Abstract
Dendrobium officinale Kimura et Migo is of great importance as a traditional Chinese herb due to its abundant metabolites. The family of basic helix-loop-helix (bHLH) transcription factors widely exists in plants and plays an essential role in plant growth and development, secondary metabolism as well as responses to environmental changes. However, there is limited information on bHLH genes in D. officinale. In the present study, a total of 98 putative DobHLH genes were identified at the genomic level, which could be classified into 18 clades. Gene structures and conserved motifs in DobHLH genes showed high conservation during their evolution. The conserved amino acids and DNA bindings of DobHLH proteins were predicted, both of which are pivotal for their function. Furthermore, gene expression from eight tissues showed that some DobHLH genes were ubiquitously expressed while other DobHLH genes were expressed in the specific tissues. Expressional changes of DobHLH genes under MeJA and ABA treatments were detected by qRT-PCR. The protein–protein interactions between DobHLHs were predicted and several interactions were confirmed by yeast two hybrid. Therefore, our results here contribute to the understanding of bHLH genes in D. officinale and lay a foundation for the further functional study of its biological processes.
Keywords: Dendrobium officinale, basic helix-loop-helix transcription factors, genome-wide identification, expression analysis, protein–protein interaction, skin care
1. Introduction
Dendrobium officinale Kimura et Migo (or known as D. catenatum) is a perennial herbal plant, belonging to the family Orchidaceae. D. officinale plants are widely distributed across southern provinces in China, with limited spread to southeast Asian countries [1]. D. officinale is of great importance due to its medicinal and ornamental uses. D. officinale ranks first among the nine Chinese herbs for longevity and has a documented use in folk medicine for over 1300 years [2]. In the past decades, about 190 compounds have been identified and isolated from D. officinale plants including phenanthrenes, bibenzyls, saccharides and glycosides, alkaloids, essential oils, and others such as phenols, acids, esters, and amides [3]. Among them, polysaccharides, bibenzyls, alkaloids, and flavones have been confirmed as the major and bioactive ingredients [3,4]. The abundance of polysaccharides in its stems and the specific alkaloids make D. officinale have anticancer [1], anti-oxidant [5], and anti-inflammatory effects [1,6] as well as support immune modulation [7,8], hepatop protection, and hypoglycemia [8,9]. Due to its multiple health benefits, demand for high-quality D. officinale is increasing across medicine, food, and skin care.
As an epiphytic plant, wild D. officinale prefers to grow in a warm and humid environment. In nature, D. officinale inevitably suffers environmental stresses such as high temperature and drought, slowing growth, and lowering yield [10]. Despite abundant polysaccharides, the contents of other active components in D. officinale are low [11,12,13]. At present, many potential genes involved in the biosynthesis of polysaccharides, alkaloids, and flavonoids have been identified in D. officinale through transcriptome and genomic sequencings [4,14]. In D. officinale, polysaccharides in the stem, leaves, and flowers are one of the major active ingredients, and sucrose synthase (Susy) and two gene families (β-galactosidase and galacturonosyltransferase) are related to the contents and richness, respectively [2]. In addition, 10 sucrose phosphate synthase and 15 Susy genes showed marked expansion [2]. The upstream pathways of alkaloid biosynthesis in plants are conserved and clear, which are responsible for the formation of strictosidine. The downstream steps of the alkaloid biosynthesis in D. officinale are modification of strictosidine including cytochrome P450s (CYP450s)-mediated oxidation and hydroxylation reactions [15], which needs further investigation. However, transcription regulation of the biosynthesis has been largely unknown. As an important switch of gene expression, transcription factor (TF) can activate or repress the expression of specific target genes through interaction with cis-elements in the gene promoter region, which moderates various biological processes such as growth, response to stresses, and the biosynthesis of secondary metabolites [16].
Basic helix-loop-helix (bHLH) TFs are widespread in the eukaryotes and also are the second largest family of TFs in plants [17,18,19,20]. bHLH TFs are named due to the presence of a highly conserved bHLH domain, and the bHLH domain of 50–60 amino acids contains one basic region and one HLH region [21,22]. The basic region of 15 amino acids is at the N-terminus of the bHLH domain, while the HLH region of 40–50 amino acids is at the C-terminus of the bHLH domain including two alpha helices and one less conserved loop with variable lengths. The bHLH domain can specifically recognize and bind a conserved cis-element CANNTG, named E-box [23,24]. The two helices from the same bHLH TF or different bHLH TFs can interact to form a homodimer or a heterodimer, which then binds different regions in the promoter to regulate the expression of their target genes [25]. The other sequences of bHLH TFs are variable and less conserved.
The bHLH TF was first found in murine muscle [21], and later in animals [23] and plants [22,26]. Based on differences in the sequences, DNA binding, bHLH domain and functions, the bHLH TF family in animals is divided into six lineages (group A–F) [23]. In plants, most bHLH TFs share a similar structure with that of group B from animals, which can bind the G-box (CACGTG), although they also bind E-box [16]. Unlike in animals, Arabidopsis bHLH TFs are divided into many more clades [22]. The classification of plant bHLHs is gradually being clarified and most plants contain 15–26 clades due to their identification in more plants including tomato [27], maize [28], grape [29], cotton [30], jujube [25], and Brachypodium distachyon [31].
bHLH TFs play important roles in plant secondary metabolites. The first bHLH protein identified in plant was R gene in maize, which can regulate the expression of at least two genes in the flavonoid/anthocyanin pathway [32]. In Arabidopsis, the three bHLH proteins Glabra3 (GL3), Enhancer of Glabra3 (EGL3), and Transparent Testa8 (TT8) can interact with the myeloblastosis (MYB) transcription factor and WD40 protein to form a MYB–bHLH–WD complex, activating multiple genes in the biosynthesis of anthocyanin and resulting in anthocyanin accumulation [33]. An increasing number of research is reporting that bHLH TFs also participate in the regulation of other secondary metabolites including flavonols [33], terpenoid indole alkaloids [34], and tanshinone [35]. Recently a bHLH TF was found to be responsible for the specific anthocyanin of lip tissues in Dendrobium hybrids [36]. Identification of bHLH TFs in D. officinale is the first step to understanding their roles in the accumulation of bioactive compounds.
In addition, bHLH TFs participate in plant growth and development including photomorphogenesis, light signal transduction, flowering time, and the development of various tissues such as flower and root [16]. For example, two Arabidopsis phytochrome interacting factors (AtPIF4 and PIF3) can interact with phytochrome to control the expression of the genes involved in the regulation of light response [22,37]. A large number of bHLH TFs play a vital role in the response to environmental stresses such as salt, drought, and high temperature. Inducer of CBF expression 1 (ICE1), belonging to the III b subclade of Arabidopsis bHLH family, can activate downstream genes through the mediation of C-repeat binding factor (CBF) in response to freezing stress [38].
Although there has been much research on bHLH TFs in various plants, a comprehensive investigation of the bHLH family in D. officinale has yet not been carried out. The competence of genome sequencing for D. officinale makes the search for potential genes associated with important traits possible [2,39]. In the present study, we first identified a total of 98 candidate DobHLH members. Their phylogenetic relationship, conserved bHLH domain and motifs, gene structures, and cis-elements were characterized. The expression patterns in eight tissues were analyzed from the RNA-Seq data. The expression levels of some DobHLH genes under hormone treatments were detected by quantitative real time-polymerase chain reaction (qRT-PCR). The possible protein–protein interactions were predicted, and some interactions were confirmed by yeast two hybrid. Accordingly, our results provide information on the bHLH family in D. officinale and lay the foundation for further investigation into its function in its biological process.
2. Results
2.1. Genome-Wide Identification of DobHLH Members in Dendrobium officinale
After the hidden markov model (HMMER) search and confirmation of the bHLH domain presences with online CD-search tool and SMART, a total of 98 members in D. officinale were considered DobHLH candidates. They were named from DobHLH1 to DobHLH98 according to their relationship with AtbHLH proteins. The basic information of DobHLH members is shown in Table 1 and Table S1. The DobHLH proteins contain 85–662 AAs with molecular weights of 9.53–74.54 kDa and isoelectric points of 4.41–10.78. The subcellular localization of 98 DobHLH proteins were predicted by WOLF PSORT. The results showed that most of the predicted proteins (89) were localized in the nucleus, and five and four DobHLH proteins were in the cytoplasmic and mitochondrial, respectively. The 98 DobHLH genes were distributed in 85 scaffolds. The CDS lengths of the DobHLH genes varied from 258 bp (DobHLH95 and DobHLH96) to 1989 bp (DobHLH24) while the lengths of genomic DNA were from 496 bp (DobHLH79) to 26,581 bp (DobHLH58).
Table 1.
The list of 98 DobHLH genes in D. officinale.
| Gene Name | Gene ID |
NCBI References |
Clade | Gene Name | Gene ID |
NCBI References |
Clade |
|---|---|---|---|---|---|---|---|
| DobHLH1 | LOC110116721 | XP_020706080.1 | 1 | DobHLH50 | LOC110116300 | XP_020705489.1 | 10 |
| DobHLH2 | LOC110115756 | XP_020704772.1 | 1 | DobHLH51 | LOC114579352 | XP_028549432.1 | 10 |
| DobHLH3 | LOC110107102 | XP_020692919.1 | 1 | DobHLH52 | LOC110095032 | XP_020676060.1 | 11 |
| DobHLH4 | LOC110096422 | XP_020678032.1 | 1 | DobHLH53 | LOC110113329 | XP_020701532.1 | 11 |
| DobHLH5 | LOC110116474 | XP_020705700.1 | 1 | DobHLH54 | LOC110113204 | XP_020701328.1 | 11 |
| DobHLH6 | LOC110098508 | XP_020681021.1 | 1 | DobHLH55 | LOC110114625 | XP_020703217.1 | 12 |
| DobHLH7 | LOC110111619 | XP_020699225.1 | 1 | DobHLH56 | LOC110106166 | XP_020691604.1 | 12 |
| DobHLH8 | LOC110112576 | XP_020700508.1 | 1 | DobHLH57 | LOC110112285 | XP_020700113.1 | 12 |
| DobHLH9 | LOC110108487 | XP_020694816.2 | 1 | DobHLH58 | LOC110108563 | XP_020694908.1 | 12 |
| DobHLH10 | LOC110110298 | XP_020697356.1 | 2 | DobHLH59 | LOC110101829 | XP_020685553.1 | 13 |
| DobHLH11 | LOC110093619 | XP_020674221.1 | 2 | DobHLH60 | LOC110095526 | XP_020676768.1 | 13 |
| DobHLH12 | LOC110115484 | XP_020704390.1 | 2 | DobHLH61 | LOC110111433 | XP_028555769.1 | 13 |
| DobHLH13 | LOC110093741 | XP_020674407.1 | 3 | DobHLH62 | LOC110093428 | XP_020673967.1 | 13 |
| DobHLH14 | LOC110102219 | XP_020686105.1 | 3 | DobHLH63 | LOC110104725 | XP_020689612.1 | 14 |
| DobHLH15 | LOC110098909 | XP_020681520.1 | 3 | DobHLH64 | LOC110102342 | XP_028552167.1 | 14 |
| DobHLH16 | LOC110107046 | XP_028551992.1 | 3 | DobHLH65 | LOC110107722 | XP_020693735.1 | 14 |
| DobHLH17 | LOC110096269 | XP_020677790.1 | 3 | DobHLH66 | LOC110100649 | XP_020683910.1 | 14 |
| DobHLH18 | LOC110116147 | XP_020705280.1 | 4 | DobHLH67 | LOC110114258 | XP_028550306.1 | 14 |
| DobHLH19 | LOC110094813 | XP_028551573.1 | 4 | DobHLH68 | LOC110114447 | XP_028556766.1 | 14 |
| DobHLH20 | LOC110107826 | XP_020693884.2 | 4 | DobHLH69 | LOC110109277 | XP_028547820.1 | 14 |
| DobHLH21 | LOC110094726 | XP_020675682.1 | 4 | DobHLH70 | LOC110096494 | XP_020678137.1 | 14 |
| DobHLH22 | LOC110114654 | XP_020703261.1 | 5 | DobHLH71 | LOC110111863 | XP_028551171.1 | 14 |
| DobHLH23 | LOC110111891 | XP_020699606.1 | 5 | DobHLH72 | LOC110107433 | XP_028552737.1 | 14 |
| DobHLH24 | LOC110097687 | XP_020679864.1 | 5 | DobHLH73 | LOC110114740 | XP_020703378.2 | 14 |
| DobHLH25 | LOC110103241 | XP_020687528.1 | 6 | DobHLH74 | LOC110095482 | XP_020676694.1 | 14 |
| DobHLH26 | LOC110094094 | XP_020674906.1 | 6 | DobHLH75 | LOC110099461 | XP_020682274.1 | 14 |
| DobHLH27 | LOC110116682 | XP_020706025.1 | 6 | DobHLH76 | LOC110099214 | XP_028548138.1 | 14 |
| DobHLH28 | LOC110099101 | XP_020681804.1 | 6 | DobHLH77 | LOC110098199 | XP_020680601.1 | 15 |
| DobHLH29 | LOC110113808 | XP_020702166.2 | 6 | DobHLH78 | LOC110098270 | XP_020680696.1 | 15 |
| DobHLH30 | LOC110094435 | XP_020675326.1 | 6 | DobHLH79 | LOC110094287 | XP_020675140.1 | 15 |
| DobHLH31 | LOC110116479 | XP_020705705.1 | 6 | DobHLH80 | LOC110113891 | XP_020702260.1 | 15 |
| DobHLH32 | LOC110114462 | XP_020703009.1 | 6 | DobHLH81 | LOC110112441 | XP_020700324.1 | 15 |
| DobHLH33 | LOC110092865 | XP_020673218.1 | 6 | DobHLH82 | LOC110101026 | XP_020684454.1 | 15 |
| DobHLH34 | LOC110109085 | XP_020695652.1 | 7 | DobHLH83 | LOC110111081 | XP_020698439.1 | 15 |
| DobHLH35 | LOC110096203 | XP_020677673.1 | 7 | DobHLH84 | LOC110104845 | XP_020689772.1 | 15 |
| DobHLH36 | LOC110112097 | XP_020699850.1 | 7 | DobHLH85 | LOC110095998 | XP_020677402.1 | 15 |
| DobHLH37 | LOC110107930 | XP_020694038.1 | 8 | DobHLH86 | LOC110110399 | XP_020697513.2 | 15 |
| DobHLH38 | LOC110115493 | XP_020704404.1 | 8 | DobHLH87 | LOC110109507 | XP_028547425.1 | 15 |
| DobHLH39 | LOC110094861 | XP_020675853.1 | 8 | DobHLH88 | LOC114578594 | XP_028547764.1 | 15 |
| DobHLH40 | LOC110112072 | XP_020699818.1 | 8 | DobHLH89 | LOC110105526 | XP_020690722.2 | 15 |
| DobHLH41 | LOC110114469 | XP_028556744.1 | 9 | DobHLH90 | LOC110100961 | XP_020684346.1 | 15 |
| DobHLH42 | LOC110107963 | XP_020694091.1 | 9 | DobHLH91 | LOC110112336 | XP_020700191.1 | 16 |
| DobHLH43 | LOC110106259 | XP_028550041.1 | 9 | DobHLH92 | LOC110110400 | XP_020697514.1 | 17 |
| DobHLH44 | LOC110107318 | XP_028547365.1 | 10 | DobHLH93 | LOC110112399 | XP_028555091.1 | 17 |
| DobHLH45 | LOC110103817 | XP_028551326.1 | 10 | DobHLH94 | LOC110108630 | XP_020695011.1 | 17 |
| DobHLH46 | LOC110114754 | XP_020703396.1 | 10 | DobHLH95 | LOC110110529 | XP_020697710.1 | 17 |
| DobHLH47 | LOC110107832 | XP_020693899.1 | 10 | DobHLH96 | LOC110108826 | XP_020695307.1 | 17 |
| DobHLH48 | LOC110107031 | XP_020692824.1 | 10 | DobHLH97 | LOC110105593 | XP_020690812.1 | 17 |
| DobHLH49 | LOC110098511 | XP_020681024.1 | 10 | DobHLH98 | LOC110100228 | XP_020683310.1 | 18 |
Compared with the numbers of bHLH members in other plant species, the number in D. officinale was similar to that found in grape [29] and Chinese jujube [25] and was less than that found in most other species with over 100 bHLH members such as Arabidopsis (166), rice [40], Brachypodium distachyon [31], and tomato [27]. Some important crops had up to 200 or even many more bHLH members including maize [28], poplar [41], wheat [42], cotton [30], and Brassica napus [43]. The density of bHLH members in the D. officinale genome was approximately 0.28, which was similar to peach, but higher than that found in the genomes of the two lower plants Volvox carteri (0.024) and Chlorella vulgaris (0.081) [44]. These results showed that the number of bHLH genes in different species could be connected with the genome size.
2.2. Phylogenetic Relationship of DobHLH Proteins
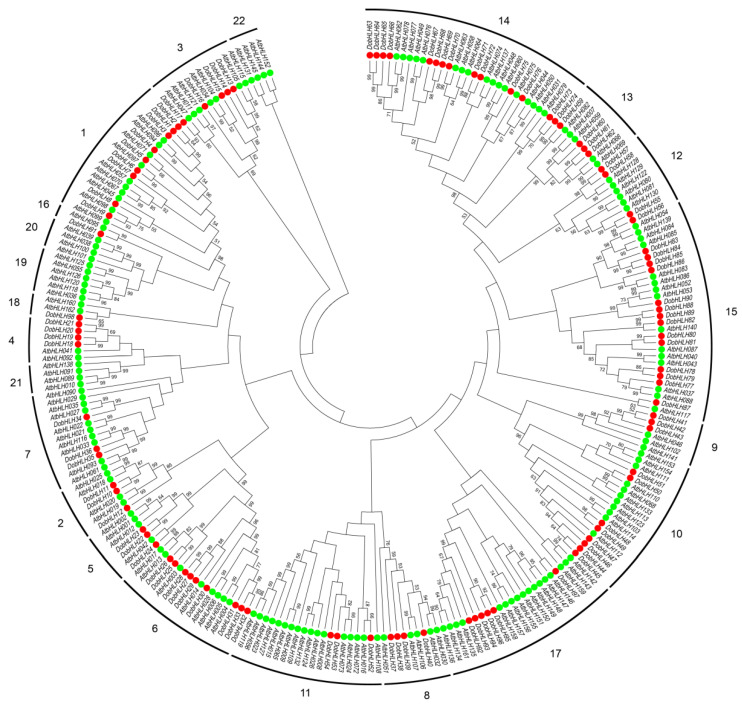
To reveal the evolution of bHLH members, an unrooted-tree was constructed using MEGA 7.0 with the neighbor-joining method including 98 DobHLHs and 166 AtbHLHs (Figure 1). The phylogenetic tree could be divided into 22 clades, and each clade included two to 31 members. Ninety-eight DobHLHs were distributed in 18 clades (clade 1 to clade 18) and four clades (clade 19, 20, 21, 22) contained bHLH proteins only from Arabidopsis, without proteins from D. officinale, which suggested that gene deletion could occur during the evolution of D. officinale. In most clades, there were fewer bHLH proteins from D. officinale than from Arabidopsis. Clade 4 and 6 had more DobHLHs than AtbHLHs and clade 9 and 16 had the same number of bHLH members (three and one).
Figure 1.
Phylogenetic analysis of bHLH proteins from D. officinale and Arabidopsis. A total of 98 DobHLH proteins and 162 AtbHLH from Arabidopsis (Arabidopsis thaliana) were used to construct the unrooted neighbor-joining (NJ) tree with a bootstrap of 1000 replicates. The bHLH proteins are grouped into 22 clades, and marked in different colors: AtbHLH genes are in green while DobHLH genes are in red.
Moreover, an unrooted tree was constructed with bHLH proteins from D. officinale and rice (Figure S1). For as many as 15 clades, there were fewer bHLH proteins from D. officinale than from rice. Clades 6 and 17 contained more bHLH proteins from D. officinale than from rice. There were more bHLH proteins in the rice genome than in D. officinale, which could result from genomic or random duplication after differentiation with D. officinale [40].
2.3. Gene Structures and Conserved Motifs of DobHLH Genes
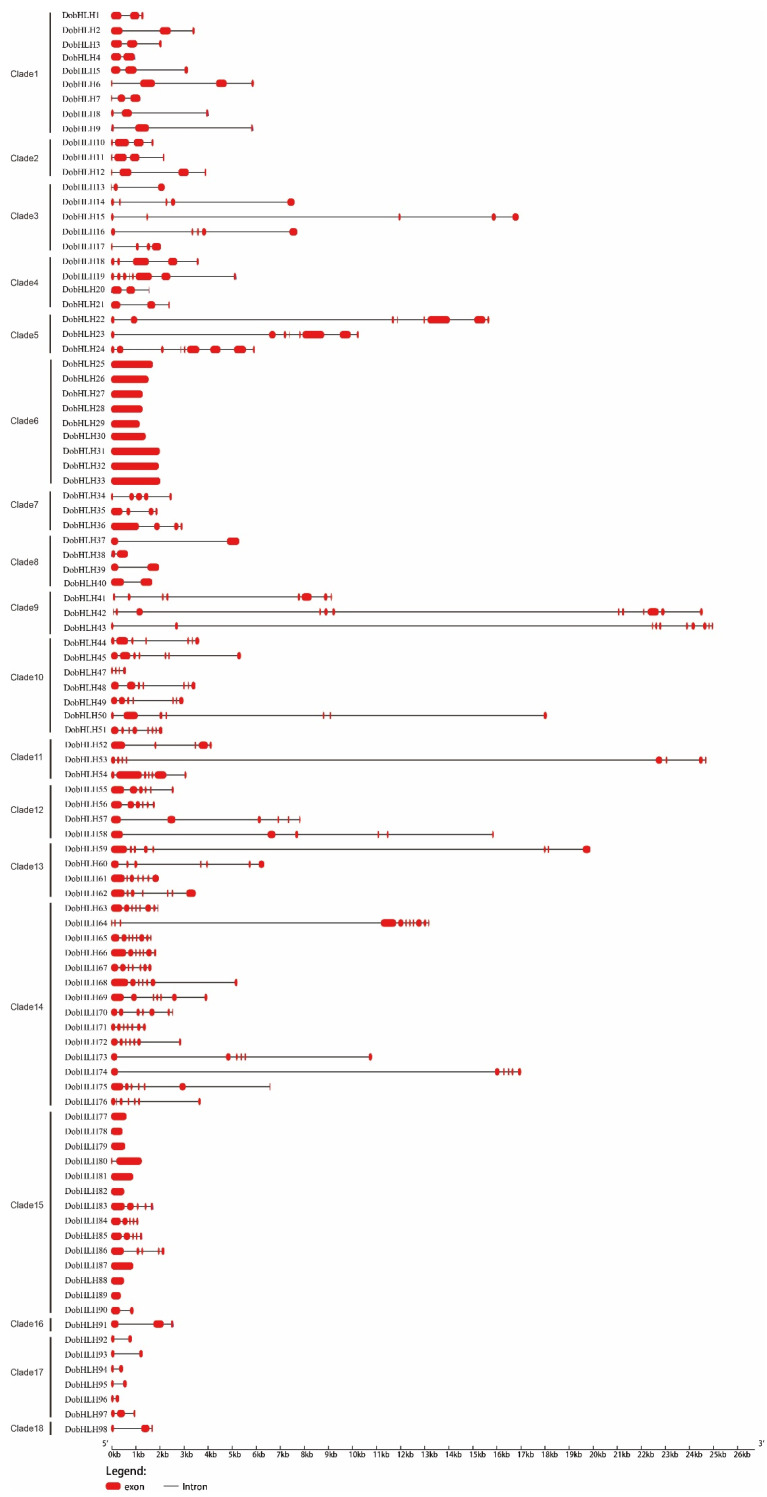
To reveal the evolution of DobHLH genes, the exon-intron structures were investigated by comparing CDS sequences with genomic sequences. The results showed that the intron numbers varied greatly among the DobHLH family members, from some completely lacking intron to some having up to 11 introns (Figure 2). Most of the members within the same clade shared the same number of introns. For example, clades 1, 2, 8, 10, 12, 16, 17 had two, three, one, six, five, two, and one intron, respectively, while all members in clade 6 and eight members in clade 15 had no introns. Appropriately 25% of the members (25) had one or two introns, mainly including members from clades 1, 5, 8, 16, 17 and 18. Most members from clades 3, 5, 7, 10, 11, 12, 13, and 14 including 43 DobHLH genes contained three to seven introns. Four genes (DobHLH24, 42, 43, and 64) had more than seven introns. The lengths of introns imbedded in the DobHLH genes ranged from 44 bp to 21,971 bp, with the longest intron in the DobHLH53 gene.
Figure 2.
The gene structures of DobHLH genes in D. officinale. The exon-intron structures of 98 DobHLH genes were obtained with GSDS 2.0. The red rectangles and black lines indicate exons and introns, respectively, according to their lengths.
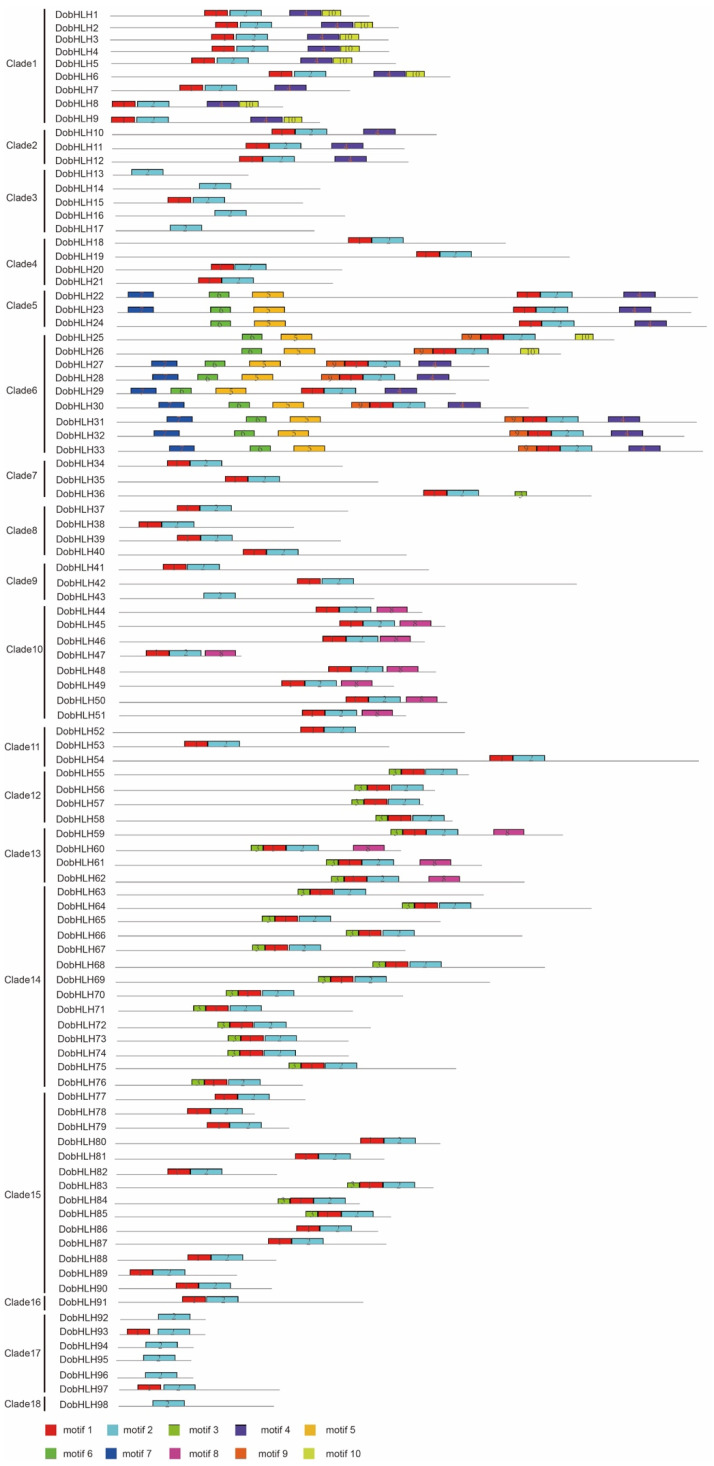
To further understand the structure of DobHLH proteins, we searched 10 conserved motifs in DobHLHs with MEME software. As shown in Figure 3, motif 1 and motif 2 were the two most conserved motifs, which were widely present in DobHLH members. The latter ten residues in the basic region and the first helix were motif 1, while the second helix was the main part of motif 2. Motif 1 was present in 88 DobHLH proteins while motif 2 was present in all the 98 DobHLH proteins. Ten DobHLH proteins (including DobHLH13, 14, 16, 17, 43, 92, 94, 95, 96, 98) had motif 2 only and lacked the basic region of bHLH domain. The other 88 DobHLH proteins had both motif 1 and motif 2, and 30 DobHLH members contained only motif 1 and motif 2, mainly from clades 4, 7, 8, 9, 11, 15, 16, and 17. Clades 5 and 6 contained the most motifs (five to seven). Motif 3 was present in all the members from clades 12, 13, 14 and some members from clade 15. Motif 3 was distributed in clade 1, 2, 5, and 6, with the exception of DobHLH25 and DobHLH26. Motif 5 and motif 6 were both present in the genes of clade 5 and clade 6. Motif 7 was limited in two members from clade 5 (DobHLH22 and DobHLH23) and seven members from clade 6. Motif 8 was present in all genes from clade 10 and clade 13. Motif 9 was limitedly present in the members of clade 6 without DobHLH29. Motif 10 was present in nine members from clades 1 and 6 (DobHLH1-6, 8, 9, 25, 26). There were similar motifs in most members from each clade. Eight of nine DobHLH members from clade 1 contained four motifs (motif 1, 2, 4, 10), and three members from clade 2 had motif 1, 2, and 4. Four of five members in clade 3 included motif 2. Ten DobHLH members from clade 10 had motifs 1, 2, and 8 while 14 members from clade 14 included motifs 1, 2, and 3.
Figure 3.
The conserved motifs of DobHLH proteins predicted by MEME. The colored boxes with numbers represent 10 conserved motif and the grey lines indicate non-conserved lines.
2.4. Conserved Amino Acids in the DobHLH Domains and DNA-Binding Ability
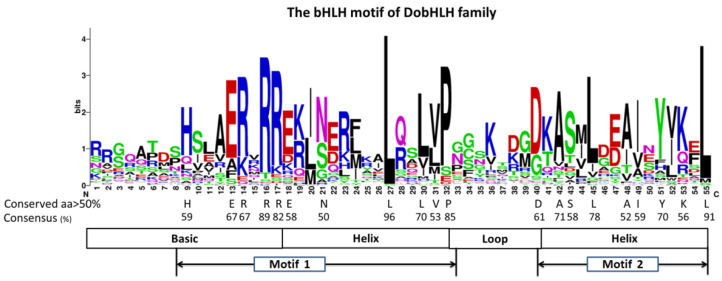
As the bHLH domain composed of motif 1 and motif 2 is especially important for bHLH TF to function, we analyzed the amino acids in bHLH domains from 98 DobHLH proteins (Figure 4). It was found that the D. officinale bHLH domain included 55 AAs, which was similar in Arabidopsis (56, [22]) but shorter than in tomato (61, [27]). The basic region in the D. officinale bHLH domain included 17 AAs and five AAs, showing over 50% identity in sequences. The first helix and the second helix both contained 15 AAs and six and eight AAs had more than 50% identity, respectively. The loop in the D. officinale bHLH domain had AAs and only Asp-40 was conserved with an identity of 60%.
Figure 4.
Analysis of the conserved amino acids in all DobHLH domains. The overall height of each amino acid represents the sequence conservation at that position. The conserved AAs and the consensus of more than 50% among 98 DobHLH domains are indicated in black letters.
As seen in Table S2, there were 20 AAs in the D. officinale bHLH domain that showed over 50% conservation and three residues (Arg-16, Leu-27, and Leu-55) were highly conserved in D. officinale, up to 89%, 96% and 91%, respectively. It has been found that these three residues were conserved both in plants and animals [22], suggesting their core sites in the bHLH domain. Moreover, seven residues including Ile-20, Leu-24, Gln-28, Met-44, Ile-49, Val-52, and Leu-55 were more conserved in plants such as D. officinale and Arabidopsis than in animals, which was also observed in eggplant [22]. This could imply their important role in plants. Compared with residues in tomato and Arabidopsis, Glu-13, Arg-14, Gln-28, Lys-36, Ala-48, and Ile-52 were less conserved.
As TFs, the most important function for bHLH proteins is to bind the promoter region of their target genes. Based on previous studies on the ability to bind DNA, we distinguished the types of DobHLH proteins (Table 2). It was found that 98 DobHLH proteins could be divided into two types: 22 non-DNA binding DobHLHs and 76 DNA binding DobHLHs. The 22 non-DNA binding DobHLHs mainly included members from clades 2, 10, 12, 15, and 17, and there were less than six residues in the basic region of 17 AAs in the bHLH domain. According to differences in the DNA binding sequences, 76 DNA binding DobHLHs could be further classified into 65 E-box binders and 11 non-E-box binders. When the 13 and 16 residues in the basic region are Glu and Arg, respectively, the bHLH protein has the capacity to bind E-box. Otherwise, the DobHLH protein belongs to non-E-box binders. Eleven non-E-box binders were from clades 10, 15, 17, and 18. The E-box binders could be further discriminated into G-box binders and non-G-box binders. When the 9, 13, and 17 residues in the basic region are His/Lys, Glu and Arg, respectively, the bHLH protein is considered to bind the classic G-box. In D. officinale, there were four DobHLH proteins that belong to non-G-box binders including DobHLH34, 35, and 36 from clade 7 and DobHLH97 from clade 17. The remaining 61 DobHLH proteins were G-box binders, accounting for 62.24% of all the DobHLH members.
Table 2.
The predicted types of DNA-binding based on DobHLH domain sequences.
| Predicted DNA-Binding Types Based on the bHLH Domain | Number of DobHLHs |
The Ratio |
|---|---|---|
| DNA binding | ||
| E-box | 65 | 62.25% |
| G-box | 61 | 62.24% |
| Non-G-box | 4 | 4.08% |
| Non-E-box | 11 | 11.22% |
| Total | 76 | 77.55% |
| Non-DNA binding | 22 | 22.45% |
2.5. Cis-Elements in the Promoter Regions of DobHLH Genes
Due to functional divergence, the members in the same gene family could show diverse expression patterns. To investigate regulatory patterns, we detected the cis-elements in the promoter regions of DobHLH genes (Table S3). Many cis-elements related to responses to stresses were predicted by PlantCARE. The most frequent cis-element was G-box, which was present in 72 DobHLH genes, suggesting that a light signal could be vital in transcriptionary regulation. Other cis-elements such as the CGTCA-motif (or TGACG-motif), ABRE, and ERE were distributed in 65, 69, and 78 DobHLH genes, respectively, which are responsible for responses to methyl jasmonate (MeJA), abscisic acid (ABA), and ethylene, respectively. There were 166 CGTCA-motifs (or TGACG-motifs), 201 ABRE cis-elements, and 188 ERE cis-elements. The cis-elements related to stress responses were also found in DobHLH gene promoter regions such as TC-rich repeats in 47 DobHLH genes, WUN-motif in 50 DobHLH genes, LTR in 39 DobHLH genes, and MBS in 42 DobHLH genes. These cis-elements can respond to defense and stress, wounding, low temperature, and drought. Other cis-elements involved in plant growth and development were observed in some DobHLH genes including ARE, CAT-box, and O2-Site.
2.6. Expression Patterns of DobHLH Genes in Eight Tissues
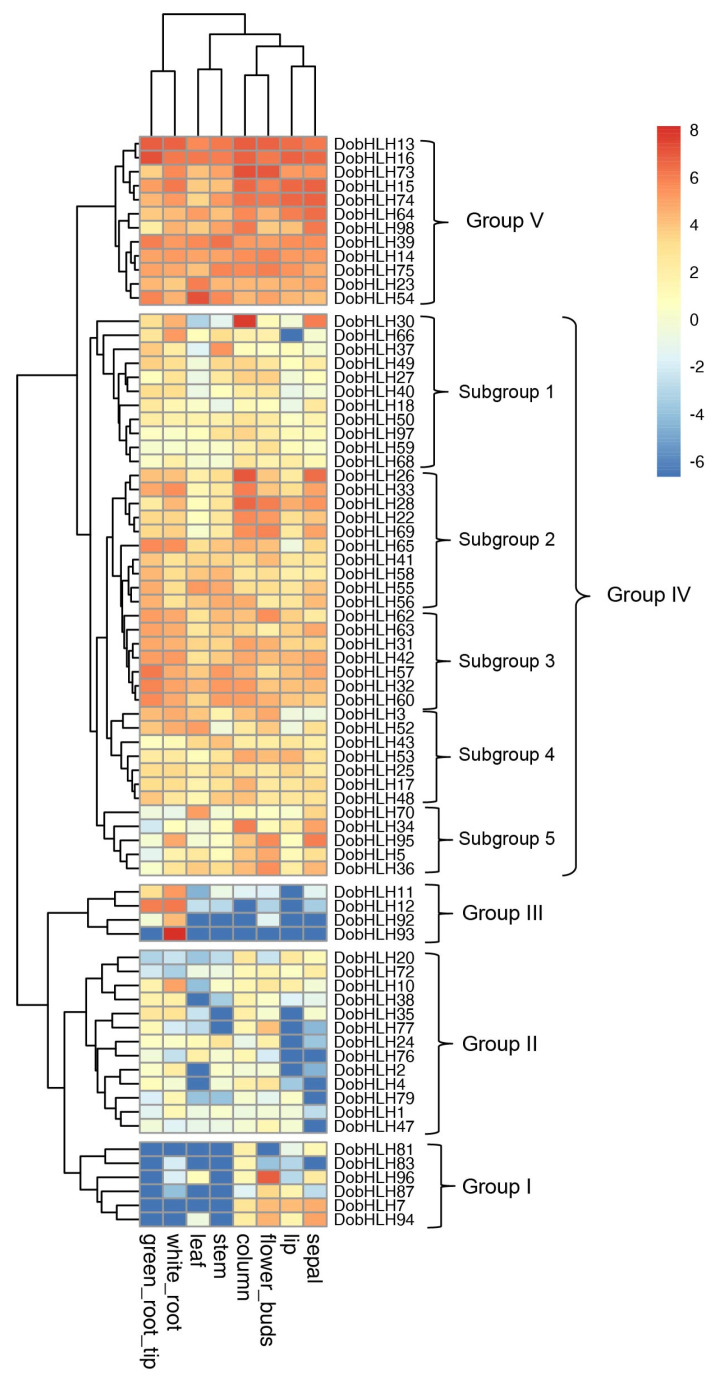
The expression patterns of DobHLH genes were analyzed based on the transcriptomic data of eight tissues including column, flower buds, lip, sepal, leaf, stem, white root, and green root tip. There were 23 DobHLH genes that were hardly detected in any of the eight tissues The expression of 23 DobHLH genes was not detected in any of the eight tissues (when FPKM values were < 2, the gene was considered to be not expressed). Those 23 DobHLH genes were members from clades 1, 4, 10, 14, and 15. The other 75 DobHLH genes were expressed in at least one tissue tested with FPKM > 2. A heatmap of the expression patterns including 75 DobHLH genes was constructed by R, as shown in Figure 5. The genes could be classified into five groups.
Figure 5.
Expression patterns of 75 DobHLH genes in eight tissues. The expression levels of 75 DobHLH genes were from the RNA-Seq data. The eight samples included the column, flower buds, lip, sepal, leaf, stem, white root and green root tip. The color scale represents the values of Log2(FPKM + 0.01).
Group I included six genes (DobHLH7, DobHLH81, DobHLH83, DobHLH87, DobHLH94, and DobHLH96). Their transcripts could not be detected across four vegetative tissues (leaf, stem, white root, and green root tip) and were expressed with various levels in four flower tissues. DobHLH81 and DobHLH83 were lowly expressed only in the column. DobHLH87 was moderately expressed in flower buds while the transcripts of DobHLH96 were abundant in flower buds with FPKM of 124.68. DobHLH7 and DobHLH87 were expressed in four flower tissues with different levels.
Group II included 13 genes (DobHLH1, 2, 4, 10, 20, 24, 35, 38, 47, 72, 76, 77, and 79). Only two genes (DobHLH10 and DobHLH77) were moderately expressed in certain tissues (white root and flower buds, respectively). The other genes were not expressed in most tissues and expressed only in one or two tissues with low FPKM values (<7.50).
Group III included four genes (DobHLH11, DobHLH12, DobHLH92, DobHLH93) that showed a strong tissue-specific expression pattern. These four genes were low expressed in root tissues. DobHLH11 and DobHLH12 were expressed in both white root and green root tip. DobHLH12 was highly expressed in both white root and green root tip, while DobHLH11 was highly expressed in the white root, but low in the green root tip. The transcripts of DobHLH92 and DobHLH93 were limitedly detected in the white root, but not the green root tip. DobHLH93 showed the highest expression level in the white root with FPKM of up to 286.63.
Group IV contained 40 DobHLH genes. According to their expression differences, these genes could be further divided into five subgroups. Subgroup 1 included 11 genes (DobHLH18, 27, 28, 30, 37, 40, 49, 50, 59, 66, 68, and 97). DobHLH30, 37, and 66 were not expressed or lowly expressed in most tissues and highly expressed in some tissues. For example, DobHLH30 showed the highest expression with FPKM of 231.28 in the column, followed by FPKM of 61.66 in the sepal. DobHLH37 was the most abundant with FPKM of 42.56 in the stem while DobHLH66 was expressed with the highest FPKM of 40.61 in the white root. The remaining eight DobHLH genes were not expressed or expressed with low FPKM values. Subgroup 2 contained five members (DobHLH22, DobHLH26, DobHLH28, DobHLH33, and DobHLH69). With the exception of DobHLH28, which was moderately expressed in the lip, all these genes were not expressed or lowly expressed in the leaf, lowly expressed in the stem and lip (5 < FPKM < 10), moderately expressed in the white root and green root tip, and moderately or abundantly expressed in the column, flower buds, and sepal. Subgroup 3 contained 12 genes (DobHLH31, 32, 41, 42, 55, 56, 57, 58, 60, 62, 63,and 65) that were widely expressed in all tested tissues with various levels. All these genes were moderate or abundant with FPKM values of more than 14.68 in the green root tip. For six genes (DobHLH32, 42, 57, 60, 63, and 65), there were more transcripts in the white root and green root tip than in other tissues tested. Except for DobHLH42, other genes had higher expression levels in the green root tip than in the white root. DobHLH31, 41, 55, 56, 58, 62, and 63 genes showed the highest expression levels in non-root tissues. Subgroup 4 contained seven genes (DobHLH3, 17, 25, 43, 48, 52, and 53). Most of them were ubiquitously expressed in eight tissues tested with low or moderate levels. DobHLH52 showed the highest expression level with FPKM of 35.73 in the leaf. Subgroup 5 contained five genes (DobHLH5, 34, 36, 70, and 95). These genes showed no or low expression levels in the lip, leaf, stem, white root, and green root tip, exhibiting moderate levels in the column, flower buds, and sepal tissues. DobHLH95 showed the highest level with FPKM of 64.94 in the sepal and DobHLH34 contained a maximum FPKM of 63.41 in the column.
Group V contained 12 genes (DobHLH13, 14, 15, 16, 23, 39, 54, 64, 73, 74, 75, and 98). Only DobHLH98 was expressed in the green root tip with a low level. All other genes were moderately or highly expressed in all tested tissues with FPKM of more than 11.89, suggesting their various roles in the eight tissues. DobHLH13 and DobHLH16 had the most abundant transcripts in the green root tip with FPKM values of 122.75 and 140.77, respectively. DobHLH73 was highly expressed in the column and flower buds. DobHLH54 had the most transcripts in the leaf with FPKM of 151.78, while DobHLH39 showed the highest expression level in the stem with FPKM of 80.19.
To further confirm the expression profiles of DobHLH obtained from RNA-Seq data, eight genes (including DobHLH11-16, 23, and 52) were randomly selected for the performance of qRT-PCR assays. The results showed that most of the genes exhibited similar expression patterns to RNA-Seq data (Figure S2). For example, DobHLH11 and DobHLH12 genes were expressed highest in roots and DobHLH13, 14, 15, and 16 were expressed in the root, stem, and leaf tissues, consistent with RNA-Seq data. However, the expression pattern of DobHLH23 showed a slight difference from the RNA-Seq data.
2.7. Expressional Changes of DobHLH Genes Concerned with ABA and JA Signals under MeJA and ABA Treatments
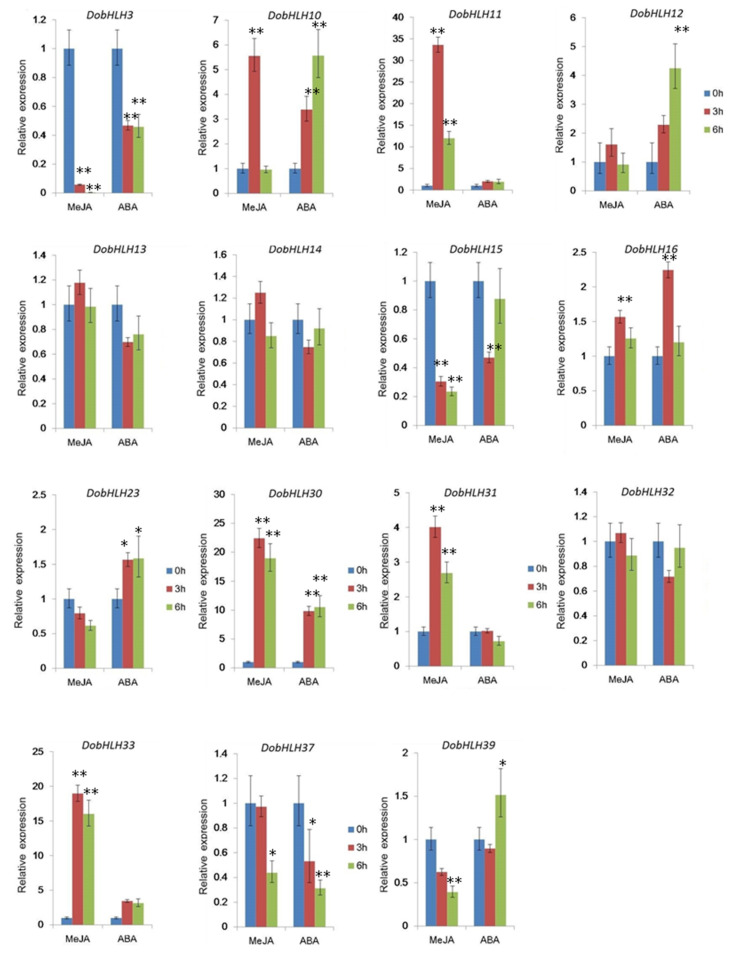
To investigate whether DobHLH genes that contain stress-related regulatory cis-elements could respond to different stresses, we simulated the stress treatment experiments using exogenous MeJA and ABA treatments and inspected the expressional changes of 16 DobHLH genes concerned with ABA and JA signals by the qRT-PCR technique. As shown in Figure 6, eight (including DobHLH 3, 10, 15, 16, 23, 31, 33, and 37) of 12 DobHLH genes that contained JA-responsive elements (CGTCA-motif cis-element) exhibited expressional changes. In contrast to the controls, four DobHLH genes (including DobHLH12, 13, 14 and 32) exhibited no expressional changes in response to MeJA treatments. Similarly, with ABA treatment, 11 (including DobHLH 3, 11, 12, 15, 16, 23, 30, 32, 33, 37, and 39) of 13 DobHLH genes that contained ABA-responsive elements (ABRE-motif cis-element) exhibited expressional changes compared to the controls, whereas two DobHLH genes (DobHLH13, 14) exhibited no expressional changes in response to ABA treatments. In particular, we noted that six DobHLH genes (DobHLH 3, 15, 16, 23, 33, and 37) of the ten DobHLH genes (including DobHLH 3, 12, 13, 14, 15, 16, 23, 32, 33, and 37) contained both JA and ABA-responsive elements, which showed expressional responses to both ABA and MeJA treatments. However, DobHLH13 and 14 did not respond to both ABA and MeJA treatments at the transcription level. These results suggest that most of the DobHLH genes might function in respond to stress signals at the transcription level, likely through specific cis-elements in promoter regions, though the processes of regulatory transcription might be complex.
Figure 6.
Expression patterns of 15 stress-related DobHLH genes under exogenous MeJA and ABA treatments by the qRT-PCR technique. After exogenous MeJA and ABA treatments in leaf tissues, the relative expression levels at 0, 3, and 6 h were tested with three biological duplicates. The relative expression levels at the beginning (0 h) were applied as the control. The bars denote the standard deviation. The star indicates the significance difference (* at p < 0.05, ** at p < 0.01) with the Student’s t-test.
2.8. Protein–Protein Interaction Networks
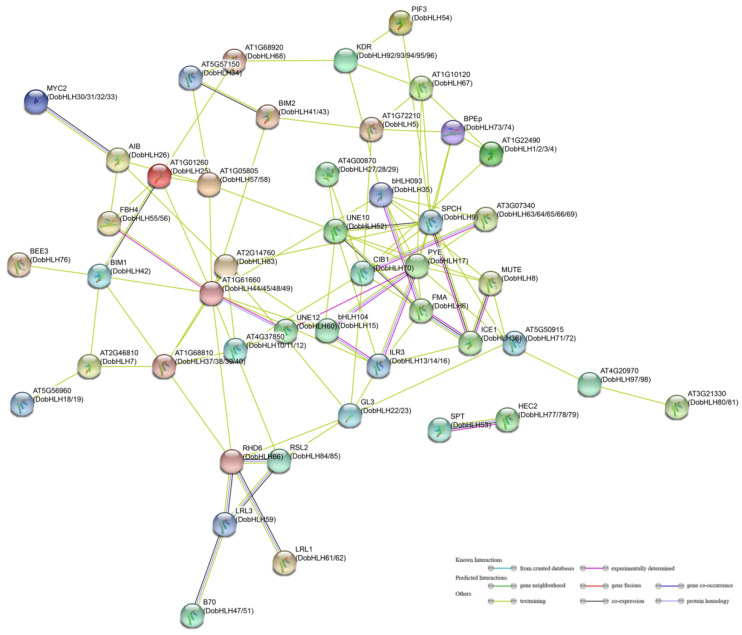
It has been reported than bHLH proteins can form a homodimer or a heterodimer to bind DNA sequences in the promoter region to regulate the transcription of the target gene. Therefore, interactions between bHLH proteins play an important role in their function. The predicted protein–protein interaction of DobHLH proteins based on homologs with Arabidopsis bHLH proteins is shown in Figure 7. It was observed than most DobHLH proteins could interact with one or more DobHLH proteins. For example, it was predicted that DobHLH17 can interact with multiple DobHLH proteins including DobHLH6, 8, 9, 13, 14, 15, 16, and 60. The homolog of DobHLH17 in Arabidopsis is POPEYE (PYE), which is involved in the regulation of responses to iron deficiency in Arabidopsis roots. It has been demonstrated that PYE can interact with multiple proteins such as UNE12, AtbHLH104, ILR3, FMA, MUTE, and SPCH. ILR3 plays a role in the development of root hair. The interaction between DobHLH17 and DobHLH13 or DobHLH14 could have a similar function in D. officinale. Moreover, FMA, SPCH, and MUTE together regulated the stomata formation in Arabidopsis and ICE1 could interact with FMA, SPCH, and MUTE [25], implying that their homologs in D. officinale could potentially participate in the regulation of stomatal differentiation. HEC2 and SPT both play a role in floral development [25], and interactions between their homologs in D. officinale (DobHLH77, 78, 79, and DobHLH53) could influence pistil development through the control of hormones. These predicted interactions suggest that DobHLH proteins could function together to regulate plant growth, development, and other biological processes that merit further investigation.
Figure 7.
The predicted network of protein–protein interactions between DobHLHs by STRING. The different colors represent different types of interactions. Arabidopsis bHLH names are marked while their homologs in D. officinale are in parentheses.
To further verify protein–protein interactions in the predicted network, we randomly selected 12 DobHLH genes to investigate their interactions using the Y2H technique in yeast. Based on their co-expressions in a given tissue, we made different combinations in Y2H experiments. The selected DobHLH genes were cloned into AD and BD vectors, respectively. As shown in Figure 8, none of the DobHLH proteins could interact with the empty BD vector, while three DobHLH proteins (DobHLH36, 48, and 70) could interact with the empty AD on SD/-Leu-Trp-His-Ade medium, suggesting their self-activation. In the combination of DobHLH proteins, it was found that DobHLH25 and DobHLH26 showed an interaction on the SD/-Leu-Trp-His medium and DobHLH60 and DobHLH17 showed a strong interaction on the SD/-Leu-Trp-His-Ade medium. Other protein–protein combinations had no interaction. These results can partially confirm the predicted interaction networks and indicate that they could play similar functions in D. officinale.
Figure 8.

The protein–protein interactions between DobHLHs detected by Y2H. The sequences of DobHLH25, 30, 32, 55, 56, and 60 genes were ligated to the GAL4 activation domain (AD) while the sequences of DobHLH17, 26, 36, 48, and 70 genes were fused to the GAL4 DNA-binding domain (BD). SD-LT, SD-LTH, and SD-LTHA represent the SD-LeuTrp, SD-Leu-Trp-His, and SD-Leu-Trp-His-Ade medium, respectively.
3. Discussion
TFs can bind cis-element sequences in downstream genes to activate or repress their expression, which confers vital roles in plant growth, development, and responses to stresses [16]. The genome sequences of D. officinale have been reported [2,39], which makes it possible to identify the TF family at the genome level. The bHLH family is the second largest family of TFs and plays an essential role in plants. In the present study, based on the published genome data, bHLH family members were first identified in D. officinale, which provides valuable information to dissect their potential function in molecular and physiological processes.
A total of 98 DobHLH members were identified according to the genome data [39]. In previous transcriptome data, some bHLH members were predicted [13,14,15,39,45] and the number of bHLH members varied amongst these publications. The number of bHLH members identified by the genome in the present study was greater than the numbers by the transcriptome data. This was because some bHLH genes were limitedly expressed in the specific tissues and could not be detected in the specific tissues used for RNA-Seq. Only in three transcriptomes from the root, stem, and leaf were there as many as 140 bHLH transcripts, which could result from alternative splicing events [13]. Compared with other TF families identified in D. officinale, the number of bHLH family members ranked second, after the MYB family [46], and was greater than other families such as GRAS [10] and MADS [47]. The number of bHLH members in D. officinale was similar to that in jujube [25] and peach [44] and was more than in Carthamus tinctorius [48]. It has been found that many crops contain more than 150 bHLH members including maize [28], wheat [42,49], and Panax ginseng [50].
The bHLH members in plants could be divided into different clades (or subfamilies) and the numbers of clades in different species varied from 15 to 25 [22]. In the present study, the 98 DobHLH genes in D. officinale could be classified into 18 clades. We found that four clades in Arabidopsis were not present in D. officinale. The similar results were found in other plants such as C. tinctorius [48], apple [51], and wheat [42], which suggests those genes were lost during their evolution. Clades 14 and 15 of the bHLH family in D. officinale contained 14 and 15 members, respectively, which accounted for as many as 28.57% members of the family. The expansion of two clades in D. officinale could potentially imply their important functions. However, the numbers of members from clade 7 and 11 in D. officinale were lower than those in Arabidopsis and rice, which suggests that these clades shrunk in D. officinale.
It was found that the bHLH family in D. officinale showed conservation in both gene structure and protein motif. The gene structure of bHLH family in D. officinale showed highly conserved patterns. Most members in the same clade had the same number of introns and all numbers from clade 6 had no intron. Similar results were observed in Nelumbo nucifera [52], apple [51], cotton [30] and rice [53], which further suggested conserved gene organization during the evolution of different species. However, there were some exceptions where some members in the same clade contained variable intron numbers such as clade 14, which had five, six, seven, or 10 introns. The events of exon loss and gain occurred during the evolution of these genes, which could result in functional diversities between closely-related genes.
The most conserved motifs in the DobHLH proteins were motif 1 and motif 2, which consisted of the bHLH domain. As a core domain for the bHLH family, the bHLH domain was also highly conserved in other species such as tomato [27], wheat [42], and maize [28]. In the bHLH domain of D. officinale, the most conservative residues were Leu-27 and Leu-54, which were in consistence with the residues in Arabidopsis, tomato, and rice [22,27,53]. These results suggest that the two residues may be essential for their functions, especially for the formation of dimers [27]. Moreover, most residues in the domain had similar conservation as in Arabidopsis and tomato, while some residues in D. officinale showed less conservation than those in Arabidopsis and tomato including Glu-13, Arg-14, Gln-28, Lys-36, Ala-48, and Ile-52.
As TFs, their most important function is to bind the specific DNA sequences to regulate the gene expression. According to previous reports and the bHLH domain, the potential types of DNA-binding for DobHLHs were predicted in D. officinale. Compared with predicted DNA-binding types in Arabidopsis and tomato, the number of DNA bindings (77.55%) in D. officinale was lower than that in Arabidopsis (81.63%), but higher than in tomato (69.18%). Among them, the number of G-box DNA binding bHLHs (61/98, 62.24%) in D. officinale was similar to that in Arabidopsis (89/147, 60.54%), which was much higher than that in tomato (72/159, 45.28%). The number of the non-E-box binders in D. officinale was slightly higher than that in Arabidopsis as well as in tomato. The comparative results suggest that a large number of bHLH proteins in D. officinale could bind DNA similar to other species. Notably, most members from the same clade showed the same type of DNA-binding, which was also observed in other species such as Arabidopsis and tomato [27]. Furthermore, the bHLH proteins from D. officinale and Arabidopsis in the same clade exhibited the same type of DNA-binding, which confirmed their conservation during evolution.
The cis-regulatory elements are located at the 5′ upstream of the gene and are responsible for gene transcription, which plays an essential role in the regulatory network and control of plant growth and development. Many cis-elements related to the development, responses to stress as well as hormone were predicted in the promoter regions of DobHLH genes, which suggest their potential roles in the responding conditions. The most common cis-element observed in DobHLH gene promoters was the G-box, which can respond to light. Similar results were found in other species such as C. tinctorius [48] and maize [28]. The second most common types of cis-elements in DobHLH genes were the hormone-responsive motifs that responded to MeJA, ABA, and ethylene. Among 98 DobHLH genes, there were as many as 201 ABRE elements in 69 DobHLH genes, suggesting their putative roles in response to ABA. Notably, a total of 188 ethylene-responsive ERE elements were predicted in 78 DobHLH genes, while few ERE elements were observed in bHLH genes from other plants. The rest of the cis-elements including TC-rich repeats, LTR element, and MBS were also predicted in DobHLH gene promoters, which were related to abiotic stresses such as low temperature and drought. The results showed that bHLH genes can respond to various environmental changes through their cis-elements.
There has almost been no report about bHLH functions in D. officinale despite many reports on other plants such as Arabidopsis and rice. The potential gene function could be predicted based on its expression pattern. In the present study, the comprehensive expression patterns of DobHLH genes were detected in eight tissues according to the previous RNA-Seq data, which strongly showed tissue-specific expression. A total of 23 DobHLH genes were barely expressed in these tissues. Similarly, some bHLH genes from other species were also not detected in any tissue [49]. For example, there were 30 bHLH genes in potato that were not expressed in any of the 12 tested tissues [54]. However, some bHLH genes could widely be expressed in various tissues such as the 12 bHLH genes in D. officinale including DobHLH13 and DobHLH16. These ubiquitous and abundant transcripts of bHLH genes could be found in jujube [25] and potato [54]. The widespread expression of bHLH genes in multiple tissues suggests their multiple roles in these tissues.
The strongly tissue-specific expression patterns of 75 DobHLH genes were observed. A group of bHLH genes in C. tinctorius was found to exhibit high transcripts in petals [48]. During the flower development of jujube, ZjbHLH62 and ZjbHLH53 genes were stably expressed while the expression levels of ZjbHLH4, 12, 23, 78, and 87 genes decreased [25]. There were seven bHLH genes with high expression levels in the petals of wheat [49]. DobHLH96 was specifically and abundantly expressed in the flower bud, which suggests its potential role in the development of the flower bud. Moreover, the expression levels of DobHLH74 in flower organs were much higher than that in the vegetative tissues. Its homolog in Arabidopsis is AtbHLH31 (At1G59640, BIGPETAL, BPE), which participated in the control of petal size and under the regulation of APETALA3, PISTILLATA, APETALA1, PISTILLATA3, and AGAMOUS [55].
Some bHLH genes could be highly expressed in roots such as DobHLH12 and DobHLH93. In other species, some bHLH genes were also found to show abundant expression levels in roots such as five bHLH genes in wheat [49] and as many as 78 bHLH genes in maize [28]. These results suggest bHLH could play a conserved role in root development. Moreover, we found that some bHLH genes were abundant in the green root tip, implying that it could function to maintain root apical meristem. Some DobHLH genes had abundant transcripts in leaf. For example, DobHLH54 had three cis-elements in its promoter region and may play a role in the process of photosynthesis.
MYC2 is an important regulator in the JA signaling pathway. In the present study, four putative MYC2 homologs were identified. A homolog of MYC2 (DobHLH33) in D. officinale was reported [56], which could be induced by MeJA and was involved in the biosynthesis of terpenoid indole alkaloids through the key enzyme genes in the MVA pathway. Moreover, four putative MYC2 homologs were expressed in roots as well as other tissues, suggesting their various functions in multiple tissues. Notably, DobHLH30 had the highest expression in the column (231.28). MYC2 (DobHLH33) also had abundant expression in flower organs, which could have implications for functioning in flower development.
Usually, bHLH proteins function through the formation of homodimers or heterodimers to co-bind with their target genes to regulate their expressions. Many studies have reported the interactions of bHLH proteins to regulate the expressions of targeted genes [25,31,42,48,57]. Our current study found that many DobHLH proteins did have potential interactions via model prediction. In addition, we verified protein interactions between several DobHLH proteins such as DobHLH 25 and 26, and DobHLH17 and 60. We verified protein interactions between limited DobHLH proteins in this study. Many DobHLH proteins may function together to regulate plant growth and development, responses to environmental changes, and secondary metabolite processes in D. officinale. Additionally, we noted that DobHLH17 and DobHLH60 are homologous to UNE12 and PYE in Arabidopsis, and to ZmbHLH114 and ZmbHLH163 in maize, whose interactions have been verified in previous studies [28,57]. It is likely that protein interactions between given bHLHs proteins might be, at least partially, conserved in plants.
4. Materials and Methods
4.1. Plant Materials
The seedling plants of D. officinale in vitro (provided by Prof. Shibao Zhang’s group) were well grown in the tissue culture room of the Kunming Institute of Botany, Chinese Academy of Sciences (Kunming, Yunnan, China). The medium was Murashige and Skoog (MS) medium with 30 g/L sucrose, and the growth was 25 °C with a constant photoperiod (12 h light/12 h dark) with an illumination of 40 µmol quanta/(m2 s). The fresh and healthy leaves, stem, and roots were harvested, immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
Three individual D. offcinale plants of six months old in vitro were used for each treatment. MeJA (Coolaber, China) and ABA (Sangon, China) solutions were prepared with ethanol and water. The treated plants were sprayed with 100 µM MeJA or 100 µM ABA for three or six hours while the control plants were sprayed with ethanol and water. The leaves were harvested and quickly frozen at −80 °C for further RNA extraction.
4.2. Identification of the bHLH TF Family in Dendrobium officinale
Although two research groups reported the genome of D. officinale (D. catenatum), respectively [2,39], at present only one genome (PRJNA262478) is available. The protein data of D. officinale were downloaded from NCBI. HMMER was used to identify the potential bHLH proteins with the bHLH domain (PF00010) [58]. The presence of the bHLH domain was manually confirmed by CDD and SMART. After repeated or uncomplete proteins were discarded, the remaining proteins with the bHLH domain were considered as the bHLH candidate members in D. officinale. The isoelectric point and molecular weight of candidate DobHLH proteins were analyzed with the ProtParam tool [59]. The subcellular localization of DobHLH proteins was predicted by WOLF PSORT (https://www.genscript.com/psort.html).
4.3. Phylogenetic Analysis of DobHLH Proteins
The AtbHLH proteins of Arabidopsis were downloaded from TAIR. The proteins of rice bHLHs were also downloaded [40]. The sequences of bHLH proteins from these species were aligned with ClustalW software [60], and then an un-rooted tree was obtained with MEGA 7.0 software using a neighbor-joining method with a bootstrap of 1000 replicates [61].
4.4. Gene Structure and Conserved Motif Analysis of DobHLHs
The gene structures of DobHLH genes were analyzed by comparing cDNA sequences with the correspondent genomic DNA sequences with GSDS [62]. MEME was used to search 10 conserved motifs in DobHLH proteins [63] and the InterPro database was used to further analyze those motifs [64].
4.5. Expression Patterns of DobHLH Genes in Different Tissues
The previously reported RNA-Seq data of eight D. officinale tissues were used to analyze the expression patterns of DobHLH genes. First, the raw sequencing reads (PRJNA348403) were downloaded from the NCBI [65]. After low-quality reads and adapter sequences were filtered, all the clean reads were mapped to the reference genome of D. officinale, resulting in unique reads using HISAT2 software [66]. Then, transcript abundances were estimated with the FPKM method using Stringtie software [66]. The heatmap of DobHLH genes was obtained by the R package pheatmap, v1.0.10 (https://rdrr.io/cran/pheatmap/).
4.6. Predicted Protein–Protein Interaction of DobHLH Members
The potential protein–protein interactions were predicted with the online STRING server (https://string-db.org). The protein sequences of 98 DobHLHs were uploaded into the server and Arabidopsis thaliana was chosen as the comparative organism. After the BLAST analysis was finished, the interaction internet of DobHLHs was generated with genes showing the highest bitscore.
4.7. Cis-Element Analysis in Promoter Regions of DobHLH Genes
The 2000 bp sequences at the upstream of the transcription start site for each DobHLH gene were downloaded from the NCBI. Cis-regulatory elements were analyzed with PlantCARE.
4.8. RNA Extraction and cDNA Preparation
The total RNA was extracted with the RNAprep Pure Plant Plus Kit (Cat DP441, Tiangen, China) according to the manufacturer’s protocol. The concentration and quality of RNA samples were detected on a Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA). cDNA was prepared with the one-step cDNA Synthesis Kit (Cat AT311, Transgen, China), which was used for DobHLH gene amplification in yeast two hybrid assays. A sample of 200 ng of total RNA in a reaction system of 20 μL was converted to cDNA with TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Cat AT341, Transgen, China), which was used for qRT-PCR assays.
4.9. Yeast Two Hybrid Assays
The CDS sequences of 11 DobHLH genes were amplified with PCR using cDNA prepared as templates. The primers used are shown in Table S4. All the sequences were confirmed to be correct by sequencing. Then, the sequences of DobHLH25, 30, 32, 55, 56, and 60 genes were cloned into the pDest22 construct (Invitrogen) with the GAL4 activation domain (AD) while the sequences of DobHLH17, 25, 26, 36, 48, and 70 genes were cloned into pDest32 (Invitrogen) with the GAL4 DNA-binding domain (BD) through recombination reaction. Yeast two hybrid (Y2H) was performed as previously described [67].
4.10. qRT-PCR Assays
One μL of prepared cDNA mixture was used for qRT-PCR in a 20 μL volume with SuperReal PreMix Plus SYBR Green Kit (TIANGEN Biotech, China), according to the manufacturer’s instructions. The reaction was carried out on an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System (ThemoFisher Scientific, USA) and the cycling conditions was as follows: first at 95 °C for 15 min, then 40 cycles of at 95 °C for 10 s, at 60 °C for 20 s, and finally at 72 °C for 32 s. The melt curves were performed at 95 °C for 15 s, at 60 °C for 1 min, and at 95 °C for 15 s. The 2-ΔΔCT method was used to calculate Log2fold change with DoActin gene as an internal reference [13,68]. CT values stand for the average of cycle times from three technical replicates. The primer sequences used for qRT-PCR are listed in Table S4.
5. Conclusions
This research presents a genome-wide identification and characterization of the bHLH TF family in D. officinale. The phylogenetic analysis found that 98 DobHLH members were classified into 18 clades, which showed conserved gene structures and motifs. The characteristics of the DobHLH domains for their function were predicted. Furthermore, the transcript data of DobHLH genes revealed their universal or specific expression profiles, which may contribute to understanding their function in D. officinale. Therefore, the present study provides additional insights into the bHLH TF family and will be helpful for the further investigation of DobHLH functions in D. officinale.
Acknowledgments
We are grateful to senior engineer Hua Wang of the Kunming Institute of Botany, Chinese Academy of Sciences for his kind donation of in vitro D. officinale materials.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/8/1044/s1, Figure S1: The phylogenetic tree of bHLH proteins from D. officinale and rice. Figure S2: Expression patterns of eight DobHLH genes in leaf, stem, and root tissues using the qRT-PCR technique. The relative expression levels were obtained from the mean of three biological duplicates. The bars denote the standard deviation. Table S1: The additional information on 98 DobHLH genes in D. officinale. Table S2: The consensus amino acids of bHLH domains in different species. Table S3: The predicted cis-elements in DobHLH genes. Table S4: The primer sequences used in the study.
Author Contributions
Conceptualization, A.L.; Data curation, Y.W.; Investigation, Y.W.; Methodology, Y.W.; Supervision, A.L.; Writing–original draft, Y.W.; Writing–review & editing, A.L. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was financially supported by the National Natural Science Foundation of China (grant number 31701465) and the Yunnan Provincial Science and Technology Department (grant number 2016FB060) as well as Beijing DR PLANT Biotechnology Co. Ltd. (E0514832C1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ng T.B., Liu J., Wong J.H., Ye X., Sze S.C.W., Tong Y., Zhang K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012;93:1795–1803. doi: 10.1007/s00253-011-3829-7. [DOI] [PubMed] [Google Scholar]
- 2.Yan L., Wang X., Liu H., Tian Y., Lian J., Yang R., Hao S., Wang X., Yang S., Li Q., et al. The Genome of Dendrobium officinale Illuminates the Biology of the Important Traditional Chinese Orchid Herb. Mol. Plant. 2015;8:922–934. doi: 10.1016/j.molp.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Tang H., Zhao T., Sheng Y., Zheng T., Fu L., Zhang Y. Dendrobium officinale Kimura et Migo: A Review on Its Ethnopharmacology, Phytochemistry, Pharmacology, and Industrialization. Evid. Based Complement. Alternat. Med. 2017;2017:7436259. doi: 10.1155/2017/7436259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen C., Guo H., Chen H., Shi Y., Meng Y., Lu J., Feng S., Wang H. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 2017;7:187. doi: 10.1038/s41598-017-00292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing S., Zhang X., Ke H., Lin J., Huang Y., Wei G. Physicochemical properties of polysaccharides from Dendrobium officinale by fractional precipitation and their preliminary antioxidant and anti-HepG2 cells activities in vitro. Chem. Cent. J. 2018;12:100. doi: 10.1186/s13065-018-0468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J., Wu Y., Yuan H., Yang Y., Xiong Q., Liang C., Li Z., Li C., Zhang G., Lai X., et al. Dendrobium officinale polysaccharides attenuate learning and memory disabilities via anti-oxidant and anti-inflammatory actions. Int. J. Biol. Macromol. 2019;126:414–426. doi: 10.1016/j.ijbiomac.2018.12.230. [DOI] [PubMed] [Google Scholar]
- 7.Tao S.C., Lei Z.X., Huang K.W., Li Y.R., Ren Z.Y., Zhang X.F., Wei G., Chen H.M. Structural characterization and immunomodulatory activity of two novel polysaccharides derived from the stem of Dendrobium officinale Kimura et Migo. J. Funct. Foods. 2019;57:121–134. doi: 10.1016/j.jff.2019.04.013. [DOI] [Google Scholar]
- 8.Kuang M.T., Li J.Y., Yang X.B., Yang L., Xu J.Y., Yan S., Lv Y.F., Ren F.C., Hu J.M., Zhou J. Structural characterization and hypoglycemic effect via stimulating glucagon-like peptide-1 secretion of two polysaccharides from Dendrobium officinale. Carbohydr. Polym. 2020;241:116326. doi: 10.1016/j.carbpol.2020.116326. [DOI] [PubMed] [Google Scholar]
- 9.Pan L.H., Li X.F., Wang M.N., Zha X.Q., Yang X.F., Liu Z.J., Luo Y.B., Luo J.P. Comparison of hypoglycemic and antioxidative effects of polysaccharides from four different Dendrobium species. Int. J. Biol. Macromol. 2014;64:420–427. doi: 10.1016/j.ijbiomac.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X., Ling H., Chen X., Guo S. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum (Orchidaceae) Gene. 2019;705:5–15. doi: 10.1016/j.gene.2019.04.038. [DOI] [PubMed] [Google Scholar]
- 11.Huang K., Li Y., Tao S., Wei G., Huang Y., Chen D., Wu C. Purification, Characterization and Biological Activity of Polysaccharides from Dendrobium officinale. Molecules. 2016;21:701. doi: 10.3390/molecules21060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z., Liao Y., da Silva J.A.T., Yang Z., Duan J. Differential Accumulation of Anthocyanins in Dendrobium officinale Stems with Red and Green Peels. Int. J. Mol. Sci. 2018;19:2857. doi: 10.3390/ijms19102857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y., Zhang J., Liu X., Meng M., Wang J., Lin J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics. 2020;112:1781–1794. doi: 10.1016/j.ygeno.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Lei Z., Zhou C., Ji X., Wei G., Huang Y., Yu W., Luo Y., Qiu Y. Transcriptome Analysis Reveals genes involved in flavonoid biosynthesis and accumulation in Dendrobium catenatum From Different Locations. Sci. Rep. 2018;8:6373. doi: 10.1038/s41598-018-24751-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X., Li Y., Li C., Luo H., Wang L., Qian J., Luo X., Xiang L., Song J., Sun C., et al. Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene. 2013;527:131–138. doi: 10.1016/j.gene.2013.05.073. [DOI] [PubMed] [Google Scholar]
- 16.Sun X., Wang Y., Sui N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018;503:397–401. doi: 10.1016/j.bbrc.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 17.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pires N., Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore A.W., Barbel S., Jan L.Y., Jan Y.N. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc. Natl. Acad. Sci. USA. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 21.Murre C., McCaw P.S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 22.Toledo-Ortiz G., Huq E., Quail P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atchley W.R., Terhalle W., Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 1999;48:501–516. doi: 10.1007/PL00006494. [DOI] [PubMed] [Google Scholar]
- 24.Massari M.E., Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Gao W., Xue C., Zhang Y., Liu Z., Zhang Y., Meng X., Liu M., Zhao J. Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube. BMC Genom. 2019;20:568. doi: 10.1186/s12864-019-5936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey P.C., Martin C., Toledo-Ortiz G., Quail P.H., Huq E., Heim M.A., Jakoby M., Werber M., Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H., Fan H.J., Ling H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015;16:9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T., Lv W., Zhang H., Ma L., Li P., Ge L., Li G. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 2018;18:235. doi: 10.1186/s12870-018-1441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P., Su L., Gao H., Jiang X., Wu X., Li Y., Zhang Q., Wang Y., Ren F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018;9:64. doi: 10.3389/fpls.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu R., Zhang J., Liu D., Wei Y.L., Wang Y., Li X.B. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum) BMC Plant Biol. 2018;18:304. doi: 10.1186/s12870-018-1523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu X., Guan Y., Chen S., Li H. Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genom. 2017;18:619. doi: 10.1186/s12864-017-4044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig S.R., Wessler S.R. Maize R Gene Family: Tissue-Specific Helix-Loop-Helix Proteins. Cell. 1990;62:849–851. doi: 10.1016/0092-8674(90)90259-H. [DOI] [PubMed] [Google Scholar]
- 33.Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Van Moerkercke A., Steensma P., Schweizer F., Pollier J., Gariboldi I., Payne R., van den Bossche R., Miettinen K., Espoz J., Purnama P.C., et al. The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. USA. 2015;112:8130–8135. doi: 10.1073/pnas.1504951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Luo H., Xu Z., Zhu Y., Ji A., Song J., Chen S. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 2015;5:11244. doi: 10.1038/srep11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C., Qiu J., Ding L., Huang M., Huang S., Yang G., Yin J. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 2017;112:335–345. doi: 10.1016/j.plaphy.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 38.Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G.Q., Xu Q., Bian C., Tsai W.C., Yeh C.M., Liu K.W., Yoshida K., Zhang L.S., Chang S.B., Chen F., et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016;6:19029. doi: 10.1038/srep19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao K., Li S., Yao W., Zhou B., Li R., Jiang T. Characterization of the basic helix-loop-helix gene family and its tissue-differential expression in response to salt stress in poplar. PeerJ. 2018;6:e4502. doi: 10.7717/peerj.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X.J., Wang J.R. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol. 2017;17:90. doi: 10.1186/s12870-017-1038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen W., Cui X., Li H., Teng R.-M., Wang Y.-X., Liu H., Zhuang J. Genome-wide identification and analyses of bHLH family genes in Brassica napus. Can. J. Plant Sci. 2019;99:589–598. doi: 10.1139/cjps-2018-0230. [DOI] [Google Scholar]
- 44.Zhang C., Feng R., Ma R., Shen Z., Cai Z., Song Z., Peng B., Yu M. Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS ONE. 2018;13:e0195974. doi: 10.1371/journal.pone.0195974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Wang Y., Lyu P., Chen L., Shen C., Sun C. Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 2019;132:419–429. doi: 10.1007/s10265-019-01099-6. [DOI] [PubMed] [Google Scholar]
- 46.He C., da Silva J.A.T., Wang H., Si C., Zhang M., Zhang X., Li M., Tan J., Duan J. Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci. Rep. 2019;9:1381. doi: 10.1038/s41598-019-49812-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y., Shen Q., Lyu P., Lin R., Sun C. Identification and expression profiling of selected MADS-box family genes in Dendrobium officinale. Genetica. 2019;147:303–313. doi: 10.1007/s10709-019-00071-5. [DOI] [PubMed] [Google Scholar]
- 48.Hong Y., Naveed A., Tian Y., Liu J., Wang L., Wang G., Liu X., Dong Y., Wang F., Liu W., et al. Genome-Wide Identification, Expression Analysis, and Subcellular Localization of Carthamus tinctorius bHLH Transcription Factors. Int. J. Mol. Sci. 2019;20:3044. doi: 10.3390/ijms20123044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L., Xiang L., Hong J., Xie Z., Li B. Genome-wide Analysis of bHLH Transcription Factor Family Reveals Their Involvement in Biotic and Abiotic Stress Responses in Wheat (Triticum aestivum L.) 3 Biotech. 2019;9:236. doi: 10.1007/s13205-019-1742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu Y., Xiao S., Su H., Liao B., Zhang J., Xu J., Chen S. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharm. Sin. B. 2018;8:666–677. doi: 10.1016/j.apsb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., Gao M., Huang L., Wang Y., van Nocker S., Wan R., Guo C., Wang X., Gao H. Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 2017;7:28. doi: 10.1038/s41598-017-00040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao T.Y., Liu Y.Y., Zhu H.H., Zhang J., Yang J.X., Fu Q., Wang N., Wang Z. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ. 2019;7:e7153. doi: 10.7717/peerj.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang R., Zhao P., Kong N., Lu R., Pei Y., Huang C., Ma H., Chen Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes. 2018;9:54. doi: 10.3390/genes9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varaud E., Brioudes F., Szécsi J., Leroux J., Brown S., Perrot-Rechenmann C., Bendahmane M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell. 2011;23:973–983. doi: 10.1105/tpc.110.081653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y., Meng C., Zhu L., Li D., Jin Q., Song C., Cai Y., Fan H., Lin Y. Cloning and characterization of DoMYC2 from Dendrobium officinale. Plant Cell Tiss. Organ. Cult. 2017;129:533–541. doi: 10.1007/s11240-017-1198-3. [DOI] [Google Scholar]
- 57.Braun P., Carvunis A.-R., Charloteaux B., Dreze M., Ecker R.J., Hill E.D., Roth P.F., Vidal M., Galli M., Balumuri P. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 60.McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang G.Q., Liu K.W., Li Z., Lohaus R., Hsiao Y.Y., Niu S.C., Wang J.Y., Lin Y.C., Xu Q., Chen L.J., et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y., Xu W., Chen Z., Han B., Haque M.E., Liu A. Gene structure, expression pattern and interaction of Nuclear Factor-Y family in castor bean (Ricinus communis) Planta. 2018;247:559–572. doi: 10.1007/s00425-017-2809-2. [DOI] [PubMed] [Google Scholar]
- 68.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.