Abstract
Cells detect external chemical stimuli by directly binding a signaling molecule, but the strategies used by cells to detect and respond to non-chemical cues have been mysterious. Recent work suggests that a bacterial protein detects changes in environmental temperature by physically measuring membrane thickness.
The ability to sense environmental conditions and mount an adaptive transcriptional response is conserved in all domains of life. Moreover, reversible protein phosphorylation is a widespread mechanistic strategy for the intracellular transduction of extracellular signals. In bacteria, a paradigmatic signal transduction mechanism (appropriately termed a “two-component system”) utilizes just two proteins [1]. The first is a transmembrane protein called a “sensor kinase” that detects an environmental signal and transmits the information to the interior of the cell. Sensor kinases have two functional domains: a signal recognition domain (often extracellular) that may physically bind a ligand, and an intracellular autokinase domain that contains a characteristic histidine residue [2]. Detection of an environmental signal activates the autokinase domain and results in phosphorylation of this histidine. The second component of the system is canonically a soluble transcription factor called a “response regulator”, which is activated when it accepts the phosphoryl group from its cognate sensor kinase. The response regulator then typically activates the transcription of appropriate genes that respond to the extracellular stress. In this model, chemical factors such as salts, nutrients, and signaling peptides may be detected by direct binding of these ligands to the sensor kinase which leads to its activation by autophosphorylation (Fig 1A). How, though, can a sensor kinase detect a more abstract, non-chemical environmental cue such as temperature?
Figure 1. Temperature sensing by measuring membrane thickness.
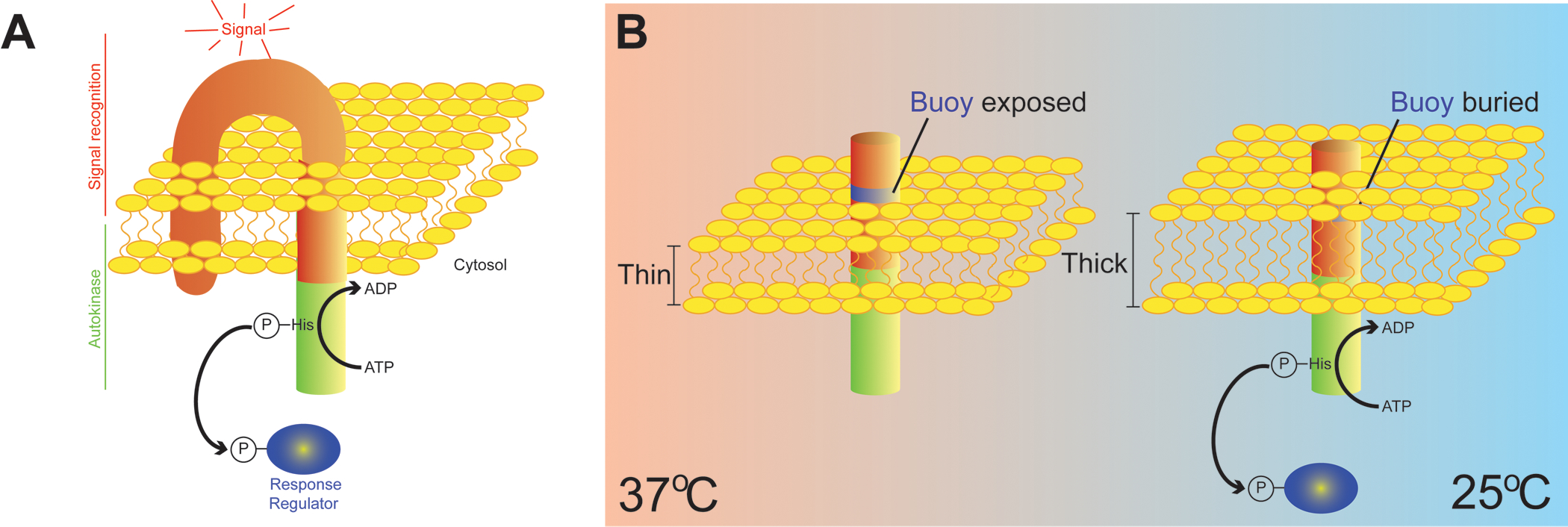
(A) Depiction of a canonical bacterial two component regulatory system. A transmembrane sensor kinase is shown with an extracytoplasmic signal recognition domain (red) and an intracellular autokinase domain (green). Sensor kinases often have multiple membrane-spanning domains, but for simplicity, only two membrane-spanning domains are depicted. Detection of an activating signal results in autophosphorylation of a characteristic histidine residue in the autokinase domain and subsequent transfer of the phosphoryl group onto a cognate response regulator (blue), typically a transcription factor. (B) Depiction of an engineered minimal signal sensing domain of DesK, a sensor kinase in Bacillus subtilis that detects changes in environmental temperature. The minimal thermo-sensor harbors a cluster of hydrophilic amino acids (blue ring) near its amino terminus (extracytoplasmic) that floats like a buoy near the lipid/water interface at high temperatures when the membrane is thinner (left). At lower temperatures (right), an increase in lipid ordering results in a thicker membrane, forcing the “buoy” into the hydrophobic lipid bilayer, thereby activating the autokinase domain. DesK therefore responds to changes in temperature by directly measuring membrane thickness.
In this issue, Cybulski et al. [3] report their studies on the sensor kinase DesK in the bacterium Bacillus subtilis, which is activated in response to reduced temperature [4]. Specifically, the authors wondered if there is a physical feature of the cell which changes in response to fluctuations in temperature that DesK may physically detect and exploit as an indirect measure for temperature. The plasma membrane of the bacterium was a top candidate for harboring this physical feature for two reasons. First, DesK is a polytopic membrane protein [4]. Second, previous studies had demonstrated that increasing the fluidity of the plasma membrane by increasing the incorporation of branched-chain fatty acids decreased the activation of DesK, suggesting that DesK directly responds to a physical property of the plasma membrane [5]. In this study, the authors propose a model in which DesK detects an increase in the thickness of the plasma membrane upon a drop in temperature, resulting in DesK activation.
The authors began by performing a systematic deletion analysis of the unwieldy five-pass integral membrane protein and discovered that deletion of just the first transmembrane region (TM1) abolished the ability of DesK to respond to lower temperature and resulted in a constitutively active protein, suggesting that TM1 harbored a temperature-sensing motif. Meanwhile, previous structural studies had indicated that the last transmembrane helix of DesK (TM5) is attached to the autokinase domain through a two helical coiled-coil motif that appeared to be critical for regulation of DesK activity [6]. The authors envisioned a model in which TM1 would detect a drop in temperature and transmit that information to TM5, which would then activate DesK. They therefore created a chimeric transmembrane region, consisting of N-terminal residues of TM1 and C-terminal residues of TM5, and fused it to the autokinase domain. Remarkably, this simplified DesK, harboring a single engineered transmembrane segment (the minimal thermo-sensor), worked almost as well as the wild type DesK harboring five membrane-spanning helices.
Curiously, the N-terminus of the minimal thermo-sensor harbored a cluster of hydrophilic amino acids near the lipid/water interface. At high temperatures, when lipids are disordered and the bilayer is thinner, the authors reasoned that these residues could “float” on the membrane surface like a buoy while being tethered to the membrane itself by the hydrophobic residues of the transmembrane region (Fig 1B). In this conformation, the authors supposed that DesK autokinase activity would be low. A drop in temperature, conversely, would increase lipid ordering, resulting in a thicker plasma membrane. In this scenario, the authors predicted that these hydrophilic residues would become forcibly buried into the hydrophobic lipid bilayer, resulting in a conformational change that would activate the autokinase activity of DesK. To test this, the authors lengthened the transmembrane region of the thermo-sensor so that the “buoy” was farther away from the surface of the membrane and would not be pulled into the bilayer when the membrane thickened. As expected, the lengthened version of the minimal thermo-sensor was unable to activate DesK at lower temperature; that is, it was unable to detect the thickening of the membrane. In complementary biochemical studies, the authors reconstituted DesK harboring the minimal thermo-sensor into membrane vesicles made of phosopholipids of varying fatty acyl chain length (and therefore varying membrane thicknesses). As predicted, autokinase activity of DesK was higher at lower temperature when DesK was reconstituted into vesicles made from long chain phospholipids (thick membranes). In the presence of short chain phospholipids (thin membranes), DesK autokinase activity was diminished, presumably because the “buoy” was too far away from the surface of the membrane to accurately measure its expansion and contraction. Taken together, the data suggested a model in which the cluster of hydrophilic residues of DesK that form the buoy actually functions like the end of a ruler that physically measures membrane thickness as an indication of environmental temperature.
As the authors mentioned, DesK now joins a list of proteins, with diverse cellular functions, that harbor domains that act as molecular rulers. Tail lengths of bacteriophages and needle lengths of bacterial type III protein secretion machines are precisely determined by a single molecular ruler protein: deletion or addition of amino acid residues to the middle of these proteins results in the proportional decrease or increase in length of the tail or needle [7, 8]. In yeast, during the biosynthesis of very long chain fatty acids, shortening the distance between a particular amino acid residue and the active site of a component of the biosynthetic machinery results in the production of fatty acids with shorter chain lengths [9]. Beyond rulers, several proteins of diverse structure and function have even been described that are equipped with “molecular protractors” that measure degrees of membrane curvature when carrying out their function [10–12]. A longstanding challenge in studying two component regulatory systems has been to understand the input signals that actually activate sensor kinases. Could physical cues, like membrane thickness, curvature, viscosity, or tension, be activators of other transmembrane sensor kinases for whom chemical activating signals have been difficult identify? Although the exploitation of tools such as molecular rulers and protractors have more often been described for morphogenetic processes like the assembly of structures and the localization of proteins, perhaps future studies will reveal that other systems like gene regulatory pathways also often measure and respond not just to chemical signals, but to physical cues like geometry.
Acknowledgments
Funding provided by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
References
- 1.Hoch JA (2000). Two-component and phosphorelay signal transduction. Current opinion in microbiology 3, 165–170. [DOI] [PubMed] [Google Scholar]
- 2.Dutta R, Qin L, and Inouye M (1999). Histidine kinases: diversity of domain organization. Molecular microbiology 34, 633–640. [DOI] [PubMed] [Google Scholar]
- 3.Cybulski LE, Martin M, Mansilla MC, Fernandez A, and de Mendoza D (2010). Membrane Thickness Cue for Cold Sensing in a Bacterium. Curr Biol. [DOI] [PubMed] [Google Scholar]
- 4.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, and de Mendoza D (2001). Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. The EMBO journal 20, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cybulski LE, Albanesi D, Mansilla MC, Altabe S, Aguilar PS, and de Mendoza D (2002). Mechanism of membrane fluidity optimization: isothermal control of the Bacillus subtilis acyl-lipid desaturase. Molecular microbiology 45, 1379–1388. [DOI] [PubMed] [Google Scholar]
- 6.Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, and Buschiazzo A (2009). Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proceedings of the National Academy of Sciences of the United States of America 106, 16185–16190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsura I (1987). Determination of bacteriophage lambda tail length by a protein ruler. Nature 327, 73–75. [DOI] [PubMed] [Google Scholar]
- 8.Journet L, Agrain C, Broz P, and Cornelis GR (2003). The needle length of bacterial injectisomes is determined by a molecular ruler Science (New York, N.Y: 302, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 9.Denic V, and Weissman JS (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677. [DOI] [PubMed] [Google Scholar]
- 10.Bigay J, Casella JF, Drin G, Mesmin B, and Antonny B (2005). ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. The EMBO journal 24, 2244–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, and McMahon HT (2004). BAR domains as sensors of membrane curvature: the amphiphysin BAR structure Science (New York, N.Y: 303, 495–499. [DOI] [PubMed] [Google Scholar]
- 12.Ramamurthi KS, Lecuyer S, Stone HA, and Losick R (2009). Geometric cue for protein localization in a bacterium Science (New York, N.Y: 323, 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]


