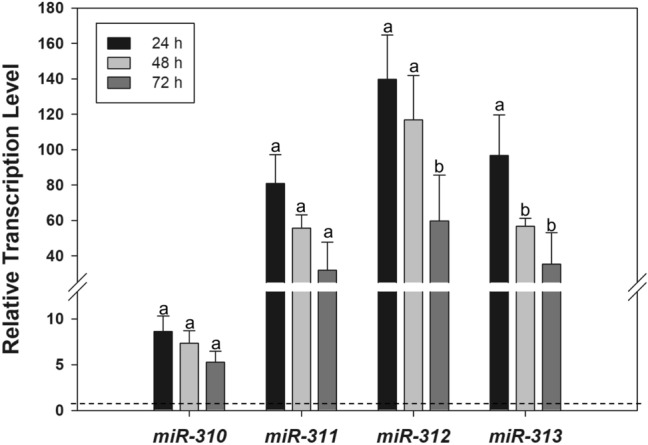
Relative transcription levels of miR-310, miR-311, miR-312, and miR-313 after microinjection of the miR-310s mimics, negative control siRNA (NCsiRNA) mimic, and DEPC-water into 91-R female at 24 h, 48 h, and 72 h post-injection. All levels of miR-310s are given relative to the transcription levels of NC and DEPC-water which were ascribed an arbitrary value of 1 (dashed line). There was no difference between the DEPC and NCsiRNA microinjection groups (p = 0.229–0.943). Different letters on the bars indicate that the means are significantly different across three time points within each miRNA (p < 0.05).
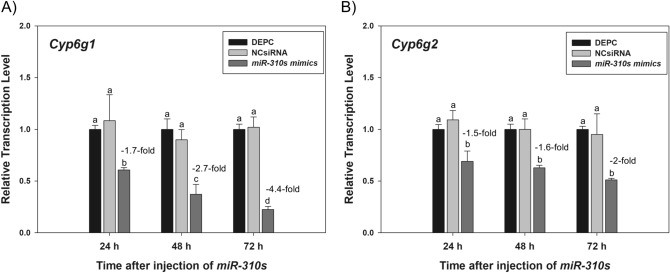
The cellular levels among the putatively targeted endogenous Cyp6g1 and Cyp6g2 transcripts were evaluated in response to miR-310s mimics injections in 91-R strain. Specifically, flies in the miR-310s mimics-injected treatment groups showed a reduction in the levels of Cyp6g1 and Cyp6g2 transcripts as compared to NCsiRNA and DEPC-injected control (Fig. 2). The relative proportion of reduction in Cyp6g1 transcript levels were 1.7-, 2.7-, and 4.4-fold at 24 h, 48 h, and 72 h after mimics injections, respectively (F = 73.4, df = 8, p < 0.05 for all comparisons), as compared to the NCsiRNA and DEPC-injected control (Fig. 2A).
Figure 2.
Relative transcription levels of (A) Cyp6g1 and (B) Cyp6g2 after microinjection of the miR-310s mimics, negative control siRNA (NCsiRNA) mimic, and DEPC-water into 91-R female flies at 24 h, 48 h, and 72 h post-injection. There was no difference between the DEPC-water and NC microinjection groups across three time points for Cyp6g1 (p = 0.55) and Cyp6g2 (p = 0.953). Different letters on the bars indicated that the means were significantly different across three time points (p < 0.05).
Analogously, the relative Cyp6g2 transcript levels were decreased by 1.5-, 1.6-, and 2-fold at 24 h, 48 h, and 72 h after injection of mimics, respectively (F = 16.1, df = 8, p < 0.05 for all comparisons), as compared to the NCsiRNA and DEPC-injected control (Fig. 2B). The Cyp6g1 transcripts showed the lowest relative level at 72 h post miR-310s injection as compared to 24 h and 48 h post injection (p < 0.05). However, there was no significant difference in the expression of Cyp6g2 transcripts across three time points in the mimic-injected group (p = 0.879–0.998).
Validation of miR-310s-mediated regulation of Cyp6g1 and Cyp6g2 transcripts in vitro
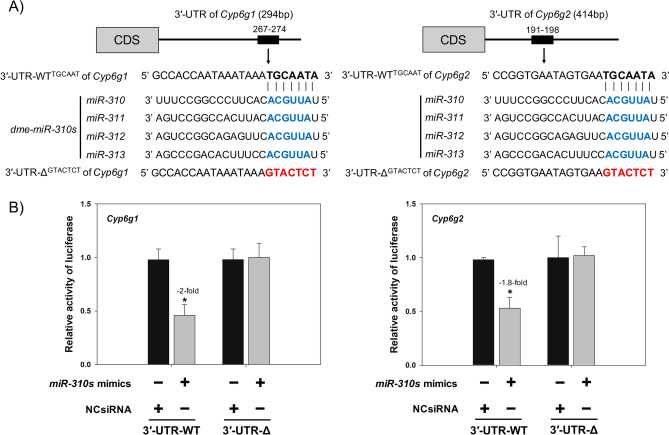
The miR-310s cluster shares an identical seed sequence among miR-310, miR-311, miR-312, and miR-313 within the 3ʹ-UTRs of both Cyp6g1 and Cyp6g2 (Fig. 3A). Specifically, the seed sequences for miR-310s, ACGUUA, were located at positions of 267 to 274 and 191 to 198 of the 3ʹ-UTR for Cyp6g1 and Cyp6g2, respectively. The putative transcript target sites showed 100% complementarity to the seed regions, ACGUUA, for all endogenous and mimic miR-310s sequences. Assays from S2 cells co-transfected with dual reporter psiCHECK-2-3ʹ-UTR-WTTGCAAT constructs + miR-310s mimics resulted in a decrease in luciferase activity; specifically, assays using wild-type 3ʹ-UTRs (3ʹ-UTR-WTTGCAAT) of Cyp6g1 and Cyp6g2, respectively, showed an approximate reduction of 2-fold (F = 8.2, df = 1, p < 0.05) and 1.8-fold (F = 125.5, df = 1, p < 0.05) as compared to the NCsiRNA treatment (Fig. 3B). However, no significant change in luciferase activity was detected for experiments that were co-transfected with the miR-310s mimics along with the constructs containing the mutant 3ʹ-UTR-ΔGTACTCT seed regions from Cyp6g1 (F = 0.01, df = 1, p = 0.918) or Cyp6g2 (F = 0.167, df = 1, p = 0.704) as compared with the NCsiRNA treatment (Fig. 3B).
Figure 3.
Validation of miR-310s -mediated regulation of Cyp6g1 and Cyp6g2 transcripts in vitro. (A) Predicted target binding site (3′-UTR-WTTGCAAT) and mutated target binding site (3′-UTR-ΔGTACTCT) of miR-310s within the 3′-UTR of the putative target Cyp6g1 and Cyp6g2 from 91-R strain. (B) Relative luciferase activity in S2 cells co-transfected with miR-310s or negative control mimics and the wide- or mutant-type luciferase reporter vectors (3′-UTR-WTTGCAAT and 3′-UTR-ΔGTACTCT). An asterisk (*) indicate a difference across the treatment groups at p < 0.05.
Impact of miR-310s modulation on DDT-induced mortality
To validate the putative involvement of increased levels of miR-310s mimics in DDT resistance in the 91-R strain, mortality bioassays were performed at 24 h post-injection of adults with miR-310s mimics. Probit analyses demonstrated that the miR-310s mimics injected flies exhibited a LT50 value of 17.0 h (χ2 = 81.1, df = 2, p < 0.01), which was a shorter time span as compared to the NCsiRNA treatment (28.5 h; χ2 = 64.9, df = 2, p < 0.01) and the DEPC-water control (29.1 h; χ2 = 59.1, df = 2, p < 0.01) (Table 1). However, evidenced by the overlapping 95% CL, the NCsiRNA injected flies did not exhibit any statistically significant difference for their LT50 value as compared to the DEPC-water control treatment.
Table 1.
Evaluation of DDT sensitivity in Drosophila 91-R strain after miR-310s mimics injection using a topical exposure to DDT (0.5 µg/fly).
| Treatment | LT50a (hour) | 95% C.L.b | χ2 (df) | Slope ± SE | P value |
|---|---|---|---|---|---|
| miR-310s mimics | 17.0 | 12.0–21.3 | 81.1 (2) | 2.2 ± 0.41 | < 0.01 |
| NCsiRNA | 28.6 | 22.1–37.3 | 65 (2) | 2.0 ± 0.44 | < 0.01 |
| DEPC-water control | 29.1 | 21.6–38.8 | 59.1 (2) | 1.98 ± 0.5 | < 0.01 |
aLethal Time 50 (hour) that killed 50% of the flies.
b95% Confidence limit.
Using the Fisher F-test, we observed that the regression lines between the miR-310s mimics injection group and NCsiRNA group were significantly different (F = 14.5, df = 3, p < 0.01; Fig. 4). Analogously, the estimated regression line for the miR-310s mimics injection treatment were different than the DEPC-water control treatment (F = 12.1, df = 3, p < 0.0001; Fig. 4). In contrast, results of the F-test predicted the relative equality between regression lines between the NCsiRNA and DEPC-water control groups (F = 1.4, df = 3, p = 0.263; Fig. 4). Additionally, the acetone-only negative control showed no fly mortality (data not shown).
Figure 4.
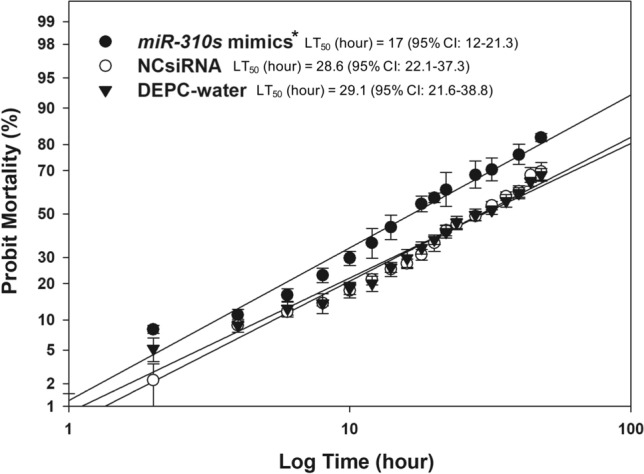
Comparative DDT (0.5 μg/fly) induced mortality of microinjected 91-R female flies with miR-310s mimics, negative control siRNA mimic (NCsiRNA), and DEPC-water. There was no difference between the DEPC-water and NCsiRNA microinjection groups (p = 0.263). The Fisher F-test was performed to verify if the regression lines were equal to each other. An asterisk (*) indicates a difference across the treatment groups at p < 0.05. 95% CI: 95% Confidence limit.
Polymorphisms in TF-biding motif putatively associated with expression level of miR-310s
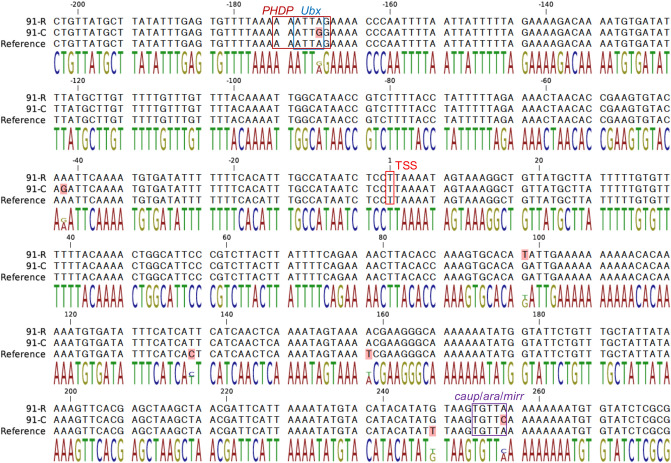
The predicted transcriptional start site (TSS) of the miR-310s cluster was located 268 bp upstream of the first miRNA, miR-313, but no other TSSs were observed within the cluster. Using the JASPAR database, a total of 216 different putative TF-binding motifs were predicted in the cis-regulatory region of the miR-310s cluster with relative profile score threshold 95% (Supplemental Table S1). A comparison of nucleotide sequences from this region, derived from short read alignments between DDT-resistant 91-R and DDT-susceptible 91-C, predicted that two mutation sites were within three putative TF-binding motifs upstream of the miR-310s cluster. Specifically, predicted binding motifs for PHPD and Ubx contain a single nucleotide polymorphism (A for 91-R and G for 91-C), respectively (Fig. 5). Additionally, caup/ara/mirr binding motif contains a change in single nucleotide from A for 91-R to C for 91-C (Fig. 5).
Figure 5.
The position of putative transcription factor binding sites within 200 bp up-stream and 280 bp down-stream of the predicted miR-310s cluster transcription start site (TSS). The transcription factor binding sites are boxed. The polymorphisms among three strains are shaded in red.
Discussion
Arthropods continue to damage agricultural commodities and vector diseases threatening human welfare due to ongoing challenges in pest control that arise from instances where selection has caused high levels of resistance following repeated exposures to insecticidal agents. The involvement of miRNAs in the regulation of metabolic resistance to insecticides has been suggested via comparative and correlative studies for pyrethroids29 and ryanoids30, but strong evidence for miRNAs as causative factors among these resistant phenotypes is arguably lacking. The present study demonstrated that members for the miR-310s cluster interact with target binding sites within 3ʹ-UTR sequences of cytochrome P450, thereby regulating transcript levels in vivo. Specifically, we showed here that transcript from two cytochrome P450 genes, Cyp6g1 and Cyp6g2, are up-regulated in the DDT-resistant strain 91-R as compared to the -susceptible strains 91-C and Canton-S, reconfirming our previously published results12,31.
Previous studies provide supporting evidence that high to moderate level DDT resistance is polygenic in Drosophila, with multiple resistance genes, including P450s, associated with the DDT resistance phenotype8–10. The Cyp6 subfamily has been associated with DDT resistance and cross-resistance to neonicotinoid insecticides32,33. For example, statistically significant differences in expression were documented for Cyp6a2, Cyp6a8, Cyp6g1, Cyp6g2, and Cyp6w1 in DDT-resistant 91-R and Wisconsin strains as compared to DDT- susceptible Canton-S and 91-C strains; relatedly, previous evidence of significant changes in expression of Cyp6 subfamily is functionally involved in DDT resistance in Drosophila12,19,34. Moreover, overexpression of the CYP6W1-Ala370 allele in transgenic Drosophila was sufficient to confer low levels of DDT tolerance relative to CYP6W1-Val370 and CYP6W1-Gly37016. Many of these genes, however, were not found to be differentially expressed in field-derived resistance strains35. The one exception was found in prior experiments that genetically mapped the positioned locus of major effect on DDT resistance within a genome region encoding Cyp6g118,33. These authors also documented that Cyp6g1 was up-regulated in resistant adult Drosophila flies18. Furthermore, expression of the Cyp6g1 enzyme in a heterologous system was capable of carrying out a dichlorination step in the metabolism of DDT36, indicating a likely role of the translated protein in the cellular detoxification mechanism.
In keeping with these previous studies, our results support the hypothesis that the miR-310s cluster mediate the regulation of Cyp6g1 and Cyp6g2 transcript levels, as well as part of the DDT resistance phenotype in the 91-R strain. These results advance research based on our previous study that reported the miR-310s are down-regulated in the DDT-resistant 91-R strain as compared to its susceptible counterpart 91-C21. Equally, the expression of the miR-310s was inversely correlated with the up-regulation of a number of detoxification genes (P450s, GSTs, and esterases) that also had computationally-predicted miR-310s target binding sites in their 3ʹ-UTRs21. Specifically, validation experiments here showed a reduction in Cyp6g1 and Cyp6g2 transcript levels following injection of miR-310s mimics into adult 91-R females (Figs. 1, 2), hypothetically via target-specific degradation via the RISC pathway37. Although a range of other target genes could be influenced by miR-310s mimic injection, the resulting reduction in Cyp6g1 and Cyp6g2 transcript levels and concomitant more rapid mortality (median lethal time; LT50) within the miR-310s injected group as compared to the control-injected group from strain 91-R (Fig. 4) suggests changes in one or both Cyp6g1 and Cyp6g2 transcripts contributes to a portion of the observed changes in DDT susceptibility.
However, we cannot firmly establish any direct involvement of miR-310-mediated regulation of Cyp6g2 on the DDT resistance phenotype in 91-R. Specifically, although the role of Cyp6g1 in DDT resistance is established, neither is any direct role of Cyp6g2 in DDT resistance yet reported from Drosophila, nor is there direct functional evidence for enzymatic products of Cyp6g2 in DDT detoxification. However, a number of previous studies suggested that the overexpression of Cyp6g2 gene was associated with resistance to several insecticides such as imidacloprid, ivermectin, and diazinon in Drosophila35,38,39. Moreover, the significant overexpression of Cyp6g2 was associated with the DDT-resistant 91-R compared to susceptible strains Canton-S and 91-C12,19,31. Additionally, the current study provides empirical evidence demonstrating a direct impact of miR-310s on corresponding transcript levels of Cyp6g1 and Cyp6g2 and DDT resistance in 91-R. Regardless, disentangling the independent effects of Cyp6g1 and Cyp6g2 on DDT resistance remains unresolved. Expression of Cyp6g1 and Cyp6g2 is not only highly correlated, which was suggested to be a consequence of physically linked allelic variants38,40 but also shown to results from impacts of the miR-310 cluster. Therefore, it remains plausible that Cyp6g2 associations with DDT may be a consequence of genomic proximity to and recent co-ancestry with Cyp6g1. The independent role of Cyp6g2 in DDT resistance remains to be investigated through future functional experiments.
Evidence from our microinjection experiments suggested a direct involvement of miR-310s in regulation of Cyp6g1 and Cyp6g2, and concomitant mediation of at least a portion of the DDT resistance phenotype in female 91-R Drosophila. Previously, Chung, et al.41 demonstrated that the up-regulation of Cyp6g1 is associated with the insertion of an Accord retrotransposon within the upstream region of the Cyp6g1 in a resistant field strain. Moreover, Schmidt et al. also found that Cyp6g1 increased expression was influenced by copy number variation42 and that these structural variants are an outstanding feature in Cyp6g1 associated with DDT resistance43. Additionally, a functional Nrf2/Maf (NF-E2-related factor 2/Muscle aponeurosis fibromatosis) transcription factor can enhance the constitutive up-regulation of Cyp6a2 and Cyp6a8 transcription, which also was associated with DDT resistance in Drosophila44,45. Furthermore, a nucleotide mutation in the estrogen-related receptor (ERR) gene led to over-expression of Cyp12d1 and Cyp6g2 in Drosophila46. These aforementioned lines of evidence demonstrate that the regulation of transcript levels for P450s, as exemplified with Cyp6g1 and Cyp6g2, may involve multiple regulatory factors; moreover, the genetic background of the flies may influence which one (or several) of these factors cause constitutive over-transcription. Current evidence supports the hypothesis that miR-310s may play a role in post-transcriptional longevity of Cyp6g1 and Cyp6g2 transcripts in DDT-resistant 91-R strain and the corresponding higher turnover of these transcripts in susceptible counterparts 91-C and Canton-S strains.
Despite documenting that miR-310s are likely causative of a portion of the DDT resistance phenotype in 91-R, demonstration of a putative miR-310s seed sequence dependence of transcript quantities in vitro were lacking. In order to address this shortfall, a luciferase assay verified that an intact miR-310s target binding sites in the 3ʹ-UTR of both Cyp6g1 and Cyp6g2 is necessary and sufficient for the degradation of corresponding transcript levels within S2 cells. Specifically, we revealed that the relative luciferase activity of wild type 3ʹ-UTR (WTTGCAAT) for both Cyp6g1 and Cyp6g2 was reduced when co-expressed with miR-310s mimics, but this effect was not seen for the mutant version, ΔGTACTCT (Fig. 3). This suggested that the functional target binding sites in 3 ʹ -UTRs of Cyp6g1 and Cyp6g2 mRNAs likely facilitate complementary base pairing with miR-310s and, in turn, may impact stability or degradation of Cyp6g1 and Cyp6g2 mRNAs.
The role of miRNAs in regulating P450 gene expression has also been investigated in several other insect species. Prior work supports the hypothesis that miR-2b-3p regulates the expression of two P450s, Cyp9f2 and Cyp307a1, putatively involved in deltamethrin resistance in Plutella xylostella29. Let-7 and miR-100 have been proposed to modulate the post-transcriptional regulation of Cyp6cy3 gene to alter the tolerance of Myzus persicae nicotianae to nicotine47. The upregulation of miR-285 is thought to increase resistance to deltamethrin in Culex pipiens pallens by binding to 3′-UTR region of Cyp6n23 resulting in changing in expression of Cyp6n2348, but direct evidence was not provided. In this study, we provide functional evidence demonstrating a likely role of miR-310s species in the binding and subsequent stability of putatively targeted cytochrome P450s and is the first known instance of making such a connection for a gene shown to have a role in mediating the expression of a DDT resistance trait.
The molecular genetic or biochemical basis of resistance has been resolved in cases where traits are monogenic or share a conserved mechanism across genera49,50. However, phenotypes showing a high level of resistance based on polygenic mechanisms have been more difficult to elucidate. The laboratory DDT-selected Drosophila strain 91-R has served as a model system to understand the molecular mechanisms underlying polygenic pesticide resistance14,31,51,52. Thus, the increased levels of Cyp6g1 and Cyp6g2 transcripts, and assumed relative increases in corresponding protein levels, mediated by reduced degradation by down-regulated miR-310s likely contributes partially to the DDT resistance phenotype in 91-R. This conclusion is based on the phenotypic response of 91-R following injection of miR-310s mimics (Fig. 4), as well as evidence that other genes are functionally involved, such as the phase III detoxification ATP binding cassette transporter, Mdr4913. Furthermore, RNAi mediated knockdown of several phase I (Cyp6g1, Cyp12d1, and Cyp4g1), phase III (Mdr49, Mdr50, Mdr65, and Mrp1), and other cuticular (Lcp1) genes documented their capacity to contribute to DDT resistance in 91-R8. Moreover, DDT resistance in 91-R is dependent upon genetic factors located within 13 major and 3 minor genome regions affected by selective sweeps14, as well as changing in multiple pathways that impact neuronal function and cell stress response31. Our DDT bioassay results support the hypothesis that miR-310s -mediated expression of Cyp6g1 and Cyp6g2 contributes to the polygenic DDT resistant phenotype in 91-R.
The possibility that the reduced expression of miR-310s in 91-R may impact a range of other genes cannot be ruled out. Computational predictions suggest putatively regulated transcripts encode a broad range of proteins, including a subset with functional annotation suggesting roles in xenobiotic detoxification21. Thus, the role of miR-310s in the DDT resistance phenotype may be more complex, and a future genome-wide transcriptome analysis of miR-310s mimics injected Drosophila samples could potentially provide a precise and unbiased measure of impact on target transcripts.
The impact of genetic variants on miRNA expression and function still remain unclear. A number of studies analyzing genome-wide nucleotide and miRNA expression variation identified putative polymorphisms significantly associated with the regulation of miRNA expression53,54. In this study, nucleotide mutations were identified in the cis-regulatory region of putative TF-binding motifs from the miR-310s cluster. However, the impact of these alterations on transcript expression levels of the miR-310s cluster and their contribution to DDT resistance phenotypes in Drosophila remain to be investigated.
Regardless, despite the other unknown molecular function(s) of miR-310s, the current study establishes a role of miR-310s in modulating cellular levels of Cyp6g1 and Cyp6g2 transcripts. Due to the co-repression of these two transcripts via miR-310 mimics, their independent effects on subsequent DDT resistance levels cannot be disentangled, such that the novel possibility of Cyp6g2 involvement in the resistance phenotype of 91-R could not be dissected using our methodology. Furthermore, since there is a paucity of in vivo and in vitro functional studies that aim to uncover protein function or the complex regulatory networks interconnecting miRNA and target mRNAs, the identification of novel genes and pathways (and both their direct and indirect consequences) are needed. Such studies will undoubtedly assist in the elucidation of relationships for miRNA-based gene regulation and DDT resistance in Drosophila and potentially shed light on analogous mechanisms across arthropods.
Materials and methods
Drosophila strains
The DDT-susceptible Canton-S, the low-level DDT-resistant 91-C, and the highly DDT-resistant 91-R strains of Drosophila have been maintained for almost two decades in the Pittendrigh laboratory (Michigan State University, East Lansing, MI, USA). The 91-R strain has been continually reared as described previously31, with selection maintained by growing flies in the presence of 150 mg DDT impregnated paper filter disks while Canton-S and 91-C were maintained without any exposure to DDT. The 91-R strain has been shown to be ~ 1,500-fold and ~ 107-fold resistant to DDT compared to the susceptible Canton-S and 91-C strains through the use of contact and topical bioassays31,55. In order to compare the constitutive expression of miRNAs and cytochrome P450 genes in subsequent analyses (see below), all flies were not exposed to DDT within that generation.
Microinjection of female adult Drosophila and RNA isolation
A set of mirVana mimics were synthesized for miR-310, miR-311, miR-312, and miR-313 by Invitrogen (Ambion, Life Technologies) at a concentration of 100 µM. Members of the miR-310s cluster are positioned less than 1 kb apart and show 100% homology between independent seed sequences. Clustered miRNAs are often co-expressed56,57 and co-regulate functionally related genes58,59, therefore were treated as a single functional unit of study within our experiments. One-day-old 91-R female flies were anesthetized on ice, and 69 nl of combined all four mimics, miR-310s, (final concentration 25 µM), were injected into the side of the thorax of individual Drosophila adults using 2-in. needles using a Drummond’s Nanoject II microinjector (Drummond Scientific Company, USA). AllStars Negative Control siRNA (NCsiRNA; Qiagen, Valencia, CA) were similarly injected into female flies with the same volume and concentration as the mimics of miR-310s treatments. Corresponding negative controls consisting of the diethylpyrocarbonate (DEPC)-treated water were analogously injected. After they were microinjected, the flies were immediately placed into small plastic tubes and allowed to recover at 25 °C and 50%-70% humidity with 16/8 h day-night light cycle with commercially available medium (Jazz-Mix Drosophila Food, Fischer Scientific). Each experiment was performed in triplicate with independent microinjections. Total RNA was extracted from a pool of fifteen flies from each replicate of mimic, NC-siRNA, and DEPC-water injected groups at 24 h, 48 h, and 72 h post-microinjections using the Qiagen miRNeasy Mini Kit according to the manufacturer’s instructions (Qiagen). Each sample was treated with DNase I (Zymo Research, Orange, CA) to remove contaminating genomic DNA prior to cDNA synthesis.
Gene expression by Reverse Transcriptase-quantitative PCR (RT-qPCR)
First-strand cDNA synthesis was performed using the miScript II RT kit (Qiagen). Each synthesized cDNA sample was then used as a template for RT-qPCR reactions using the miScript SYBR Green PCR Kit (Qiagen) according to the manufacturer’s instructions with miRNA-specific forward primers (Table 2). The same cDNA template was analogously used in RT-qPCR reactions primed by forward and reverse primers for corresponding putative targeted P450 transcripts (Table 2) using the Power SYBR Green PCR Master Mix according to the manufacturer’s instructions (Applied Biosystems Inc., Foster City, CA). All RT-qPCR amplification reactions were performed on a StepOnePlus Real-Time PCR system (Applied Biosystems Inc.), with three technical replicates across all biological replicates. Normalization of the relative expression levels of each miR-310s and target cytochrome P450s was made with respect to the reference genes, 5S rRNA and rp49, respectively. Normalized miRNA and target transcript expression levels were calculated using the 2−∆∆C(t) method60. Statistical analysis was performed using a one-way analysis of variance (ANOVA) by Tukey’s multiple sample comparisons using XLSTAT software (Addinsoft, NY, USA), and a significance threshold set at p < 0.05.
Table 2.
Sequences of the primers used in this study.
| miRNA or gene | Forward primers | Reverse primers | Remarks |
|---|---|---|---|
| 5S rRNA | CGACCATACCACGCTGAATA | Universal primer (supplied from miScript SYBR Green PCR Kit) | RT-qPCR |
| miR-310-3p | UAUUGCACACUUCCCGGCCUUU | Universal primer (supplied from miScript SYBR Green PCR Kit) | |
| miR-311-3p | UAUUGCACAUUCACCGGCCUGA | Universal primer (supplied from miScript SYBR Green PCR Kit) | |
| miR-312-3p | UAUUGCACUUGAGACGGCCUGA | Universal primer (supplied from miScript SYBR Green PCR Kit) | |
| miR-313-3p | UAUUGCACUUUUCACAGCCCGA | Universal primer (supplied from miScript SYBR Green PCR Kit) | |
| Rp49 | CGGATCGATATGCTAAGCTGT | GCGCTTGTTCGATCCGTA | |
| Cyp6g1 | GAATTCGCACCAAGCTGACT | TCCCAGAGTTCTTCTCTCCA | |
| Cyp6g2 | ATGTAGGTGTAGGGCGTGT | CAAGGGCATGCCCGTTTATA | |
| Cyp6g1 3′UTR | XhoI: tccgctcgagATTTGAATCGCATGAACTGTG | NotI: agaatgcggccgcATAATCGTAAAGATAGCATTT | Vector construction |
| Cyp6g2 3′UTR | XhoI: tccgctcgagAGCTGGTGTCGCATCTTAAA | NotI: agaatgcggccgcTGAGCAGCTAGCAGCTACTC |
Construction of luciferase reporter vectors and luciferase assay
The psiCHECK-2 dual fluorescent reporter system (Promega, Madison, WI, USA) was used to determine any interaction of miR-310s with putative target binding sites in Cyp6g1 and Cyp6g2. The psiCHECK-2 system incorporates Renilla luciferase as a reporter gene and firefly luciferase as a control to normalize for transfection efficiency and cell number in the reporter gene assay. Constructs in the current study were designed to contain either the 3ʹ-UTR of wild-type Cyp6g1 or Cyp6g2, or a mutant version of each, cloned upstream of the Renilla luciferase reporter gene. For this aforementioned cloning, a partial region of the wild-type 3ʹ-UTR including target binding sites of miR-310s cluster for Cyp6g1 and Cyp6g2 were PCR amplified from 91-R isolated gDNA using the primers incorporating 5ʹ extensions with XhoI and NotI restriction endonuclease recognition sites (Table 2). All PCR reactions were carried out using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) with 91-R genomic DNA as the template. Each PCR product was purified using the QIAquick PCR Purification Kit (Qiagen) and then ligated into the pGEM-T Easy Vector Systems (Promega). The subsequent transformed clones were purified with a QIAprep Miniprep kit (Qiagen). After sequence analysis, the purified plasmid products were digested using the XhoI and NotI enzymes (New England Biolabs) and then cloned into the downstream of Renilla luciferase reporter gene in psiCHECK-2 plasmid vector (Promega). The putative miR-310s binding site within Cyp6g1 and Cyp6g2 3′-UTRs, TGCAAT, was altered to GTACTCT (ΔGTACTCT) using the Q5 Site‐Directed Mutagenesis Kit (NEB). Mutant Cyp6g1 and Cyp6g2 3ʹ-UTR Renilla luciferase reporter gene psiCHECK-2 constructs were generated as described above. All the constructs were verified by Sanger sequencing in both directions.
Drosophila Schneider 2 (S2) Cells from the Drosophila Genomics Resource Center (DGRC, Bloomington, IL) were obtained, and cultured in Schneider’s Drosophila medium (GIBCO, Rockville, MD) containing 10% fetal bovine serum (FBS; VWR, Radnor, PA), 50 units/ml penicillin, and 50 μg/ml streptomycin at 27 °C. The S2 cells were seeded at a density of 4 X 105 cells per well in 6-well plates and were cultured for 24 h to 80% confluence. The cells were divided into four groups for the following four treatments: (1) psiCHECK-2_3ʹ-UTR-WTTGCAAT + miR-310s mimics; (2) psiCHECK-2_3ʹ-UTR-WTTGCAAT + NCsiRNA (control); (3) psiCHECK-2_3ʹ-UTR- ΔGTACTCT + miR-310s mimics; and, (4) psiCHECK-2_3ʹ-UTR- ΔGTACTCT + NCsiRNA (control). S2 cells were co-transfected with 0.5 μg of each reporter construct and 100 nM miRNA of the mimics or NCsiRNA per well using the FuGENE HD Transfection Reagent (Promega). The firefly and Renilla luciferase activities were assayed with the Dual-Luciferase Assay System (Promega) after 48 h according to the manufacturer’s protocols. The Renilla luciferase activity was normalized by firefly luciferase activity. All experiments were repeated at least three times. Significant differences were determined based on ANOVA followed by Tukey's HSD multiple comparison test (p < 0.05).
DDT sensitivity bioassay
One-day-old female 91-R (90 flies per each injection) were injected separately with miR-310s mimics, NCsiRNA, and DEPC-water using microinjection methods described above and allowed one day for recovery. A 2.5 mg ml−1 solution of DDT dissolved in acetone was prepared, and then 0.2 µl (0.5 µg DDT per fly) was topically applied to the pronotum of female flies using a 50 µl glass micropipette (Hamilton 705SNR, Reno, NV) fitted in a repeating dispenser (Hamilton PB-600). Treated flies were transferred to 20 ml glass vials and capped with cotton plugs moistened with 5% sucrose solution in distilled water. Acetone-only treated flies were used as a control. The effect of DDT toxicity on flies was assessed to determine if the flies were unable to fly and crawl over the inner surface of the glass vial. Flies that remained immobile on the bottom of vial with slow leg twitching were considered dead. Median lethal time (LT50) was estimated for each treatment group based on the regression of Log10 time versus PROBIT percent mortality using the statistical software XLSTAT (Addinsoft). Each bioassay was repeated three times with independent microinjections. The three logistic regression curves were examined by F-test to determine whether any differences in the resulting mortality curves from differently treated fly groups were statistically significant (p < 0.01).
Analysis of putative transcription factor (TF)-binding-motifs of miR-310s
The putative promoter regions were analysed for the potential cis-acting elements and motifs of miR-310s cluster which could potentially account for expression changes in strain 91-R as compared to 91-C. The transcription start site (TSS) of the miR-310s cluster was predicted for the Drosophila genome release 6 by McPromoter61. A 480 bp region containing 200 bp up-stream and 280 bp down-stream of the predicted miR-310s cluster transcription start site (TSS) was excised from the Drosophila reference genome assembly release 6, in which putative regulatory element motifs were predicted using the JASPAR database release 7 online query tool62 with an applied relative profile score threshold of 95%. Default parameters of the ‘Map Reads to Reference’ tool of the CLC genomic workbench v.12.0 (Qiagen) were used to map Illumina short genomic sequence reads from 91-C and 91-R [SRA accession: SRX516723 and SRX516724]14 to the excised upstream regulatory regions of miR-310s cluster. The consensus nucleotide sequence of miR-310s cluster from each 91-R and 91-C were aligned with the corresponding region of the Drosophila reference sequence release 6 using the Alignments and Tree tool of the CLC Genomic Workbench (Qiagen), and polymorphism within putative cis-regulatory TF binding sites predicted by JASPAR were identified manually.
Supplementary information
Acknowledgements
This research was primarily supported by MSU Foundation Professor and AgBioResearch funds provided to BRP (MICL02503) and was a joint contribution from the United States Department of Agriculture (USDA) Agricultural Research Service (ARS) (CRIS Project 5030-22000-018-00D), and the Iowa Agriculture and Home Economics Experiment Station, Ames, IA (Project 3543). USDA is an equal opportunity employer and provider.
Author contributions
K.M.S designed the study and performed the experiments. K.M.S. analyzed the data and interpreted the results. K.M.S. and B.S.C. wrote the manuscript. B.R.P. supervised the whole work and revised the manuscript. All authors edited and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71250-0.
References
- 1.Ripper W. Effect of pesticides on balance of arthropod populations. Annu. Rev. Entomol. 1956;1:403–438. doi: 10.1146/annurev.en.01.010156.002155. [DOI] [Google Scholar]
- 2.Govella NJ, Okumu FO, Killeen GF. Insecticide-treated nets can reduce malaria transmission by mosquitoes which feed outdoors. Am. J. Trop. Med. Hyg. 2010;82:415–419. doi: 10.4269/ajtmh.2010.09-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esu E, Lenhart A, Smith L, Horstick O. Effectiveness of peridomestic space spraying with insecticide on dengue transmission; systematic review. Trop. Med. Int. Health. 2010;15:619–631. doi: 10.1111/j.1365-3156.2010.02489.x. [DOI] [PubMed] [Google Scholar]
- 4.Mallet J. The evolution of insecticide resistance: have the insects won? Trends Ecol. Evol. 1989;4:336–340. doi: 10.1016/0169-5347(89)90088-8. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien R, Matsumura F. DDT: a new hypothesis of its mode of action. Science. 1964;146:657–658. doi: 10.1126/science.146.3644.657. [DOI] [PubMed] [Google Scholar]
- 6.Kabasenche WP, Skinner MK. DDT, epigenetic harm, and transgenerational environmental justice. Environ. Health. 2014;13:62. doi: 10.1186/1476-069X-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson TG. Drosophila melanogaster (Diptera: Drosophilidae): a model insect for insecticide resistance studies. J. Econ. Entomol. 1988;81:22–27. doi: 10.1093/jee/81.1.22. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, et al. Identification and interaction of multiple genes resulting in DDT resistance in the 91-R strain of Drosophila melanogaster by RNAi approaches. Pestic. Biochem. Physiol. 2018;151:90–99. doi: 10.1016/j.pestbp.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Seong KM, Mittapalli O, Clark JM, Pittendrigh BR. A review of DDT resistance as it pertains to the 91-C and 91-R strains in Drosophila melanogaster. Pestic. Biochem. Physiol. 2019;161:86–94. doi: 10.1016/j.pestbp.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Gellatly KJ, et al. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 2015;121:107–115. doi: 10.1016/j.pestbp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Seong KM, Coates BS, Pittendrigh BR. Impacts of sub-lethal DDT exposures on microRNA and putative target transcript expression in DDT resistant and susceptible Drosophila melanogaster strains. Front. Genet. 2019;10:45. doi: 10.3389/fgene.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seong KM, Coates BS, Berenbaum MR, Clark JM, Pittendrigh BR. Comparative CYP-omic analysis between the DDT-susceptible and-resistant Drosophila melanogaster strains 91-C and 91-R. Pest Manag. Sci. 2018;74:2530–2543. doi: 10.1002/ps.4936. [DOI] [PubMed] [Google Scholar]
- 13.Seong KM, Sun W, Clark JM, Pittendrigh BR. Splice form variant and amino acid changes in MDR49 confers DDT resistance in transgenic Drosophila. Sci. Rep. 2016;6:23355. doi: 10.1038/srep23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steele LD, et al. Selective sweep analysis in the genomes of the 91-R and 91-C Drosophila melanogaster strains reveals few of the ‘usual suspects’ in dichlorodiphenyltrichloroethane (DDT) resistance. PLoS ONE. 2015;10:e0123066. doi: 10.1371/journal.pone.0123066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuler MA, Berenbaum MR. Structure and function of cytochrome P450s in insect adaptation to natural and synthetic toxins: insights gained from molecular modeling. J. Chem. Ecol. 2013;39:1232–1245. doi: 10.1007/s10886-013-0335-7. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt JM, et al. Insights into DDT resistance from the Drosophila melanogaster genetic reference panel. Genetics. 2017;207:1181–1193. doi: 10.1534/genetics.117.300310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David J-P, Ismail HM, Chandor-Proust A, Paine MJI. Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. B. Biol. Sci. 2013;368:20120429. doi: 10.1098/rstb.2012.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daborn PJ, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 19.Pedra JH, McIntyre L, Scharf M, Pittendrigh BR. Genome-wide transcription profile of field-and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seong KM, Coates BS, Pittendrigh BR. Cytochrome P450s Cyp4p1 and Cyp4p2 associated with the DDT tolerance in the Drosophila melanogaster strain 91-R. Pestic. Biochem. Physiol. 2019;159:136–143. doi: 10.1016/j.pestbp.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Seong KM, Coates BS, Kim DH, Hansen AK, Pittendrigh BR. Differentially expressed microRNAs associated with changes of transcript levels in detoxification pathways and DDT-resistance in the Drosophila melanogaster strain 91-R. PLoS ONE. 2018;13:e0196518. doi: 10.1371/journal.pone.0196518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta. Mol. Cell Res. 1803;1231–1243:2010. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Lucas K, Raikhel AS. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013;43:24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.25Asgari, S. in Advances in Insect Physiology: Crop Protection (ed Guy Smagghe) 19–45 (Academic Press, 2018).
- 26.Wei X, et al. miR-276 and miR-3016-modulated expression of acetyl-CoA carboxylase accounts for spirotetramat resistance in Aphis gossypii Glover. Insect Biochem. Mol. Biol. 2016;79:57–65. doi: 10.1016/j.ibmb.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Guo Q, et al. The role of miR-2~13~71 cluster in resistance to deltamethrin in Culex pipiens pallens. Insect Biochem. Mol. Biol. 2017;84:15–22. doi: 10.1016/j.ibmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. A microRNA-1 gene, tci-miR-1-3p, is involved in cyflumetofen resistance by targeting a glutathione S-transferase gene, TCGSTM4, in Tetranychus cinnabarinus. Insect Mol. Biol. 2018;27:352–364. doi: 10.1111/imb.12375. [DOI] [PubMed] [Google Scholar]
- 29.Etebari K, et al. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 2018;27:478–491. doi: 10.1111/imb.12387. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Guo L, Zhou X, Gao X, Liang P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.) Sci. Rep. 2015;5:1–9. doi: 10.1038/srep14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seong KM, Coates BS, Sun W, Clark JM, Pittendrigh BR. Changes in neuronal signaling and cell stress response pathways are associated with a multigenic response of Drosophila melanogaster to DDT selection. Genome Biol. Evol. 2017;9:3356–3372. doi: 10.1093/gbe/evx252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daborn PJ, Boundy S, Yen J, Pittendrigh BR, Ffrench-Constant R. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genomics. 2001;266:556–563. doi: 10.1007/s004380100531. [DOI] [PubMed] [Google Scholar]
- 33.Brandt A, et al. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst (2) DDT locus. Insect Mol. Biol. 2002;11:337–341. doi: 10.1046/j.1365-2583.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- 34.Qiu X, et al. Genome-wide analysis of genes associated with moderate and high DDT resistance in Drosophila melanogaster. Pest Manag. Sci. 2013;69:930–937. doi: 10.1002/ps.3454. [DOI] [PubMed] [Google Scholar]
- 35.Daborn PJ, Lumb C, Boey A, Wong W, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Joußen N, Heckel DG, Haas M, Schuphan I, Schmidt B. Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1-overexpressing strains of Drosophila melanogaster, leading to resistance. Pest Manag. Sci. 2008;64:65–73. doi: 10.1002/ps.1472. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y, Kim VN. MicroRNA factory: RISC assembly from precursor microRNAs. Mol. Cell. 2012;46:384–386. doi: 10.1016/j.molcel.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Denecke S, et al. Multiple P450s and variation in neuronal genes underpins the response to the insecticide imidacloprid in a population of Drosophila melanogaster. Sci. Rep. 2017;7:11338. doi: 10.1038/s41598-017-11092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, et al. RNA interference validation of detoxification genes involved in ivermectin tolerance in Drosophila melanogaster. Insect Mol. Biol. 2018;27:651–660. doi: 10.1111/imb.12512. [DOI] [PubMed] [Google Scholar]
- 40.Battlay P, et al. Structural variants and selective sweep foci contribute to insecticide resistance in the Drosophila Genetic Reference Panel. G3 (Bethesda) 2018;8:3489–3497. doi: 10.1534/g3.118.200619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt JM, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty M, Emerson J, Macdonald SJ, Long AD. Structural variants exhibit widespread allelic heterogeneity and shape variation in complex traits. Nat. Commun. 2019;10:4872. doi: 10.1038/s41467-019-12884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes. Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan H, et al. Nrf2/Maf-binding-site-containing functional Cyp6a2 allele is associated with DDT resistance in Drosophila melanogaster. Pest Manag. Sci. 2014;70:1048–1058. doi: 10.1002/ps.3645. [DOI] [PubMed] [Google Scholar]
- 46.Sun W, et al. A glycine insertion in the estrogen-related receptor (ERR) is associated with enhanced expression of three cytochrome P450 genes in transgenic Drosophila melanogaster. PLoS ONE. 2015;10:e0118779. doi: 10.1371/journal.pone.0118779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng T, et al. Reduced abundance of the CYP6CY3-targeting let-7 and miR-100 miRNAs accounts for host adaptation of Myzus persicae nicotianae. Insect Biochem. Mol. Biol. 2016;75:89–97. doi: 10.1016/j.ibmb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Tian M, et al. MiR-285 targets P450 (CYP6N23) to regulate pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 2016;115:4511–4517. doi: 10.1007/s00436-016-5238-4. [DOI] [PubMed] [Google Scholar]
- 49.Ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993;363:449–451. doi: 10.1038/363449a0. [DOI] [PubMed] [Google Scholar]
- 50.Heckel DG. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 2012;104:103–110. doi: 10.1016/j.pestbp.2012.05.007. [DOI] [Google Scholar]
- 51.Crow JF. Genetics of insect resistance to chemicals. Annu. Rev. Entomol. 1957;2:227–246. doi: 10.1146/annurev.en.02.010157.001303. [DOI] [Google Scholar]
- 52.Dapkus D, Merrell D. Chromosomal analysis of DDT-resistance in a long-term selected population of Drosophila melanogaster. Genetics. 1977;87:685–697. doi: 10.1093/genetics/87.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huan T, et al. Genome-wide identification of microRNA expression quantitative trait loci. Nat. Commun. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borel C, et al. Identification of cis-and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011;21:68–73. doi: 10.1101/gr.109371.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strycharz JP, et al. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic. Biochem. Physiol. 2013;107:207–217. doi: 10.1016/j.pestbp.2013.06.010. [DOI] [Google Scholar]
- 56.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 58.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 59.Kim Y-K, et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucl. Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 61.Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucl. Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.