Abstract
Olive (Olea europaea L.) is a very important woody tree and favored by consumers because of the fruit’s high-quality olive oil. Chloroplast genome analysis will provide insights into the chloroplast variation and genetic evolution of olives. The complete chloroplast genomes of three accessions (O. europaea subsp. cuspidata isolate Yunnan, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. europaea var. frantoio) were obtained by next-generation sequencing technology. A total of 133 coding regions were identified in the three chloroplast genomes without rearrangement. O. europaea subsp. europaea var. sylvestris and O. europaea subsp. europaea var. frantoio had the same sequences (155,886 bp), while O. europaea subsp. cuspidata isolate Yunnan (155,531 bp) presented a large gap between rps16 and trnQ-UUG genes with six small gaps and fewer microsatellites. The whole chloroplast genomes of 11 O. europaea were divided into two main groups by a phylogenetic tree and O. europaea subsp. cuspidata formed a separate group (Cuspidata group) with the other subspecies (Mediterranean/North African group). Identification of consistency and diversity among O. europaea subspecies will benefit the exploration of domestication events and facilitate molecular-assisted breeding for O. europaea.
Keywords: Olea europaea L., comparative chloroplast genome, genetic diversity, phylogenetic analyses
1. Introduction
Olive (Olea europaea L.) is a famous woody tree in the world and has been cultivated for about five to six thousand years in Mediterranean countries [1,2,3]. Except for a few fermented table olives, most olive fruits are used for oil extraction. Because of the mechanical method, olive oil is regularly consumed in its crude form without loss of nutrients. Therefore, it is considered as “liquid gold” and popular among consumers all over the world [4,5]. The olive belongs to the O. europaea species, which comprises of six subspecies, including O. europaea subsp. europaea (Mediterranean basin), O. europaea subsp. maroccana (Macaronesia), O. europaea subsp. cerasiformis (Macaronesia), O. europaea subsp. guanchica (Macaronesia), O. europaea subsp. laperrinei (Saharan mountains), and O. europaea subsp. cuspidata (from South Africa to South Asia) [3,6,7]. For O. europaea subsp. europaea, the cultivated olive (O. europaea subsp. europaea var. europaea) and wild olive (O. europaea. subsp. europaea var. sylvestris) are differentiated. There are currently more than 2600 cultivars grown for oil extraction after a long period of domestication with biogeographic conditions and human influence [8]. Olive trees are primarily distributed in Spain, Italy, and Greece, where they enjoy the moderate temperatures and semi-arid Mediterranean climate. Nowadays, olive trees have been introduced into about 40 countries such as China, Australia, and the US [9].
Until now, more than 2000 olive accessions have been collected in the Olea databases (http://www.oleadb.it). The phenomenon of synonyms, homonyms, and unclear genetic relationship still exists among olive germplasms [10,11]. Researchers have done lots of studies on the molecular markers to distinguish different olive accessions, such as the amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and single nucleotide polymorphism (SNP) [12,13,14]. D’Agostino et al. [15] and Zhu et al. [16] conducted whole-genome level SNP exploration for 97 and 57 olive cultivars, respectively. The two studies produced high identity-by-state values between different pairs of cultivars, which had formerly been considered the same cultivar in the past years. In addition, the screening of core loci provided a more efficient and faster method for identification of different olive germplasms [16,17]. Until now, the genomic sequencing of three olive trees, O. europaea subsp. europaea cv. leccino, O. europaea subsp. europaea cv. farga, and O. europaea subsp. europaea var. sylvestris, were available [18,19,20]. More studies identifying germplasm resources at the whole-genome level and determining the mechanism of agronomic traits need to be done urgently.
Organelle DNA genomes mtDNA and cpDNA are maternally inherited and provide scientists simple and fast methods to study the different genetic backgrounds of olive germplasms [21]. Molecular markers and organelle DNA sequences are available in olive. Using lengths of restriction fragments markers, Amane et al. [22] classified the chloroplast of 72 cultivars and 101 wild olives into five chlorotypes and found that the same chlorotype was predominant over the whole geographical distributions of cultivated olive and the oleaster forms. More numerous variant chlorotypes were observed in oleasters than in cultivated olive, although they all displayed low variation [22]. With PCR-RFLP and microsatellite markers, 143 cultivated olive, 334 wild olive, 77 subspecies, and 1 outgroup (Olea woodiana Knobl.) were classified into five clades with only 15 chlorotypes [23]. Mariotti et al. [24] and Besnard et al. [21] conducted chloroplast DNA sequencing and found that the sizes of olive chloroplast DNA varied from 155,531 to 155,896 bp with low nucleotide divergence (<0.07%) among the lineages. Olive trees shared a high similarity in the europaea subspecies with more variation between different subspecies [21]. Here, we sequenced the cpDNAs of O. europaea subsp. cuspidata isolate Yunnan, O. europaea subsp. europaea var. sylvestris, which displayed significant differences from most olive cultivars in tree characteristics, fruit traits, and resistance. As a control, the cultivated olive O. europaea subsp. europaea var. frantoio was also employed to analyze genome variation and genetic association among olive chloroplasts. Through the analysis of structure comparison and evolution relation among all the O. europaea species, this study provides a better understanding of chloroplast variation and genetic evolution of olive at the whole-genome level.
2. Materials and Methods
2.1. Plant Material and DNA Extraction
Three olive accessions were collected and analyzed in this study including O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan. The first two accessions were collected from Italy and Spain, respectively, while O. europaea subsp. cuspidata isolate Yunnan was collected from China. Fresh young leaves (~100 mg) were sampled from the new shoots and frozen in liquid nitrogen for further analysis.
Total DNA was isolated with modified cetyltrimethylammonium bromide (CTAB) method as described by Murray et al. [25]. Agarose gel electrophoresis (1.2%) was used to detect DNA integrity, purity, and concentration, and a qubit fluorometer was used to determine DNA concentration.
2.2. Sequencing and Data Quality Control
Complete DNA sequencing was done using Illumina’s next-generation sequencing technology. The genome sequencing was performed on the Illumina MiSeq 2000 (Illumina Inc., San Diego, CA, USA) with paired-end methods (150 bp). The raw sequence reads were filtered using the NGSQC Tool Kit v2.3.3 as follows: (1) remove adapter sequence in the reads; (2) remove the reads whose 5’-end base was unknown; (3) remove the reads with the quality value ≤ Q20; (4) remove reads whose unknown bases ≥ 10%; (5) remove reads whose length was less than 50 bp.
2.3. Chloroplast Genome Assembly and Annotation
The quality of the raw reads was assessed by FastQC [26] and carried out by Cutadapt [27]. Clean reads were assembled into scaffolds using the de novo assembler SPAdes [28] and further assembled using Blastn and exonerated with O. europaea subsp. europaea var. manzanilla (FN996972.1) as a reference. Sequence extension, hole filling, and splicing were performed with paired-read iterative contig extension (PRICE) and MITObim (https://github.com/chrishah/MITObim). The chloroplast genes were annotated using the DOGMA and UGENE ORFs finder tool [29] and visualized with OGDraw 1.2 [30].
Each of the assembled cpDNA sequences has been submitted to GenBank and acquired the following accession numbers: MT182984 and MT182986 for O. europaea subsp. europaea var. frantoio and O. europaea subsp. europaea var. sylvestris, and MT182985 for O. europaea subsp. cuspidata isolate Yunnan.
2.4. Comparative Genomic and Repetitive Sequences Analysis
Except for the three O. europaea chloroplast genomes sequenced here, the other three subspecies genomes, including O. europaea subsp. laperrinei (MG255765.1), O. europaea subsp. guanchica (MG255764.1), O. europaea subsp. maroccana (FN998900.2), were used for comparative genomic analysis. Sequence identity and rearrangement were performed using the mVISTA program with LAGAN mode [31] and Mauve alignment [32].
Repetitive simple sequence repeat (SSR) sequences were analyzed with MISA software (http://pgrc.ipk-gatersleben.de/misa/). Four types of repeat sequences, including forward, reverse, complement, and palindrome sequences, were determined by REPuter software with a minimum repeat size of 20 bp as described by Liu et al. [33].
2.5. Phylogenetic Analysis
All of the nucleic acid sequences of 11 O. europaea chloroplast genomes were used to conduct the phylogenetic tree with Olea lancea (NC_042278.1) as the outgroup. In addition to the three O. europaea chloroplast genomes sequenced here, the other eight genomes included O. europaea subsp. laperrinei (MG255765.1), O. europaea subsp. guanchica (MG255764.1), O. europaea subsp. maroccana (FN998900.2), O. europaea subsp. europaea var. bianchera (NC_013707.2), O. europaea subsp. europaea var. manzanilla (FN996972.1), O. europaea subsp. cuspidata isolate Maui 1 (FN650747.2), O. europaea subsp. cuspidata isolate Guangzhou 1 (FN996944.1), and O. europaea subsp. cuspidata isolate Almihwit 5.1 (FN996943.2). These were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov). MAFFT 7.427 (https://www.ebi.ac.uk/Tools/msa/mafft/) and Gblocks (–t = d, –b5 = h) were used for multi-sequence alignment and editing. The phylogenetic tree was built using IQTREE 1.6.10 software (http://www.iqtree.org) with maximum likelihood method (GTR + I + G) and edited with Figtree 1.4.3 software (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. Assembly and Validation of Chloroplast Genome
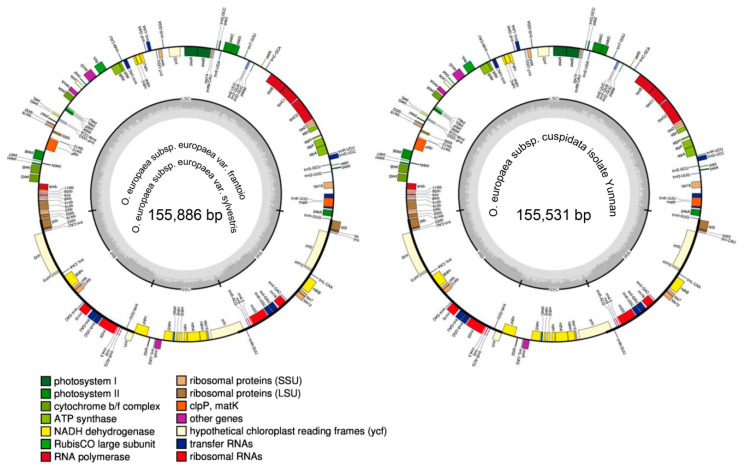
The chloroplast genome sizes of O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan were 155,886, 155,886, and 155,531 bp with 42512X, 35953X, and 48376X depth, respectively. After performing the de novo and reference-guided assembly with minor modifications, the complete chloroplast genome sequences of three O. europaea accessions were obtained (Figure 1). O. europaea subsp. europaea var. frantoio and O. europaea subsp. europaea var. sylvestris shared the completely same sequence (Figure 1; Table 1), which was consistent with O. europaea subsp. europaea var. manzanilla (FN996972.1) [21]. The genomes of all of the three O. europaea had two copies of inverted repeat (IR, 25,742 and 25,731 bp) separated by large single-copy (LSC, 86,611 and 86,279 bp) and small single-copy (SSC, 17,791 and 17,790 bp) regions (Table 1). There were 133 coding regions, including 88 protein-coding genes, 8 rRNA, and 37 tRNA (Figure 1; Table 2).
Figure 1.
Chloroplast gene maps of Olea europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan. Genes with different functions were shown in different colors. Those transcribed clockwise or counter-clockwise were shown inside or outside the circle. LSC, large single-copy region; SSC, small single-copy region; IR, inverted repeat.
Table 1.
Summary of the three chloroplast genomes sequenced in this study.
| Category | O. europaea subsp. europaea var. frantoio/O. europaea subsp. europaea var. sylvestris | O. europaea subsp. cuspidata isolate Yunnan |
|---|---|---|
| Total length | 155,886 bp | 155,531 bp |
| Length of large single copy (LSC) region | 86,611 bp | 86,279 bp |
| Length of small single copy (SSC) region | 17,791 bp | 17,790 bp |
| Length of inverted repeat (IR) region | 25,742 bp | 25,731 bp |
| GC content | 37.8% | 37.8% |
| Total number of genes | 133 | 133 |
| Number of protein encoding genes | 87 | 87 |
| Number of rRNA genes | 8 | 8 |
| Number of tRNA genes | 37 | 37 |
| Loci of JLA | 86,612 bp | 86,280 bp |
| Loci of JSA | 112,353 bp | 112,010 bp |
| Loci of JSB | 130,145 bp | 129,801 bp |
| Loci of JLB | 155,886 bp | 155,531 bp |
Table 2.
Genes identified in the chloroplast genome of olive.
| Category for Genes | Group of Genes | Name of Genes |
|---|---|---|
| Self-replication | tRNA genes | rrn4.5, rrn5, rrn16, rrn23 |
| rRNA genes | trnA-UGC†, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC†, trnH-GUG, trnI-CAU, trnI-GAU†, trnK-UUU†, trnL-CAA, trnL-UAA†, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC†, trnW-CCA, trnY-GUA | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12§, rps14, rps15, rps16†, rps18, rps19 | |
| Large subunit of ribosome | rpl2†, rpl14, rpl16†, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1†, rpoC2 | |
| Genes for photosynthesis | Subunits of NADH-dehydrogenase | ndhA†, ndhB†, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Genes for photosynthesis | Subunits of NADH-dehydrogenase | ndhA†, ndhB†, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome b/f complex | petA†, petB†, petD, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF†, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP ‡ | |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Translational initiation factor 1 | infA | |
| Genes of unknown function | ycf1, ycf2, ycf3‡, ycf4, ycf15 |
† Genes contain one intron; ‡ genes contain two introns; § genes that need trans-splicing.
3.2. Genetic Structure of the Chloroplast Genome of Olive
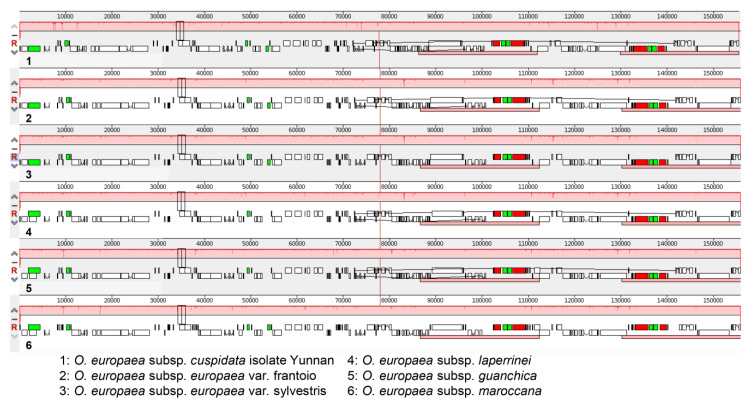
To make a comprehensive comparison of O. europaea chloroplast genomes, the gene structures of O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan were drafted with the other three O. europaea subspecies, including O. europaea subsp. laperrinei (MG255765.1), O. europaea subsp. guanchica (MG255764.1), and O. europaea subsp. maroccana (FN998900.2) obtained from NCBI (https://www.ncbi.nlm.nih.gov). The six O. europaea chloroplast genomes showed collinear gene organization with no rearrangement that occurred (Figure 2). Compared to O. europaea subsp. europaea var. sylvestris, O. europaea subsp. cuspidata isolate Yunnan had a large gap between rps16 and trnQ-UUG genes with six small gaps located in intergenic spacers (Figure 3). Furthermore, O. europaea subsp. laperrinei, O. europaea subsp. guanchica, and O. europaea subsp. maroccana had 4, 0, and 4 gaps, respectively.
Figure 2.
Synteny comparisons of six O. europaea chloroplast genomes. The chloroplast genome of O. europaea subsp. europaea var. sylvestris was used as reference sequence. Within each of the alignments, local collinear blocks were marked by the same color and connected by lines.
Figure 3.
Comparisons of six O. europaea chloroplast genomes. Chloroplast genome of O. europaea subsp. europaea var. sylvestris was used as reference sequence, and the horizontal axis indicated the coordinates with other chloroplast genomes. Gene, exon, intron, and intergenic spacer were colored.
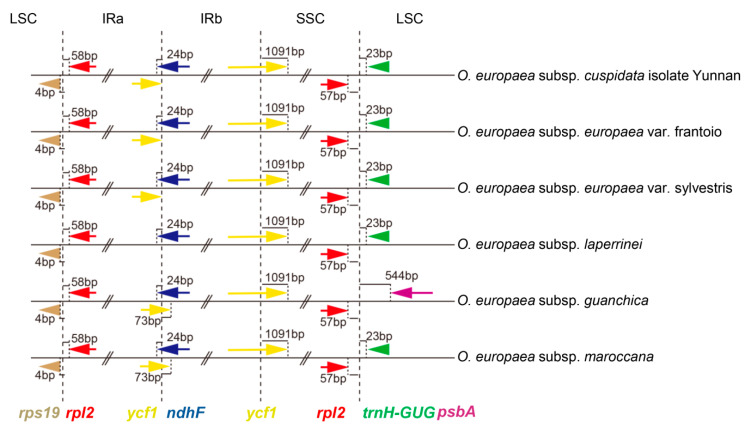
3.3. IR Expansion and Contraction
There were two significant differences of the chloroplast genomes among these six O. europaea accessions. O. europaea subsp. laperrinei and O. europaea subsp. guanchica lacked the ycf1 and the trnH-GUG gene near the IRa-SSC border and IRb-LSC border, respectively (Figure 4). While the nucleic acid sequences at the corresponding genes in these two samples were not significantly different from the other samples except for some SNPs, it was speculated that the ycf1 and the trnH-GUG genes were exhaustively annotated and existed.
Figure 4.
Border comparisons of six O. europaea chloroplast genomes. Chloroplast genome of O. europaea subsp. europaea var. sylvestris was used as reference sequence. LSC, large single-copy region; SSC, small single-copy region; IR, inverted repeat.
We also found that the ycf1 gene at the boundary between IRa and SSC had different expansion and contraction. As in Figure 4, the ycf1 gene from the three samples (O. europaea subsp. cuspidata isolate Yunnan, O. europaea subsp. europaea var. frantoio, and O. europaea subsp. europaea var. sylvestris) were right at the border of IRa and SSC, while in O. europaea subsp. guanchica and O. europaea subsp. maroccana, the ycf1 gene was located across both IRa and SSC regions.
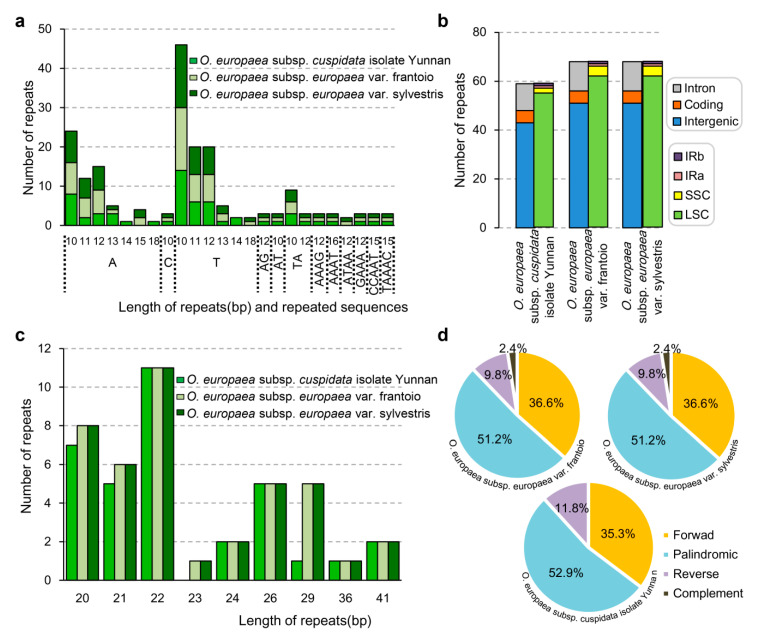
3.4. Repetitive Sequences and Hotspot Regions in Chloroplast Genomes
To further explore more differences, the microsatellites of three O. europaea chloroplast genomes were also studied. There were 68, 68, and 59 microsatellites identified in O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan, respectively (Figure 5a). For the 68 microsatellites identified from O. europaea subsp. europaea var. frantoio and O. europaea subsp. europaea var. sylvestris, 56 were mono-nucleotide, 6 were di-nucleotide, 4 were tetra-nucleotide, 2 were penta-nucleotide. No tri-nucleotide or hexa-nucleotide was found (Figure 5a). Among these microsatellites, 51, 5, and 12 microsatellites were located in the intergenic, protein-coding, and intron regions (Figure 5b). Of the 59 microsatellites identified from the O. europaea subsp. cuspidata isolate Yunnan, 48 were mono-nucleotide, 6 were di-nucleotide, 3 were tetra-nucleotide, and 2 were penta-nucleotide. No tri-nucleotide or hexa-nucleotide was found (Figure 5a). Among these microsatellites, 43, 5, and 11 microsatellites were located in the intergenic, protein-coding, and intron regions (Figure 5b).
Figure 5.
Statistical information of simple sequence repeats (SSRs) detected in O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan. (a) Distribution of SSRs in the different regions; (b) length and repeated sequences; (c) type of SSRs with 20 bp or longer; (d) statistics of SSRs with 20 bp or longer.
For microsatellites with 20 bp or longer, 41, 41, and 34 repeats were detected from O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris, and O. europaea subsp. cuspidata isolate Yunnan, respectively (Figure 5c). In detail, 20, 21, 22, 24, 26, 29, 36, and 41 bp-long repeats occurred in all of these three chloroplast genomes, while 23 bp-long repeats were only detected in O. europaea subsp. europaea var. frantoio, O. europaea subsp. europaea var. sylvestris. There were 51.2% and 52.9% considered as palindromic repeats in O. europaea subsp. europaea and O. europaea subsp. cuspidata isolate Yunnan (Figure 5d). No complement repeats were identified in O. europaea subsp. cuspidata isolate Yunnan (Figure 5d).
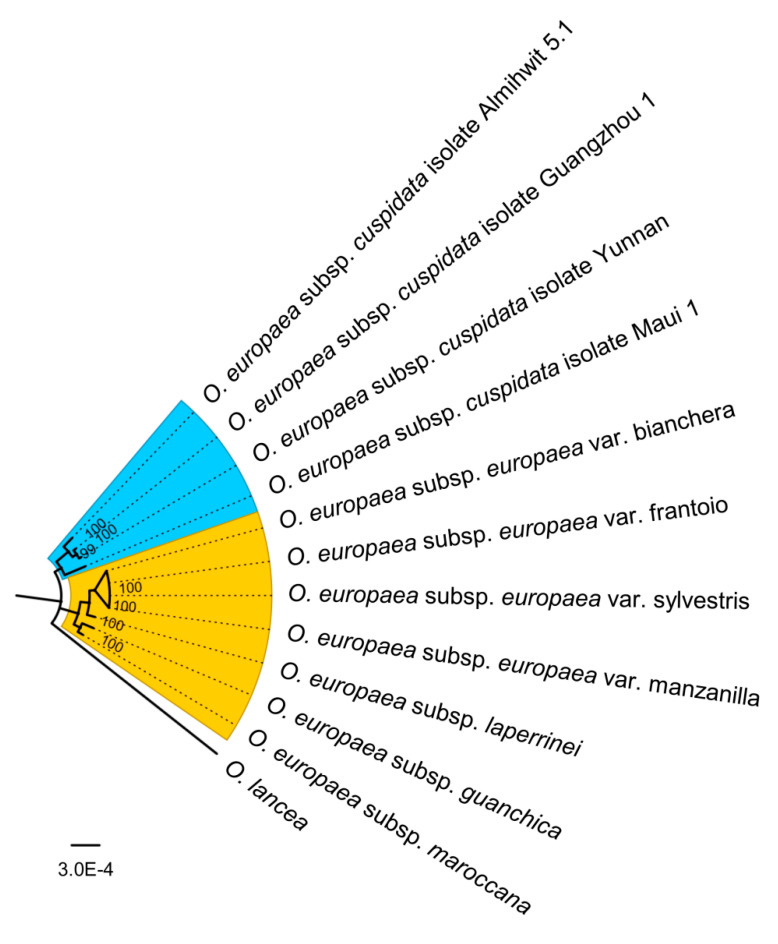
3.5. Genetic Phylogenetic Analysis
Due to the low genetic diversity, the whole chloroplast genome sequences of 11 O. europaea were constructed the genetic phylogenetic analysis based on maximum likelihood method with Olea lancea (NC_042278.1) as the outgroup (Figure 6). O. europaea chloroplast genomes were classified into two branches. O. europaea subsp. cuspidata was relatively different from the rest and grouped as an individual branch, forming the cuspidata clade as Besnard et al. [23] described. Four O. europaea subspecies O. europaea subsp. europaea, including the wild and cultivated, O. europaea subsp. laperrine, O. europaea subsp. guanchica, and O. europaea subsp. maroccana, showed closer relationships and formed the Mediterranean/North African clade.
Figure 6.
Phylogenetic analysis of O. europaea species. Whole chloroplast sequences of 11 O. europaea including O. europaea subsp. europaea var. frantoio (MT182984), O. europaea subsp. europaea var. sylvestris (MT182986), O. europaea subsp. cuspidata isolate Yunnan (MT182985), O. europaea subsp. laperrinei (MG255765.1), O. europaea subsp. guanchica (MG255764.1), O. europaea subsp. maroccana (FN998900.2), O. europaea subsp. europaea var. bianchera (NC_013707.2), O. europaea subsp. europaea var. manzanilla (FN996972.1), O. europaea subsp. cuspidata isolate Maui 1 (FN650747.2), O. europaea subsp. cuspidata isolate Guangzhou 1 (FN996944.1), O. europaea subsp. cuspidata isolate Almihwit 5.1 (FN996943.2), and Olea lancea (NC_042278.1) were used as the outgroup. Phylogenetic tree was built using IQTREE 1.6.10 software (http://www.iqtree.org) with maximum likelihood method (GTR + I + G).
4. Discussion
Six olive subspecies are recognized as before [3,6,7]. Among them, O. europaea subsp. europaea is generally considered to include two differentiated variants: The cultivated (O. europaea subsp. europaea var. europaea) and wild (O. europaea subsp. europaea var. sylvestris) olive [34,35]. The two variants show overlapping distributions in the Mediterranean basin. Although the diversity of morphology and stress physiology is clear, the botanical and genetic studies have verified that the cultivated variants are derived from wild olives [34,35,36,37]. Single or multiple independent domestication events has been a debate [38]. Here, the chloroplast genome of O. europaea subsp. europaea var. sylvestris was first sequenced and showed exactly the same as O. europaea subsp. europaea var. frantoio. They also displayed a high similarity with cultivated olives, indicating that few differentiation events were present in O. europaea subsp. europaea chloroplasts. More exploration of domestication events should be conducted to study the genome sequences.
The phylogenetic analysis based on the whole chloroplast sequences showed that O. europaea occupied two main groups, the Mediterranean/North African and the Cuspidata groups, which confirmed previous research using polymorphic sites [23,24,39]. The genetic structure and repetitive sequences displayed the divergence clearly between cuspidata and other subspecies. Although O. europaea subsp. cuspidata isolate Yunnan had the same number of coding regions without rearrangement, a large gap exists between rps16 and trnQ-UUG with six small gaps was present. Moreover, 59 microsatellites were identified from O. europaea subsp. cuspidata isolate Yunnan, compared to 68 found in O. europaea subsp. europaea. The results indicate high diversity between Cuspidata and Mediterranean/North African groups and further benefit the development of molecular markers.
In the genus Olea, only the cultivars of O. europaea are economically valuable, and O. europaea shows low genetic variation and obvious regionalization. O. europaea subsp. cuspidata has no economic value other than as an ornamental. The diversity of O. europaea subsp. europaea with other subspecies identified here could be used as an important gene resource to broaden the genetic background of olive cultivars through conventional or molecular breeding methods. They appear to be compatible using the conventional breeding methods. Ma et al. [40,41] reported that the variety Jinyefoxilan, derived from a cross between of O. europaea subsp. europaea var. frantoio and O. europaea subsp. cuspidata isolate Yunnan, had stronger abiotic stress-resistance tolerance, more vigorous vegetative growth, and a later flowering stage compared to the female parent. Our findings will provide more information on O. europaea subsp. cuspidata isolate Yunnan for molecular assisted breeding.
Author Contributions
Conceptualization, S.Z. and E.N.; sampling, E.N.; methodology, E.N. and Y.Z.; data curation, E.N.; writing—original draft preparation, E.N.; writing—review and editing, S.Z., C.J., and W.W.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Project, grant number “2019YFD1001205”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Muzzalupo I. Olive Germplasm—The Olive Cultivation, Table Olive and Olive Oil Industry in Italy. InTech Press; Rijeka, Croatia: 2012. [Google Scholar]
- 2.Zohary D., Spiegel-Roy P. Beginnings of fruit growing in the old world. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]
- 3.Sebastiani L., Busconi M. Recent developments in olive (Olea europaea L.) genetics and genomics: Applications in taxonomy, varietal identification, traceability and breeding. Plant Cell Rep. 2017;36:1–16. doi: 10.1007/s00299-017-2145-9. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Jiménez F., Ruano J., Perez-Martinez P., Lopez-Segura F., Lopez-Miranda J. The influence of olive oil on human health: Not a question of fat alone. Mol. Nutr. Food Res. 2010;51:1199–1208. doi: 10.1002/mnfr.200600273. [DOI] [PubMed] [Google Scholar]
- 5.Rigacci S., Stefani M., Stefania R., Massimo S. Nutraceutical properties of olive oil polyphenols. An itinerary from cultured cells through animal models to humans. Int. J. Mol. Sci. 2016;17:843. doi: 10.3390/ijms17060843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez C.M., Trujillo I., Martinez-Urdiroz N., Barranco D., Rallo L., Marfil P., Gaut B.S. Olive domestication and diversification in the Mediterranean Basin. New Phytol. 2015;206:436–447. doi: 10.1111/nph.13181. [DOI] [PubMed] [Google Scholar]
- 7.Besnard G., Khadari B., Baradat P., Bervillé A. Combination of chloroplast and mitochondrial DNA polymorphisms to study cytoplasm genetic differentiation in the olive complex (Olea europaea L.) Theor. Appl. Genet. 2002;105:139–144. doi: 10.1007/s00122-002-0868-6. [DOI] [PubMed] [Google Scholar]
- 8.Muzzalupo I., Vendramin G., Chiappetta A. Genetic biodiversity of Italian olives (Olea europaea) germplasm analyzed by SSR markers. Sci. World J. 2014;2014:296590. doi: 10.1155/2014/296590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaniewski D., Van Campo E., Boiy T., Terral J.F., Khadari B., Besnard G. Primary domestication and early uses of the emblematic olive tree: Palaeobotanical, historical and molecular evidences from the Middle East. Biol. Rev. Camb. Philos. Soc. 2012;87:885–899. doi: 10.1111/j.1469-185X.2012.00229.x. [DOI] [PubMed] [Google Scholar]
- 10.Bracci T., Sebastiani L., Busconi M., Fogher C. Evaluation of genetic diversity in Liguria region olive (Olea europaea L.) germplasm by SSR markers. Acta Hortic. 2008;122:209–215. doi: 10.1016/j.scienta.2009.04.010. [DOI] [Google Scholar]
- 11.Fabbri A., Lambardi M., Ozden-Tokatli Y. Olive breeding. In: Mohan Jain S., Priyadarshan P.M., editors. Breeding Plantation Tree Crops: Tropical Species. 1st ed. Springer; Boston, MA, USA: 2009. pp. 423–465. [Google Scholar]
- 12.Belaj A., Dominguez-Garcia M., Atienza S.G., Urdíroz N.M., De la Rosa R., Satovic Z., Martín A., Kilian A., Trujillo I., Valpuesta V., et al. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes. 2012;8:365–378. doi: 10.1007/s11295-011-0447-6. [DOI] [Google Scholar]
- 13.Grati-Kamoun N., Lamy-Mahmoud F., Rebaï A., Gargouri A., Panaud O., Saar A. Genetic diversity of Tunisian olive tree (Olea europaea L.) cultivars assessed by AFLP markers. Genet. Resour. Crop. Evol. 2006;53:265–275. doi: 10.1007/s10722-004-6130-0. [DOI] [Google Scholar]
- 14.Khaleghi E., Sorkheh K., Chaleshtori M.H., Ercisli S. Elucidate genetic diversity and population structure of Olea europaea L. germplasm in Iran using AFLP and IRAP molecular markers. 3 Biotech. 2017;7:71. doi: 10.1007/s13205-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.D’Agostino N., Taranto F., Camposeo S., Mangini G., Fanelli V., Gadaleta S., Miazzi M.M., Pavan S., di Rienzo V., Sabetta W., et al. GBS-derived SNP catalogue unveiled wide genetic variability and geographical relationships of Italian olive cultivars. Sci. Rep. 2018;8:15877. doi: 10.1038/s41598-018-34207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S., Niu E., Shi A., Mou B. Genetic diversity analysis of olive germplasm (Olea europaea L.) with genotyping-by-sequencing technology. Front. Genet. 2019;10:755. doi: 10.3389/fgene.2019.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu S., Niu E., Wang W., Shi A. Identification and evaluation of SNP core loci for olive germplasm (Olea europaea L.) Mol. Plant Breed. 2020;18:1548–1557. [Google Scholar]
- 18.Barghini E., Natali L., Cossu R.M., Giordani T., Pindo M., Cattonaro F., Scalabrin S., Velasco R., Morgante M., Cavallini A. The peculiar landscape of repetitive sequences in the olive (Olea europaea L.) genome. Genome Biol. Evol. 2014;6:776–791. doi: 10.1093/gbe/evu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz F., Julca I., Gómez-Garrido J., Loska D., Marcet-Houben M., Cano E., Galán B., Frias L., Ribeca P., Derdak S., et al. Genome sequence of the olive tree, Olea europaea. Gigascience. 2016;5:29. doi: 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unver T., Wu Z., Sterck L., Turktas M., Lohaus R., Li Z., Yang M., He L., Deng T., Escalante F.J., et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA. 2017;114:E9413–E9422. doi: 10.1073/pnas.1708621114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besnard G., Hernández P., Khadari B., Dorado G., Savolainen V. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biol. 2011;11:80. doi: 10.1186/1471-2229-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amane M., Lumaret R., Hany V., Ouazzani N., Debain C., Vivier G., Deguilloux M.F. Chloroplast-DNA variation in cultivated and wild olive (Olea europaea L.) Theor. Appl. Genet. 1999;99:133–139. doi: 10.1007/s001220051217. [DOI] [Google Scholar]
- 23.Besnard G., Khadari B., Baradat P., Bervillé A. Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theor. Appl. Genet. 2002;104:1353–1361. doi: 10.1007/s00122-001-0832-x. [DOI] [PubMed] [Google Scholar]
- 24.Mariotti R., Cultrera N.G., Diez C.M., Baldini L., Rubini A. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biol. 2010;10:211. doi: 10.1186/1471-2229-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 2 June 2014)]; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 28.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 30.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 31.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Wang Y., He P., Li P., Fu C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018;19:235. doi: 10.1186/s12864-018-4633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green P.S. A revision of Olea L. (Oleaceae) Kew Bull. 2002;57:91–140. doi: 10.2307/4110824. [DOI] [Google Scholar]
- 35.Gros-Balthazard M., Besnard G., Sarah G., Holtz Y., Leclercq J., Santoni S., Wegmann D., Glémin S., Khadari B. Evolutionary transcriptomics reveals the origins of olives and the genomic changes associated with their domestication. Plant J. 2019;100:143–157. doi: 10.1111/tpj.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belaj A., León L., Satovic Z., de la Rosa R. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analysed by agromorphological traits and SSR markers. Sci. Hort. 2011;129:561–569. doi: 10.1016/j.scienta.2011.04.025. [DOI] [Google Scholar]
- 37.Pritsa T.S., Voyiatzis D.G., Voyiatzi C.J., Sotiriou M.S. Evaluation of vegetative growth traits and their relation to time to first flowering of olive seedlings. Crop. Pasture Sci. 2003;54:371–376. doi: 10.1071/AR02131. [DOI] [Google Scholar]
- 38.Besnard G., Rafael R.D.C. Single vs multiple independent olive domestications: The jury is (still) out. New Phytol. 2016;209:466–470. doi: 10.1111/nph.13518. [DOI] [PubMed] [Google Scholar]
- 39.Rugini E., Baldoni L., Muleo R., Sebastiani L. The Olive Tree Genome. Springer Nature; Cham, Switzerland: 2016. [Google Scholar]
- 40.Ma T., Ning D.L., Yang W.M., Zhang Z.Z., Li Y.J., Xu T., Xiao L.J. Breeding of a new olive varieties “Jinyefoxilan”. China Fruits. 2014;6:3–4. [Google Scholar]
- 41.Ma T., Xu T., Ning D.L., Xiao L.J., Li J. Comparative study on the growth and morphology of new olive varieties “Jinyefoxilan” and its parents. South. Hortic. 2015;26:1–3. [Google Scholar]