Abstract
Weedy rice (Oryza spp.) is a worldwide weed of domesticated rice (O. sativa), considered particularly problematic due to its strong competition with the crop, which leads to reduction in yields and harvest quality. Several studies have established multiple independent origins for weedy rice populations in the United States and various parts of Asia; however, the origins of weedy rice in South America have not been examined in a global context. We evaluated the genetic variation of weedy rice populations in Colombia, as well as the contributions of local wild Oryza species, local cultivated varieties, and exotic Oryza groups to the weed, using polymorphism generated by genotyping by sequencing (GBS). We found no evidence for genomic contributions from local wild Oryza species (O. glumaepatula, O. grandiglumis, O. latifolia, and O. alta) to Colombian weedy rice. Instead, Colombian weedy rice has evolved from local indica cultivars and has also likely been inadvertently imported as an exotic pest from the United States. Additionally, weeds comprising de novo admixture between these distinct weedy populations now represent a large proportion of genomic backgrounds in Colombian weedy rice. Our results underscore the impressive ability of weedy rice to evolve through multiple evolutionary pathways, including in situ de‐domestication, range expansion, and hybridization.
Keywords: agricultural weed evolution, genotyping by sequencing, Oryza sativa, red rice
1. INTRODUCTION
The creation of the agricultural environment through crop domestication provided a new environment for opportunistic plants. Thus in the last 10,000 years, humans have had to contend with the continued evolution of agricultural weeds, which are currently a leading cause of crop losses worldwide (Oerke, 2006). Understanding how agricultural weeds arise and evolve is necessary to devise effective weed management strategies (Vigueira, Olsen, & Caicedo, 2013). Particularly intriguing from an evolutionary point of view are agricultural weeds that are related to crop species. How plant groups amenable to domestication through artificial selection can also give rise to plants that compete with crops and evade human efforts of removal has long been a question of interest, and may provide insight into mechanisms of plant adaptation (Ellstrand et al., 2010; Vigueira et al., 2013).
Among weeds related to crops, weedy rice (Oryza spp.) stands out in its worldwide occurrence and its impact on crop rice (Oryza sativa L.), which is the staple food for about half of the world's population (Prasad, Shivay, & Kumar, 2017). Weedy rice is a type of weedy Oryza that only infests cultivated rice fields, and is particularly difficult to control in systems employing direct seeding and continuous sowing. Around the world, weedy rice is known to be one among the most problematic and predominant weeds of cultivated rice (Singh et al., 2017; Watson, 2002). The negative effects of weedy rice include yield reduction, harvest quality deterioration, rice milling quality decrease, crop seed contamination, production cost increase, and land devaluation (Clavijo & Montealegre, 2010; Delouche et al., 2007). The importance and negative impact of weedy rice has been reported in many regions of the world, including Europe (Fogliatto, Vidotto, & Ferrero, 2012; Grimm, Fogliatto, Nick, Ferrero, & Vidotto, 2013; Messeguer, Marfà, Català, Guiderdoni, & Melé, 2004), Latin America (Arrieta‐Espinoza et al., 2005; Avila, Marchezan, & Menezes, 2000; Canal, Arnaude, Ortiz‐Domínguez, Valverde, & Fuentes, 2009; Clavijo & Montealegre, 2010; Federici et al., 2001), the United States (Burgos, Norman, Gealy, & Black, 2006; Delouche et al., 2007), and Asia (Cao, Li, Song, Cai, & Lu, 2007; Cao et al., 2006; Zhiwen et al., 2016). Weedy rice infestations have been found to have the capacity to reduce cultivated rice yields by up to 80% (Olsen, Caicedo, & Jia, 2007).
While it has often not been well‐defined taxonomically (Kraehmer, Jabran, Mennan, & Chauhan, 2016), in many countries weedy rice is classified in the same genus and species as cultivated rice. In fact, a distinguishing feature of weedy rice is that it presents similar morphological and physiological characteristics to those of the crop, as well as similar nutritional and agro‐climatic requirements (Ellstrand et al., 2010; Valverde, 2005). However, unlike the crop, it has negative characteristics such as seed shattering (dispersal), red pericarp and associated seed dormancy, rapid growth, and efficient resource use, which make it a very competitive and successful plant (Delouche et al., 2007).
The occurrence of weedy rice in multiple and different regions of the world suggests parallel evolutionary events with independent origins. In fact, in recent years, genomic studies have been clarifying the extent to which weedy rice is an example of convergent evolution. Thus, for example, weedy rice from Northeast Asia has been found to have evolved from japonica rice varieties, which are the typical cultivars grown in colder regions in subtropical and temperate zones (Vigueira et al., 2019). Some weedy rice populations in South Asia and in South‐East Asia have evolved from indica cultivars, which are typically cultivated in lowland tropical and subtropical regions (Huang et al., 2017; Song, Chuah, Tam, & Olsen, 2014). Other weedy populations from South Asia have been found to have origins from aus cultivated varieties, which are typically grown in Bangladesh, or from local wild rice (O. rufipogon Griff., the ancestor of cultivated rice) (Huang et al., 2017, 2018). The origin of weedy rice in different regions of the world is thus often influenced by the prevailing local variety and/or the availability of reproductively compatible wild Oryza (Cao et al., 2006; Ishikawa et al., 2005; Olsen et al., 2007).
In the world, Colombia ranks 26th among rice‐producing countries and is third in Latin America after Brazil and Peru. Rice is the fourth most important crop in Colombia, after sugarcane, oil palm, and banana. However, rice cultivation in Colombia has reported losses from 30% to 73% due to weeds (Cobb & Reade, 2010). A major weed of cultivated rice in Colombia is weedy rice, for which the presence of 24 plants per m2 in early‐growth stages (40 days after emergence) can reduce crop yields by 50% (Fischer & Ramírez, 1993). Despite its economic importance, no systematic survey of the evolutionary origins of weedy rice has yet been carried out in Colombia.
Several Oryza types could be possible sources of weedy rice in Colombia. The main varieties of cultivated rice grown are routinely released by Fedearroz (National Federation of Rice Growers) and are classified morphologically as indica. A few areas in the western part of the country, collectively known as “Bajo Cauca,” are also cultivated with traditional landraces (i.e., “criollos”), whose classification is unknown. Additionally, Colombia and other countries of Central and South America are unique in the occurrence of several native species of wild Oryza, different from those found in Asia, and these could play a role in weedy rice evolution. These wild species are as follows: O. glumaepatula Steud. (also known as O. glumipatula Steud., diploid, AA genome, 2n = 24, and included in the O. sativa complex), and O. latifolia Desv., O. grandiglumis (Döll) Prod., and O. alta Swallen (all tetraploid, with CCDD genomes, 2n = 48, and included in the O. officinalis Wall. complex.) (Veasey et al., 2011). In Colombia, native wild Oryza species grow mainly in patches of untreated vegetation bordering commercial rice crops; they are found in humid soils, wetlands, and streams, soils with high organic matter content, and can also appear in zones distant from agricultural areas (Villafañe, Estrada, Lentini, Fory, & Palacio, 2007). Oryza latifolia is the species with the greatest distribution, whereas O. grandiglumis has the most restricted range (Estrada, Villafañe, Palacio, Fory, & Lentini, 2010). As with most wild Oryza species, this South American Oryza has traits that are often found in weedy rice, such as red pericarp color, shattering, and seed dormancy.
The plethora of possible contributors to weedy rice in Colombia make the country an interesting case study to contrast with weedy rice evolution in other world regions. Understanding how weedy rice emerges in and adapts to agricultural environments is a necessary first step for designing control strategies for this pest (Vigueira et al., 2013). To determine the origins of weedy rice in Colombia, we have carried out a thorough sampling of weedy rice diversity in the country, as well as sampling of local cultivars and wild species. Here, we use genome‐level information to understand the relationships between Colombian weedy rice and local Oryza. We further examine the origins of Colombian weedy rice within an international context, to attempt to understand how the weed evolves and diversifies at a global scale.
2. MATERIALS AND METHODS
2.1. Plant material and DNA extraction
Weedy rice seeds were collected by sampling in the five principal rice production areas of the country in 2014–2015 (Figure 1). Twenty‐six accessions were collected in the Central zone, 34 in the Llanos plains, 28 in the Bajo Cauca river valley, 26 in the North coast, and 26 in the Santanderes area, for a total of 140 Colombian weedy rice (CWR) accessions. These were additionally classified according to hull color (lemma and palea), pericarp color, grain size, and awn presence, abundance, and length. Nine varieties of commercial rice were supplied by the National Federation of Rice Growers (Fedearroz), which included the following: four varieties that are currently cultivated and have been in the market for 10–17 years (F473 cultivated in the low Cauca valley area, F2000 in Santanderes and North coast zones, F174 in the Eastern plains, and F60 in the Central zone), one variety with national relevance and cultivated for approximately 17 years (F50), and four varieties that have left the market but were important over a 25‐year period (Cica 6, Cica 8 and Cica 9 and Orizica1). Additionally, 19 landraces (traditional crop with manual dry seeding system, present in the low Cauca valley area) that belong to the germplasm bank of the Fedearroz Monteria section were sampled. We also obtained seeds of 22 wild South America Oryza from the International Rice Research Institute (IRRI); these included seven O. glumaepatula, six O. grandiglumis, five O. alta, and four O. latifolia (Table S1).
FIGURE 1.
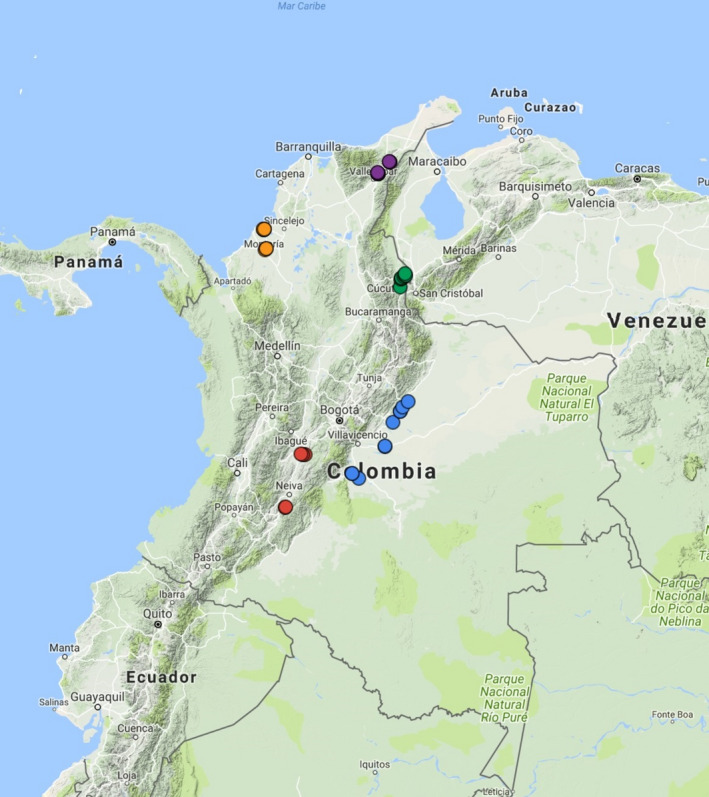
Map of Colombia showing weedy rice collection areas used in this study. Central zone (red dots), Llanos plains (blue dots), Bajo Cauca river valley (orange dots), North coast (purple dots), and Santanderes area (green dots)
Staggered sowing of all selected materials (CWR, commercial rice and landraces) was carried out in the Weed Science greenhouse at Universidad Nacional de Colombia. In order to break dormancy, weeds were exposed to 50°C for 12 hr (materials that did not germinate under this treatment were kept at 50°C for 72 hr). South American wild rice materials were planted in the Biology greenhouse at the University of Massachusetts, following a dormancy breaking treatment at 50°C for 5 days. 200 mg of young leaf tissue was used for DNA extraction with the Qiagen DNeasy® Plant Mini Kit. The concentration and purity of the DNA in the samples was quantified with a Qubit 2.0 Fluorometer; 30 μl of DNA with a concentration of 30–100 ng/μl for each sample was sent to the Cornell University Biotechnology Resource Center (BRC) for genotyping by sequencing (GBS).
2.2. GBS library preparation and sequence analysis
Genotyping by sequencing was used to detect polymorphisms distributed throughout the genome among our samples (Elshire et al., 2011). Restriction digestions were carried out with the enzyme ApeKI, and the fragments were ligated with individual barcoded and common adapters. DNA fragments were pooled for further PCR amplification to enrich the libraries. Single‐end fragments of 100 base pairs (bp) were sequenced on an Illumina HiSeq 2500 platform. Raw GBS data have been deposited in the NCBI SRA (experiment SUB6244405). Initial data processing was performed at the Biotechnology Institute of Cornell University with the TASSEL‐GBS v.3.0 pipeline (Glaubitz et al., 2014) and the MSU7 rice reference genome. The first filters removed SNPs with minimum minor allele frequency <0.01 or missing data per site >90%. Additional filters were applied to the initial VCF file with the NGSEP pipeline (Duitama et al., 2014) at the University of Massachusetts. Final SNPs were supported by at least five reads (for the analysis of Colombian material) or three reads (for the global analysis), and displayed heterozygosity levels of <50%. SNPs with more than 15% missing data and individuals with more than 70% missing data were removed. Only biallelic SNPs were retained, filtering any other type of variants. The final VCF has been deposited in DRYAD.
Data generated for Colombian and South American samples were integrated with other published GBS databases that included 128 cultivars, 173 weedy rice samples, and 53 samples of the wild ancestor of cultivated rice (O. rufipogon/O. nivara) from Asia (Huang et al., 2017; Vigueira et al., 2019); nine cultivars and 17 weedy rice samples from the United States (Burgos et al., 2014); and four out‐group samples (O. meridionalis and O. barthii) from these same datasets (NCBI Short Read Archive experiment SRX576894), for a total of 574 accessions (Table S1).
2.3. Population analyses
Population structure was analyzed using STRUCTURE (version 2.3.3, Hubisz, Falush, Stephens, & Pritchard, 2009), on the Massachusetts Green High Performance Computing Cluster (http://www.mghpcc.org/). Due to the limitation in the amount of input data that can be handled by the program (Falush, Stephens, & Pritchard, 2003; Pritchard, Stephens, & Donnelly, 2000), approximately 10,000 SNPs were randomly selected for each analysis with a roughly 15,000 base pairs (bp) spacing. Heterozygotes were recorded as “N,” and all the simulations were run with data coded as haploid, because weedy and cultivated rice are highly self‐pollinated. The program was run for an ancestral “admixture” model, with no correlated allele frequencies. Runs were carried out using K values between 1 and 15, with three replicates per K, a burn‐in period of 100,000 and 500,000 subsequent iterations. The optimal number of genetic groupings was determined using ΔK (Evanno, Regnaut, & Goudet, 2005) according to the program Structure Harvester (Earl & vonHoldt, 2012). The program CLUMPP (Jakobsson & Rosenberg, 2007) was used to obtain a single Q matrix for each K. The final matrix for each K value was visualized with Distruct (Rosenberg, 2004). For comparison, we also analyzed our complete SNP dataset for worldwide samples with the Bayesian clustering analysis fastStructure (version 1.0, Raj, Stephens, & Pritchard, 2014), without recoding heterozygotes, with no prior grouping. FastStructure runs were conducted for K from 2 to 8. The best K was determined through chooseK.py, and the POPHELPER R package was used to generate an image (Francis, 2017).
To investigate the genetic divergence among individuals for all SNPs, the program SmartPCA from the EIGENSOFT package (Patterson, Price, & Reich, 2006; Price et al., 2006) was used. Figures with eigenvalues as coordinates were generated by RStudio 1.0.143.
To infer the phylogenetic relationships among samples, RAxML (Randomized Axelerated Maximum Likelihood) version 8 (Stamatakis, 2014) was used. The RAxML HPC2 on XSEDE tool was selected in the CIPRES portal (http://www.phylo.org/), with GTRGAMMA model and 1,000 bootstraps. Because SNP data only present variable sites, ascertainment bias correction (ASC) was performed. The best phylogenetic tree result was plotted using iTol v4 (Letunic & Bork, 2016).
Genetic diversity for each population was measured by evaluating the expected heterozygosity calculated for all loci, and paired FST was used to estimate the genetic differences among populations with the software ARLEQUIN (ver 3.5.2.2., Excoffier & Lischer, 2010). Additionally, an AMOVA was performed to analyze variation among and within populations.
2.4. Shattering (sh4) and pericarp color (Rc) gene sequencing
Seventy‐one weedy rice accessions from the five rice production areas in Colombia, five commercial rice, and ten landraces, for a total of 96 accessions, were selected for candidate gene analysis. Additionally, for the Rc gene, four accessions of wild diploid materials (O. glumaepatula) were sequenced, and for sh4, publicly available sequences of O. glumaepatula (GU221016, GU221017, and DQ421813) were included for comparison.
A portion of the second exon of sh4 gene was amplified using primers designed with Primer3 (Untergasser et al., 2012), based on the PopSet by Thurber et al. (2010) (sh4_01_for: 5′‐ACGGGCACCTGACTGCTACG‐3′; sh4_01_rev: 5′‐GAGGTGGGTGGTGGTGATGG‐3′), yielding a band size of ~678 bp. PCR was performed in a 25 μl volume containing 2 μl of genomic DNA template (5 ng/μl), 4 µl of dNTPs (2.5 mM), 0.50 µl of each primer (10 µM), 12.5 µl of 2X GC Buffer I, 0.25 µl of TaKaRa LA Taq DNA polymerase (5 U/µl; Takara, Shiga, Japan), and 0.50 µl of DMSO. PCR amplification was carried out by initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, 61°C annealing for 30 s, and 72°C elongation for 2 min, and a final extension of 72°C for 5 min.
Exon 7 of Rc was amplified with primers designed off the PopSet by Gross et al. (2010), Rc_02_for (5′‐AGTGGCATCACCTGAAAATACC‐3′) y Rc_02_rev (5′‐CGGCTTTATAGAAATAGAGGGAGT‐3′), yielding a band size of ~509 bp. PCR amplifications were carried out in a 20 μl reaction mixture containing 2 μl of template DNA (5 ng/µl), 1.6 μl of dNTPs (2.5 mM), 1 μl of each primer (10 µM), 2 μl of 10X Ex Taq Buffer, and 0.1 μl of TaKaRa Ex Taq (5 U/µl). The PCR program consisted of an initial denaturing step at 95°C for 3 min, followed by 30 cycles of denaturation 95°C for 1 min, annealing at 54°C for 30 s, and extension at 68°C for 30 s, with a final extension at 72°C for 10 min.
Sanger sequencing was carried out by either GENEWIZ or Beckman. DNA sequences were aligned and edited with the software Geneious (ver 9.1.3., http://www.geneious.com, Kearse et al., 2012). Sequence alignments have been deposited as PopSets in NCBI GenBank. Rc sequences were examined for the presence/absence of the 14‐bp deletion present in exon 7 associated with loss of pericarp color; sh4 sequences were examined for the occurrence of the G/T single nucleotide substitution in exon 2 associated with loss of shattering during domestication. Gene trees were constructed under the Tamura‐Nei genetic distance model and the neighbor‐joining (NJ) method in the program Geneious. For the sh4 gene, three O. glumaepatula DNA sequences deposited in the GenBank, accessions GU221016 and GU221017 reported by Thurber et al. (2010) and DQ421813 reported by Li, Zhou, and Sang (2006), were included. Trees were plotted in iTol v4 (Letunic & Bork, 2016).
Seed shattering and pericarp color were assessed for sequenced accessions. The determination of pericarp color was visually performed at the mature stage of the grain taking into account the intensity of the color. When there were doubts about the color due to the differences in weedy rice maturity in the field, the 2% potassium hydroxide (KOH) test was performed (Louisiana State Seed Testing Laboratory, 1980; Rosta, 1975), immersing dehulled seeds in this solution for 10–20 min. After this time, the KOH solution turns red or dark orange if the seeds are red, while the white seed shows a light golden yellow color or remains colorless. Shattering measures were taken 30 days after flowering using a digital force gauge to measure breaking tensile strength (BTS), and averaging values for 10 seeds in the same panicle (Thurber et al., 2010). Statistical analysis was performed by analysis of variance (ANOVA) of one factor, using the statistical package SPSS version 23. The comparison of means was performed by the Tukey test at p < .05.
3. RESULTS
3.1. Colombian weedy rice origins
A total of 47,200 high‐quality SNPs were obtained from GBS for the 190 Colombian and wild South American Oryza samples. When all materials are included, a population structure analysis based on 11,808 SNPs suggests the presence of two differentiated populations (highest ΔK at K = 2; Evanno et al., 2005; Figure 2a; Tables S2 and S3). One population includes all Colombian weedy rice (CWR) and commercial rice varieties; the second corresponds to native tetraploid wild Oryza species (O. grandiglumis, O. alta, and O. latifolia). Landraces and O. glumaepatula appear as a mixture of these populations. These results are congruent with a principal component analysis (PCA) on all SNPs, which shows separate clusters for tetraploid wild Oryza species with CCDD genome (O. grandiglumis, O. alta, and O. latifolia), the diploid wild species (O. glumaepatula), and CWR with commercial rice and landraces (Figure 2b, Table S4).
FIGURE 2.
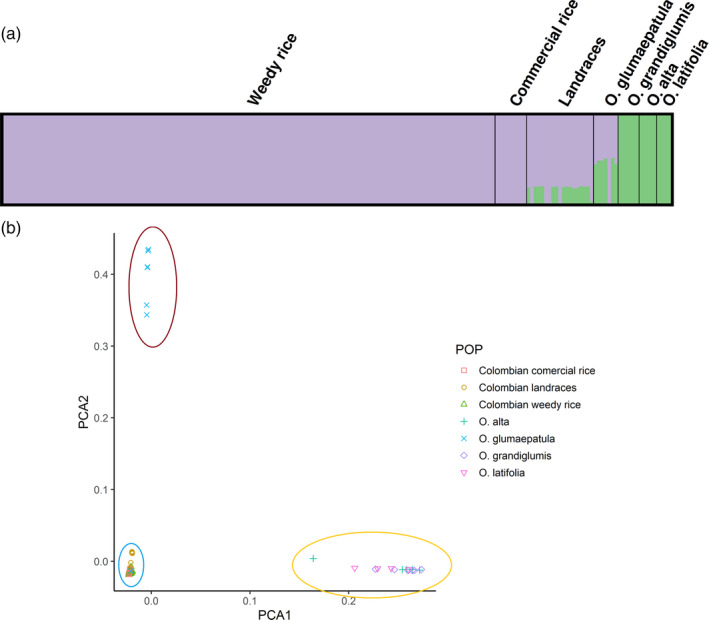
Population analysis of 190 Oryza accessions (140 Colombian weedy rice, 9 commercial rice lines from Colombia, 19 landraces from Colombia, 7 O. glumaepatula, 6 O. grandiglumis, 5 O. alta, and 4 O. latifolia). (a) Population structure, each individual is represented by a color bar, with color partitions that reflect the relative proportion of genetic membership in a given group (K = 2). (b) Principal component analysis
Both population structure and PCA suggest no contribution of native wild Oryza to CWR; thus, we excluded these groups and re‐analyzed population structure with the available 10,923 SNPs. Three ΔK peaks were observed, at K = 2, K = 5, and K = 7, with the highest occurring at K = 7 (Figure 3, Tables S5 and S6). Under the progressive models, Colombian landraces are highly differentiated from CWR and commercial rice, and the commercial varieties make contributions to a heterogeneous collection of CWR samples. These analyses suggest that the sources of remaining CWR genomic heterogeneity are from unsampled material, perhaps from sources outside of Colombia.
FIGURE 3.
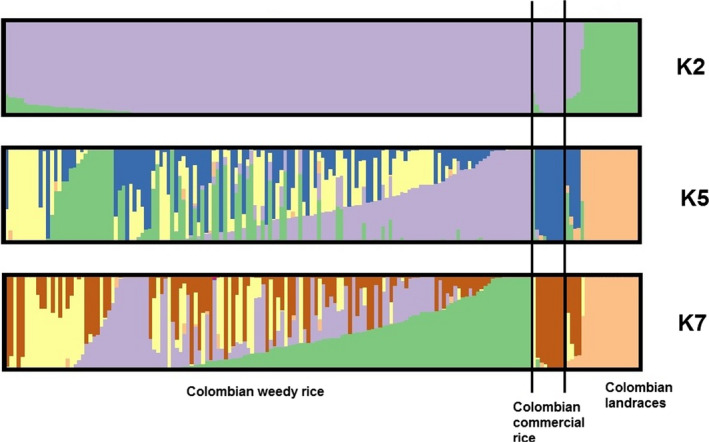
Population structure of 168 Colombian Oryza accessions (140 weedy rice, 9 commercial rice, and 19 landraces). Each individual is represented by a color bar, with color partitions that reflect the relative proportion of genetic membership in a given group (K = 2, K = 5, and K = 7)
To further investigate the origin of CWR, we obtained high‐quality SNPs from merged data from this study and from published GBS data of global samples of weedy rice, cultivated rice, and the wild ancestor of cultivated rice, a species complex known as O. rufipogon/nivara, and carried out a population structure analysis on 12,284 SNPs. We excluded wild rice from South America and out‐groups. The highest ΔK value was obtained for K = 2, with also substantially high values for K = 3–4 (Figure 4a, Tables S7 and S8). At K = 2, indica + aus and japonica cultivated lineages are clearly differentiated, showing that CWR and Colombian commercial varieties most resemble indica cultivars, a common cultivar group in the tropical lowlands of Asia, while most Colombian landraces used in the last 400 years resemble japonica. Because the Evanno method can underestimate K when there is a hierarchical structure of the population (Waples & Gaggiotti, 2006), we examined models with K = 3 and K = 4. These models are consistent population structure detected for US and Asian weedy rice (Huang et al., 2017; Reagon et al., 2010; Vigueira et al., 2019) and readily distinguish the three main groups of cultivated groups of rice, indica, aus, and japonica, with K = 4 also distinguishing the wild ancestor of cultivated rice (Figure 4a). With this higher resolution, we see that CWR has admixture, with contributions primarily from indica and aus cultivated varieties (and very occasionally from japonica; Figure 4), and shares resemblance with other weed groups around the world. A fastStructure run based on 21,664 SNPs, for which a K value of 5 maximized the marginal likelihood, gave congruent results (Fig. S1, Table S9). Based on these results, we classified all CWR samples according to predominant ancestry (>80% contribution from a single group), resulting in 5 aus‐like individuals, 85 indica‐like individuals, and 50 admixed indica‐aus individuals. Interestingly, these CWR types seem distributed in all the rice‐growing regions of Colombia (Figure 4b).
FIGURE 4.
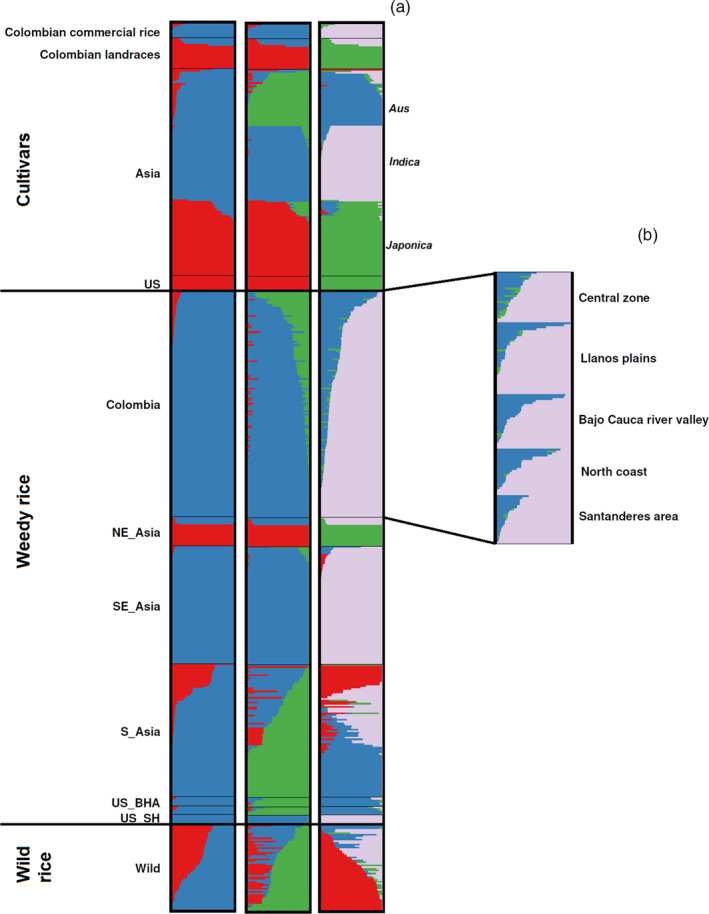
Population structure of 548 Colombian and global Oryza accessions. Each individual is represented by a colored bar, with color partitions that reflect the relative proportion of genetic membership in a given group. (a) Results of the analysis of Colombian accessions (140 weedy rice, 9 commercial rice, and 19 landraces), Asian (128 cultivars, 173 weedy rice, and 53 wild rice), and United States (9 cultivars and 17 weedy rice) at K = 2, K = 3, and K = 4 populations. (b) Population structure of Colombian weedy rice according to the rice geographic areas at K = 4
The ancestral contributions in CWR could stem directly from local (Colombian) cultivars that have undergone de‐domestication, or from dispersal of weedy rice from other world regions into Colombia. We constructed a phylogenetic tree with our global set of samples, including out‐groups, and note CWR falling in different portions of the tree (Figure 5 and Figure S2). In particular, CWR classified as indica‐like based on our structure analysis is related to world weedy rice with known indica origins (e.g., US SH weeds, South Asian weeds) and to known indica cultivars. However, the closest relationship is with the Colombian commercial rice varieties, suggesting that local de‐domestication events gave rise to indica‐like weedy rice in Colombia. Of the five accessions of CWR identified as >80% aus‐like based on our population structure analysis, two are nested within the BHA clade, which is a monophyletic group of US weeds with known aus origin (Figure 5). Two others cluster close by, but with low bootstrap support, and one clusters with indica‐like weeds from South Asia, also with low bootstrap support. This suggests that aus‐like weeds in Colombia are more likely to have originated directly from US weeds and perhaps were inadvertently imported, although an importation event from Asia cannot be completely discounted for some accessions. The low incidence of non‐admixed aus‐like weeds in Colombia compared to the United States makes an origin in Colombia with export to the United States unlikely. CWR samples identified as admixed in our population structure analyses fall between known indica and aus cultivars and do not group with aus‐like weeds from South Asia, consistent with their likely origins though hybridization events between aus‐like and indica‐like weeds in Colombia (Figure 5).
FIGURE 5.
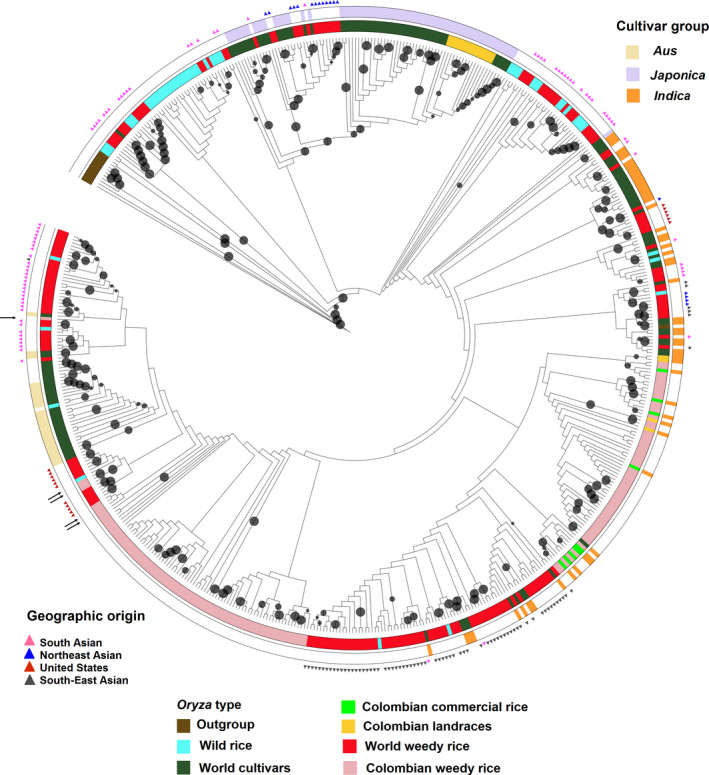
Maximum‐likelihood tree of 559 Colombian and global Oryza accessions and out‐groups. Circles represent the level of bootstrap support of each branch (only bootstrap values >70% are shown). Inner ring colors indicate Oryza type groups, as indicated in the figure key. Outer ring colors represent cultivar groups of O. sativa. Colored triangles at the tip of each branch represent the geographic origin for world weedy rice accession. Arrows show accessions of CWR identified as >80% aus‐like. Individual accession IDs can be found in Figure S1
3.2. Genetic diversity in Colombian weedy rice
We examined levels of genetic diversity in CWR compared to other Oryza groups. Colombian weedy rice as a whole has lower levels of expected heterozygosity than any cultivar group examined (Table 1). Among CWR groups, aus‐like weeds present a greater genetic diversity, despite their low numbers, than indica‐like or weeds of admixed origins (Table 1). When genetic diversity was analyzed for the different rice‐growing regions in Colombia, we did not see a substantial difference among regions (Table 1).
TABLE 1.
Genetic diversity according to Oryza groups, Colombian weedy rice ancestry, and geographic area
| Oryza groups | H e | Ancestry | H e | Geographic area | H e |
|---|---|---|---|---|---|
| Colombian weedy rice (CWR) | 0.146 | indica | 0.170 | Central | 0.188 |
| aus | 0.367 | Llanos plains | 0.168 | ||
| Admixed indica‐aus | 0.209 | Bajo Cauca river valley | 0.198 | ||
| North coast | 0.199 | ||||
| Santanderes | 0.182 | ||||
| Landraces | 0.197 | – | – | – | – |
| Commercial rice | 0.263 | – | – | – | – |
| japonica | 0.161 | – | – | – | – |
| indica | 0.188 | – | – | – | – |
| aus | 0.212 | – | – | – | – |
H e: expected heterozygosis calculated for all loci.
We assessed levels of genetic differentiation between CWR groups and major cultivated rice groups, which confirmed the very low F ST values for indica‐like weeds and commercial rice from Colombia (F ST = 0.093), which is of indica type, followed by an also modest F ST for indica‐like CWR and indica (F ST = 0.162) (Table S10). Additionally, the lowest F ST values for Colombian aus‐like weeds and crop groups were with aus cultivars (F ST = 0.232). Comparisons of CWR with weedy rice groups distributed worldwide revealed that the lowest levels of differentiation for CWR aus‐like weeds were with US weeds of aus origin (BHA; F ST = 0.14; Table S11). This FST value was lower than that between aus‐like weeds and aus, supporting a direct origin from US weeds. Interestingly, the lowest FST values for admixed CWR were in comparison with indica‐like CWR, supporting the greater genetic contribution of this group to these admixed weeds.
3.3. Shattering and the sh4 gene
Seed shattering was recorded for 86 accessions of CWR and cultivated varieties (commercial and landraces) as reported in Hoyos, Plaza, and Caicedo (2019). In general terms, weeds have lower BTS (equivalent to greater shattering) necessary to separate grains from the panicle with a general average of 4.6 gf (±8.3), followed by Colombian commercial varieties with an average of 25.5 gf (±3.7), and finally landraces with 30.8 gf (±13.1) (Figure 6a). Shattering averages were significantly different (Tukey's test [HSD] at the 0.05 probability level) across CWR ancestral groups (Table S12), with indica‐like weeds displaying less seed shattering (BTS 6.9 gf ± 10.0), and admixed indica‐aus weeds showing the greatest shattering (BTS 1.4 gf ± 3.5) (Figure 6b).
FIGURE 6.
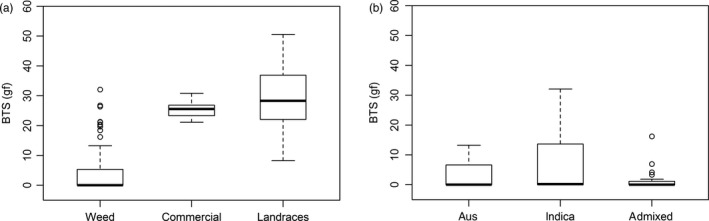
Seed shattering phenotype. Distributions are the mean values of breaking tensile strength (BTS) expressed in grams‐force (gf). The black line represents the median of each distribution. (a) Shattering accessions of Colombian weedy rice, commercial varieties, and landraces. (b) Shattering accessions of weedy rice according to their ancestry
Sequencing of 628 bp within the second exon of sh4 for 87 accessions (69 weedy rice, 5 commercial varieties, 10 landraces, and 3 O. glumaepatula) revealed that all CWR samples, regardless of origin, showed the presence of the T allele associated with the weakening of the gene function and loss of shattering during domestication (Figure S3). The fixation of the derived T allele occurs despite the primarily shattering phenotype displayed by CWR (Figure S3, Table S13).
3.4. Pericarp color and the Rc gene
We recorded pericarp color for 95 accessions, 70 Colombian weedy rice, 5 commercial varieties, 10 landraces, and 4 O. glumaepatula (Figure 7a, Table S14). All domesticated rice was found to have white pericarps, whereas all wild O. glumaepatula have red pericarps. In contrast, 77% of CWR samples had red pericarps, with the remainder being white. Additionally, we observed variation in the extent of pericarp pigmentation among weeds, with light red being present in 52% of these samples. The extent of pericarp coloration in CWR was correlated with ancestry, with 100% of aus‐like weeds possessing red pericarps, whereas indica and admixed weeds have some samples with white pericarps (33% and 11%, respectively) (Figure 7b).
FIGURE 7.

Pericarp color phenotype. (a) Percentage of individuals of Colombian wild rice, weedy rice, commercial varieties, and landraces with white and red colors. (b) Percentage of weedy rice accessions according to their ancestry with white and red colors
We sequenced 421 bp spanning the seventh exon of the Rc locus for phenotyped accessions. As expected, all accessions with white pericarp (both weeds and cultivated rice) possessed the 14‐bp deletion, which has been shown to be responsible for the loss of color in most cultivated rice (Furukawa et al., 2007; Sweeney, Thomson, Pfeil, & McCouch, 2006; Figure S4). For accessions with red pericarps, wild diploid samples of O. glumaepatula lacked the deletion, as expected, as did most of the red pericarp CWR, independent of their ancestry. However, seven weedy rice accession (mainly of indica origin) also with red pericarps carried the 14 bp deletion as well. We examined for the presence of the two additional mutations which have been reported to reverse the effect of the 14 bp deletion on Rc function, re‐establishing the red colored pericarp (Brooks, Yan, Jackson, & Deren, 2008; Lee, Lupotto, & Powell, 2009), but neither were found in the accessions evaluated. Our results suggest that in a subset of weeds, other loci are having an effect on pericarp color.
4. DISCUSSION
4.1. Colombian weedy rice has multiple origins
Understanding the origin and evolution, genetic diversity, and structure of weedy rice populations is basic knowledge that is necessary for the design of effective methods of weed control (He, Kim, & Park, 2016; He et al., 2014; Vigueira et al., 2013), as well as preventing weedy rice adaptation to crop fields (Huang et al., 2017). Three main hypotheses have at various times been put forward for the evolution and origins of weedy rice: (a) direct colonization and adaptation of wild rice groups to the cultivated environment; (b) "de‐domestication" of cultivated rice varieties that become weedy; and (3) gene flow between cultivated rice and reproductively compatible wild populations, leading to weedy rice of hybrid origins (Vigueira et al., 2013). In studies of weedy rice around the world, examples of all of these origins have been found (e.g., Huang et al., 2017; Li, Li, Jia, Caicedo, & Olsen, 2017; Reagon et al., 2010; Song et al., 2014; Vigueira et al., 2019), underscoring the ease with which weedy rice can repeatedly arise in diverse cultivated environments.
As a continent with a unique set of wild Oryza species, some of which are reproductively compatible with O. sativa, South America presents an interesting case study for the origins of local weedy rice. In Colombia, many of these wild Oryza species can be found growing at the borders of rice fields. Our population structure and PCA results, however, show no contribution of wild South American species nor of wild Asian species (O. rufipogon or O. nivara) to Colombian weedy rice. These results are consistent with microsatellite‐based reports that weedy rice from southern Brazil groups separately from O. glumaepatula, which is native to South America, and also from O. rufipogon, O. longistaminata, and O. glaberrima, which are wild AA species from Asia (Goulart, Borba, Menezes, & Merotto, 2014). The difference in ploidy among wild Oryza of the CCDD type and cultivated rice with an AA genome likely creates a genetic barrier that reduces the possibility of gene flow between most South American wild species and cultivated rice (Oka & Chang, 1961). The remaining wild species, O. glumaepatula, has the same AA genome as cultivated rice, and there are reports of it hybridizing with O. sativa (Espinoza & Lentini, 2005). However, there is no evidence for hybridization in our study, and we suggest that in several rice‐growing regions of Colombia, O. glumaepatula is too geographically limited to have an effect. Although this species grows in Colombia in non‐intervention areas bordering commercial rice crops (Estrada et al., 2010), it has a low potential distribution in rice‐growing regions of the Caribbean and inter‐Andean valleys (Villafañe et al., 2007). Early studies with microsatellites in Colombia reported that some CWR accessions from Tolima (the Central zone) with black hulls and awns showed high genetic similarity with O. rufipogon (González et al., 2003, 2007); given the lack of evidence for contribution of this Asian species to CWR, we suggest that this was an erroneous conclusion likely due to the lack of inclusion of the diversity present in rice cultivars, in particular exclusion of aus varieties.
Multiple studies have supported de‐domestication as the principal route for weedy rice origins around the world, for example, in Bhutan (Ishikawa et al., 2005), Brazil (Goulart et al., 2014), United States (Li et al., 2017; Reagon et al., 2010), China (Cao et al., 2006; Li et al., 2017; Qiu et al., 2014; Zhang et al., 2008; Zhang, Dai, Wu, Song, & Qiang, 2012), Malaysia (Song et al., 2014), Korea (He et al., 2016), and South‐East Asia (Vigueira et al., 2019). De‐domestication is an evolutionary process that involves a loss of some traits added under domestication, and likely begins with the formation of a "volunteer weed," a crop that acquires a trait by mutation or hybridization, which provides an advantage in the environment (Ellstrand et al., 2010; Gressel, 2005).
In Colombia, two types of cultivated rice are planted, either of which could potentially give rise to weedy rice through de‐domestication. Landraces, commonly known as “criollos,” are unimproved varieties cultivated in the department of Chocó and the Low Cauca area that are planted manually in small plots mainly for self‐consumption (Espinal, Martínez, & Acevedo, 2005; Pérez, Saavedra, Chimá, & Toro, 2005). Our results are consistent with prior results from chloroplast DNA polymorphisms (Villafañe, Bocanegra, & Moreno, 2010) in finding that most Colombian landraces are japonica cultivars. The influence of japonica in the country has a possible origin with the European colonization of the Americas and the introduction of rice; some historians believe that Christopher Columbus brought this crop on his second trip in 1,493 to Central and South America (GRiSP, 2013). However, these japonica landraces are not the source of weedy rice in Colombia.
Commercial cultivars in Colombia have long been presumed to be indica, which is confirmed by our results. These cultivars also seem to be the largest source of weedy rice in Colombia, as most of our sampled CWR was found to be indica‐like, clustered close to Colombian cultivars in our tree, and harbored very little genetic differentiation with Colombian commercial cultivars based on F ST. Our results support growing evidence that in many parts of the world, the origins of this weed are often related to local crop varieties. However, research on weedy rice in the United States has also shown that de‐domesticated weeds do not necessarily always evolve from local cultivars, as SH and BHA weedy rice descend from indica and aus cultivars, respectively, neither of which are cultivated commercially in the United States (Gealy, Agrama, & Eizenga, 2009; Londo & Schaal, 2007; Reagon et al., 2010). Such exotic origins of de‐domesticated weeds are also evident in Colombia, which harbors a small proportion of weedy rice that is aus‐like in genomic background. Yet, there are no records of aus cultivars ever having been grown in Colombia, and its cultivation is restricted to the center and Northeast of India and Bangladesh (Garris, Tai, Coburn, Kresovich, & Mccouch, 2005; Londo, Chiang, Hung, Chiang, & Schaal, 2006), which are the centers of origin of this cultivar (Civáň, Craig, Cox, & Brown, 2015).
The most likely explanation for the contribution of aus cultivars to CWR is through an involuntary introduction of BHA weedy rice from the United States. This is supported by the phylogenetic clustering of most aus‐like CWR and US BHA weeds, and the low genetic differentiation (as measured by F ST) between CWR and US weeds with the same ancestry. To our knowledge, rice seeds in Colombia have not been sourced from Asia historically, and another possibility that BHA weeds arose from Colombian aus‐like weedy rice is unlikely given that Colombia has not exported rice to the United States or contributed to its breeding program. BHA weedy rice was possibly introduced to the United States as a stock seed contaminant or as escaped breeding material (Delouche et al., 2007; Reagon et al., 2010). Similarly, it has been previously suggested that weedy rice in Latin America and the Caribbean was introduced as a contaminant in rice seeds imported from the United States (Hernandez, 1971; Ortiz‐Dominguez, 1999). While this is an unlikely explanation for indica‐like CWR, it is the most likely explanation for the occurrence of aus‐like CWR.
The last, and second most common, group of CWR seems to be an admixture of indica and aus backgrounds. Given the occurrence of both indica‐like weeds and aus‐like weeds in the country, the most likely explanation for the origin of these admixed weeds is hybridization between weedy groups in Colombian fields. Admixed CWR is thus the product of a complex evolutionary scenario consisting of three parts: (a) a de novo de‐domestication event (local indica cultivars giving rise to indica‐like CWR), (b) exotic gene flow (exotic de‐domesticated aus‐like weed arrives from elsewhere), and (c) a hybridization event (indica‐aus weeds arise through hybridization between the two weedy rice groups). In hybridizing, a widely successful new type of weed, admixed indica‐aus CWR, has emerged, which is now found in all rice‐growing regions of Colombia, and which rivals its progenitor weeds in abundance.
That all three groups of Colombian weedy rice have managed to adapt to the principal rice‐growing regions in Colombia is shown by the lack of geographic structure in the weeds. This adaptation has occurred, in spite of what are relatively lower levels of expected heterozygosity than observed in the commercial and landrace varieties used in the country. The rice‐growing regions in Colombia vary in environmental conditions such as temperature, altitude, and rainfall, and rice is planted at different times throughout the year. However, there is great homogeneity in the provenance of the seeds used, because approximately 80% of the commercial varieties sown in the country are supplied by a single company, FEDEARROZ, and in some areas, this percentage exceeds 90% (personal communication FEDEARROZ 2017). Additionally, the exchange of contaminated seeds among farmers from different regions could be another factor that affects homogeneity in the regions. For the year 2016, only 58.8% of the planted area in Colombia was sown with weed‐free‐certified seed (DANE & FEDEARROZ, 2017). This implies that the remaining areas are being planted with seed that may not be weed free and that often stems from farmers exchanging seed saved from their fields, providing a route for weedy rice to expand its range.
4.2. The source of weedy traits in Colombian weedy rice
As for all de‐domesticated weeds, a major question about weedy rice is how it has acquired the traits that enable weediness, which are not typical of their crop ancestors. Two particularly characteristic traits of weedy rice are the dispersal of seeds through shattering and the red pericarp color, which is associated with seed dormancy. As expected, Colombian weeds showed high levels of seed shattering in comparison with cultivated varieties, but also high phenotypic variability in this trait, similar to other weed rice studies (Markus, Merotto, Barcelos, & Dalazen, 2013; Nunes, Delatorre, & Merotto, 2014; Nunes, Markus, Delatorre, & Merotto, 2014; Thurber et al., 2010). The greatest shattering levels were observed in admixed indica‐aus, while the lowest values were found in indica‐like weeds. Similar results were found in South Asian weeds, where indica‐like weeds showed lower shattering (higher BTS) compared to aus‐like or wild type (Huang et al., 2017).
All CWR groups carry the domesticated allele of the gene sh4, which contains the non‐shattering T substitution that was fixed during rice domestication. Sh4 is involved in the formation and function of the abscission zone from which a mature grain separates from the pedicel (Li et al., 2006). While there is evidence that cultivated alleles of sh4 are not always sufficient for loss of shattering and are likely dependent on genetic background (Zhu, Ellstrand, & Lu, 2012), sh4 remains among the most significant genes to have experienced selection by humans during domestication (Li et al., 2006; Wang et al., 2018; Zhang et al., 2009). Similar sh4 allele frequencies have been observed for other de‐domesticated weedy rice in the United States (Thurber et al., 2010), Japan (Akasaka, Konishi, Izawa, & Ushiki, 2011), other parts of Asia (Huang et al., 2018; Zhu et al., 2012), and Italy (Grimm, 2014). Thus, the results for CWR add to mounting evidence that de‐domesticated weeds evolve the shattering trait through novel genetic mechanisms rather than reversals to ancestral alleles. Preliminary mapping studies in SH and BHA weedy rice groups suggest that these novel loci may not necessarily be shared among independently arisen weedy rice groups (Qi et al., 2015; Thurber, Jia, Jia, & Caicedo, 2013). Given that a subset of CWR shares ancestry with BHA weeds from the United States, however, it is possible that the same loci are involved in reacquisition of shattering in BHA as in Colombian aus‐like and indica‐aus admixed weeds groups.
The proanthocyanidins responsible for pericarp are believed to play an important role in seed dormancy and in the protection against bacteria and fungi in the soil (Sweeney et al., 2007). In weedy rice, seed dormancy allows the seeds to remain in the soil for long periods of time, guaranteeing the permanence of the group (Grimm, 2014). As is common in weedy rice groups (Ziska et al., 2015), most of Colombian weedy rice accessions (77%) have reddish pericarp coloration. These reddish colorations in the pericarp are present in all types of Colombian weedy rice (aus, indica and admixed). In contrast, weeds with white pericarps, which are a minority, only occur in indica‐like or admixed weeds, supporting the perceived advantage of red pericarps in weedy rice.
As expected based on prior studies of Rc (Furukawa et al., 2007; Sweeney et al., 2007), the red pericarp in most CWR can be explained by the presence of the functional ancestral Rc alleles that lack the 14‐bp deletion that knocks out the gene and leads to white pericarps. Rc is a pleiotropic gene that affects both pericarp coloration and dormancy through promotion of expression of genes involved in the biosynthesis of abscisic acid, which is a dormancy‐inducing hormone, and through activation of a conserved network of eight genes involved in synthesis of flavonoids, which are pigments (Gu et al., 2011). The occurrence of wild functional Rc alleles in what is a de‐domesticated weed is most likely explained through direct inheritance of this low‐frequency allele from domesticated ancestors. Although most cultivated rice has a white pericarp and the 14‐bp deletion in Rc, cultivars with red pericarps and ancestral alleles also exist, and this standing variation in crop ancestors could have been selectively favored in weedy rice. Although no red pericarp cultivars occur in Colombian commercial rice, we cannot be absolutely sure these have not been used for breeding purpose in the past.
A startling find is that a small subset of weedy rice display red pericarps despite having the 14‐bp Rc deletion. Restorer mutations in this gene have previously been reported in some cultivars, such as Rc‐r (Lee et al., 2009) and Rc‐g (Brooks et al., 2008), but we did not find these. Our data are insufficient to explain whether undiscovered restorer mutations have occurred or if other loci are involved in this red coloration, but this finding again underscores the selective advantage that red pericarps must confer on weedy rice.
5. CONCLUSIONS
Since the beginning of agriculture, weedy plants have evolved to flourish in crop fields with detrimental effects on agricultural production. Understanding the source of such plants and how they adapt to agricultural conditions are important pre‐requisites for ameliorating their impact crop yields. As studies of weedy rice, a major weed of cultivated rice, accumulate, it has become evident that this weed has had multiple independent origins, most often from domesticated ancestors. Our results from Colombian weedy rice support this growing evidence that the origins of weedy rice are influenced more by domesticated groups than by wild rice, even when local wild Oryza species exist, as several do in South America. De‐domestication has additionally been increasingly recognized as a significant mechanism for agricultural weed origins beyond the weedy rice system (Ellstrand et al., 2010), as evidenced by recent studies showing crop origins for Tibetan semi‐wild wheat and weedy rye (Burger & Ellstrand, 2014; Burger, Holt, & Ellstrand, 2007; Jiang et al., 2019).
An important consequence of this study is the highlighting of the need to consider weedy rice evolution in any locality within a global context. Although indica‐like weedy rice in Colombia most likely has origins in de‐domestication from the local indica crop, our data suggest that aus‐like weeds are likely exotic introductions, a finding that would have been impossible to discern if we had not compared our Colombian weeds with the range of worldwide diversity in weedy and cultivated Oryza. Our study also underscores how dynamic the evolution of weedy rice can be: Given a local weedy rice origin and an exotic introduction, a new category of admixed weedy rice has successfully emerged in the country. We propose that continued use of a global perspective in weedy rice evolution studies, as well as in other weed evolution studies, will enhance our understanding of the facility with which agricultural weeds can evolve and the mechanisms they use to adapt to the agricultural environment.
CONFLICT OF INTEREST
No conflicts of interest have been declared.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S14
ACKNOWLEDGEMENTS
This project was funded, in part, by a Developing Country Collaboration supplement to U.S. National Science Foundation (NSF) grant IOS‐1032023 and the Universidad Nacional de Colombia, Bogotá, Special thanks to Fedearroz staff for their help in obtaining cultivated rice accessions and Sherin Perera for help with genotyping. We also thank Jacob Barnett for help with scripts.
Hoyos V, Plaza G, Li X, Caicedo AL. Something old, something new: Evolution of Colombian weedy rice (Oryza spp.) through de novo de‐domestication, exotic gene flow, and hybridization. Evol Appl. 2020;13:1968–1983. 10.1111/eva.12955
DATA AVAILABILITY STATEMENT
Raw genotyping‐by‐sequencing data have been deposited at the NCBI Short Read Archive (experiment BioProjectIDPRJNA616397). DNA sequences obtained for this study have been deposited in GenBank under accessions MT089735—MT089829 for Rc gene and MT089830—MT089913 for sh4 gene. VCF files have been submitted to DRYAD (10.5061/dryad.sf7m0cg2q).
REFERENCES
- Akasaka, M. , Konishi, S. , Izawa, T. , & Ushiki, J. (2011). Histological and genetic characteristics associated with the seed‐shattering habit of weedy rice (Oryza sativa L.) from Okayama. Japan. Breeding Science, 61(2), 168–173. 10.1270/jsbbs.61.168 [DOI] [Google Scholar]
- Arrieta‐Espinoza, G. , Sánchez, E. , Vargas, S. , Lobo, J. , Quesada, T. , & Espinoza, A. M. (2005). The weedy rice complex in Costa Rica. I. Morphological study of relationships between commercial rice varieties, wild Oryza relatives and weedy types. Genetic Resources and Crop Evolution, 52(5), 575–587. 10.1007/s10722-004-6109-x [DOI] [Google Scholar]
- Avila, L. A. D. , Andres, A. , Marchezan, E. , & Menezes, V. G. (2000). Banco de sementes de arroz vermelho em sistemas de semeadura de arroz irrigado. Ciencia Rural, 30(5), 773–777. 10.1590/S0103-84782000000500005 [DOI] [Google Scholar]
- Brooks, S. A. , Yan, W. , Jackson, A. K. , & Deren, C. W. (2008). A natural mutation in rc reverts white‐rice‐ pericarp to red and results in a new, dominant, wild‐type allele: Rc‐g. Theoretical and Applied Genetics, 117, 575–580. 10.1007/s00122-008-0801-8 [DOI] [PubMed] [Google Scholar]
- Burger, J. C. , & Ellstrand, N. C. (2014). Rapid evolutionary divergence of an invasive weed from its crop ancestor and evidence for local diversification. Journal of Systematics and Evolution, 52(6), 750–764. 10.1111/jse.12111 [DOI] [Google Scholar]
- Burger, J. C. , Holt, J. M. , & Ellstrand, N. C. (2007). Rapid phenotypic divergence of feral rye from domesticated cereal rye. Weed Science, 55(3), 204–211. 10.1614/ws-06-177.1 [DOI] [Google Scholar]
- Burgos, N. R. , Norman, R. J. , Gealy, D. R. , & Black, H. (2006). Competitive N uptake between rice and weedy rice. Field Crops Research, 99(2–3), 96–105. 10.1016/j.fcr.2006.03.009 [DOI] [Google Scholar]
- Burgos, N. R. , Singh, V. , Tseng, T. M. , Black, H. , Young, N. D. , Huang, Z. , … Caicedo, A. L. (2014). The impact of herbicide‐resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiology, 166(3), 1208–1220. 10.1104/pp.114.242719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canal, R. , Arnaude, O. , Ortiz‐Domínguez, A. , Valverde, B. , & Fuentes, C. (2009). Caracterización morfológica de poblaciones de arroz maleza, en el distrito de riego del río zulia, Norte de Santander. Colombia. Agronomia Tropical, 59(4), 387–400. [Google Scholar]
- Cao, Q. J. , Li, B. , Song, Z. P. , Cai, X. X. , & Lu, B.‐R. (2007). Impact of weedy rice populations on the growth and yield of direct‐seeded and transplanted rice. Weed Biology and Management, 7(2), 97–104. 10.1111/j.1445-6664.2007.00241.x [DOI] [Google Scholar]
- Cao, Q. , Lu, B. , Xia, H. , Rong, J. , Sala, F. , Spada, A. , & Grassi, F. (2006). Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in North‐eastern China revealed by Simple Sequence Repeat (SSR) markers. Annals of Botany, 98(6), 1241–1252. 10.1093/aob/mcl210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň, P. , Craig, H. , Cox, C. J. , & Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nature Plants, 1(11), 15164 10.1038/nplants.2015.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo, J. , & Montealegre, F. A. (2010). Arroz rojo en Colombia: Comportamiento y riesgos In Degiovanni V. B., Martinez C. P. R., & Motta F. O. (Eds.), Produccion eco‐eficiente del arroz en América latina (pp. 413–430). Cali, Colombia: Centro Internacional de Agricultura Tropical. [Google Scholar]
- Cobb, A. H. , & Reade, J. P. H. (2010). Herbicides and Plant Physiology. Oxford, UK: Wiley‐Blackwell. [Google Scholar]
- DANE & FEDEARROZ (2017). 4° Censo Nacional Arrocero Año 2016 (180 p). Bogotá D.C.: Departamento Administrativo Nacional de Estadística (DANE) y Federación Nacional de Arroceros (FEDEARROZ). [Google Scholar]
- Delouche, J. C. , Burgos, N. R. , Gealy, D. R. , Zorrilla, G. , Labrada, R. , Larinde, M. , & Rosell, C. (2007). Weedy rices – Origin, biology, ecology and control. Rome, Italy: FAO Plant Production and Protection Paper 188. [Google Scholar]
- Duitama, J. , Quintero, J. C. , Cruz, D. F. , Quintero, C. , Hubmann, G. , Foulquié‐Moreno, M. R. , … Tohme, J. (2014). An integrated framework for discovery and genotyping of genomic variants from high‐throughput sequencing experiments. Nucleic Acids Research, 42(6), 1–13. 10.1093/nar/gkt1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ellstrand, N. C. , Heredia, S. M. , Leak‐Garcia, J. A. , Heraty, J. M. , Burger, J. C. , Yao, L. I. , … Ridley, C. E. (2010). Crops gone wild: Evolution of weeds and invasives from domesticated ancestors. Evolutionary Applications, 3, 494–504. 10.1111/j.1752-4571.2010.00140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire, R. J. , Glaubitz, J. C. , Sun, Q. , Poland, J. A. , Kawamoto, K. , Buckler, E. S. , & Mitchell, S. E. (2011). A robust, simple genotyping‐by‐sequencing (GBS) approach for high diversity species. PLoS ONE, 6(5), 1–10. 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinal, C. , Martínez, H. , & Acevedo, X. (2005). La cadena del arroz en Colombia: una mirada global de su estructura y dinámica 1991‐2005. Min. Agricultura y Desarrollo Rural, Obs. Agrocadenas Colombia. Retrieved from http://www.agronet.gov.co/www/docs_agronet/2005112145659_caracterizacion_cacao.pdf [Google Scholar]
- Espinoza, A. M. , & Lentini, Z. (2005). Coexistence of weedy rice and rice in Tropical America — Gene flow and genetic diversity In Gressel J. (Ed.), Crop ferality and volunteerism (1st Editio, pp. 305–322). Boca Raton, FL: CRC Press. [Google Scholar]
- Estrada, O. A. , Villafañe, C. , Palacio, J. D. , Fory, L. F. , & Lentini, Z. (2010). Caracterización de especies silvestres del género Oryza recolectadas en las principales zonas arroceras de Colombia, mediante marcadores moleculares In X Congreso Latinoamericano de Botánica. “Conservación y uso sustentable de la flora nativa latinoamericana”. La Serena: Asociación Latinoamericana de Botánica. [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3), 564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. , & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164(4), 1567–1587. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici, M. T. , Vaughan, D. , Tomooka, N. , Kaga, A. , Wang, X. W. , Doi, K. , … Saldain, N. (2001). Analysis of Uruguayan weedy rice genetic diversity using AFLP molecular markers. Electronic Journal of Biotechnology, 4(3), 130–145. [Google Scholar]
- Fischer, A. J. , & Ramírez, A. (1993). Red rice (Oryza sativa): Competition studies for management decisions. International Journal of Pest Management, 39(2), 133–138. 10.1080/09670879309371777 [DOI] [Google Scholar]
- Fogliatto, S. , Vidotto, F. , & Ferrero, A. (2012). Morphological characterisation of Italian weedy rice (Oryza sativa) populations. Weed Research, 52(1), 60–69. 10.1111/j.1365-3180.2011.00890.x [DOI] [Google Scholar]
- Francis, R. M. (2017). POPHELPER: An R package and web app to analyse and visualize population structure. Molecular Ecology Resources, 17(1), 27–32. 10.1111/1755-0998.12509 [DOI] [PubMed] [Google Scholar]
- Furukawa, T. , Maekawa, M. , Oki, T. , Suda, I. , Iida, S. , Shimada, H. , … Kadowaki, K.‐I. (2007). The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant Journal, 49, 91–102. 10.1111/j.1365-313X.2006.02958.x [DOI] [PubMed] [Google Scholar]
- Garris, A. J. , Tai, T. H. , Coburn, J. , Kresovich, S. , & Mccouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics, 169, 1631–1638. 10.1534/genetics.104.035642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealy, D. R. , Agrama, H. A. , & Eizenga, G. C. (2009). Exploring genetic and spatial structure of U. S. weedy red rice (Oryza sativa) in relation to rice relatives worldwide. Weed Science, 57, 627–643. 10.1614/WS-09-018.1 [DOI] [Google Scholar]
- Glaubitz, J. C. , Casstevens, T. M. , Lu, F. , Harriman, J. , Elshire, R. J. , Sun, Q. , & Buckler, E. S. (2014). TASSEL‐GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE, 9(2), e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, E. , Fory, L. F. , Arcia, K. , Blanco, A. E. , Ortiz, A. , Pérez, I. , … Lentini, Z. (2007). Genetic diversity analysis of weedy rice and wild Oryza species collected in crop‐natural ecosystems contact zones. Cali, Colombia: Centro Internacional de Agricultura Tropical. [Google Scholar]
- González, E. , Fory, L. F. , Vásquez, J. J. , Ruiz, P. , Mora, A. , Silva, J. , … Lentini, Z. (2003). Molecular characterization of rice and wild / weedy relatives by microsatellites and their use to assess gene flow in the Neo‐Tropics. Cali, Colombia: Centro Internacional de Agricultura Tropical. [Google Scholar]
- Goulart, I. C. G. R. , Borba, T. C. O. , Menezes, V. G. , & Merotto, A. Jr (2014). Distribution of weedy red rice (Oryza sativa) resistant to imidazolinone herbicides and its relationship to rice cultivars and wild Oryza species. Weed Science, 62(02), 280–293. 10.1614/WS-D-13-00126.1 [DOI] [Google Scholar]
- Gressel, J. (2005). Introduction – The challenges of ferality In Gressel J. (Ed.), Crop ferality and volunteerism (pp. 1–7). Boca Raton, FL: CRC Press. [Google Scholar]
- Grimm, A. (2014). Characterization of weedy rice groups in Northern Italy and investigation of their evolutionary origins. Karlsruhe: Karlsruher Institut für Technologie (KIT) – Universitätsbereich; https://publikationen.bibliothek.kit.edu/1000040430 [Google Scholar]
- Grimm, A. , Fogliatto, S. , Nick, P. , Ferrero, A. , & Vidotto, F. (2013). Microsatellite markers reveal multiple origins for Italian weedy rice. Ecology and Evolution, 3(14), 4786–4798. 10.1002/ece3.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRiSP (Global Rice Science Partnership) (2013). Rice almanac: Source book for one of the most economic activities on Earth (4th editio). Los Baños, Philippines: International Rice Research Institute. [Google Scholar]
- Gross, B. L. , Reagon, M. , Hsu, S. C. , Caicedo, A. L. , Jia, Y. , & Olsen, K. M. (2010). Seeing red: The origin of grain pigmentation in US weedy rice. Molecular Ecology, 19(16), 3380–3393. 10.1111/j.1365-294X.2010.04707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X.‐Y. , Foley, M. E. , Horvath, D. P. , Anderson, J. V. , Feng, J. , Zhang, L. , … Chen, Z. (2011). Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics, 189(4), 1515–1524. 10.1534/genetics.111.131169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q. , Kim, K. , & Park, Y. (2016). Population genomics identifies the origin and signatures of selection of Korean weedy rice. Plant Biotechnology Journal, 1, 1–10. 10.1111/pbi.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z. , Jiang, X. , Ratnasekera, D. , Grassi, F. , Perera, U. , & Lu, B. R. (2014). Seed‐mediated gene flow promotes genetic diversity of weedy rice within populations: Implications for weed management. PLoS ONE, 9(12), 1–21. 10.1371/journal.pone.0112778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, A. (1971). Arroz rojo amenaza de las variedades comerciales en Sinaloa (2da edició). Culiacán, Sin. Mexico: Secretaria de Agricultura y Ganadería, Instituto Nacinal de Investigaciones Agrícolas, Centro de Investigaciones Agrícolas de Sinaloa. [Google Scholar]
- Hoyos, V. , Plaza, G. , & Caicedo, A. L. (2019). Characterization of the phenotypic variability in Colombian weedy rice (Oryza spp.). Weed Science, 67(4), 441–452. 10.1017/wsc.2019.18 [DOI] [Google Scholar]
- Huang, Z. , Kelly, S. , Matsuo, R. , Li, L. , Li, Y. , Olsen, K. M. , … Caicedo, A. L. (2018). The role of standing variation in the evolution of weediness traits in South Asian Weedy Rice (Oryza spp.). G3: Genes, Genomes, Genetics, 8, 3679–3690. 10.1534/g3.118.200605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z. , Young, N. D. , Reagon, M. , Hyma, K. E. , Olsen, K. M. , Jia, Y. , & Caicedo, A. L. (2017). All roads lead to weediness: Patterns of genomic divergence reveal extensive recurrent weedy rice origins from South Asian Oryza. Molecular Ecology, 26(12), 3151–3167. 10.1111/mec.14120 [DOI] [PubMed] [Google Scholar]
- Hubisz, M. J. , Falush, D. , Stephens, M. , & Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources, 9(5), 1322–1332. 10.1111/j.1755-0998.2009.02591.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, R. , Toki, N. , Imai, K. , Sato, Y. I. , Yamagishi, H. , Shimamoto, Y. , … Sato, T. (2005). Origin of weedy rice grown in Bhutan and the force of genetic diversity. Genetic Resources and Crop Evolution, 52(4), 395–403. 10.1007/s10722-005-2257-x [DOI] [Google Scholar]
- Jakobsson, M. , & Rosenberg, N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23(14), 1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Chen, Q. , Wang, Y. , Guo, Z. , Xu, B. , Zhu, J. , … Qi, P. (2019). Re‐acquisition of the brittle rachis trait via a transposon insertion in domestication gene Q during wheat de‐domestication. New Phytologist, 224(2), 961–973. 10.1111/nph.15977 [DOI] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , … Drummond, A. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraehmer, H. , Jabran, K. , Mennan, H. , & Chauhan, B. S. (2016). Global distribution of rice weeds – A review. Crop Protection, 80, 73–86. 10.1016/j.cropro.2015.10.027 [DOI] [Google Scholar]
- Lee, D. , Lupotto, E. , & Powell, W. (2009). G‐string slippage turns white rice red. Genome, 52, 490–493. 10.1139/G09-025 [DOI] [PubMed] [Google Scholar]
- Letunic, I. , & Bork, P. (2016). Interactive Tree Of Life (iTOL) v3: An online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 44(1), W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhou, A. , & Sang, T. (2006). Rice domestication by reducing shattering. Science, 311, 1936–1939. 10.1126/science.1123604 [DOI] [PubMed] [Google Scholar]
- Li, L. , Li, Y. , Jia, Y. , Caicedo, A. L. , & Olsen, K. M. (2017). Signatures of adaptation in the weedy rice genome. Nature Genetics, 49(5), 811–814. 10.1038/ng.3825 [DOI] [PubMed] [Google Scholar]
- Londo, J. P. , Chiang, Y. , Hung, K. , Chiang, T. , & Schaal, B. A. (2006). Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa . Proceedings of the National Academy of Sciences of the United States of America, 103(25), 9578–9583. 10.1073/pnas.0603152103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo, J. P. , & Schaal, B. A. (2007). Origins and population genetics of weedy red rice in the USA. Molecular Ecology, 16, 4523–4535. 10.1111/j.1365-294X.2007.03489.x [DOI] [PubMed] [Google Scholar]
- Louisiana State Seed Testing Laboratory (1980). A potassium hydroxide test for confirmation of red rice (Oryza sativa L.) seed. Association of Official Seed Analysts Newsletter, 54(1), 68–69. [Google Scholar]
- Markus, C. , Merotto, A. , Barcelos, J. A. N. , & Dalazen, G. (2013). Expressão de genes envolvidos na formação da parede celular e sua relação com o degrane em arroz vermelho In VIII Congresso brasileiro de Arroz Irrigado2 (pp. 363–366). Santa Maria, CA: Sociedade Sul‐Brasileira de Arroz Irrigado (SOSBAI) and Universidade Federal de Santa Maria (UFSM) http://cbai2013.web2265.uni5.net/cdonline/docs/trab-2261-591.pdf [Google Scholar]
- Messeguer, J. , Marfà, V. , Català, M. M. , Guiderdoni, E. , & Melé, E. (2004). A field study of pollen‐mediated gene flow from Mediterranean GM rice to conventional rice and the red rice weed. Molecular Breeding, 13(1), 103–112. 10.1023/B:MOLB.0000012285.39859.9d [DOI] [Google Scholar]
- Nunes, A. L. , Delatorre, C. A. , & Merotto, A. Jr (2014). Gene expression related to seed shattering and the cell wall in cultivated and weedy rice. Plant Biology, 16(5), 888–896. 10.1111/plb.12133 [DOI] [PubMed] [Google Scholar]
- Nunes, A. L. , Markus, C. , Delatorre, C. A. , & Merotto, A. Jr (2014). Nucleotide variability and gene expression reveal new putative genes related to seed shattering in weedy rice. Annals of Applied Biology, 2, 1–9. 10.1111/aab.12159 [DOI] [Google Scholar]
- Oerke, E. C. (2006). Crop losses to pests. Journal of Agricultural Science, 144(1), 31–43. 10.1017/S0021859605005708 [DOI] [Google Scholar]
- Oka, H. I. , & Chang, W. T. (1961). Hybrid swarms between wild and cultivated rice species, Oryza perennis and O. sativa . Evolution, 21(1), 418–430. [Google Scholar]
- Olsen, K. M. , Caicedo, A. L. , & Jia, Y. (2007). Evolutionary genomics of weedy rice in the USA. Journal of Integrative Plant Biology, 49(6), 811–816. 10.1111/j.1744-7909.2007.00506.x [DOI] [Google Scholar]
- Ortiz‐Dominguez, A. (1999). Situación del arroz rojo en Venezuela In FAO (Ed.), Global workshop on red rice control/Taller global de control de arroz rojo (pp. 59–65). Rome, Italy: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Patterson, N. , Price, A. L. , & Reich, D. (2006). Population structure and eigenanalysis. PLoS Genetics, 2(12), 2074–2093. 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez, C. R. , Saavedra, E. , Chimá, C. J. , & Toro, C. G. (2005). El cultivo del arroz secano manual o a chuzo en el caribe húmedo. Arroz, 53(454), 4–13. [Google Scholar]
- Prasad, R. , Shivay, Y. S. , & Kumar, D. (2017). Current status, challenges, and opportunities in rice production In Chauhan B. S., Jabran K., & Mahajan G. (Eds.), Rice production worldwide (pp. 1–32). Cham, Switzerland: Springer International Publishing AG. [Google Scholar]
- Price, A. L. , Patterson, N. J. , Plenge, R. M. , Weinblatt, M. E. , Shadick, N. A. , & Reich, D. (2006). Principal components analysis corrects for stratification in genome‐wide association studies. Nature Genetics, 38(8), 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Liu, Y. , Vigueira, C. C. , Young, N. D. , Caicedo, A. L. , Jia, Y. , … Olsen, K. M. (2015). More than one way to evolve a weed: Parallel evolution of US weedy rice through independent genetic mechanisms. Molecular Ecology, 24(13), 3329–3344. 10.1111/mec.13256 [DOI] [PubMed] [Google Scholar]
- Qiu, J. , Zhu, J. , Fu, F. , Ye, C.‐Y. , Wang, W. , Mao, L. , … Fan, L. (2014). Genome re‐sequencing suggested a weedy rice origin from domesticated indica ‐ japonica hybridization: A case study from southern China. Planta, 240, 1353–1363. 10.1007/s00425-014-2159-2 [DOI] [PubMed] [Google Scholar]
- Raj, A. , Stephens, M. , & Pritchard, J. K. (2014). FastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics, 197(2), 573–589. 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagon, M. , Thurber, C. S. , Gross, B. L. , Olsen, K. M. , Jia, Y. , & Caicedo, A. L. (2010). Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice. BMC Evolutionary Biology, 10, 180 10.1186/1471-2148-10-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A. (2004). DISTRUCT: A program for the graphical display of population structure. Molecular Ecology Notes, 4(1), 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Rosta, K. (1975). Variety determination in rice (Oryza sativa L.). Seed Science and Technology, 3(1), 161–169. [Google Scholar]
- Singh, V. , Burgos, N. , Singh, S. , Gealy, D. , Gbur, E. , & Caicedo, A. L. (2017). Impact of volunteer rice infestation on yield and grain quality of rice Running title: Volunteer rice affects rice grain quality Vijay Singh. Pest Management Science, 73(3), 604–615. [DOI] [PubMed] [Google Scholar]
- Song, B.‐K. , Chuah, T.‐S. , Tam, S. M. , & Olsen, K. M. (2014). Malaysian weedy rice shows its true stripes: Wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. Molecular Ecology, 23, 5003–5017. 10.1111/mec.12922 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, M. T. , Thomson, M. J. , Cho, Y. G. , Park, Y. J. , Williamson, S. H. , Bustamante, C. D. , & Mccouch, S. R. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genetics, 3(8), e133 10.1371/journal.pgen.0030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, M. T. , Thomson, M. J. , Pfeil, B. E. , & Mccouch, S. (2006). Caught red‐handed: Rc encodes a basic helix‐loop‐helix protein conditioning red pericarp in rice. The Plant Cell, 18, 283–294. 10.1105/tpc.105.038430.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber, C. S. , Jia, M. H. , Jia, Y. , & Caicedo, A. L. (2013). Similar traits, different genes? Examining convergent evolution in related weedy rice populations. Molecular Ecology, 22(3), 685–698. 10.1111/mec.12147 [DOI] [PubMed] [Google Scholar]
- Thurber, C. S. , Reagon, M. , Gross, B. L. , Olsen, K. M. , Jia, Y. , & Caicedo, A. L. (2010). Molecular evolution of shattering loci in U.S. weedy rice. Molecular Ecology, 19(16), 3271–3284. 10.1111/j.1365-294X.2010.04708.x.Molecular [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser, A. , Cutcutache, I. , Koressaar, T. , Ye, J. , Faircloth, B. C. , Remm, M. , & Rozen, S. G. (2012). Primer3‐new capabilities and interfaces. Nucleic Acids Research, 40(15), 1–12. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, B. E. (2005). The damage by weedy rice — Can feral rice remain undetected? In Gressel, J. (Ed.), Crop ferality and volunteerism (pp. 279–294). Boca Raton, FL: CRC Press. [Google Scholar]
- Veasey, E. A. , Bressan, E. D. A. , Zucchi, M. I. , Vencovsky, R. , Cardim, D. C. , & Silva, R. M. D. (2011). Genetic diversity of American wild rice species. Scientia Agricola, 68(4), 440–446. 10.1590/S0103-90162011000400008 [DOI] [Google Scholar]
- Vigueira, C. C. , Olsen, K. M. , & Caicedo, A. L. (2013). The red queen in the corn: Agricultural weeds as models of rapid adaptive evolution. Heredity, 110(4), 303–311. 10.1038/hdy.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigueira, C. C. , Qi, X. , Song, B. K. , Li, L. F. , Caicedo, A. L. , Jia, Y. , & Olsen, K. M. (2019). Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia. Evolutionary Applications, 12, 93–104. 10.1111/eva.12581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafañe, C. , Bocanegra, J. L. , & Moreno, R. (2010). Diversidad genotípica de un grupo de variedades tradicionales de arroz (Oryza sativa L.) del Chocó colombiano. In Orjuela‐R M. A. & Moreno R. V. (Eds.), Fortalecimiento de capacidades para la implementación del Protocolo de Cartagena en Colombia: Sector Ambiente (pp. 26–32). Bogotá, Colombia: Ministerio de Ambiente, Vivienda y Desarrollo Territorial e Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. [Google Scholar]
- Villafañe, C. , Estrada, O. A. , Lentini, Z. , Fory, L. F. , & Palacio, J. D. (2007). Diagnóstico y fortalecimiento de la línea base del conocimiento del género Oryza (arroz) en Colombia, un aporte para la toma de decisiones en el ámbito de bioseguridad In Desarrollo de capacidades para evaluación y gestión de riesgos y monitoreo de organismos genéticamente modificados (OGM) Tomo I Resultados de proyectos específicos (pp. 33–49). Bogotá D.C.: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. [Google Scholar]
- Wang, W. , Mauleon, R. , Hu, Z. , Chebotarov, D. , Tai, S. , Wu, Z. , … Leung, H. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature, 557(7703), 43–49. 10.1038/s41586-018-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples, R. S. , & Gaggiotti, O. (2006). What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology, 15(6), 1419–1439. 10.1111/j.1365-294X.2006.02890.x [DOI] [PubMed] [Google Scholar]
- Watson, A. K. (2002). Flooding (weed control in rice) In Pimentel D. (Ed.), Encyclopedia of pest management (1st editio, pp. 283–285). Boca Raton, FL: CRC Press. [Google Scholar]
- Zhang, J. , Burgos, N. R. , Ma, K. , Zhou, Y. , Geng, R. , & Yu, L. (2008). Genetic diversity and relationship of weedy rice in Taizhou City, Jiangsu Province, China. Rice Science, 15(4), 295–302. 10.1016/S1672-6308(09)60007-X [DOI] [Google Scholar]
- Zhang, L. , Dai, W. , Wu, C. , Song, X. , & Qiang, S. (2012). Genetic diversity and origin of Japonica‐ and Indica‐like rice biotypes of weedy rice in the Guangdong and Liaoning provinces of China. Genetic Resources and Crop Evolution, 59, 399–410. 10.1007/s10722-011-9690-9 [DOI] [Google Scholar]
- Zhang, L.‐B. , Zhu, Q. , Wu, Z. , Ross‐ibarra, J. , Gaut, B. S. , Ge, S. , & Sang, T. (2009). Selection on grain shattering genes and rates of rice domestication. New Phytologist, 184(3), 708–720. 10.1111/j.1469-8137.2009.02984.x [DOI] [PubMed] [Google Scholar]
- Zhiwen, L. , Shiqi, Z. , Dong, Z. , Fang, Y. , Zhaoxia, J. , & Wenfu, C. (2016). Molecular phylogeny of the Chinese weedy rice based on nrDNA ITS and two cpDNA sequences. Indian Journal of Genetics and Plant Breeding, 76(1), 57–63. 10.5958/0975-6906.2016.00008.0 [DOI] [Google Scholar]
- Zhu, Y. , Ellstrand, N. C. , & Lu, B. (2012). Sequence polymorphisms in wild, weedy, and cultivated rice suggest seed‐shattering locus sh4 played a minor role in Asian rice domestication. Ecology and Evolution, 2(9), 2106–2113. 10.1002/ece3.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziska, L. H. , Gealy, D. R. , Burgos, N. , Caicedo, A. L. , Gressel, J. , Lawton‐Rauh, A. L. , … Merotto, A. (2015). Weedy (Red) rice: An emerging constraint to global rice production. Advances in Agronomy, 129, 181–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S14
Data Availability Statement
Raw genotyping‐by‐sequencing data have been deposited at the NCBI Short Read Archive (experiment BioProjectIDPRJNA616397). DNA sequences obtained for this study have been deposited in GenBank under accessions MT089735—MT089829 for Rc gene and MT089830—MT089913 for sh4 gene. VCF files have been submitted to DRYAD (10.5061/dryad.sf7m0cg2q).


