Abstract
To our best knowledge this is the first study characterizing fish pathogens isolated from marine plastics from the West coast of Norway for their potential for pathogenicity using whole genome sequencing. Marine plastic polymers identified as polyethylene, polyethylene/ethylene vinyl acetate copolymer and polypropylene, yielded a total of 37 bacterial isolates dominated by Pseudomonas spp. (70%). Six isolates representing either fish pathogens or opportunistic human pathogens were selected for whole genome sequencing (WGS). These included four isolates belonging to Aeromonas spp., one Acinetobacter beijerinckii isolate and one Morganella morganii isolate. Three Aeromonas salmonicida isolates were potentially virulent and carried virulence factors involved in attachment, type II and type VI secretion systems as well as toxins such as aerA/act, ahh1, ast, hlyA, rtxA and toxA. A. salmonicida and Acinetobacter beijerinckii carried new variants of antibiotic resistance genes (ARGs) such as β-lactamases and chloramphenicol acetyltransferase (catB), whereas Morganella morganii carried several clinically relevant ARGs. Our study shows that marine plastics carry not only potentially virulent fish pathogens but also multidrug resistant opportunistic human pathogens like M. morganii and may serve as vectors for transport of these pathogens in the marine environment.
Keywords: marine plastics, microplastics, fish pathogens, antibiotic resistance, Norway, Aeromonas sp.
1. Introduction
Plastics are synthetic, anthropogenic materials made from a wide range of organic polymers such as polyethylene (PE), polypropylene (PP), polystyrene (PS), polyvinyl chloride (PVC), polyurethane (PUR), polyethylene terephthalate (PET), polybutylene terephthalate (PBT) and polyamide (PA), commonly known as nylon [1]. Plastic materials enter the marine ecosystem in many different ways, such as being dumped, lost [2], carelessly handled [2,3] or left [2,4]. Plastics found in the environment are mostly classified into three different categories based on their sizes: nanoplastic (<100 nm), microplastic (100 nm–5 mm) and macroplastic (>5 mm) [5,6,7].
Pollution with plastics and accumulation of plastics in the oceans has a big impact on the marine ecosystem [8,9,10]. Ingestion of plastic by marine organisms can disturb their energy balance, affect behavior and sometimes block the intestinal tract leading to sublethal effect or, in the worst-case, cause death [11,12]. Small microplastic (<10 µm) and nanoplastic particles appear to have an increased plastic particle toxicity to aquatic life [13]. Plastic can also be toxic due to the breakdown products as well as due to the presence of chemicals absorbed or released from plastics, such as plasticizers [14,15,16]. Although some polymers are biodegradable, albeit at a very slow rate, others are not [1] and thus plastics stay in the environment for a long time.
Plastic provides a hydrophobic surface for the attachment of microbes and hence different microbes colonize plastics in the marine environment [5,17,18]. Therefore, marine plastics may function as vectors for the transport of bacteria, including fish pathogens, in the oceans [19]. Colonization of plastics by bacteria can also facilitate horizontal gene transfer of antibiotic resistance genes (ARGs), owing to biofilm formation [20]. Accordingly, a study has shown presence of multidrug resistant bacteria on plastics from marine environments [5].
Plastic pollution has been reported in the marine environment in Norway [21]. To date, Norway is a country with a low burden of antibiotic resistance in clinics and in the environment [22]. Although this is true, there is lack of knowledge about microbiota associated with plastic found in the marine environment in Norway as well as associated ARGs. The aim of our study was to investigate the presence of fish pathogens and antibiotic resistant bacteria on marine plastics collected from the West coast of Norway. We isolated fish pathogens and opportunistic human pathogens from plastic surfaces, analyzed whole genome sequences of these isolates and report their potential for pathogenicity as well as new variants of ARGs.
2. Materials and Methods
2.1. Sample Collection
Macroplastic samples (n = 7), with different sizes ranging from one centimeter to 15 cm, were collected from the intertidal zone at beaches in Øygarden, Vestland county, Norway. The coordinates for the sampling areas are 60°29′57.6″ N 4°55′05.3″ E and 60°29′54.5″ N 4°54′52.6″ E, respectively. The samples were collected in either sterile 50 mL tubes or sterile plastic bags. These samples were then stored at 4 °C and transported back to the lab for analysis.
2.2. Isolation and Identification of Bacterial Strains
Plastic samples were carefully washed with sterile phosphate buffer saline (PBS) before making suspensions. Suspensions were made by adding 10 mL of sterile PBS to each plastic sample in 50 mL sterile tubes. Four sterile glass beads (4 mm diameter) were added to the tubes, followed by vortexing for 90 s at maximum speed. Serial dilutions of the suspension were prepared in sterile PBS and spread on Mueller–Hinton agar plates and MacConkey agar plates, both media containing either ampicillin (100 µg/mL), cefotaxime (10 µg/mL) or no antibiotics. Mueller–Hinton agar plates were incubated at 25 °C for 24–48 h, while MacConkey agar plates were incubated at 37 °C for 24–48 h. A total of 37 colonies were picked and purified by restreaking on Mueller–Hinton agar with ampicillin. Fish pathogens like Aeromonas spp. and Vibrio spp. are intrinsically resistant to penicillins, thus, ampicillin facilitated selection of fish pathogens [23,24]. All isolates were stored as glycerol stocks at −80 °C for further use. For identification, isolates were grown on Mueller–Hinton agar with ampicillin at 30 °C for 24 h. The strains were typed using a Bruker MALDI Biotyper at Veterinær instituttet (Bergen, Norway) using the MALDI Biotyper database.
2.3. Genomic DNA Extraction and Illumina Sequencing
Genomic DNA was extracted from isolates cultivated overnight on Mueller–Hinton agar with ampicillin using QlAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. NanoDrop 1000 and Qubit 2.0 (Thermo Scientific, Waltham, MA, USA) were used to quantify the extracted DNA. Extracted DNA was sent for sequencing to the Norwegian Sequencing Centre (Oslo University Hospital, Ullevål, Oslo, Norway). Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA, USA) was used for preparing sequencing libraries. Sequencing was performed on an Illumina MiSeq platform (Illumina, San Diego, CA, USA), using 2 × 300 bp chemistry.
2.4. Genome Sequence Assembly and Screening for ARGs
Adapters were removed from the obtained raw reads and the reads were quality filtered using BBDuk (version 38.75; https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/). Sequences were assembled in SPAdes (version 3.13.0) [25] using the default parameters; spades.py −1 xx_R1_trim.fastq −2 xx_R2_trim.fastq --careful --only-assembler --cov-cutoff auto -o output_name. Genome annotation was performed using National Center for Biotechnology Information (NCBI) Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) [26]. The genome sequences were screened for antibiotic resistance genes using the CARD database (version 3.0.7) [27] and ResFinder 3.2 database [28]. The virulence genes were identified using VFanalyzer and the virulence factor database (VFDB) [29]. Genome sequences have been submitted to GenBank under the following genome accession numbers: JAACGC000000000, JAACGB000000000, JAACGA000000000, JAACFZ000000000, JAACFY000000000, JAACFX000000000 and JAACFW000000000, respectively.
2.5. Phylogenetic Analysis
Amino acid sequences for CphA, RtxA and AerA were extracted from the genome sequences of Aeromonas spp. Additional sequences for respective proteins were downloaded from GenBank (www.ncbi.nlm.nih.gov/genbank). The sequences were aligned using clustalX version 2.1 [30]. The phylogenetic tree was generated by the neighbor joining method using MEGA-X with bootstrap analysis (1000 replicates) [31].
2.6. Antibiotic Susceptibility Testing
Isolates were grown on Mueller–Hinton agar with ampicillin at 30 °C overnight and were used for making suspensions for determination of minimum inhibitory concentration (MIC). MICs were determined for cefotaxime (CT), tetracycline (TC), ciprofloxacin (CI), ampicillin (AM), meropenem (MP), streptomycin (SM), trimethoprim (TR), gentamicin (GM), imipenem (IP) and chloramphenicol (CL) using E-test according to manufacturer’s protocol (bioMérieux, Paris, France). Escherichia coli strain CCUG 17,620 was used as a control strain for E-test quality check.
2.7. Identifying Plastic Polymer Type
The polymer type for plastic samples was identified by attenuated total reflection infrared spectroscopy (Cary670 FTIR spectrometer, Agilent Technologies) equipped with a monolithic diamond crystal unit (GladiATR™, PIKE Technologies, Madison, WI, USA). Spectra were collected over the wavenumber range 440–4000 cm−1. The samples were analyzed by 32 co-added scans, with a resolution of 8 cm−1. The spectra were compared to spectra of known standards using a library of environmental relevant synthetic and natural polymers [32], and commercial libraries for polymers (Bio-Rad Sadtler) in the KnowitAll Informatics System software (BioRad Laboratories, Hercules, CA, USA). Each polymer type was determined by a combination of the best-fitted spectra and the expertise of the operator on interpreting polymer IR spectra.
3. Results
3.1. Identification of the Plastic Polymers and Bacterial Strains
The plastic polymers were identified as polyethylene, polyethylene/ethylene vinyl acetate copolymer or polypropylene (Figure 1). A total of 37 isolates were obtained from the samples (Table S1). Only two isolates were obtained from polypropylene, while the rest were found on polyethylene or polyethylene/ethylene vinyl acetate copolymer. Pseudomonas spp. dominated the isolates (26 of 37). Four isolates representing fish pathogens Aeromonas spp. and two isolates representing opportunistic human pathogens (Morganella morganii and Acinetobacter beijerinckii, respectively) were chosen for characterization with whole genome sequencing (WGS). Genome sequence assembly statistics and GenBank accession number for the genome sequences of the isolates are represented in Table S2. Sequencing coverage ranged from 55× to 110×.
Figure 1.
Images of samples collected in this study. (A,C,D,F,G) are polyethylene; (B) is polyethylene/ethylene vinyl acetate copolymer; (E,F) are polypropylene.
3.2. Antibiotic Resistance and ARGs
All three A. salmonicida isolates were resistant to ampicillin. M. morganii and Ac. beijerinckii were multidrug resistant, with resistance against at least three different classes of antibiotics (Table 1). Class C β-lactamases and chloramphenicol acetyltransferase (catB) were detected in all six isolates. Class B2 metallo-β-lactamase cphA was detected in three A. salmonicida isolates, with a new variant of cphA (≤98% identity) present in two of the three A. salmonicida isolates (Table 2 and Figure S1). Along with cphA variants all Aeromonas isolates carried qnrA gene. Isolate 2HC4 (Ac. beijerinckii) carried new variants of four different ARGs including a class A and a class C β-lactamase, aminoglycoside acetyltransferase and chloramphenicol acetyltransferase. M. morganii carried several resistance genes with 100% homology to previously described ARGs. The list of ARGs detected in the genome sequences of these isolates is presented in Table 2.
Table 1.
Minimum inhibitory concentration (MIC) of different antibiotics.
| Isolate | Name | MIC (µg/mL) | Isolation Source | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT | TC | CI | AM | MP | SM | TR | GM | IP | CL | |||
| 2MA1 | Morganella morganii | 0.023 | 256 | 0.008 | >256 | 0.032 | 4 | 0.125 | 0.75 | 0.50 | 24 | Polyethylene/Ethylene vinyl acetate copolymer |
| 5HA1 | Aeromonas salmonicida | 0.023 | 0.25 | 0.003 | >256 | 0.032 | 4 | 0.032 | 0.75 | 0.25 | 0.38 | Polyethylene |
| 2HA2 | Aeromonas salmonicida | 0.047 | 0.50 | 0.002 | >256 | 0.023 | 8 | 0.25 | 1.5 | 0.25 | 1.0 | Polyethylene/Ethylene vinyl acetate copolymer |
| 2MA4 | Aeromonas salmonicida | 0.094 | 0.38 | 0.003 | >256 | 0.032 | 8 | 0.50 | 1.5 | 0.38 | 0.75 | Polyethylene/Ethylene vinyl acetate copolymer |
| 1HA1 | Aeromonas popoffii | 0.064 | 0.19 | <0.002 | >256 | 0.004 | 6 | 0.125 | 0.75 | 0.047 | 0.38 | Polyethylene |
| 2HC4 | Acinetobacter beijerinckii | 12 | 1.5 | 0.094 | >256 | 0.50 | 2 | 6 | 1.0 | 0.19 | 24 | Polyethylene/Ethylene vinyl acetate copolymer |
Legend: CT (cefotaxime), TC (tetracycline), CI (ciprofloxacin), AM (ampicillin), MP (meropenem), SM (streptomycin), TR (trimethoprim), GM (gentamicin), IP (imipenem) and CL (chloramphenicol).
Table 2.
Overview of the different antibiotic resistance genes (ARGs) detected in whole genome sequence of the isolates.
| Isolate | Name | Gene | Closest Blast Hit | Percent Identity (Amino Acid) |
|---|---|---|---|---|
| 2MA1 | Morganella morganii | bla DHA | DHA family class C β-lactamase | 100.00% |
| tet(D) | tetracycline efflux MFS transporter Tet(D) | 100.00% | ||
| aac(3) | aminoglycoside 3-N-acetyltransferase | 100.00% | ||
| catB | antibiotic acetyltransferase | 100.00% | ||
| 5HA1 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 98.43% |
| bla OXA | class D β-lactamase | 100.00% | ||
| cphA | CphA family subclass B2 metallo-β-lactamase | 97.64% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 98.64% | ||
| 2HA2 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 97.90% |
| cphA | CphA family subclass B2 metallo- β-lactamase | 99.61% | ||
| bla OXA | class D β-lactamase | 100.00% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 98.64% | ||
| 2MA4 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 96.33% |
| bla OXA | class D β-lactamase | 99.24% | ||
| cphA | CphA family subclass B2 metallo-β-lactamase | 96.85% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 99.09% | ||
| 1HA1 | Aeromonas popoffii | bla OXA | class D β-lactamase | 99.62% |
| ampC | FOX/MOX family class C β-lactamase | 98.95% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 99.10% | ||
| 2HC4 | Acinetobacter beijerinckii | ampC | class C β-lactamase | 96.46% |
| bla | class A β-lactamase | 96.11% | ||
| aac(6′)-I | AAC(6′)-Ighjkrstuvwx family aminoglycoside N-acetyltransferase | 97.93% | ||
| catB | antibiotic acetyltransferase | 96.24% |
3.3. Virulence Factors
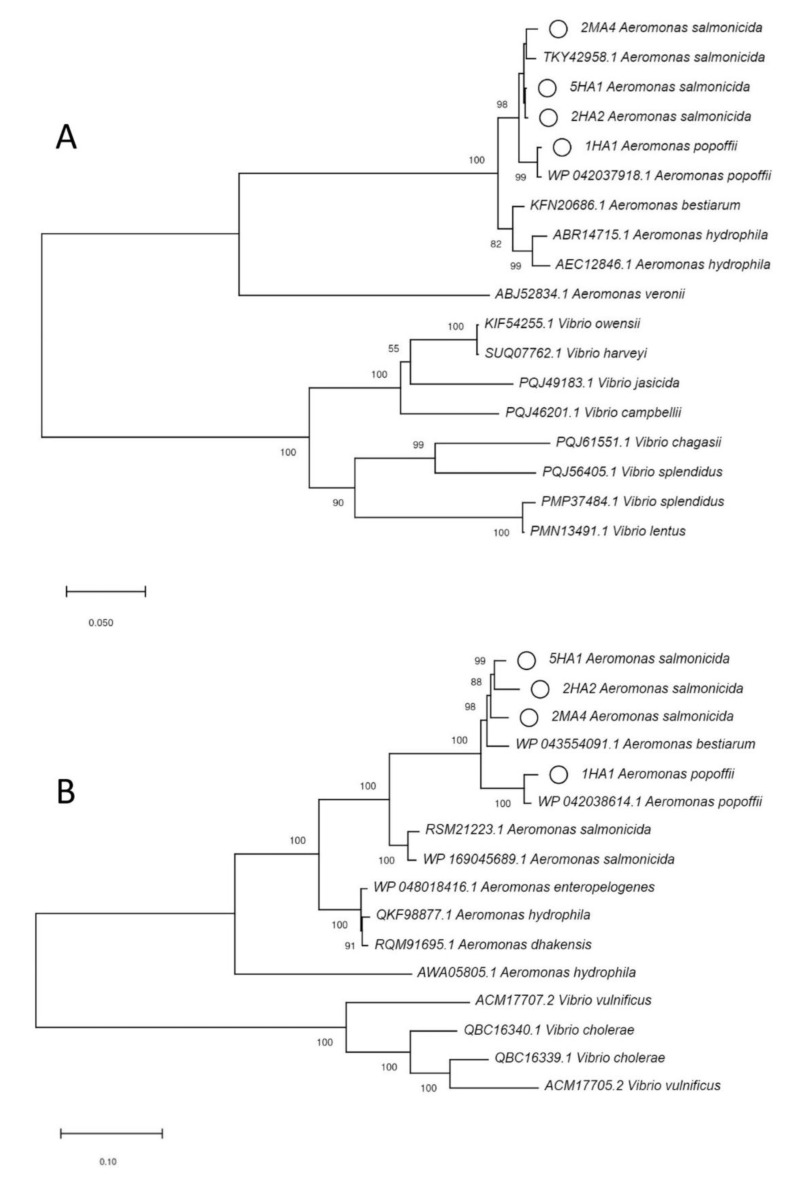
All three A. salmonicida isolates had genes for virulence factors involved in attachment, type II (T2SS) and type VI (T6SS) secretion systems as well as toxins such as aerA/act, ahh1, ast, hlyA, rtxA, rtxB, rtxC, rtxD, rtxE, rtxH and toxA. This suggests that they have potential for causing infection. M. morganii carried genes involved in acid resistance, genes involved in iron uptake, and type six secretion system proteins. A phylogenetic tree of RtxA and AerA proteins from A. salmonicida isolates and closely related proteins from other species is presented in Figure 2. A list of virulence genes detected in all sequenced isolates is presented Table S3.
Figure 2.
(A) Phylogenetic tree of AerA based on amino acid sequences. (B) Phylogenetic tree of RtxA based on amino acid sequences. The proteins highlighted with small circles are found in A. salmonicida isolates in this study. Accession numbers and host organisms are presented for sequences downloaded from GenBank. Numbers at nodes are bootstrap values (%) based on 1000 resampled datasets; only values >50% are given.
4. Discussion
To our best knowledge, this is the first study analyzing genome sequences of multidrug resistant bacteria, including fish pathogens, associated with plastic found in the marine environment in Norway. Using 16S rDNA clone libraries, Viršek et al. [33] suggested that microplastics might serve as a vector for the transport of pathogenic bacteria, especially fish pathogen A. salmonicida. Our study strengthens the notion that fish pathogens as well as other opportunistic human pathogens are present on marine plastics. We further show the presence of virulence genes and new variants of antibiotic resistance genes in A. salmonicida isolated from these plastics.
Most Aeromonas spp. are opportunistic pathogens and are widespread in the marine environments [34,35]. Aeromonas spp. are known to cause infection in both humans and animals [36]. A. salmonicida is one of the most important fish pathogens [37], causing disease in healthy wild and cultured stocks of salmon and other fish species [36]. All A. salmonicida isolates in this study carried virulence factors like the rtxA toxin. Repeats in toxins (RTX) rtxA is a cytotoxin that is an important virulence factor for many fish pathogens, including Vibrio spp. and Aeromonas spp. It disrupts host cells membranes, aiding pathogenesis [38,39]. Along with rtxA, other cytotoxins like aerolysin aerA [40], extracellular hemolysin ahh1, heat-stable cytotonic enterotoxin ast, hemolysin III and hylA were detected in these isolates [41]. A study suggests that the presence of both aerA and ahh1 represents the most cytotoxic genotype in A. hydrophila [41]. In addition, Aeromonas isolates also carried Exotoxin-A (toxA), which is a major virulence factor in Pseudomonas aeruginosa [42]. Two A. salmonicida isolates (2HA2 and 2MA4) carried genes involved in immune evasion (capsular polysaccharide), phagocytosis prevention (rmlC, wbfU, wbfY) and endotoxin from Bordetella. In addition to these, several others virulence factors like adhesion factors, T2SS [43,44] and T6SS [45,46] were detected. The presence of virulence factors in the genome does not necessarily guarantee expression of these virulence factors and further in-depth studies are needed to establish the virulence of these isolates. Nevertheless, the presence of these virulence genes indicates that these isolates have potential for causing infections. This emphasizes the risk posed by plastics as vectors for the transport of potentially virulent fish pathogens in the marine environment, especially in Norway where aquaculture is one of the major activities.
All three isolates of A. salmonicida in our study carried resistance genes like ampC, blaOXA, cphA, qnrA and catB. New variants of cphA (Class B2 metallo-β-lactamase) were present in the two isolates (5HA1 and 2MA4) of A. salmonicida whereas new variants of FOX/MOX family class C β-lactamase (96.3% to 98.4% identity) were detected in all three isolates. Marine bacteria have been shown to be a source of ARGs found in the clinics [47,48,49]. A recent study by Ebmeyer et al. [50] showed that Aeromonas spp. are the origin of several clinically important β-lactamases like CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9. Similarly, another study from Ebmeyer et al. [51] showed that FOX AmpC β-lactamases originated in A. allosaccharophila. They proposed that the mobilization and fixation of these genes may be recent and may have happened during the antibiotic era. Plastics provide a hydrophobic surface for the attachment of microbes promoting colonization and biofilm formation [52]. In accordance, an increased frequency of resistance plasmid transfer in bacteria associated with microplastics was observed [20]. Plastics also absorb a range of pollutants including antibiotic and heavy metals that are known to be drivers of antibiotic resistance [5,53,54,55]. This may create selection pressure, aiding transfer and/or mobilization of ARGs in the surface associated microbiota on marine plastics [56]. In order to better understand the role of marine plastics in mobilization and selection of ARGs, more research is thus warranted.
We detected opportunistic human pathogens like M. morganii [57,58,59], Ac. beijerinckii [60] and A. popoffii [61] on plastic surfaces. M. morganii previously belonged to family Enterobacteriaceae, which also consists of classical human pathogens like Escherichia coli, Salmonella typhi and Klebsiella pneumoniae. It was recently reclassified to be included in its own family Morganellaceae [62]. Although reclassified, M. morganii is an important emerging opportunistic human pathogen causing a variety of infections ranging from wound infections, urinary tract infections to meningitis [57,58,59]. M. morganii isolate (2MA1) carried virulence genes involved in acid resistance (ureB, ureG), genes involved in iron uptake, and T6SS proteins. Further, this isolate was resistant to tetracycline, chloramphenicol and β-lactams as well as carried several clinically important resistance genes like tetD, aac3 and catB [59]. The presence of multi-drug resistant human associated bacteria carrying multiple ARGs suggests that plastic may serve as vectors for transport of not only fish pathogens but also opportunistic human pathogens in the marine environment. Our results are in accordance with a recent study that showed presence of potentially pathogenic bacteria on plastic debris from Guanabara Bay in Brazil [63].
Aquaculture is both historically and economically important for Norway. Norway is considered the world’s second largest exporter of seafood after China and delivers fish to more than 100 countries [64,65]. In 2018, more than 1.35 million ton of fish, mostly salmon (Salmo salar) (1.28 million ton) and rainbow trout (Oncorhynchus mykiss) (68,345 ton) were farmed in Norway, with a first-hand value of 67.8 billion NOK (8.34 billion USD) [66,67]. The presence and spread of potentially virulent fish pathogens on marine plastics, could have a major impact on aquaculture in Norway. Hence, more research on understanding of the role of plastic surface-associated microbial communities and their biogeochemical functions in the marine environment is needed, especially in Norway.
5. Conclusions
Our study demonstrates the presence of fish pathogens and human associated bacteria on marine plastics from Norway. We show the potential for pathogenicity of A. salmonicida isolates obtained from marine plastics, with presence of genes encoding toxins, hemolysins and adhesion factors in their genome sequences, as well as describe new variants of ARGs carried by plastic associated bacteria. Our study strengthens the notion that plastic debris may serve as vectors for transport for fish pathogen as well as other opportunistic human pathogens in the marine environment. Marine plastics colonized by potentially virulent fish pathogens may impact aquaculture. Hence, in-depth follow-up studies for better understanding the role of plastic in the spread of antibiotic resistant pathogens in the marine environment are needed.
Acknowledgments
The authors acknowledge Didrik Hjertaker Grevskott from Institute of Marine Research (IMR) for help with sample collection. We acknowledge Ørjan Bjorøy from IMR for providing technical support and Julia Storesund from IMR for discussions regarding bioinformatics analysis. The authors also acknowledge Monica Sanden, Cecilie Smith Svanevik and Bjørn Tore Lunestad from IMR for supporting the work. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/8/1200/s1, Figure S1: Phylogenetic tree for cphA β-lactamases based on the amino acid sequence. The proteins highlighted with circles are new variants found in A. salmonicida isolates in this study. Accession numbers are presented for sequences downloaded from GenBank. PFM1 (QDC33502.1) representing subclass B2 metallo-beta-lactamase; PFM-1 from Pseudomonas fluorescens is used as an out group for construction of phylogenetic tree. Numbers at nodes are bootstrap values (%) based on 1000 resampled datasets; only values >50% are given. Table S1: Identification and isolation source of the isolates obtained in the study. Table S2: Genome sequence assembly statistics for the whole genome sequences of the six isolates. Table S3: A list of virulence genes detected in the genome sequence of the isolates.
Author Contributions
N.P.M. conceived the study and designed the experiment. V.R. and P.S.N. performed the experiments and bioinformatic analysis with assistance from N.P.M., N.P.M. and V.R. analyzed the data. A.M.B. helped with plastic analysis. N.P.M. and V.R. drafted the manuscript. P.S.N. and A.M.B. provided critical inputs. N.P.M. acquired the project funding and was responsible for the overall direction of the project. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by Institute of Marine research, under Ocean Health programme (project number-15495).
Conflicts of Interest
The authors have no conflict of interest.
Availability of Data
Genome sequences have been submitted to GenBank under the following genome accession numbers: JAACGC000000000, JAACGB000000000, JAACGA000000000, JAACFZ000000000, JAACFY000000000, JAACFX000000000 and JAACFW000000000.
References
- 1.Shah A.A., Hasan F., Hameed A., Ahmed S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008;26:246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Derraik J.G. The pollution of the marine environment by plastic debris: A review. Mar. Pollut. Bull. 2002;44:842–852. doi: 10.1016/S0025-326X(02)00220-5. [DOI] [PubMed] [Google Scholar]
- 3.Wilber R.J. Plastic in the North Atlantic. Oceanus. 1987;30:61–68. [Google Scholar]
- 4.Pruter A.T. Sources, quantities and distribution of persistent plastics in the marine environment. Mar. Pollut. Bull. 1987;18:305–310. doi: 10.1016/S0025-326X(87)80016-4. [DOI] [Google Scholar]
- 5.Yang Y., Liu G., Song W., Ye C., Lin H., Li Z., Liu W. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 2019;123:79–86. doi: 10.1016/j.envint.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 6.Blair R.M., Waldron S., Phoenix V., Gauchotte-Lindsay C. Micro-and nanoplastic pollution of freshwater and wastewater treatment systems. Springer Sci. Rev. 2017;5:19–30. doi: 10.1007/s40362-017-0044-7. [DOI] [Google Scholar]
- 7.Koelmans A.A., Besseling E., Shim W.J. Marine Anthropogenic Litter. Springer; Cham, Switzerland: 2015. Nanoplastics in the Aquatic Environment. Critical Review; pp. 325–340. [Google Scholar]
- 8.Avio C.G., Gorbi S., Regoli F. Plastics and microplastics in the oceans: From emerging pollutants to emerged threat. Mar. Environ. Res. 2017;128:2–11. doi: 10.1016/j.marenvres.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Moore C.J. Synthetic polymers in the marine environment: A rapidly increasing, long-term threat. Environ. Res. 2008;108:131–139. doi: 10.1016/j.envres.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 10.Gregory M.R. Environmental implications of plastic debris in marine settings entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos. Trans. R. Soc. B. 2009;364:2013–2025. doi: 10.1098/rstb.2008.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jepsen E.M., de Bruyn P.J.N. Pinniped entanglement in oceanic plastic pollution: A global review. Mar. Pollut. Bull. 2019;145:295–305. doi: 10.1016/j.marpolbul.2019.05.042. [DOI] [PubMed] [Google Scholar]
- 12.Kühn S., Bravo Rebolledo E.L., van Franeker J.A. Deleterious Effects of Litter on Marine Life. In: Bergmann M., Gutow L., Klages M., editors. Marine Anthropogenic Litter. Springer International Publishing; Cham, Switzerland: 2015. pp. 75–116. [DOI] [Google Scholar]
- 13.Kögel T., Bjorøy Ø., Toto B., Bienfait A.M., Sanden M. Micro- and nanoplastic toxicity on aquatic life: Determining factors. Sci. Total Environ. 2020;709:136050. doi: 10.1016/j.scitotenv.2019.136050. [DOI] [PubMed] [Google Scholar]
- 14.Li W.C., Tse H.F., Fok L. Plastic waste in the marine environment: A review of sources, occurrence and effects. Sci. Total Environ. 2016;566–567:333–349. doi: 10.1016/j.scitotenv.2016.05.084. [DOI] [PubMed] [Google Scholar]
- 15.Zarfl C., Matthies M. Are marine plastic particles transport vectors for organic pollutants to the Arctic? Mar. Pollut. Bull. 2010;60:1810–1814. doi: 10.1016/j.marpolbul.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Schneider F., Parsons S., Clift S., Stolte A., McManus M.C. Collected marine litter—A growing waste challenge. Mar. Pollut. Bull. 2018;128:162–174. doi: 10.1016/j.marpolbul.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Bryant J.A., Clemente T.M., Viviani D.A., Fong A.A., Thomas K.A., Kemp P., Karl D.M., White A.E., DeLong E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. MSystems. 2016;1:e00024-16. doi: 10.1128/mSystems.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberbeckmann S., Loeder M.G.J., Gerdts G., Osborn A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014;90:478–492. doi: 10.1111/1574-6941.12409. [DOI] [PubMed] [Google Scholar]
- 19.Eriksen M., Lebreton L.C., Carson H.S., Thiel M., Moore C.J., Borerro J.C., Galgani F., Ryan P.G., Reisser J. Plastic pollution in the world’s oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS ONE. 2014;9:e111913. doi: 10.1371/journal.pone.0111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias-Andres M., Klümper U., Rojas-Jimenez K., Grossart H.-P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018;237:253–261. doi: 10.1016/j.envpol.2018.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Booth A., Kubowicz S., Beegle-Krause C., Skancke J., Nordam T., Landsem E., Throne-Holst M., Jahren S. Microplastic in Global and Norwegian Marine Environments: Distributions, Degradation Mechanisms and Transport. Miljødirektoratet M-918. SINTEF; Trondheim, Norway: 2017. pp. 1–147. [Google Scholar]
- 22.Jensen S., Lindbaek M., Gradmann C., Lie A.K. Reflections about antibiotic resistance history in Norway. Erindr. Antibiot. Hist. Nor. 2010;130:2494. doi: 10.4045/tidsskr.10.0283. [DOI] [PubMed] [Google Scholar]
- 23.Amalina N.Z., Santha S., Zulperi D., Amal M.N.A., Yusof M.T., Zamri-Saad M., Ina-Salwany M.Y. Prevalence, antimicrobial susceptibility and plasmid profiling of Vibrio spp. isolated from cultured groupers in Peninsular Malaysia. BMC Microbiol. 2019;19:251. doi: 10.1186/s12866-019-1624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron S., Granier S.A., Larvor E., Jouy E., Cineux M., Wilhelm A., Gassilloud B., Le Bouquin S., Kempf I., Chauvin C. Aeromonas diversity and antimicrobial susceptibility in freshwater—An attempt to set generic epidemiological cut-off values. Front. Microbiol. 2017;8:503. doi: 10.3389/fmicb.2017.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Ciufo S., Li W. The NCBI Handbook [Internet] 2nd ed. Bethesda, US: Center for Biotechnology Information; Rockville, MD, USA: 2013. Prokaryotic Genome Annotation Pipeline; pp. 1–12. [Google Scholar]
- 27.Alcock B.P., Raphenya A.R., Lau T.T., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primpke S., Wirth M., Lorenz C., Gerdts G. Reference database design for the automated analysis of microplastic samples based on Fourier transform infrared (FTIR) spectroscopy. Anal. Bioanal. Chem. 2018;410:5131–5141. doi: 10.1007/s00216-018-1156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viršek M.K., Lovšin M.N., Koren Š., Kržan A., Peterlin M. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar. Pollut. Bull. 2017;125:301–309. doi: 10.1016/j.marpolbul.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Parker J.L., Shaw J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011;62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Igbinosa I.H., Igumbor E.U., Aghdasi F., Tom M., Okoh A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012;2012:1–13. doi: 10.1100/2012/625023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reith M.E., Singh R.K., Curtis B., Boyd J.M., Bouevitch A., Kimball J., Munholland J., Murphy C., Sarty D., Williams J. The genome of Aeromonas salmonicida subsp. salmonicida A449: Insights into the evolution of a fish pathogen. BMC Genom. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin B., Austin D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish. Springer; London, UK: 1999. [Google Scholar]
- 38.Wiles T.J., Mulvey M.A. The RTX pore-forming toxin α-hemolysin of uropathogenic Escherichia coli: Progress and perspectives. Future Microbiol. 2013;8:73–84. doi: 10.2217/fmb.12.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suarez G., Khajanchi B.K., Sierra J.C., Erova T.E., Sha J., Chopra A.K. Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene. 2012;506:369–376. doi: 10.1016/j.gene.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bücker R., Krug S.M., Rosenthal R., Günzel D., Fromm A., Zeitz M., Chakraborty T., Fromm M., Epple H.-J., Schulzke J.-D. Aerolysin from Aeromonas hydrophila Perturbs Tight Junction Integrity and Cell Lesion Repair in Intestinal Epithelial HT-29/B6 Cells. J. Infect. Dis. 2011;204:1283–1292. doi: 10.1093/infdis/jir504. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen-Ivey C.R., Figueras M.J., McGarey D., Liles M.R. Virulence Factors of Aeromonas hydrophila: In the Wake of Reclassification. Front. Microbiol. 2016;7:1337. doi: 10.3389/fmicb.2016.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wedekind J.E., Trame C.B., Dorywalska M., Koehl P., Raschke T.M., McKee M., FitzGerald D., Collier R.J., McKay D.B. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 2001;314:823–837. doi: 10.1006/jmbi.2001.5195. [DOI] [PubMed] [Google Scholar]
- 43.Li G., Howard S.P. ExeA binds to peptidoglycan and forms a multimer for assembly of the type II secretion apparatus in Aeromonas hydrophila. Mol. Microbiol. 2010;76:772–781. doi: 10.1111/j.1365-2958.2010.07138.x. [DOI] [PubMed] [Google Scholar]
- 44.Schoenhofen I.C., Stratilo C., Howard S.P. An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: Effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 1998;29:1237–1247. doi: 10.1046/j.1365-2958.1998.01011.x. [DOI] [PubMed] [Google Scholar]
- 45.Suarez G., Sierra J.C., Sha J., Wang S., Erova T.E., Fadl A.A., Foltz S.M., Horneman A.J., Chopra A.K. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 2008;44:344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sha J., Rosenzweig J.A., Kozlova E.V., Wang S., Erova T.E., Kirtley M.L., van Lier C.J., Chopra A.K. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology. 2013;159:1120. doi: 10.1099/mic.0.063495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cattoir V., Poirel L., Mazel D., Soussy C.-J., Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 2007;51:2650–2651. doi: 10.1128/AAC.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirel L., Rodriguez-Martinez J.-M., Mammeri H., Liard A., Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potron A., Poirel L., Nordmann P. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob. Agents Chemother. 2011;55:4405–4407. doi: 10.1128/AAC.00681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebmeyer S., Kristiansson E., Larsson D.G.J. The mobile FOX AmpC beta-lactamases originated in Aeromonas allosaccharophila. Int. J. Antimicrob. Agents. 2019;54:798–802. doi: 10.1016/j.ijantimicag.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 51.Ebmeyer S., Kristiansson E., Larsson D.J. CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three aeromonas species. J. Antimicrob. Chemother. 2019;74:1202–1206. doi: 10.1093/jac/dkz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dang H., Lovell C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016;80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J., Zhang K., Zhang H. Adsorption of antibiotics on microplastics. Environ. Pollut. 2018;237:460–467. doi: 10.1016/j.envpol.2018.02.050. [DOI] [PubMed] [Google Scholar]
- 54.Huijbers P.M., Blaak H., de Jong M.C., Graat E.A., Vandenbroucke-Grauls C.M., de Roda Husman A.M. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015;49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 55.Caruso G. Microplastics as vectors of contaminants. Mar. Pollut. Bull. 2019;146:921–924. doi: 10.1016/j.marpolbul.2019.07.052. [DOI] [PubMed] [Google Scholar]
- 56.Jutkina J., Marathe N.P., Flach C.F., Larsson D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018;616–617:172–178. doi: 10.1016/j.scitotenv.2017.10.312. [DOI] [PubMed] [Google Scholar]
- 57.Tucci V., Isenberg H.D. Hospital cluster epidemic with Morganella morganii. J. Clin. Microbiol. 1981;14:563–566. doi: 10.1128/JCM.14.5.563-566.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastroianni A., Coronado O., Chiodo F. Morganella morganii meningitis in a patient with AIDS. J. Infect. 1994;29:356–357. doi: 10.1016/S0163-4453(94)91450-8. [DOI] [PubMed] [Google Scholar]
- 59.Liu H., Zhu J., Hu Q., Rao X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016;50:10–17. doi: 10.1016/j.ijid.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 60.Visca P., Seifert H., Towner K.J. Acinetobacter infection—An emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 61.Hua H.T., Bollet C., Tercian S., Drancourt M., Raoult D. Aeromonas popoffii urinary tract infection. J. Clin. Microbiol. 2004;42:5427–5428. doi: 10.1128/JCM.42.11.5427-5428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adeolu M., Alnajar S., Naushad S., Gupta R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 63.Silva M.M., Maldonado G.C., Castro R.O., de Sá Felizardo J., Cardoso R.P., dos Anjos R.M., de Araújo F.V. Dispersal of potentially pathogenic bacteria by plastic debris in Guanabara Bay, RJ, Brazil. Mar. Pollut. Bull. 2019;141:561–568. doi: 10.1016/j.marpolbul.2019.02.064. [DOI] [PubMed] [Google Scholar]
- 64.Stévant P., Rebours C., Chapman A. Seaweed aquaculture in Norway: Recent industrial developments and future perspectives. Aquac. Int. 2017;25:1373–1390. doi: 10.1007/s10499-017-0120-7. [DOI] [Google Scholar]
- 65.Fisheries F. The State of World Fisheries and Aquaculture 2010. Food and Agriculture Organization of the United Nations; Rome, Italy: 2016. [Google Scholar]
- 66.Sentralbyrå S. Akvakultur, 2018, Årlig Endelige Tall. [(accessed on 5 May 2020)]; Available online: https://www.ssb.no/jord-skog-jakt-og-fiskeri/statistikker/fiskeoppdrett/aar.
- 67.FAO Globefish highlights—A quarterly update on world seafood markets—April 2019 issues, with Jan.–Dec. 2018 statistics. Globefish Highlights. 2019;2:1–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences have been submitted to GenBank under the following genome accession numbers: JAACGC000000000, JAACGB000000000, JAACGA000000000, JAACFZ000000000, JAACFY000000000, JAACFX000000000 and JAACFW000000000.