Abstract
Cytopathological analyses of bronchial washings (BWs) collected during fibre‐optic bronchoscopy are often inconclusive for lung cancer diagnosis. To address this issue, we assessed the suitability of conducting molecular analyses on BWs, with the aim to improve the diagnosis and outcome prediction of lung cancer. The methylation status of RASSF1A, CDH1, DLC1 and PRPH was analysed in BW samples from 91 lung cancer patients and 31 controls, using a novel two‐colour droplet digital methylation‐specific PCR (ddMSP) technique. Mutations in ALK, BRAF, EGFR, ERBB2, KRAS, MAP2K1, MET, NRAS, PIK3CA, ROS1 and TP53 and gene fusions of ALK, RET and ROS1 were also investigated, using next‐generation sequencing on 73 lung cancer patients and 14 tumour‐free individuals. Our four‐gene methylation panel had significant diagnostic power, with 97% sensitivity and 74% specificity (relative risk, 7.3; odds ratio, 6.1; 95% confidence interval, 12.7–127). In contrast, gene mutation analysis had a remarkable value for predictive, but not for diagnostic, purposes. Actionable mutations in EGFR, HER2 and ROS1 as well as in other cancer genes (KRAS, PIK3CA and TP53) were detected. Concordance with gene mutations uncovered in tumour biopsies was higher than 90%. In addition, bronchial‐washing analyses permitted complete patient coverage and the detection of additional actionable mutations. In conclusion, BWs are a useful material on which to perform molecular tests based on gene panels: aberrant gene methylation and mutation analyses could be performed as approaches accompanying current diagnostic and predictive assays during the initial workup phase. This study establishes the grounds for further prospective investigation.
Keywords: early diagnosis, liquid biopsy, lung cancer, molecular test, therapeutic decision‐making
Diagnostic and predictive assessment remains major challenges in lung cancer management. In this work, we investigate the importance of molecular investigations applied to bronchial washings collected during fibre‐optic bronchoscopy for lung lesions. Using a cancer‐specific DNA methylation panel and gene mutations analyses, we demonstrated that bronchial washing represents a useful material for performing molecular tests in lung cancer patients.
Abbreviations
- Adks
adenocarcinomas
- BAL
bronchoalveolar lavage
- BW
bronchial washing
- ddMSP
digital methylation‐specific PCR
- ddPCR
droplet digital PCR
- FOB
fibre‐optic bronchoscopy
- LOD
limit of detection
- SqK
squamous carcinoma
- Ta
annealing temperature
1. Introduction
Lung carcinoma is the deadliest cancer worldwide (Siegel et al., 2016), with a 5‐year survival rate of 10–15%. Detection of early‐stage tumours is important for reducing mortality. However, most lung cancers are asymptomatic in the early stages, and even in more advanced disease, symptoms are nonspecific and cannot be distinguished from other, nontumour lung diseases (Rivera and Mehta, 2007). Lung cancer is therefore often detected late in its development.
Fibre‐optic bronchoscopy (FOB) is usually the first invasive procedure employed for the diagnosis of lung lesions, allowing physicians to exclude or confirm the presence of synchronous lesions in the airways and other mediastinal structures. Endoscopically, lung cancer lesions can be classified into two categories: visible (central or near‐the‐hilum cancer) and not visible (peripheral bronchoalveolar tumours or paratracheal tumours). FOB allows diagnosis of malignancy in 90% of visible lesions, in 60% of distal lesions and in < 30% of lesions < 2 cm in diameter (Ofiara et al., 2012; Thiberville and Salaun, 2010).
Bronchial washing (BW) is a safe, well‐tolerated procedure performed during FOB that allows the harvesting of cytological and histological samples for diagnostic typing. Unfortunately, it is not always possible to reach a definitive diagnosis with this technique, forcing patients to undergo further, potentially more invasive, biopsies.
Several studies have identified lung cancer‐specific DNA alterations in blood, serum, plasma, exhaled breath condensate, bronchoalveolar lavage (BAL) specimens and sputum (Belinsky et al., 2005; Han et al., 2009; Topaloglu et al., 2004). Indeed, aberrant DNA methylation was reported to be a suitable lung cancer biomarker in sputum (Belinsky et al., 2006) and in BAL specimens (Topaloglu et al., 2004). However, aberrant DNA methylation generally does not affect actionable genes, so the search for clinically actionable alterations has become part of the routine diagnostic workup essential for treatment decisions in lung cancer patients. On this point, liquid biopsy is clinically a highly valuable assay for the assessment of tumour heterogeneity, minimal residual disease and response to therapy in lung cancer (Molina‐Vila et al., 2016; Yoneda et al., 2019). Analysis of cell‐free DNA for the detection of EGFR, KRAS or TP53 mutations in BAL fluid has been reported (Li et al., 2014; Park et al., 2017). However, current guidelines do not recommend BAL as a routine approach for the diagnosis of peripheral lung lesions, given that the procedure may not be tolerated by all patients and may lead to complications during the process, so impairing the diagnostic efficacy of procedures with higher diagnostic power, such as transbronchial needle aspiration or biopsy. Conversely, BW is a routinely employed, less‐invasive procedure. To our knowledge, there are no studies on the detection of cancer‐gene mutations from BWs.
Thus, we investigated the suitability of using BW fluid in molecular analyses of lung cancer via the detection of cancer‐specific alterations of DNA methylation and gene mutations. To this end, we developed a novel, sensitive method based on droplet digital PCR (ddPCR) to detect traces of altered cancer‐specific DNA methylation; given the importance of identifying sensitive and specific diagnostic tools for nonvisible lung lesions, we assessed its ability to detect distal, or peripheral, lesions. Moreover, a panel of 12 cancer genes was used for the identification of actionable mutations, which is needed for therapy selection in advanced lung adenocarcinomas (Adks).
2. Methods
2.1. Patient cohort
One hundred twenty‐nine consecutive BWs were collected at the University Hospital of Ferrara, Italy, from patients undergoing FOB for suspected lung cancer. Almost all patients were current or former smokers. The study protocol was approved by the local ethics committee and the study methodologies conformed to the standards set by Declaration of Helsinki. All study participants provided a written informed consent for the use of their sample for research purposes. Clinical features of the cohort are reported in Table 1. The 31 controls were defined as benign cases based on several criteria, which included not only negative FOB, but also definitive pathological diagnosis from other more invasive samplings, as well as proven lesion reduction/resolution or lesion stability for at least 12 months upon clinical/radiological follow‐up. Seven metastatic patients were excluded from further statistical analyses and not included in Table 1.
Table 1.
Clinicopathological features of patients enrolled in the BW study. ND, not determined; NSCLC, nonsmall cell lung cancer; P/Y, packs of cigarettes per year; SCLC, small cell lung cancer.
| Cancer patients | Control patients | |
|---|---|---|
| Patients | ||
| Total a | 91 | 31 |
| Male | 60 | 21 |
| Female | 31 | 10 |
| Median age (range) | 71 (47–85) | 66 (42–86) |
| Smoke habits | ||
| Current smoker | ||
| > 10 P/Y | 25 | 8 |
| < 10 P/Y | 0 | 4 |
| Former smoker | ||
| > 10 P/Y | 42 | 11 |
| < 10 P/Y | 5 | 0 |
| Nonsmoker | 10 | 1 |
| Not known | 9 | 7 |
| Diagnosis | ||
| NSCLC: adenocarcinoma | 41 | |
| NSCLC: squamous cell carcinoma | 32 | |
| SCLC | 11 | |
| Lung cancer: undefined | 7 | |
| Inflammation | 18 | |
| Hyperplasia | 4 | |
| Squamous metaplasia | 3 | |
| Sarcoidosis | 2 | |
| Pneumonia | 1 | |
| Pleurisy | 1 | |
| Tuberculosis | 2 | |
| Stage | ||
| 1 | 13 | |
| 2 | 7 | |
| 3 | 25 | |
| 4 | 43 | |
| ND | 3 | |
Seven patients with metastases were excluded from the analyses.
2.2. Fibre‐optic bronchoscopy
During FOB, one or more BW samples were collected for cytological investigation. FOB was performed for diagnostic purposes based on the results of chest computer tomography and/or total body positron emission tomography. The endoscopic procedure was performed by introducing a bronchoscope (FB15V, Fb18V; Pentax Corporation, Tokyo, Japan) nasally while patients were in the supine position. Patients were sedated with midazolam (0.035 mg·kg−1 IV, with incremental doses of 1 mg being given as needed), and multiparameter monitoring (pulse oximetry, heart rate, blood pressure) conducted (Contoli et al., 2013). Local anaesthesia of the upper and lower respiratory tracts was achieved with lidocaine (10% spray and 2% solution). During all FOBs, BW fluid was collected in a trap by aspiration through the operating channel after instilling 20–40 mL of isotonic saline solution. In the case of visible lesions, the bronchoscope was positioned next to the tumour, wedging the tip of the bronchoscope into the segment where the lesion was located (van der Drift et al., 2005; Du Rand et al., 2013); here, an endobronchial biopsy was also taken. For nonvisible lesions, transbronchial needle aspiration, transbronchial lung biopsy or both, were performed under fluoroscopic guidance with the C‐arm system (Archovis Ing. Burgatti S.p.A, Bologna, Italy); biopsies obtained from these three procedures and the collected BW samples were eligible for pathological diagnosis.
2.3. DNA and RNA isolation
After collection, about 10 mL of the BW samples was immediately centrifuged. Cell pellets were stored at −80°C in the homogenization solution of the Maxwell miRNA Tissue kit (Promega, Madison, WI, USA) for days to a maximum of 8 weeks. DNA and RNA were isolated using the automated Maxwell system (miRNA Tissue kit; Promega), according to the manufacturer's instructions. DNAse was not added during the extraction, allowing us to obtain DNA and RNA from the same starting sample. Nucleic acids were quantified using the Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.4. DNA bisulphite conversion
DNA samples (500 ng) were preliminarily modified by chemical treatment with sodium bisulphite (Herman et al., 1996), using the EZ DNA Methylation Gold kit (Zymo Research, Irvine, CA, USA), according to the manufacturer's instructions. If the amount of DNA was < 500 ng, salmon sperm DNA was added up to a total of 500 ng. All bisulphite‐converted DNA was purified and collected in 40 μL of Tris–EDTA solution.
2.5. Droplet digital methylation‐specific PCR
Duplex ddPCR was designed for each gene locus by including two double‐quenched hydrolysis probes: one FAM‐labelled probe to recognize the nonmethylated sequence; and one HEX‐labelled probe to recognize the methylated sequence. Both probes were designed to anneal the same promoter region of each gene. To improve precision, double‐quenched probes (having a 3'IBFQ quencher and an internal ZEN quencher) were produced. Oligonucleotide and probe sequences are listed in Table S1 along with amplification conditions. Twenty microliter was used as the final volume per reaction. For each reaction, 10 µL of 2× Probes ddPCR Supermix (No dUTP; Bio‐Rad Laboratories, Hercules, CA, USA), 0.4 µL of primers (final concentration of 400 nm each), 0.2 µL of probes (final concentration of 200 nm each), and 1 µL of bisulphite‐treated DNA were used.
Each assay was performed in duplicate. The ddPCR assays were performed on a QX‐200 ddPCR System (Bio‐Rad Laboratories) as previously described (Ferracin et al., 2016). The amplification conditions were as follows: 95 °C for 10 min, 40 cycles at 94 °C for 30 s, annealing temperature (T a) for 1 min (see Table S1 for the T a of each locus‐specific reaction), followed by 10 min at 98 °C and a final hold at 4 °C. All ramping rates were reduced to 2 °C per second. After the amplification, the ddPCR assay was assessed by the droplet reader (Bio‐Rad Laboratories to analyse each droplet using a two‐colour detection system. The number of positive and negative droplets for each fluorophore (FAM and HEX) in each sample was counted with quantasoft software (Bio‐Rad Laboratories). Poisson distribution of the positive droplets was used to determine the absolute quantification of samples targets (initial copy number of each methylated gene) expressed as copies·µL−1. A result was considered positive when both duplicates exhibited the presence of HEX‐positive droplets.
Since DNA isolated from BW samples largely originates from exfoliated normal cells, the quantification of nonmethylated DNA by FAM‐labelled probes represented an internal quality control check and a quantification of the isolated DNA. The detection of at least 200 FAM‐positive droplets was required for the further analysis of the sample. The eventual absence of HEX‐labelled droplets was considered truly negative only in the presence of > 200 FAM‐positive droplets (limit of detection, LOD = 0.5%). In most cases, we obtained 1000–2000 FAM‐positive droplets (LOD = 0.05–0.1%; an example is shown in Fig. S1), which allowed us to readily assess the presence/absence of aberrant methylation at the RASSF1A, PRPH, DLC1, and CDH1 loci.
2.6. Cancer‐gene mutation analyses by NGS
Amplicon libraries were prepared using the Oncomine Lung Cell‐Free Total Nucleic Acid assay (Thermo Fisher Scientific) from 20 to 50 ng of total DNA/RNA isolated from BW samples. To identify the sample, each library was barcoded with a unique oligonucleotide identifier, according to the manufacturer's instructions. Libraries were pooled together in groups of 24/chip (Ion 540) and sequenced on the Ion S5 System (Thermo Fisher Scientific), achieving an average sequencing depth of 7000× (molecular coverage) and average reads number of 3 500 000 for sample. Sequencing raw‐data analysis was performed using torrent suite v. 5.10.1 and ion reporter 5.10.5 (Thermo Fisher Scientific). Briefly, low‐quality reads were removed, adapter sequences trimmed and samples sequence aligned against a reference genome (hg19) using the Torrent Mapping Alignment Program (Thermo Fisher, Carlsbad, CA, USA). Subsequently, the aligned BAM files were uploaded to Ion Reporter and processed using the ad hoc Oncomine TagSeq Lung v2 Liquid Biopsy w2.1 (Thermo Fisher)—Single Sample workflow. Each sample was analysed for mutations in the ALK, BRAF, EGFR, ERBB2, KRAS, MAP2K1, MET, NRAS, PIK3CA, ROS1, and TP53 genes and for ALK, ROS1, and RET gene fusions. Next‐generation sequencing (NGS) raw data are available in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the accession number PRJEB38273.
2.7. Statistical analyses
The presence or absence of methylation (discrete category variables) was analysed with Fisher's exact test. A P‐value < 0.05 was considered significant. Diagnostic accuracy of each marker, alone or in combination with others, was also evaluated. The diagnostic value of a marker was calculated as the number of correct answers versus the total number of samples analysed. Specifically, the value was obtained from the ratio between the true positives (lung cancer patients positive for methylation) and true negatives (noncancer patients negative for methylation) out of the total cases analysed.
3. Results
3.1. DNA methylation biomarkers for lung cancer in bronchial washings
After investigating tens of potential methylation biomarkers in lung cancer samples (data not shown), we selected the CpG islands of the CDH1, RASSF1A, PRPH and DLC1 gene promoters, because they displayed aberrant methylation in more than 50% of lung cancer samples and no aberrant methylation in normal tissues. We examined the methylation status of these CpG islands in BWs from 129 nonselected consecutive patients undergoing FOB.
Malignant cancer was diagnosed in 98 patients: 91 had a lung carcinoma and seven had lung metastases due to other types of neoplasm (mostly colorectal cancer). Aberrant DNA methylation was detected in six of the seven BW samples from patients with metastatic lesions, and albeit of potential interest, they formed a group that was too small to reach any significant conclusion. In addition, since the study was designed to investigate lung cancer, data from patients with metastases in the lung were excluded from the final statistical analyses to avoid the risk of potential data distortion. The remaining 31 patients had nonmalignant disorders, so were used as negative controls (see Table 1): the definitive diagnoses for these negative controls were obtained by histo‐pathological assessments as well as by proven lesion reduction/resolution or stability for at least 12 months upon clinical/radiological follow‐up.
Methylation status was analysed by two‐colour ddMSP. Aberrant methylation in at least one marker was present in 88 of 91 (97%) lung cancer patients. In samples from noncancer patients, RASSF1A, PRPH and DLC1 exhibited excellent specificity, whereas methylation at CDH1 was detected in eight of 29 (28%) cases (Fig. 1).
Fig. 1.
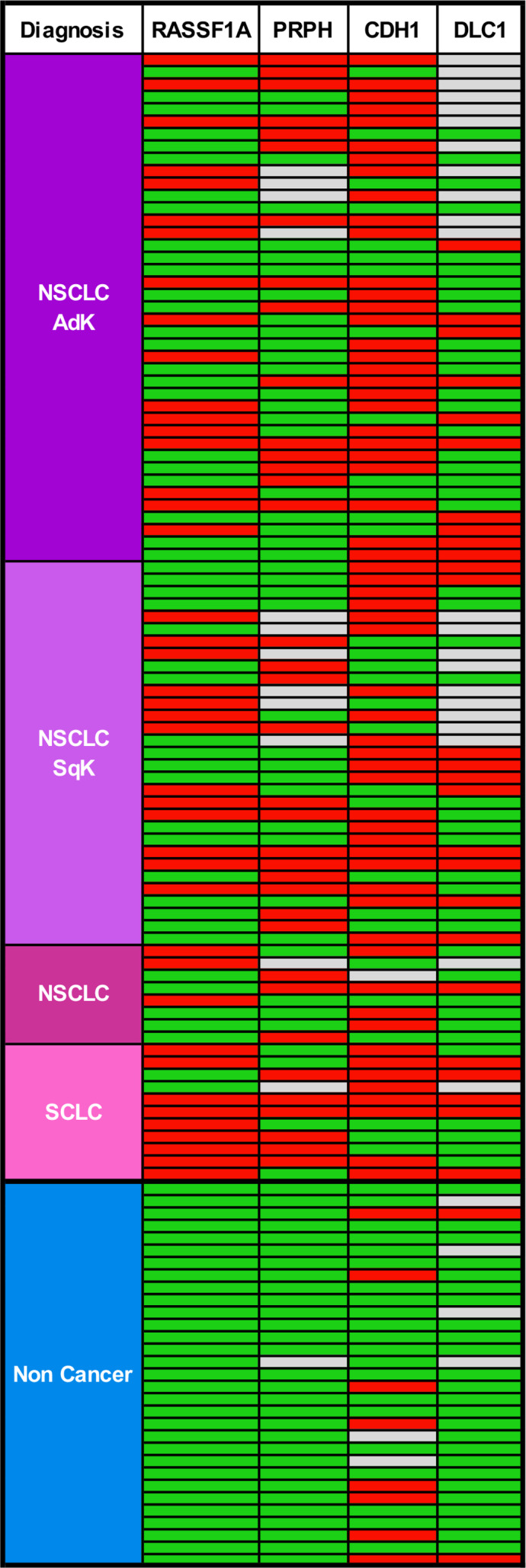
Biomarker methylation profile in BW fluid. Red boxes indicate the presence of methylated DNA, green boxes indicate nonmethylated DNA, and grey boxes indicate either not analysed samples or uninformative results. noncancer, samples from control patients; NSCLC, nonsmall cell lung cancer; SCLC, small cell lung cancer.
3.2. Diagnostic value of our four‐gene DNA methylation panel
By matching methylation results with final diagnoses, aberrant methylation at each locus was significantly associated with cancer (Table 2). For diagnostic purposes, the best result was achieved by the inclusion of RASSF1A, PRPH, DLC1 and CDH1 in a panel. The panel had a sensitivity of 97% and a specificity of 74% (overall diagnostic accuracy, 0.88). Positivity of the panel conferred a relative risk of 7.3, with a diagnostic odds ratio of 76.1 (95% CI, 18.6–312; Table 2). To perform receiver operating characteristic (ROC) curve analysis, we assigned a specific strength to each methylation marker on the basis of its own specificity (RASSF1A = 1.00; PRPH = 1.00; DLC1 = 0.94; CDH1 = 0.74), and then for each patient, the diagnostic strength was calculated as the sum of the four‐gene panel results. ROC curve analysis on these data gave an area under the curve of 0.93 and confirmed the excellent diagnostic power of the four‐gene panel (Fig. 2A).
Table 2.
Assessment of diagnostic value of DNA methylation markers. CI, confidence interval.
| Test | Sample type | Disease + (n) | Controls (n) | Fisher's exact test P‐value (two‐sided) | Sensitivity | Specificity | Relative risk (95% CI) a | Diagnostic odds ratio (95% CI) a | Diagnostic accuracy (%) | Positive predictive value (95% CI) | Negative predictive value (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytology | BW | 90 | 31 | < 0.0001 | 0.59 (0.49–0.69) | 1.00 (0.89–1.00) | 1.84 (1.48–2.28) | 89.9 (5.3–1516) | 69 | 1.00 (0.93–1.00) | 0.46 (0.33–0.58) |
| CDH1 | BW | 90 | 31 | 0.0001 | 0.64 (0.54–0.74) | 0.74 (0.55–0.88) | 1.51 (1.19–1.92) | 5.2 (2.1–13.0) | 68 | 0.88 (0.77–0.94) | 0.42 (0.29–0.56) |
| DLC1 | BW | 69 | 27 | 0.0027 | 0.37 (0.26–0.48) | 0.94 (0.79–0.99) | 1.48 (1.22–1.79) | 8.4 (1.9–37.8) | 53 | 0.94 (0.79–0.99) | 0.37 (0.26–0.48) |
| PRPH | BW | 79 | 30 | < 0.0001 | 0.40 (0.30–0.51) | 1.00 (0.89–1.00) | 1.57 (1.34–1.85) | 42.2 (2.5–712) | 56 | 1.00 (0.90–1.00) | 0.36 (0.27–0.48) |
| RASSF1A | BW | 91 | 31 | < 0.0001 | 0.46 (0.35–0.56) | 1.00 (0.89–1.00) | 1.63 (1.37–1.94) | 52.8 (3.1–890) | 60 | 1.00 (0.91–1.00) | 0.39 (0.28–0.50) |
| RASSF1A/ CDH1/ PRPH/ DLC1 | BW | 91 | 31 | < 0.0001 | 0.97 (0.91–0.99) | 0.74 (0.55–0.88) | 7.33 (2.54–21.2) | 76.1 (18.6–312) | 88 | 0.92 (0.84–0.96) | 0.87 (0.68––0.97) |
Since the values of the contingency table included a zero, the odds ratio was calculated by adding 0.5 to each value of the table.
Fig. 2.
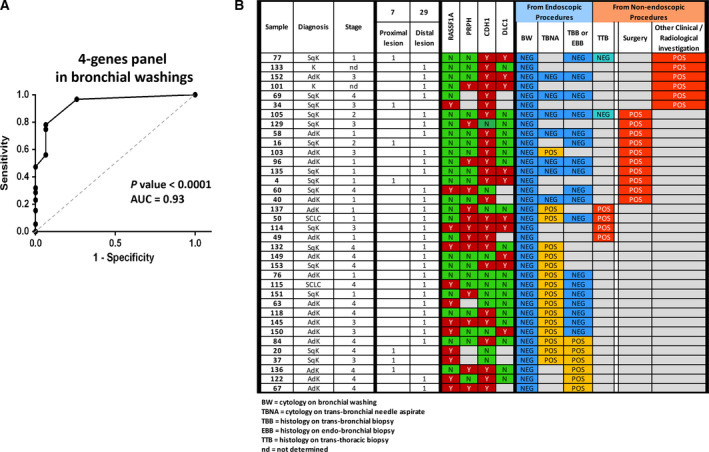
Diagnostic performance of the four‐gene panel in BW samples. (A) Receiver operating characteristic curve analysis, as described in the text. AUC, area under curve. The P‐value of the ROC curve was automatically calculated by the prism software (www.graphpad.com) to assess how close to 0.50, the null hypothesis, is the AUC. (B) The 36 lung cancer lesions missed by cytological analysis of BW samples. Twenty‐nine were from peripheral lesions. In these cases, the definitive diagnosis was achieved by transbronchial needle aspiration (TBNA) biopsy, transbronchial or endobronchial biopsy (TBB or EBB), transthoracic biopsy (TTB), or surgical intervention. In six cases, the diagnosis was based on other medical evidence. In 35 (97%) of these BW samples, the 4‐gene panel assay was positive for at least one of the markers (Y = positive; N = negative; empty (grey box) = not tested or uninformative results).
Bronchial washing samples, especially in the case of distal or peripheral lesions, are often inadequate for efficient diagnosis: indeed, we were not able to reach a definitive diagnosis in 36 patients (about 40% of our lung cancer patients, and 29 of whom had peripheral lesions) through cytological analysis of their BWs (Fig. 2B). To reach definitive diagnosis in these patients, more‐invasive and potentially risky procedures had to be performed (Fig. 2B). In contrast, analysis of methylation markers produced positive results in 35 of the 36 BW samples (Fig. 2B).
3.3. Predictive value of gene mutations assessed on BWs
We analysed DNA and RNA from 73 BW samples from cancer patients and 14 samples from noncancer patients. Results are summarized in Table 3 (details in Table S2). Mutations on EGFR, ERBB2 mutations and ROS1 fusions were found in patients diagnosed with AdKs, whereas KRAS, PIK3CA and TP53 mutations were detected in AdK as well as in squamous carcinoma (SqK) patients. Atypical mutations were found on BRAF (p.G469V) in one AdK patient and on ALK (p.R1275Q) in one AdK and two SqK patients. Analysis of the 14 noncancer patients revealed the presence of mutations on KRAS, PIK3CA and TP53 in nine individuals. The frequent mutation detection in noncancer patients revealed the low specificity of this approach for diagnostic purposes.
Table 3.
Detection of gene mutations in BWs.
| EGFR | ERBB2/HER2 | ROS1 fusions | ROS1 mutations | ALK fusions | ALK mutations a | KRAS | PIK3CA | TP53 | BRAF b | MET | MAP2K1 | NRAS | RET | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NSCLC | AdK | tot | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 |
| mut | 4 | 1 | 2 | 0 | 0 | 1 | 15 | 13 | 22 | 1 | 0 | 0 | 0 | 0 | ||
| % | 7.7 | 1.9 | 3.8 | 0.0 | 0.0 | 1.9 | 28.8 | 25.0 | 42.3 | 1.9 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| SqK | tot | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | |
| mut | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 9 | 14 | 0 | 0 | 0 | 0 | 0 | ||
| % | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.5 | 14.3 | 42.9 | 66.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Non tumour patients | tot | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | |
| mut | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | ||
| % | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.1 | 14.3 | 28.6 | 28.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
ALK mutation = p.R1275Q.
BRAF mutation = p.G469V.
In contrast, predictive value was valuable. To assess the predictive value of mutation analyses, we compared the results obtained from BWs with those from tumour biopsies. The comparison was only possible in the 52 AdK patients, since, as required by clinical guidelines, mutations in tumour samples are only investigated in that histological subtype. Details of tissue versus BWs are reported in Table S3 and summarized in Table 4. Where mutation data from tissue samples were available, the data from BWs revealed to be in good concordance with the matched tumour samples. The few differences included an EGFR mutation and an ALK fusion detected by IHC on tissue biopsies but missed in BWs, and two KRAS mutations detected in BWs but missed in tissue biopsies. A number of mutation analyses could only be performed on BWs, evidencing the presence of two additional EGFR mutations, seven KRAS mutations, and two different ROS1 fusions. Several mutations were also found on PIK3CA and TP53. These findings indicate that detection of actionable mutations is feasible for BWs. In addition to the above‐described concordance between BW analysis and tumour tissue mutational results, NGS on BWs has the advantage that it can be performed on all patients, including those for whom tumour tissue was missed, allowing the discovery of additional mutations eventually on actionable genes.
Table 4.
Concordance of mutations between tissue and BW in adenocarcinoma patients.
| Tissue | BW | Concordance (%) | Additional BWs | ||
|---|---|---|---|---|---|
| ALK fusions | Mut | 1 | 0 | 96 | 0 |
| WT | 27 | 28 | 24 | ||
| BRAF V600E | Mut | 0 | 0 | 100 | 1 |
| WT | 7 | 7 | 44 | ||
| EGFR | Mut | 3 | 2 | 97 | 2 |
| WT | 29 | 30 | 18 | ||
| ERBB2/HER2 | Mut | 1 | 1 | 100 | 0 |
| WT | 5 | 5 | 46 | ||
| KRAS | Mut | 5 | 7 | 90 | 7 |
| WT | 15 | 13 | 25 | ||
| MET | Mut | 0 | 0 | 100 | 0 |
| WT | 6 | 6 | 46 | ||
| ROS1 fusions | Mut | 0 | 0 | 100 | 2 |
| WT | 15 | 15 | 35 | ||
4. Discussion
At initial diagnosis, endoscopic FOB is the routinely used approach to obtain the biological material needed to perform cyto‐ or histo‐pathological analyses. Although BW is a low‐risk method for the collection of such samples, depending on the location of the primary tumour this technique is not always adequate for cytological diagnostic purposes. Thus, in about 40% of cases, more‐invasive approaches are needed to obtain biopsy material. To address this issue, molecular investigations on BWs could represent useful alternatives or additions for improving diagnostic performance and/or reducing the need of potentially risky procedures. Li and collaborators analysed mutations in BALs, finding that the combined detection of mutations on KRAS and TP53 yielded a sensitivity of 66% for the diagnosis of peripheral nonsmall cell lung cancer (Li et al., 2014). However, current guidelines do not recommend BAL as a routine approach for the diagnosis of peripheral lung lesions. Conversely, BW is a routinely employed, less‐invasive procedure. Thus, in the present work, we focused on BW, which, despite theoretically having a lower power for diagnosing peripheral lesions, is a procedure better tolerated by all patients. Founded on the diagnostic yield of methylation assays and the high concordance rate of mutational analysis as compared with pathology results, our findings demonstrate that BW specimens are suitable for diagnostic purposes, even for peripheral lesions.
In the present study, we investigated the use of DNA and RNA isolated from BW samples, evaluating diagnostic potential and predictive value. Our results indicate that BWs collected during FOB are excellent sample materials on which to generate useful information for diagnostic workup and predictive therapy indications based on molecular examinations. The employed methodological approaches, based on either ddMSP or NGS, exhibited an analytical sensitivity of 0.1% or lower, which is adequate to recognize the traces of nucleic acids originating from cancer cells.
For diagnostic purposes, we analysed tumour‐specific aberrant DNA methylation with a 4‐gene panel made up of CDH1, PRPH, RASSF1A and DLC1. Methylation at RASSF1A and CDH1 loci has been previously investigated in human lung cancer, either for their clinical pathological significance (Brock et al., 2008; Yu et al., 2015) or for their potential use as tumour biomarkers (Baryshnikova et al., 2008; Han et al., 2009; Hubers et al., 2014a; Hubers et al., 2014b; Hubers et al., 2015). In particular, RASSF1A methylation has been utilized because of its high specificity and early appearance in tumour development. By correlating the methylation results with final diagnoses, each locus exhibited very good specificity. Aberrant methylation on RASSF1A or PRPH exhibited 100% specificity, and on DLC1 an excellent 94% specificity. Only CDH1 exhibited less‐than‐optimal specificity with a value around 74%. Thus, our findings show that the assessment of aberrant DNA methylation on BW fluids could be a useful aid for initial lung cancer diagnosis. Nevertheless, although the RASSF1A, PRPH, CDH1 and DLC1 DNA methylation panel is highly sensitive and specific, it requires further improvement with the addition of other highly specific biomarkers.
More importantly, the four‐gene panel achieved excellent diagnostic accuracy: overall, the panel exhibited 88% diagnostic accuracy, 97% sensitivity and, largely because of CDH1, 74% specificity (RR, 7.3; diagnostic OR, 76.1). This diagnostic accuracy is greater than that obtained through traditional cytological analyses. Significantly, for the 36 BW samples from cancer patients that were inadequate for reaching a cytologically based diagnosis, results from methylation analysis provided a diagnostic indication of cancer in 35 (97%) of them, a result that could have potentially avoided more‐invasive interventions for these patients.
Most of the panel genes are not only tumour biomarkers, but are also pathogenically important. Loss of expression of CDH1‐, RASSF1A‐ and DLC1‐encoded proteins is relevant for pathogenic mechanisms that promote cell motility and metastatic potential. CDH1 encodes a calcium‐dependent cell adhesion protein [cadherin 1, type 1, E‐cadherin (epithelial)] whose loss can contribute to the metastatic potential of cancer cells (Kim et al., 2016). The protein encoded by RASSF1A binds to activated RAS to mediate apoptosis (Donninger et al., 2007; Gordon and Baksh, 2011; Grawenda and O'Neill, 2015). The protein encoded by DLC1 interacts with the DNA repair proteins XPA and BRCA2; it also inhibits the accumulation of cyclin D1 and induces cell cycle arrest. DLC1 (deleted in liver cancer 1) encodes for a GTPase‐activating protein (GAP) member of the rho‐GAP family of proteins: it can stop the signalling of RHOA, RHOB, RHOC and CDC42. Loss of DLC1 expression results in the constitutive activation of the rho‐GTPases, which promote increased cell motility and changes in morphology (Barras and Widmann, 2014; Popescu and Goodison, 2014). The role of PRPH in cancer is less clear: it encodes the cytoskeletal protein peripherin, a type III intermediate filament with homology to other cytoskeletal proteins such as desmin, and that is found in neurons of the peripheral nervous system. To date, aberrant PRPH methylation has been reported in two studies on liver cancer and neuroblastoma (Decock et al., 2012; Revill et al., 2013).
We also give evidence on the value of DNA/RNA analysis of BW specimens to direct therapeutic decisions. The use of highly sensitive approaches, such as ddPCR or NGS, is already being employed to detect traces of nucleic acids originating from tumour cells in liquid biopsies: they are performed mainly on plasma samples to monitor the patient's response to therapy. The present study highlights the possible use of BWs to provide predictive indications during a very early diagnostic phase. However, whether anticipating the start of treatment translates into clinically measurable benefits cannot be assessed here, as this would require an ad hoc trial.
When BW results were matched to molecular data obtained from AdK samples, the concordance was almost complete, with only one EGFR mutation and an ALK fusion missed. Most notably, in cases in which tumour tissues could not be analysed, additional mutations affecting the actionable genes EGFR and ROS1 were detected in BW samples. We also detected the atypical lung cancer mutations BRAF (p.G469V) and ALK (p.R1275Q), whose clinical significance should be further evaluated. Although at present the significance of these mutations in terms of response to target therapies is not known, the ALK mutation c.3824G>A (p.R1275Q), albeit infrequent in lung cancer, is commonly found in neuroblastoma, where this mutation covers about one third of all ALK mutations, which affect about 8–9% of the cases (Tate et al., 2019). The response to crizotinib or other ALK inhibitors is presently unknown, but preclinical studies have shown that R1275Q cell lines are sensitive to crizotinib (Bresler et al., 2014; Schonherr et al., 2011), and clinical trials investigating crizotinib in neuroblastoma are underway. Taken together, the findings of this study on aberrant methylation and cancer‐gene mutations strongly suggest that molecular analysis of BWs can indeed be part of the complete diagnostic and predictive workup in the very early phases of patient management.
In addition, although not specifically investigated in this study, the described approaches are potentially relevant for the early diagnosis of lung cancer. Early diagnosis, along with measures of primary prevention, could be effective in improving prognosis and reducing mortality due to lung cancer (Fleischhacker et al., 2013; Hubers et al., 2013; Langevin et al., 2015). A number of guidelines and recommendations for lung cancer screening exist (Bach et al., 2012; Jaklitsch et al., 2012; Smith et al., 2015). Individuals at risk of lung cancer have been recognized: these are aged 55–74 years old with a ≥30 packs/year smoking history, either currently smoking or who have quit within the past 15 years. However, no safe screening programme for the early detection of lung cancer is widely available yet (Kubik and Polak, 1986; Melamed et al., 1984). Low‐dose spiral computerized tomography has been suggested as a modality for lung cancer screening (Aberle et al., 2011), but limitations have been reported (Aberle et al., 2013). Thus, the need for more‐sensitive and specific approaches still exists. Can molecular investigations find application in screening programmes for the early detection of lung cancer? A number of studies have suggested the use of circulating microRNAs (Montani et al., 2015; Sozzi et al., 2014; Wang et al., 2015; Wozniak et al., 2015). Here, we show evidence that the analysis of the methylation status of a small gene panel has a very high diagnostic potential and could be used on BWs obtained periodically from individuals at risk.
Surprisingly, the present study identified mutations in cancer genes in several cancer‐free individuals. This finding is in line with recent studies reporting the presence of cancer‐gene mutations in different noncancer tissues (Lee‐Six et al., 2019; Martincorena and Campbell, 2015; Martincorena et al., 2018). Here, we identified mutations on PIK3CA, TP53, KRAS and ALK in individuals without cancer. ALK R1275Q, as discussed above, has an uncertain pathogenic significance in lung cancer. Similarly, the pathogenic significance of PIK3CA is also uncertain, judging from the unusually infrequent detection of mutations in lung cancer (2% in adenocarcinoma and 5% in SqKs). Conversely, TP53 is very frequently affected by mutations (40% in adenocarcinoma, 60% in SqK and 63% in small cell lung cancer), but it generally requires a double hit to become oncogenic. In our case series, most patients are or were heavy smokers, and mutations could have randomly accumulated on several genes in several cells. It should also be noted that these mutations are generally detected at a very low level, suggesting that they occur in a few disparate cells of the lung. Especially for TP53 mutations, the requirement of two inactivating mutations in the same cell suggests that the detected mutations might represent single hits. We speculate that these single‐hit mutations might confer an increased risk of developing cancer, but are insufficient to promote cancer initiation. Mutations in cancer genes, such as TP53, possibly represent molecular evidence of the increased risk of cancer that exists in all smokers. From a practical point of view, our findings indicate that the detection of point mutations on PIK3CA, TP53, and ALK in BWs have limited diagnostic power for lung cancer.
There were also two cases with KRAS mutations: a p.G12V mutation detected at 0.16% in patient C_059 and a p.G12D mutation at 3.3% in patient R_023. The first patient never developed a malignant lung lesion, suggesting that no additional alteration able to cooperate with the activated RAS was acquired. It is well established that multiple genetic/epigenetic changes are needed to promote a malignant phenotype. In the second patient, who incidentally exhibited a higher level of the mutant KRAS allele, a lung metastasis from a colon adenocarcinoma was diagnosed 3 years later. These findings suggest that the level of variants detected in cancer genes, especially if higher than 1%, should not be disregarded and a surveillance programme should be considered for these patients.
Alterations in certain other genes, namely EGFR, HER2 and ROS1, were instead only found in patients with cancer. Albeit relatively infrequent, these mutations might have diagnostic value. In support of this hypothesis, we observed that patient B_203 developed an lung adenocarcinoma about 1 year after an initially negative diagnosis, but a mutation on EGFR was already detected in the initial BW analysis. Although just a single case, this example suggests that the presence of mutations in cancer genes might identify individuals carrying a different risk of developing lung cancer, which puts forward the case, as indicated above, for the development of specific surveillance programmes for these individuals. To investigate this hypothesis, the present study suggests that molecular analyses conducted on BWs taken from at‐risk individuals should be thoroughly investigated through trials based on extensive case studies and accurate follow‐up.
5. Conclusions
Our study demonstrates that the use of BWs for molecular analyses is feasible. Methylation and gene mutation analyses could be performed to support and complete the current clinical diagnostic/predictive strategies.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
RR and LL performed targeted sequencing experiments and edited the manuscript; EM performed methylation experiments and statistical analyses; ES, SM, LM, DR provided clinical samples and data; CB performed bioinformatics analyses; DR, EC, VC, RR, GL, RG, AF provided clinical data and pathology review of the samples; AP, SS, FR, GLC and MN performed the study design, wrote and proofed the manuscript.
Supporting information
Fig. S1. Typical 1D and 2D plots for analysis of droplet fluorescence in RASSF1A methylation assay.
Table S1. ddPCR oligonucleotide and probe sequences, annealing temperatures and size products.
Table S2. Mutation analyses in bronchial washings from cohort study patients.
Table S3. Mutation analyses in bronchial washings and tissue biopsies from NSCLC adenocarcinoma patients.
Acknowledgements
We wish to thank Dr. Fernanda Mora for her excellent technical support. The work was supported by funding from the University of Ferrara and from the Italian Ministry of Health (Grant n. RF‐2016‐02363927) (to MN)
Roberta Roncarati, Laura Lupini and Elena Miotto equally contributed to this work.
Contributor Information
Silvia Sabbioni, Email: sbs@unife.it.
Massimo Negrini, Email: ngm@unife.it.
Data accessibility
All data are available in supplementary files. NGS row data will be uploaded in European Nucleotide Archive (ENA).
References
- Aberle DR, Abtin F and Brown K (2013) Computed tomography screening for lung cancer: has it finally arrived? Implications of the national lung screening trial. J Clin Oncol 31, 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD (2011) Reduced lung‐cancer mortality with low‐dose computed tomographic screening. N Engl J Med 365, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR et al (2012) Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 307, 2418–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras D and Widmann C (2014) GAP‐independent functions of DLC1 in metastasis. Cancer Metastasis Rev 33, 87–100. [DOI] [PubMed] [Google Scholar]
- Baryshnikova E, Destro A, Infante MV, Cavuto S, Cariboni U, Alloisio M, Ceresoli GL, Lutman R, Brambilla G, Chiesa G et al (2008) Molecular alterations in spontaneous sputum of cancer‐free heavy smokers: results from a large screening program. Clin Cancer Res 14, 1913–1919. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, Crowell RE, Karp DD, Stidley CA and Picchi MA (2005) Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res 11, 6505–6511. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y et al (2006) Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high‐risk cohort. Cancer Res 66, 3338–3344. [DOI] [PubMed] [Google Scholar]
- Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, Laudenslager M, Rappaport EF, Wood AC, McGrady PW et al (2014) ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 26, 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota‐Machida E, Han Y, Guo M, Ames S, Glockner S, Piantadosi S, Gabrielson E, Pridham G et al (2008) DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Contoli M, Gnesini G, Artioli D, Ravenna C, Sferra S, Romanazzi C, Marangoni E, Guzzinati I, Pasquini C, Papi A et al (2013) Midazolam in flexible bronchoscopy premedication: effects on patient‐related and procedure‐related outcomes. J Bronchology Interv Pulmonol 20, 232–240. [DOI] [PubMed] [Google Scholar]
- Decock A, Ongenaert M, Hoebeeck J, De Preter K, Van Peer G, Van Criekinge W, Ladenstein R, Schulte JH, Noguera R, Stallings RL et al (2012) Genome‐wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol 13, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donninger H, Vos MD and Clark GJ (2007) The RASSF1A tumor suppressor. J Cell Sci 120, 3163–3172. [DOI] [PubMed] [Google Scholar]
- Du Rand IA, Blaikley J, Booton R, Chaudhuri N, Gupta V, Khalid S, Mandal S, Martin J, Mills J, Navani N et al (2013) British Thoracic Society guideline for diagnostic flexible bronchoscopy in adults: accredited by NICE. Thorax 68(Suppl 1), i1–i44. [DOI] [PubMed] [Google Scholar]
- Ferracin M, Salamon I, Lupini L, Miotto E, Sabbioni S and Negrini M (2016) Circulating MicroRNA quantification using DNA‐binding dye chemistry and droplet digital PCR. J Vis Exp e54102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker M, Dietrich D, Liebenberg V, Field JK and Schmidt B (2013) The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med 7, 363–383. [DOI] [PubMed] [Google Scholar]
- Gordon M and Baksh S (2011) RASSF1A: not a prototypical Ras effector. Small GTPases 2, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawenda AM and O'Neill E (2015) Clinical utility of RASSF1A methylation in human malignancies. Br J Cancer 113, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Wang T, Reilly AA, Keller SM and Spivack SD (2009) Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir Res 10, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD and Baylin SB (1996) Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers AJ, Brinkman P, Boksem RJ, Rhodius RJ, Witte BI, Zwinderman AH, Heideman DA, Duin S, Koning R, Steenbergen RD et al (2014a) Combined sputum hypermethylation and eNose analysis for lung cancer diagnosis. J Clin Pathol 67, 707–711. [DOI] [PubMed] [Google Scholar]
- Hubers AJ, Heideman DA, Burgers SA, Herder GJ, Sterk PJ, Rhodius RJ, Smit HJ, Krouwels F, Welling A, Witte BI et al (2015) DNA hypermethylation analysis in sputum for the diagnosis of lung cancer: training validation set approach. Br J Cancer 112, 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers AJ, Prinsen CF, Sozzi G, Witte BI and Thunnissen E (2013) Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer 109, 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers AJ, van der Drift MA, Prinsen CF, Witte BI, Wang Y, Shivapurkar N, Stastny V, Bolijn AS, Hol BE, Feng Z et al (2014b) Methylation analysis in spontaneous sputum for lung cancer diagnosis. Lung Cancer 84, 127–133. [DOI] [PubMed] [Google Scholar]
- Jaklitsch MT, Jacobson FL, Austin JH, Field JK, Jett JR, Keshavjee S, MacMahon H, Mulshine JL, Munden RF, Salgia R et al (2012) The American Association for Thoracic Surgery guidelines for lung cancer screening using low‐dose computed tomography scans for lung cancer survivors and other high‐risk groups. J Thorac Cardiovasc Surg 144, 33–38. [DOI] [PubMed] [Google Scholar]
- Kim SA, Inamura K, Yamauchi M, Nishihara R, Mima K, Sukawa Y, Li T, Yasunari M, Morikawa T, Fitzgerald KC et al (2016) Loss of CDH1 (E‐cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br J Cancer 114, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik A and Polak J (1986) Lung cancer detection. Results of a randomized prospective study in Czechoslovakia. Cancer 57, 2427–2437. [DOI] [PubMed] [Google Scholar]
- Langevin SM, Kratzke RA and Kelsey KT (2015) Epigenetics of lung cancer. Transl Res 165, 74–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee‐Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, Georgakopoulos N, Torrente F, Noorani A, Goddard M et al (2019) The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537. [DOI] [PubMed] [Google Scholar]
- Li J, Hu YM, Wang Y, Tang XP, Shi WL and Du YJ (2014) Gene mutation analysis in non‐small cell lung cancer patients using bronchoalveolar lavage fluid and tumor tissue as diagnostic markers. Int J Biol Markers 29, e328–e336. [DOI] [PubMed] [Google Scholar]
- Martincorena I and Campbell PJ (2015) Somatic mutation in cancer and normal cells. Science 349, 1483–1489. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR et al (2018) Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed MR, Flehinger BJ, Zaman MB, Heelan RT, Perchick WA and Martini N (1984) Screening for early lung cancer. Results of the Memorial Sloan‐Kettering study in New York. Chest 86, 44–53. [DOI] [PubMed] [Google Scholar]
- Molina‐Vila MA, Mayo‐de‐Las‐Casas C, Gimenez‐Capitan A, Jordana‐Ariza N, Garzon M, Balada A, Villatoro S, Teixido C, Garcia‐Pelaez B, Aguado C et al (2016) Liquid biopsy in non‐small cell lung cancer. Frontiers in medicine 3, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, Bertolotti R, Bellomi M, Rampinelli C, Maisonneuve P et al (2015) miR‐Test: a blood test for lung cancer early detection. J Natl Cancer Inst 107, djv063. [DOI] [PubMed] [Google Scholar]
- Ofiara LM, Navasakulpong A, Ezer N and Gonzalez AV (2012) The importance of a satisfactory biopsy for the diagnosis of lung cancer in the era of personalized treatment. Curr Oncol 19, S16–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Hur JY, Lee KY, Lee JC, Rho JK, Shin SH and Choi CM (2017) Assessment of EGFR mutation status using cell‐free DNA from bronchoalveolar lavage fluid. Clin Chem Lab Med 55, 1489–1495. [DOI] [PubMed] [Google Scholar]
- Popescu NC and Goodison S (2014) Deleted in liver cancer‐1 (DLC1): an emerging metastasis suppressor gene. Mol Diagn Ther 18, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM and Powers S (2013) Genome‐wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 145, 1424–1435, e1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MP and Mehta AC (2007) Initial diagnosis of lung cancer: ACCP evidence‐based clinical practice guidelines. Chest 132, 131S–148S. [DOI] [PubMed] [Google Scholar]
- Schonherr C, Ruuth K, Yamazaki Y, Eriksson T, Christensen J, Palmer RH and Hallberg B (2011) Activating ALK mutations found in neuroblastoma are inhibited by Crizotinib and NVP‐TAE684. Biochem J 440, 405–413. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD and Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66, 7–30. [DOI] [PubMed] [Google Scholar]
- Smith RA, Manassaram‐Baptiste D, Brooks D, Doroshenk M, Fedewa S, Saslow D, Brawley OW and Wender R (2015) Cancer screening in the United States, 2015: a review of current American cancer society guidelines and current issues in cancer screening. CA Cancer J Clin 65, 30–54. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N et al (2014) Clinical utility of a plasma‐based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 32, 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E et al (2019) COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiberville L and Salaun M (2010) Bronchoscopic advances: on the way to the cells. Respiration 79, 441–449. [DOI] [PubMed] [Google Scholar]
- Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D and Moon CS (2004) Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res 10, 2284–2288. [DOI] [PubMed] [Google Scholar]
- van der Drift MA, van der Wilt GJ, Thunnissen FB and Janssen JP (2005) A prospective study of the timing and cost‐effectiveness of bronchial washing during bronchoscopy for pulmonary malignant tumors. Chest 128, 394–400. [DOI] [PubMed] [Google Scholar]
- Wang C, Ding M, Xia M, Chen S, Van Le A, Soto‐Gil R, Shen Y, Wang N, Wang J, Gu W et al (2015) A Five‐miRNA panel identified from a multicentric case‐control study serves as a novel diagnostic tool for ethnically diverse non‐small‐cell lung cancer patients. EBioMedicine 2, 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MB, Scelo G, Muller DC, Mukeria A, Zaridze D and Brennan P (2015) Circulating microRNAs as non‐invasive biomarkers for early detection of non‐small‐cell lung cancer. PLoS One 10, e0125026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda K, Imanishi N, Ichiki Y and Tanaka F (2019) A liquid biopsy in primary lung cancer. Surg Today 49, 1–14. [DOI] [PubMed] [Google Scholar]
- Yu Q, Guo Q, Chen L and Liu S (2015) Clinicopathological significance and potential drug targeting of CDH1 in lung cancer: a meta‐analysis and literature review. Drug design, development and therapy 9, 2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Typical 1D and 2D plots for analysis of droplet fluorescence in RASSF1A methylation assay.
Table S1. ddPCR oligonucleotide and probe sequences, annealing temperatures and size products.
Table S2. Mutation analyses in bronchial washings from cohort study patients.
Table S3. Mutation analyses in bronchial washings and tissue biopsies from NSCLC adenocarcinoma patients.
Data Availability Statement
All data are available in supplementary files. NGS row data will be uploaded in European Nucleotide Archive (ENA).


