Abstract
Soft tissue sarcoma (STS) is a heterogeneous group of rare malignancies with a five-year survival rate of approximately 50%. Reliable molecular markers for risk stratification and subsequent therapy management are still needed. Therefore, we analyzed the prognostic potential of miR-155-5p and miR-203a-3p expression in a cohort of 79 STS patients. MiR-155-5p and miR-203a-3p expression was measured from tumor total RNA by qPCR and correlated with the demographic, clinicopathological, and prognostic data of the patients. Elevated miR-155-5p expression was significantly associated with increased tumor stage and hypoxia-associated mRNA/protein expression. High miR-155-5p expression and low miR-203a-3p expression, as well as a combination of high miR-155-5p and low miR-203a-3p expression, were significantly associated with poor disease-specific survival in STS patients in the Kaplan–Meier survival analyses (p = 0.027, p = 0.001 and p = 0.0003, respectively) and in the univariate Cox regression analyses (RR = 1.96; p = 0.031; RR = 2.59; p = 0.002 and RR = 4.76; p = 0.001, respectively), but not in the multivariate Cox regression analyses. In conclusion, the oncomiR miR-155-5p and the tumor suppressor-miR miR-203a-3p exhibit an association with STS patient prognosis and are suggested as candidates for risk assessment.
Keywords: soft tissue sarcoma, prognosis, miR-155-5p, miR-203a-3p
1. Introduction
Soft tissue sarcoma (STS) comprises approximately 50 subtypes of tumors characterized by their mesenchymal origin and heterogeneous phenotype [1]. Although STS is a rare tumor entity, with an incidence rate of only 3.6 per 100,000 [2], the low five-year survival rate of 50% is a major issue [3]. Therefore, there is still an urgent need for reliable, specific, and easily measurable prognostic markers for risk stratification and subsequent therapy management.
MicroRNAs are 18–25 nucleotide long, noncoding RNAs that post-transcriptionally regulate gene expression by repressing the translation of their respective target genes through RNA-induced silencing complex (RISC)-mediated binding to the 3′-UTR and subsequent inhibition of proper ribosomal assembly [4]. Frequently, microRNA expression patterns are specific for physiologic tissues, but also systemic diseases or malignancies. In this context, sarcomas have been subject to a characterization of the individual microRNA patterns putatively associated with their heterogeneous appearance and tumor-biological behavior (reviewed in [5]).
Furthermore, there is a multitude of studies showing a considerable impact of the dysregulation of certain microRNAs in all major malignancies on the prognosis of patients. Previously, we were able to demonstrate an association between increased miR-155, miR-210, or miR-203 expression and a significantly worsened prognosis in pancreatic adenocarcinoma [6]. An intermediate miR-210 level was also associated with worsened survival in STS patients [7].
MiR-155-5p (henceforth designated miR-155) and its complementary strand miR-155-3p are processed from the B-cell integration cluster (BIC) gene located on chromosome 21, which is alternatively designated the MIR155 host gene (MIR-155HG). Although miR-155 plays a role in hematopoiesis, inflammation, and immune responses, it may act as an oncogenic miRNA. It is not only upregulated in different leukemias and lymphomas, but also in solid tumors such as cancer of the breast, colon, lung, pancreas, and thyroid (reviewed in [8]). Furthermore, miR-155 was reported to be overexpressed in liposarcoma (LS)-derived cell lines or liposarcoma biopsies in comparison to normal fat tissue [9,10] and contribute to LS progression by targeting the central Wnt pathway component casein kinase 1α (CK1α) [9]. Interestingly, the Kaposi sarcoma-associated herpesvirus genome encodes an ortholog of miR-155 [11], and overexpression of this viral-encoded microRNA is associated with the suppression of cell growth control [12]. MiR-155 is also overexpressed in osteosarcoma samples in comparison to corresponding normal tissue and targets the Wnt pathway repressor HMG-box transcription factor 1 (HBP1), thereby driving osteosarcoma cell proliferation by enhancing Wnt pathway action [13]. Further important cellular pathways regulated by miR-155 in osteosarcoma include the PTEN/AKT/mTOR pathway [14] and the NFκB pathway [15].
The miR-203a gene is located on chromosome 14q32.33 [16]. MiR-203a-3p (henceforth designated as miR-203) is expressed aberrantly in different cancers, such as gastric, pancreatic, colon and esophageal cancer, compared to normal tissue (reviewed in [17,18]), which suggests its expression to serve as a diagnostic marker. MiR-203 was identified to be downregulated both in rhabdomyosarcoma (RMS) cell lines and in rhabdomyosarcoma patient samples due to promoter hypermethylation. Re-induction of miR-203 expression by demethylating agents downregulates p63 and leukemia inhibiting factor receptor (LIFR) [19]. Furthermore, miR-203 is significantly downregulated in osteosarcoma samples and cell lines compared to controls, while its target gene RAS-related protein Rab22A (RAB22A) is overexpressed. Conversely, the overexpression of miR-203 blocks osteosarcoma growth and migration [20].
In this study, we aimed to analyze the impact of miR-155 and miR-203 on the diagnostics as well as the correlation with clinical characteristics and with the prognosis of STS patients.
2. Results
2.1. miR-155 and miR-203 Expression in STS Specimens
MiR-155 and miR-203 were detectable in all analyzed STS specimens, with median expression values of 16.7 and 1.9, respectively. The distribution of miR-155 expression values ranged from 0.3–189.5, while miR-203 expression values ranged from 0.003 to 14358.7.
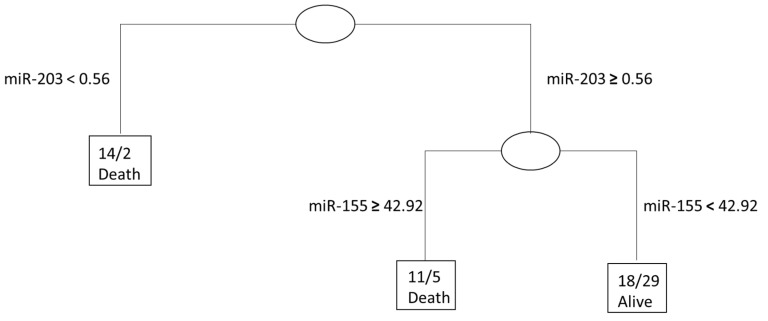
By using recursive partitioning methods, we established the influence and optimized cut-offs of each microRNA when analyzing the combinational effect on STS patient survival. With an expression of miR-203 ≥ 0.56 and miR-155 ≥ 42.92, there was a 68% correct classification rate of tumor-related death in the STS patients (see Figure 1).
Figure 1.
Classification tree establishing the influence of each microRNA in a combinational approach towards risk stratification.
Optimized cut-off values were also defined by assessing the value with the highest Youden index in receiver operating characteristics (ROC) analyses, testing the sensitivity and specificity of each microRNA in the tumor-related death of the patients (see Figure 2).
Figure 2.
ROC analyses for miR-155 (green), miR-203 (blue) and a combination of miR-155 + miR-203 (model in red) and their prognostic application. Both microRNAs exhibited fair area under the curve values (AUCs; miR-155: AUC 0.62 (0.49–0.74), p = 0.067; miR-203: AUC 0.67 (0.55–0.79), p = 0.011). MiR-155 or miR-203 expression with the highest Youden index according to single ROC analyses was chosen as the cut-off for subsequent survival analyses. A fitted linear regression model that combined miR-155 and miR-203 also exhibited only a fair AUC of 0.68 (0.56–0.80), p = 0.006.
The optimal cut-off was 18.0 for miR-155 and 0.9 for miR-203. When assessing low and high expression of the microRNAs with respect to age, gender, tumor stage, resection type, tumor localization, and histological subtypes of the respective tumor, we detected a significant association between increased miR-155 expression and increased tumor stage (p = 0.01, Kruskal–Wallis test, see Figure 3).
Figure 3.
The distribution of miR-155 expression in relation to tumor stage. MiR-155 expression was significantly associated with tumor stage (p = 0.01, Kruskal-Wallis test).
Regarding the histological subtype, especially liposarcoma and neuronal sarcoma exhibited a lower miR-155 expression in comparison to the other sarcoma entities (see Supplementary Figure S1). These differences were significant (p = 0.001, Kruskal–Wallis test). However, this was based on relatively low case numbers (n = 7–21 per category).
2.2. Bivariate Correlation Analyses
We analyzed the molecular biological factors associated with miR-155 and miR-203 expression in soft tissue sarcoma. When considering all significant correlations with rs > 0.25 or rs < −0.25, we retrieved eight and five factors associated with miR-155 and miR-203, respectively (see Table 1).
Table 1.
Bivariate correlation analyses (Spearman-Rho) with miR-155 or miR-203 and different molecular-pathological factors in soft tissue sarcoma.
| Parameters | rs | p | n | Reference | |
|---|---|---|---|---|---|
| miR-155 | p53 mutations | 0.260 | 0.040 | 63 | [21] |
| OPN protein | 0.361 | 0.002 | 72 | [22] | |
| uPA protein | 0.604 | <0.001 | 79 | [23] | |
| uPAR protein | 0.440 | <0.001 | 79 | [23] | |
| PAI-1 protein | 0.347 | 0.002 | 79 | [23] | |
| EFNA3 mRNA | −0.378 | 0.001 | 75 | n.p. | |
| LGR5 mRNA | −0.389 | 0.001 | 69 | [24] | |
| miR-210 | 0.349 | 0.002 | 78 | [7] | |
| miR-203 | miR-210 | 0.265 | 0.019 | 78 | [7] |
| OPN-a mRNA | −0.267 | 0.021 | 74 | [25] | |
| OPN-b mRNA | −0.324 | 0.005 | 74 | [25] | |
| OPN-c mRNA | −0.260 | 0.025 | 74 | [25] | |
| pAKT473 protein | −0.434 | <0.001 | 73 | [26] |
Abbreviations: rs—correlation factor according to Spearman-Rho; n.p.—not published own data.
Both miR-155 and miR-203 expression levels were significantly correlated with miR-210 expression. Moreover, miR-155 expression was significantly correlated with the expression of proteins of the urokinase-type plasminogen activator (uPA) system (rs = 0.347–0.604, Spearman-Rho) and the tumor tissue level of osteopontin (OPN) in the tumors of STS patients (rs = 0.361). Additionally, miR-155 expression was significantly associated with ephrin-A3 (EFNA3) mRNA expression (rs = −0.378) and LGR5 mRNA expression (rs = −0.389). MiR-203 was significantly inversely associated with OPN mRNA expression (splice variants a-c: rs = −0.26–−0.324) and pAKT tumor tissue level (rs = −0.434).
2.3. Survival Analyses
Finally, we tested the association of miR-155 and miR-203 expression with the prognosis of STS patients. In Kaplan–Meier survival analyses, elevated miR-155 expression (≥18.0) was significantly associated with worsened tumor-related survival (median survival time 25 months in comparison to 118 months; see Figure 4a). On the other hand, lower miR-203 expression (<0.9) was significantly associated with worsened tumor-related survival in the STS patients (median survival time 18 months vs. 86 months, see Figure 4b).
Figure 4.
Kaplan-Meier survival analyses of the prognostic impact of miR-155 (a) and miR-203 (b) expression or a combination of both microRNAs (c) on soft tissue sarcoma patient survival. Below the figures, the number of patients at risk at specific time points is given.
Furthermore, when combining the expression of both microRNAs, the unfavorable combination miR-155 high + miR-203 low was significantly associated with a worsened survival of the patients in comparison to the miR-155 low + miR-203 high combination in tumor tissues (p < 0.001, log-rank-test, see Figure 4c).
In the univariate Cox regression analyses, increased miR-155 expression (p = 0.03; RR = 1.96) and decreased miR-203 expression (p = 0.002; RR = 2.59) were also associated with a higher risk of tumor-related death (see Table 2). When combining both microRNAs, the less favorable combination miR-155 high + miR-203 low exhibited a significant 4.76-fold increased risk of tumor-related death (p = 0.001). However, although the estimated risk ratios remained numerically equivalent in multivariate Cox regression analyses adjusted for tumor stage, tumor localization, resection type, and STS sub-entity, the association between miR-155 or the combination of miR-155 and miR-203 was no longer an independent predictor of STS patient survival, probably because of the interdependence of increased miR-155 expression and a higher tumor stage. Furthermore, the relapse-free survival of the patients was not associated with miR-155 or miR-203 expression in uni- or multivariate Cox regression analyses (see Table 2).
Table 2.
Univariate and multivariate Cox regression survival analyses of the impact of miR-155 and miR-203 on soft tissue sarcoma patient survival. Multivariate Cox regression analyses adjusted for tumor stage, resection type, tumor localization and tumor sub-entity. RR = relative risk with 95% confidence interval in brackets.
| Parameters | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | |||||
|---|---|---|---|---|---|---|---|
| n | p | RR | n | p | RR | ||
| miR-155 low | 45 | Reference | 45 | Reference | Tumor-specific survival | ||
| miR-155 high | 34 | 0.031 | 1.96 (1.06–3.61) | 34 | 0.766 | 1.13 (0.52–2.46) | |
| miR-203 low | 29 | 0.002 | 2.59 (1.41–4.76) | 29 | 0.077 | 1.99 (0.93–4.25) | |
| miR-203 high | 50 | Reference | 50 | Reference | |||
| miR-155 low + miR-203 high | 29 | Reference | 29 | Reference | |||
| miR-155 high + miR-203 low | 13 | 0.001 | 4.76 (1.89–12.05) | 13 | 0.21 | 4.05 (0.46–35.7) | |
| miR-155 low | 45 | Reference | 45 | Reference | Relapse-free survival | ||
| miR-155 high | 34 | 0.985 | 1.01 (0.5–2.01) | 34 | 0.512 | 1.32 (0.34–1.73) | |
| miR-203 low | 29 | 0.189 | 1.61 (0.79–3.26) | 29 | 0.405 | 1.60 (0.21–1.90) | |
| miR-203 high | 50 | Reference | 50 | Reference | |||
| miR-155 low + miR-203 high | 29 | 0.455 | 1.5 (0.23–1.93) | 29 | Reference | ||
| miR-155 high + miR-203 low | 13 | Reference | 13 | 0.247 | 3.04 (0.46–19.99) | ||
Significant values are in bold face.
.
Furthermore, when analyzing the effect of miR-155 or miR-203 on the patients survival in distinct histological subtypes, it became evident that in myogenic sarcomas (rhabdo- and leiomyosarcoma) a lowered miR-155 expression as well as a lowered miR-203 expression was significantly associated to a worsened survival of the STS patients (See Table S1).
3. Discussion
We measured miR-155 and miR-203 expression in 79 soft tissue sarcoma (STS) specimens. Both microRNAs were detectable in all samples to a different extent, with higher expression of miR-155 than miR-203. This is consistent with the general theory of miR-155 being an oncomiR (cancer-associated microRNA) defined by its general upregulation in lymphomas, leukemias, and solid tumors [8,27,28], and miR-203 being a tumor suppressor miR due to its general downregulation in many tumor entities [20,29,30]. Interestingly, several groups report miR-155 to be overexpressed, especially in liposarcoma in comparison to normal fat tissue [9,10]. We did not observe any association between the STS sub-entities present in our cohort and altered miR-155 expression; however, we could not monitor normal tissue corresponding to our tumor specimens. On the other hand, increased miR-155 expression was significantly associated with elevated tumor stage in our STS cohort. Higher expression of miR-155 in connection with advanced tumor stages was also observed in breast cancer [31], laryngeal squamous cell carcinoma [32], or chordoma [33]. In contrast, miR-155 expression in pediatric Wilms tumors was lower in advanced tumor stages and increased due to chemotherapy [34].
MiR-155 and miR-203 expression was correlated with several–mainly hypoxia-associated–molecular factors in our STS cohort. MiR-155 was significantly positively correlated with the hypoxia-dependent microRNA miR-210 and inversely correlated with the mRNA expression of the known miR-210 target gene ephrin-A3 (EFNA3) [6,35]. There is a negative feed-back-loop between miR-155 and von-Hippel-Lindau factor (VHL) [36]. On one hand, the VHL protein suppresses miR-155 and miR-210 expression. On the other hand, Kong and colleagues demonstrated the pivotal role of miR-155 in tumor angiogenesis of breast cancer tissue through targeting VHL and subsequent HIF-1α stabilization [36]. They suggest that miR-155 regulates the VHL/HIF pathway to induce tumor angiogenesis and metastasis in triple-negative breast cancer. Furthermore, miR-210 is upregulated by HIF-1α and therefore it is possible that miR-155 indirectly increases miR-210 by stabilizing hypoxia inducible factor 1 subunit alpha (HIF1α). In this way, increasing miR-210 levels can reduce mRNA levels of its target gene EFNA3.
Additionally, we found a correlation between miR-155 expression and p53 mutation status. This is in accordance with previous findings demonstrating that miR-155 can be downregulated by wild-type p53, p63 and breast cancer type 1 susceptibility protein (BRCA1). However, an upregulation of miR-155 through mutant p53 or mutant BRCA1 has been reported (reviewed in [8,37]).
Moreover, we detected a significant association between increased miR-155 expression and several components of the uPA system, namely, uPA, uPAR, and PAI-1. The expression of miR-155 is also positively correlated with the expression of serpine1/PAI-1 in breast cancer [37]. Wild-type p53 represses uPA transcription by repressing the uPA promoter and enhancer [38]. However, oncogenic p53 mutants exhibit a significant loss or total ablation of this repressing activity [38]. Altogether, the correlation of miR-155 expression with the whole uPA system warrants further investigation.
In addition, the miR-155 level is negatively correlated with the mRNA expression level of the stem cell marker leucine rich repeat containing G protein-coupled receptor 5 (LGR5) [24]. However, LGR5 does not appear to be a direct target of miR-155 (http://mirtar.mbc.nctu.edu.tw/human/). Interestingly, LGR5-induced p53 degradation was described for hepatocellular carcinoma cells, which was also associated with therapeutic resistance against doxorubicin [39]. It appears that there is a LGR5/p53/miR-155 axis, but this needs further investigation.
MiR-203 is negatively associated with the expression of several osteopontin splice variants. Interestingly, neither the existing literature nor available databases (TargetScan.org) hint at the direct targeting of OPN by miR-203. However, a reduction in OPN mRNA/protein under hypoxia has been reported in normal human cells [40]. Therefore, it remains to be elucidated whether there is a direct or indirect regulation of this gene by miR-203. Furthermore, a negative correlation between miR-203 expression and the protein level of phosphorylated AKT (pAKT) was observed in our study. This is in agreement with the findings of Yang and coworkers [41]. They identified phosphatidylinositol 4,5-bisphosphate 3-kinase (PIK3CA) as a direct target gene of miR-203. Inhibition of miR-203 resulted in increased levels of pAKT and pPIK3CA. Therefore, they suggest that miR-203 targets the PI3K/Akt signaling pathway [41].
In our cohort, increased miR-155 expression exhibited a significant negative impact on STS patient survival, which was demonstrated for different STS entities for the first time. In this way, our results support the findings of Kapodistrias and colleagues, who showed that increased miR-155 expression is an independent indicator of unfavorable prognosis in liposarcoma [42]. In non-small-cell lung cell carcinoma, the effect of miR-155 expression on patient survival was dependent on the histological subtype, with negative effects of increased miR-155 expression in adenocarcinoma and decreased miR-155 expression in squamous cell carcinoma [43]. However, in general, increased miR-155 expression is associated with worsened survival in tumor patients, as seen in pancreatic ductal adenocarcinoma [6], colorectal carcinoma [44], hepatocellular carcinoma [45], breast cancer [46], glioma [47], osteosarcoma [13], or leukemia [48].
On the other hand, we identified, for the first time in STS, a significant association between a lower miR-203 expression and a worsened STS patient outcome. The role of miR-203 in tumor patient survival is multifaceted, with increased miR-203 being a negative predictor of patient survival in pancreatic adenocarcinoma [6,49], epithelial ovarian cancer [50], breast cancer [51], colon/colorectal cancer [52,53], and ependymoma [54], but being a positive predictor for patient survival in glioma [55], hepatocellular carcinoma [56], epithelial ovarian cancer [57], and esophageal cancer [17]. MiR-203 has been characterized to act as a tumor suppressor microRNA in laryngeal carcinoma by targeting survivin [58] and in lung cancer by targeting v-src avian sarcoma viral oncogene (SRC) and protein kinase C alpha (PKCα/PRKCA) [57,59]. Furthermore, a tumor suppressive role of miR-203 was shown in prostate cancer [60]. MiR-203 levels are downregulated in clinical samples of primary prostate cancer and further reduced in metastatic prostate cancer [60]. Transfection with miR-203 precursors resulted in a reduction of phosphorylated EGFR and phosphorylated ERK1/2 in DU145 derivative PCa cells. Siu et al suggested that the loss of miR-203 is a molecular link in the progression of prostate cancer metastasis and tyrosine kinase inhibitor (TKI) resistance characterized by high EGFR ligand output and antiapoptotic protein activation [60]. Interestingly, hypoxia can upregulate phosphorylated ERK1/2 in normal human cells [40], and a reduction in miR-203 level is associated with increased phosphorylated ERK1/2 as well [60]. Therefore, both hypoxia and a reduction in miR-203 level may trigger activation of MAPK (ERK1/2)/AKT signaling, which also plays a role in different sarcomas [61].
Altogether, increased miR-155 is associated with more aggressive behavior and poor prognosis in many different cancers, but for miR-203, a controversial picture can be drawn depending on the tumor entity. In this way, miR-155 appears to be a potential target for miRNA-based therapy, as suggested seven years ago [8]. However, the physiological role of miR-155 in myelopoiesis and erythropoiesis should also be considered [8]. Since that time, miRNA therapies have undergone evaluation in several recently undertaken clinical trials where results are eagerly awaited [62]. MiR-155 inhibition by MRG106/cobomarsen was recently tested in a clinical phase 1 study in different leukemia and lymphoma patients (NCT02580552) and in two clinical phase 2 studies in cutaneous T-cell lymphoma/mycosis fungoides patients (NCT03837457, NCT03713320). Our results suggest that inhibition of miR-155 could also have potential as a future treatment option for STS patients.
4. Materials and Methods
4.1. Patients and Specimens
The tumor specimens analyzed in this study originated from 79 adult patients from a previously described STS cohort [63]. The study was approved by the ethics committee of Martin Luther University Halle-Wittenberg (date of approval: 24 January 2007). All patients gave written informed consent. The demographic and clinicopathological details of the patients are summarized in Table 3.
Table 3.
MiR-155 and miR-203 expression in relation to clinical and histopathological characteristics in soft tissue sarcoma patients.
| Parameters | miR-155 Low (<18.0) | miR-155 High (≥18.0) |
Chi2 Test (p-Value) | miR-203 Low (<0.9) | miR-203 High (≥0.9) | Chi2 Test (p-Value) | |
|---|---|---|---|---|---|---|---|
| age | <60 years | 23 | 20 | n.s. | 15 | 28 | n.s. |
| >60 years | 22 | 14 | 14 | 22 | |||
| gender | female | 24 | 12 | n.s. | 16 | 20 | n.s. |
| male | 21 | 22 | 13 | 30 | |||
| patients status | alive | 25 | 11 | 0.04 | 7 | 29 | 0.004 |
| deceased | 20 | 23 | 22 | 21 | |||
| tumor stage a | I | 12 | 1 | 0.011 | 3 | 10 | n.s. |
| II | 19 | 12 | 10 | 21 | |||
| III | 11 | 15 | 11 | 15 | |||
| IV | 3 | 6 | 5 | 4 | |||
| resection | radical (R0) | 32 | 21 | n.s. | 17 | 36 | n.s. |
| not radical (R1) | 13 | 13 | 12 | 14 | |||
| tumor localization | extremities | 29 | 23 | n.s. | 18 | 34 | n.s. |
| trunk wall | 3 | 2 | 2 | 3 | |||
| head/neck | 2 | 0 | 1 | 1 | |||
| abdomen/peritoneum | 9 | 9 | 6 | 12 | |||
| multiple locations | 2 | 0 | 2 | 0 | |||
| histological subtypes | LS | 12 | 9 | 0.011 | 10 | 11 | n.s. |
| FS + NOS | 12 | 8 | 4 | 16 | |||
| NS | 4 | 3 | 3 | 4 | |||
| RMS + LMS | 11 | 10 | 8 | 13 | |||
| other | 6 | 4 | 4 | 6 | |||
| tumor size | T1 | 6 | 5 | n.s. | 3 | 8 | n.s. |
| T2 | 39 | 29 | 26 | 42 | |||
| number of relapses | 0 | 28 | 20 | n.s. | 15 | 33 | n.s. |
| 1 | 8 | 5 | 6 | 7 | |||
| >2 | 9 | 9 | 8 | 10 |
a Union for International Cancer Control Guidelines; Abbreviations: LS—liposarcoma; FS—fibrosarcoma; RMS—rhabdomyosarcoma; LMS—leiomyosarcoma; NS—neuronal sarcoma; Syn—synovial sarcoma; NOS—not otherwise specified; T1—≤5 cm in diameter; T2—>5 cm in diameter; n.s.—not significant.
4.2. RNA Isolation and cDNA Synthesis
Total RNA was isolated by phenol/chloroform extraction using TRIzol reagent (Invitrogen, Karlsruhe, Germany) from 10–30 tumor slides of 10 μm thickness. Briefly, tumor tissue was suspended in TRIzol reagent and vortexed vigorously, and then chloroform (Roth, Karlsruhe, Germany) was added and mixed. The suspension was centrifuged for 10 min at 6000× g, and the aqueous phase was mixed with isopropanol (Roth). After precipitation overnight, the RNA pellet was washed twice in ice-cold ethanol (96% and 70%, respectively; Merck, Darmstadt, Germany) and eluted in 20 μL DEPC-treated water (Invitrogen).
Ten nanograms of total RNA was introduced to cDNA synthesis of miR-155, miR-203 and U18 snoRNA (reference gene) with the stem-loop primer system (Applied Biosystems, Darmstadt, Germany). Reverse transcription reactions were carried out with the SuperScript II Reverse transcription kit (Invitrogen, Karlsruhe, Germany) according to the manufacturers’ protocols. The qPCR measurements were performed in duplicate with the HotStart Taq Polymerase Kit (Qiagen, Hilden, Germany) on the Rotorgene 3000 real-time PCR system (LTF, Wasserburg, Germany). The resulting mean CT values were analyzed in comparison to U18 snoRNA expression as a reference gene. Further, 2−ΔCT values were designated according to Livak and Schmittgen [64].
4.3. Statistical Analysis
Bivariate linear regression analysis (Spearman’s rank test) was applied to test the correlation between miR-155 or miR-203 expression and molecular biological tumor markers. Pearson’s Chi2 test and nonparametric tests (Kruskal-Wallis test) were applied to detect interdependences between microRNA expression and demographic or clinicopathological parameters. Survival analyses (Kaplan–Meier analyses, uni- and multivariate Cox regression analyses) were performed to test associations between microRNA expression and STS patient survival. All these tests were performed with SPSS 25.0 (IBM, Ehingen, Germany). p values ≤ 0.05 were considered significant. ROC analyses and recursive partitioning were applied to test the individual contributions of miR-155 and miR-203 in combined risk stratification. These tests were performed with the R statistical framework (v. 3.2.1; R foundation for statistical computing, Vienna, Austria) and the additional function libraries pROC and rpart.
5. Conclusions
We analyzed the association of miR-155 and miR-203 expression with the prognosis of soft tissue sarcoma patients. Both microRNAs were detectable in all tumor specimens to different extents and exhibited correlations with several hypoxia-associated molecular markers, such as miR-210 and, in the case of miR-155, components of the urokinase-like plasminogen activator (uPA) system and ephrin-A3 (EFNA3). Furthermore, miR-155 expression was higher in advanced tumor stages. The expression of miR-155 and miR-203 as well as the combination of both microRNAs was significantly associated with a worsened disease-specific survival in soft tissue sarcoma patients. In summary, miR-155 and miR-203 are interesting candidates for risk stratification in soft tissue sarcoma patients, and miR-155 may be an option for therapy management in the future.
Acknowledgments
We acknowledge the financial support within the funding program Open Access Publishing by the German Research Foundation (DFG).
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/8/2254/s1, Figure S1: The distribution of miR-155 expression and miR-203 in relation to the histological subtype, Table S1: Univariate and multivariate Cox regression survival analyses of the impact of miR-155 and miR-203 on soft tissue sarcoma patient survival in different histological subtypes.
Author Contributions
Conceptualization, T.G., P.W., and H.T.; Formal analysis, T.G., H.-J.H., P.W., and H.T.; Funding acquisition, H.T.; Investigation, T.G., F.K., and A.G.; Methodology, T.G., M.K., and S.W.; Project administration, H.T.; Visualization, P.W. and H.T.; Writing—original draft, T.G., S.W., and H.T.; Writing—review & editing, T.G., P.W., M.K., S.W., and H.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Taylor B.S., Barretina J., Maki R.G., Antonescu C.R., Singer S., Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducimetière F., Lurkin A., Ranchère-Vince D., Decouvelaere A.-V., Péoc’h M., Istier L., Chalabreysse P., Muller C., Alberti L., Bringuier P.-P., et al. Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS ONE. 2011;6:e20294. doi: 10.1371/journal.pone.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari A., Sultan I., Huang T.T., Rodriguez-Galindo C., Shehadeh A., Meazza C., Ness K.K., Casanova M., Spunt S.L. Soft tissue sarcoma across the age spectrum: a population-based study from the Surveillance Epidemiology and End Results database. Pediatr. Blood Cancer. 2011;57:943–949. doi: 10.1002/pbc.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Smolle M.A., Leithner A., Posch F., Szkandera J., Liegl-Atzwanger B., Pichler M. MicroRNAs in Different Histologies of Soft Tissue Sarcoma: A Comprehensive Review. Int. J. Mol. Sci. 2017;18:1960. doi: 10.3390/ijms18091960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greither T., Grochola L.F., Udelnow A., Lautenschläger C., Würl P., Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 7.Greither T., Würl P., Grochola L., Bond G., Bache M., Kappler M., Lautenschläger C., Holzhausen H.-J., Wach S., Eckert A.W., et al. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int. J. Cancer. 2012;130:1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- 8.Elton T.S., Selemon H., Elton S.M., Parinandi N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532:1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Zhang P., Bill K., Liu J., Young E., Peng T., Bolshakov S., Hoffman A., Song Y., Demicco E.G., Terrada D.L., et al. MiR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances β-catenin signaling. Cancer Res. 2012;72:1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincenzi B., Iuliani M., Zoccoli A., Pantano F., Fioramonti M., de Lisi D., Frezza A.M., Rabitti C., Perrone G., Onetti Muda A., et al. Deregulation of dicer and mir-155 expression in liposarcoma. Oncotarget. 2015;6:10586–10591. doi: 10.18632/oncotarget.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalsky R.L., Samols M.A., Plaisance K.B., Boss I.W., Riva A., Lopez M.C., Baker H.V., Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W.H., Chi J.-T.A., Braich R., Manoharan M., Soutschek J., Ohler U., et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X., Geng X., Zhang J., Zhao H., Liu Y. miR-155 promotes the growth of osteosarcoma in a HBP1-dependent mechanism. Mol. Cell. Biochem. 2015;403:139–147. doi: 10.1007/s11010-015-2344-z. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Tang B., Han H., Mao D., Chen J., Zeng Y., Xiong M. miR-155 Affects Osteosarcoma MG-63 Cell Autophagy Induced by Adriamycin Through Regulating PTEN-PI3K/AKT/mTOR Signaling Pathway. Cancer Biother. Radiopharm. 2018;33:32–38. doi: 10.1089/cbr.2017.2306. [DOI] [PubMed] [Google Scholar]
- 15.Lu S., Liao Q.-S., Tang L. MiR-155 affects osteosarcoma cell proliferation and invasion through regulating NF-κB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7633–7639. doi: 10.26355/eurrev_201811_16380. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hezova R., Kovarikova A., Srovnal J., Zemanova M., Harustiak T., Ehrmann J., Hajduch M., Svoboda M., Sachlova M., Slaby O. Diagnostic and prognostic potential of miR-21, miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell carcinoma of esophagus. Diagn. Pathol. 2015;10:42. doi: 10.1186/s13000-015-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y., Yang W., Zhu Y., Yuan Y. Prognostic role of microRNA-203 in various carcinomas: evidence from a meta-analysis involving 13 studies. Springerplus. 2016;5:1538. doi: 10.1186/s40064-016-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diao Y., Guo X., Jiang L., Wang G., Zhang C., Wan J., Jin Y., Wu Z. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J. Biol. Chem. 2014;289:529–539. doi: 10.1074/jbc.M113.494716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D., Liu G., Wang K. miR-203 Acts as a Tumor Suppressor Gene in Osteosarcoma by Regulating RAB22A. PLoS ONE. 2015;10:e0132225. doi: 10.1371/journal.pone.0132225. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Taubert H., Meye A., Würl P. Soft tissue sarcomas and p53 mutations. Mol Med. 1998;4:365–372. doi: 10.1007/BF03401743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bache M., Kappler M., Wichmann H., Rot S., Hahnel A., Greither T., Said H.M., Kotzsch M., Würl P., Taubert H., et al. Elevated tumor and serum levels of the hypoxia-associated protein osteopontin are associated with prognosis for soft tissue sarcoma patients. BMC Cancer. 2010;10:132. doi: 10.1186/1471-2407-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotzsch M., Magdolen V., Greither T., Kappler M., Bache M., Lautenschläger C., Füssel S., Eckert A.W., Luther T., Baretton G., et al. Combined mRNA expression levels of members of the urokinase plasminogen activator (uPA) system correlate with disease-associated survival of soft-tissue sarcoma patients. BMC Cancer. 2011;11:273. doi: 10.1186/1471-2407-11-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rot S., Taubert H., Bache M., Greither T., Würl P., Eckert A.W., Schubert J., Vordermark D., Kappler M. A novel splice variant of the stem cell marker LGR5/GPR49 is correlated with the risk of tumor-related death in soft-tissue sarcoma patients. BMC Cancer. 2011;11:429. doi: 10.1186/1471-2407-11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahnel A., Wichmann H., Greither T., Kappler M., Würl P., Kotzsch M., Taubert H., Vordermark D., Bache M. Prognostic impact of mRNA levels of osteopontin splice variants in soft tissue sarcoma patients. BMC Cancer. 2012;12:131. doi: 10.1186/1471-2407-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rot S., Taubert H., Bache M., Greither T., Würl P., Holzhausen H.-J., Eckert A.W., Vordermark D., Kappler M. Low HIF-1α and low EGFR mRNA Expression Significantly Associate with Poor Survival in Soft Tissue Sarcoma Patients; the Proteins React Differently. Int. J. Mol. Sci. 2018;19:3842. doi: 10.3390/ijms19123842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z., Ma T., Huang C., Hu T., Li J. The pivotal role of microRNA-155 in the control of cancer. J. Cell. Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 28.Jurkovicova D., Magyerkova M., Kulcsar L., Krivjanska M., Krivjansky V., Gibadulinova A., Oveckova I., Chovanec M. miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma. 2014;61:241–251. doi: 10.4149/neo_2014_032. [DOI] [PubMed] [Google Scholar]
- 29.Furuta M., Kozaki K.-i., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 30.Sonkoly E., Lovén J., Xu N., Meisgen F., Wei T., Brodin P., Jaks V., Kasper M., Shimokawa T., Harada M., et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis. 2012;1:e3. doi: 10.1038/oncsis.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng S.-R., Guo G.-L., Zhang W., Huang G.-L., Hu X.-Q., Zhu J., Huang Q.-D., You J., Zhang X.-H. Clinical significance of miR-155 expression in breast cancer and effects of miR-155 ASO on cell viability and apoptosis. Oncol. Rep. 2012;27:1149–1155. doi: 10.3892/or.2012.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X.-D., Zhang W., Liang H.-J., Ji W.-Y. Overexpression of miR -155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS ONE. 2013;8:e56395. doi: 10.1371/journal.pone.0056395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osaka E., Kelly A.D., Spentzos D., Choy E., Yang X., Shen J.K., Yang P., Mankin H.J., Hornicek F.J., Duan Z. MicroRNA-155 expression is independently predictive of outcome in chordoma. Oncotarget. 2015;6:9125–9139. doi: 10.18632/oncotarget.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo X., Dong J., He X., Shen L., Long C., Liu F., Liu X., Lin T., He D., Wei G. MiR-155-5p exerts tumor-suppressing functions in Wilms tumor by targeting IGF2 via the PI3K signaling pathway. Biomed. Pharmacother. 2020;125:109880. doi: 10.1016/j.biopha.2020.109880. [DOI] [PubMed] [Google Scholar]
- 35.Fasanaro P., D’Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M.C., Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong W., He L., Richards E.J., Challa S., Xu C.-X., Permuth-Wey J., Lancaster J.M., Coppola D., Sellers T.A., Djeu J.Y., et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neilsen P.M., Noll J.E., Mattiske S., Bracken C.P., Gregory P.A., Schulz R.B., Lim S.P., Kumar R., Suetani R.J., Goodall G.J., et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2013;32:2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- 38.Kunz C., Pebler S., Otte J., von der Ahe D. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 1995;23:3710–3717. doi: 10.1093/nar/23.18.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z., Guo D., Wang Q., Liu P., Xiao Y., Wu P., Wang Y., Chen B., Liu Z., Liu Q. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics. 2019;9:2967–2983. doi: 10.7150/thno.30562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Han M.-x., Li S., Xu Y., Wang L. Hypoxia regulates the proliferation and osteogenic differentiation of human periodontal ligament cells under cyclic tensile stress via mitogen-activated protein kinase pathways. J. Periodontol. 2014;85:498–508. doi: 10.1902/jop.2013.130048. [DOI] [PubMed] [Google Scholar]
- 41.Yang X., Li X., Lin Q., Xu Q. Up-regulation of microRNA-203 inhibits myocardial fibrosis and oxidative stress in mice with diabetic cardiomyopathy through the inhibition of PI3K/Akt signaling pathway via PIK3CA. Gene. 2019;715:143995. doi: 10.1016/j.gene.2019.143995. [DOI] [PubMed] [Google Scholar]
- 42.Kapodistrias N., Mavridis K., Batistatou A., Gogou P., Karavasilis V., Sainis I., Briasoulis E., Scorilas A. Assessing the clinical value of microRNAs in formalin-fixed paraffin-embedded liposarcoma tissues: Overexpressed miR-155 is an indicator of poor prognosis. Oncotarget. 2017;8:6896–6913. doi: 10.18632/oncotarget.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donnem T., Eklo K., Berg T., Sorbye S.W., Lonvik K., Al-Saad S., Al-Shibli K., Andersen S., Stenvold H., Bremnes R.M., et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J. Transl. Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibuya H., Iinuma H., Shimada R., Horiuchi A., Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 45.Han Z.-B., Chen H.-Y., Fan J.-W., Wu J.-Y., Tang H.-M., Peng Z.-H. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J. Cancer Res. Clin. Oncol. 2012;138:153–161. doi: 10.1007/s00432-011-1076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J., Wang B.-C., Tang J.-H. Clinical significance of microRNA-155 expression in human breast cancer. J. Surg. Oncol. 2012;106:260–266. doi: 10.1002/jso.22153. [DOI] [PubMed] [Google Scholar]
- 47.Sun J., Shi H., Lai N., Liao K., Zhang S., Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med. Oncol. 2014;31:911. doi: 10.1007/s12032-014-0911-x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X., Wang Y., Guo Q., Diao Y., Liu H., Song G., Wang W., Zhang Z., Yin H., Li L. Prognostic role of microRNA-155 in patients with leukemia: A meta-analysis. Clin. Chim. Acta. 2018;483:6–13. doi: 10.1016/j.cca.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 49.Ikenaga N., Ohuchida K., Mizumoto K., Yu J., Kayashima T., Sakai H., Fujita H., Nakata K., Tanaka M. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann. Surg. Oncol. 2010;17:3120–3128. doi: 10.1245/s10434-010-1188-8. [DOI] [PubMed] [Google Scholar]
- 50.Wang C., Wang X., Liang H., Wang T., Yan X., Cao M., Wang N., Zhang S., Zen K., Zhang C., et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS ONE. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madhavan D., Peng C., Wallwiener M., Zucknick M., Nees J., Schott S., Rudolph A., Riethdorf S., Trumpp A., Pantel K., et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37:461–470. doi: 10.1093/carcin/bgw008. [DOI] [PubMed] [Google Scholar]
- 52.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L.W., Au G.K.H., et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bovell L.C., Shanmugam C., Putcha B.-D.K., Katkoori V.R., Zhang B., Bae S., Singh K.P., Grizzle W.E., Manne U. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin. Cancer Res. 2013;19:3955–3965. doi: 10.1158/1078-0432.CCR-12-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa F.F., Bischof J.M., Vanin E.F., Lulla R.R., Wang M., Sredni S.T., Rajaram V., Bonaldo M.d.F., Wang D., Goldman S., et al. Identification of microRNAs as potential prognostic markers in ependymoma. PLoS ONE. 2011;6:e25114. doi: 10.1371/journal.pone.0025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He J., Deng Y., Yang G., Xie W. MicroRNA-203 down-regulation is associated with unfavorable prognosis in human glioma. J. Surg. Oncol. 2013;108:121–125. doi: 10.1002/jso.23315. [DOI] [PubMed] [Google Scholar]
- 56.Chen H.-Y., Han Z.-B., Fan J.-W., Xia J., Wu J.-Y., Qiu G.-Q., Tang H.-M., Peng Z.-H. miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med. Oncol. 2012;29:1859–1865. doi: 10.1007/s12032-011-0031-9. [DOI] [PubMed] [Google Scholar]
- 57.Wang S., Zhao X., Wang J., Wen Y., Zhang L., Wang D., Chen H., Chen Q., Xiang W. Upregulation of microRNA-203 is associated with advanced tumor progression and poor prognosis in epithelial ovarian cancer. Med. Oncol. 2013;30:681. doi: 10.1007/s12032-013-0681-x. [DOI] [PubMed] [Google Scholar]
- 58.Bian K., Fan J., Zhang X., Yang X.-W., Zhu H.-Y., Wang L., Sun J.-Y., Meng Y.-L., Cui P.-C., Cheng S.-Y., et al. MicroRNA-203 leads to G1 phase cell cycle arrest in laryngeal carcinoma cells by directly targeting survivin. FEBS Lett. 2012;586:804–809. doi: 10.1016/j.febslet.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 59.Wang N., Liang H., Zhou Y., Wang C., Zhang S., Pan Y., Wang Y., Yan X., Zhang J., Zhang C.-Y., et al. miR-203 suppresses the proliferation and migration and promotes the apoptosis of lung cancer cells by targeting SRC. PLoS ONE. 2014;9:e105570. doi: 10.1371/journal.pone.0105570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siu M.K., Abou-Kheir W., Yin J.J., Chang Y.-S., Barrett B., Suau F., Casey O., Chen W.-Y., Fang L., Hynes P., et al. Loss of EGFR signaling regulated miR-203 promotes prostate cancer bone metastasis and tyrosine kinase inhibitors resistance. Oncotarget. 2014;5:3770–3784. doi: 10.18632/oncotarget.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teicher B.A. Searching for molecular targets in sarcoma. Biochem. Pharmacol. 2012;84:1–10. doi: 10.1016/j.bcp.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Bonneau E., Neveu B., Kostantin E., Tsongalis G.J., de Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30:114–127. [PMC free article] [PubMed] [Google Scholar]
- 63.Kappler M., Köhler T., Kampf C., Diestelkötter P., Würl P., Schmitz M., Bartel F., Lautenschläger C., Rieber E.P., Schmidt H., et al. Increased survivin transcript levels: an independent negative predictor of survival in soft tissue sarcoma patients. Int. J. Cancer. 2001;95:360–363. doi: 10.1002/1097-0215(20011120)95:63.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 64.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.