Abstract
Fluoropyrimidine drugs (FPs), including 5-fluorouracil, tegafur, capecitabine, and doxifluridine, are among the most widely used anticancer agents in the treatment of solid tumors. However, severe toxicity occurs in approximately 30% of patients following FP administration, emphasizing the importance of predicting the risk of acute toxicity before treatment. Three metabolic enzymes, dihydropyrimidine dehydrogenase (DPD), dihydropyrimidinase (DHP), and β-ureidopropionase (β-UP), degrade FPs; hence, deficiencies in these enzymes, arising from genetic polymorphisms, are involved in severe FP-related toxicity, although the effect of these polymorphisms on in vivo enzymatic activity has not been clarified. Furthermore, the clinical usefulness of current methods for predicting in vivo activity, such as pyrimidine concentrations in blood or urine, is unknown. In vitro tests have been established as advantageous for predicting the in vivo activity of enzyme variants. This is due to several studies that evaluated FP activities after enzyme metabolism using transient expression systems in Escherichia coli or mammalian cells; however, there are no comparative reports of these results. Thus, in this review, we summarized the results of in vitro analyses involving DPD, DHP, and β-UP in an attempt to encourage further comparative studies using these drug types and to aid in the elucidation of their underlying mechanisms.
Keywords: fluoropyrimidine, dihydropyrimidine dehydrogenase, dihydropyrimidinase, β-ureidopropionase, genetic polymorphism
1. Introduction
Fluoropyrimidine drugs (FPs), including 5-fluorouracil (5-FU) and its oral prodrugs tegafur, capecitabine, and doxifluridine, are widely used in the treatment of solid tumors in the gastrointestinal tract, breast, liver, lung, head, and neck [1,2,3]. FP-based treatments have a narrow therapeutic index, which has led to severe adverse effects in approximately 30% of cancer patients, including mucositis, diarrhea, neutropenia, thrombocytopenia, and hand–foot syndrome [4,5,6,7,8]. Additionally, severe treatment toxicities could lead to treatment interruption, which increases the subsequent risk of therapeutic failure as well as patient death [9].
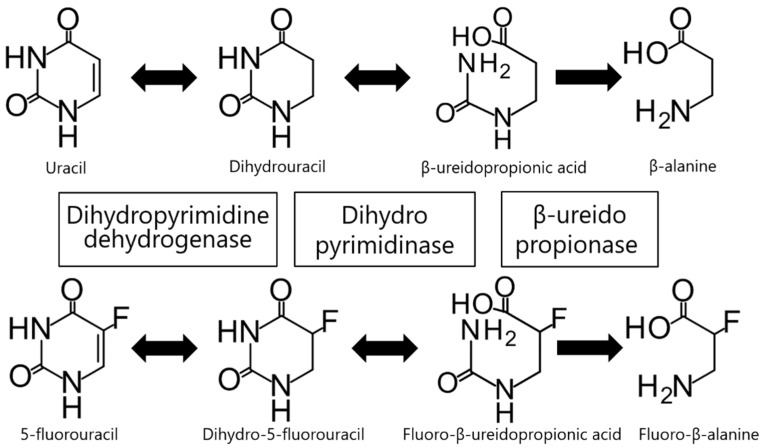
Genetic polymorphisms of thymidylate synthase (TYMS), methylene tetrahydrofolate reductase (MTHFR), and miR-27a are associated with the development of severe toxicities as well as treatment resistance; however, FP-related toxicity is mainly dependent on FP catabolism. Over 80% of an administered dose of 5-FU is rapidly degraded by three consecutive enzymes belonging to the endogenous pyrimidine, uracil, and thymine catabolic pathways (Figure 1), the only known 5-FU in vivo degradation pathway. Initially, the rate-limiting enzyme dihydropyrimidine dehydrogenase (DPD, EC 1.3.1.2), mainly found in the liver, catalyzes the reduction of 5-FU to dihydro-5-fluorouracil (FUH2). Subsequently, dihydropyrimidinase (DHP, EC 3.5.2.2) catalyzes the hydrolytic ring opening of FUH2 to form fluoro-β-ureidopropionic acid (FUPA). Even though DPD and DHP catalysis is reversible, the positive reaction is dominant in vivo [10,11,12,13,14,15]. Lastly, β-ureidopropionase (β-UP, EC 3.5.1.6) catalyzes the hydrolysis of FUPA to fluoro-β-alanine. The three enzymes (DPD, DHP, and β-UP) are encoded by the DPYD, DPYS, and UPB1 genes, respectively [16,17,18].
Figure 1.
Uracil and 5-fluorouracil degradation pathway. Uracil and 5-fluorouracil are catabolized successively by dihydropyrimidine dehydrogenase, dihydropyrimidinase, and β-ureidopropionase. β-Alanine and fluoro-β-alanine are the final metabolites in this pathway.
Decreased DPD and DHP enzymatic activities have been linked to genetic polymorphisms identified in patients with severe FP-related toxicities; for each causative polymorphism, the reduction in activity is caused mainly by the substitution or deletion of amino acids [19,20,21]. However, the relationship between β-UP activity and the development of FP-related toxicity is still unknown. To date, the specific effects of previously identified polymorphisms on enzymatic function are largely unknown. Only four DPYD variants (c.1905 + 1G > A (IVS14 + 1G > A, DPYD*2A); c.1679T > G (DPYD*13, p.I560S); c.1129 − 5923C > G /hapB3; and c.2846A > T (p.D949V)) have been characterized as predictive markers for FP-related toxicity in Caucasians [22]. However, significant racial and individual differences in polymorphism location and frequency make it challenging to safely extrapolate the clinical data and institute regional guidelines from one population to another. Thus, it is necessary to further clarify the effects of genetic polymorphisms in an attempt to establish their effect on in vivo enzymatic function. For example, before FP administration, PCR-Restriction Fragment Length Polymorphism (RFLP) analysis, Sanger sequencing, and next-generation sequencing analysis are often used for detecting genetic polymorphisms and establishing patient risk. Moreover, hepatic DPD activity, and thus DPD deficiency incidence, can be predicted by assessing peripheral blood mononuclear cell (PBMC) DPD activity. However, to date, there are no established methods to quantify DHP and β-UP activity clinically.
The most direct method to understand the effect of the genetic polymorphisms of these enzymes on FP pharmacokinetics is to measure metabolite concentrations in blood and urine from subjects with the respective genotypes after FP administration. However, in vivo testing is highly invasive due to continuous blood sampling and poses a considerable risk of FP-related toxicity. Additionally, as the variants of interest are mainly low-frequency polymorphisms, the recruitment of an adequate subject pool to obtain statistically significant data is considerably difficult. While pyrimidine metabolites in blood and urine have been previously quantified to assess enzymatic activity in vivo, these have yielded contradictory results [23,24].
In contrast, in vitro testing using heterologous expression systems has yielded reproducible results using non-invasive methods to facilitate enzymatic activity assessment [25]. Amongst these, several in vitro FP analyses using Escherichia coli or mammalian cells have been reported. While other in vitro techniques have been used to evaluate genetic polymorphisms including gene expression profiling, in this review, we focus on the in vitro analysis of the FP-metabolizing enzymes: DPD, DHP, and β-UP, thus providing further information to aid in the application of genetic testing in a clinical setting in light of recent novel insights.
2. Dihydropyrimidine Dehydrogenase (DPD)
DPD, the rate-limiting enzyme of the pyrimidine degradation pathway, catalyzes the reduction of 5-FU and uracil to FUH2 and dihydrouracil (UH2). The DPD gene (DPYD) is expressed in most human tissues, but the expression level is highest in the liver and PBMCs [26]. Located on chromosome 1p21, human DPYD is comprised of 23 exons and features a 3078 bp open reading frame, encoding a polypeptide containing 1025 amino acid residues [27].
DPD deficiency is an autosomal recessive disorder first reported in a child with neurological symptoms by Bakkeren et al., which was characterized by the accumulation of uracil and thymine in urine, blood, and cerebrospinal fluid [28]. The clinical symptoms include convulsions, autism, microcephaly, growth impairment, and intellectual disability, although asymptomatic cases have also been reported [29,30,31]. The frequency of DPD-deficient patients varies greatly across world populations. While Caucasian frequencies range from 3–5% for partial deficiency and 0.2% for complete deficiency, it is estimated to be extremely rare in Asians [32,33]. In the case of asymptomatic DPD deficiency, there is a considerable risk of FP accumulation during treatment, including 5-FU, which could lead to severe toxicity in patients [34,35,36]. Therefore, it is imperative to diagnose DPD deficiency before chemotherapy administration, even in cases with no prior clinical evidence of this condition.
Of the three metabolic enzymes, DPYD is the most studied gene. More than 500 DPYD polymorphisms to date have been identified and have been linked to FP-related toxicity in cancer patients [22,37,38,39,40,41,42,43,44,45]. Several of these variants are known to alter amino acid sequence or mRNA splicing, resulting in decreased enzymatic activity. Within the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, three variants (c.85T > C (DPYD*9A, p.C29R), c.1627A > G (DPYD*5, p.I543V), and c.2194G > A (DPYD*6, p.V732I)) are reported to have no effect on enzyme activity [46]. Four variants that cause exon 14 skipping or amino acid substitution (c.1905 + 1G > A (IVS14 + 1G > A, DPYD*2A), c.1679T > G (DPYD*13, p.I560S), c.1129 − 5923C > G/hapB3, and c.2846A > T (p.D949V)) are designated as having reduced enzymatic function and thus increase the risk of developing toxicity. Similarly, the Dutch Pharmacogenetics Working Group (DPWG) guidelines define these same four variants as risk factors for FP-related toxicity and recommend reducing treatment dosage when a patient possesses one of them [47]. Although DPYD*9A, *5, and *6 are common variants in many ethnic groups, these four risk variants have not yet been identified in Asians [48,49,50].
For most identified DPYD variants, except those mentioned above, the effect on DPD activity is unknown, and it is important to clarify the DPD phenotype [51]. The current standard to predict DPD activity measures its enzymatic activity in PBMCs, which correlates with hepatic DPD activity [52,53]. However, this method is not easily implemented in its current form in routine medical care, as it lacks solid evidence of clinical utility. Due to insufficient sensitivity, methods for quantifying pyrimidine metabolites in blood or urine might not identify patients with partial DPD deficiencies [23]. Moreover, additional studies on the clinical validity and utility of these tests are required before implementation can be justified.
In vitro testing is one of the methods used for estimating DPD phenotypes and for the functional analysis of identified non-synonymous variants [54,55,56,57,58,59,60]. Several studies of such tests using E. coli or mammalian cell expression systems have been reported (Table 1). Ogura et al. functionally analyzed two variants (G366A and T768K) identified from 150 healthy Japanese volunteers using an E. coli expression system [57]. Interestingly, while the G366A mutation produced a decreased intrinsic clearance (CLint) for 5-FU, reducing DPD activity by 50%, the T768K mutation did not. However, T768K-related activity decreased at a faster rate than that of wild-type DPD, suggesting protein instability. In a subsequent study, Offer et al. expressed 80 non-synonymous variants in HEK293T/c17 cells and measured their enzymatic activities using 5-FU as a substrate [58]. M166V, E828K, K861R, and P1023T exhibited significantly higher activity than wild-type DPD. In contrast, 31 variants, including D949V, exhibited significantly lower activity than wild-type DPD. Elraiyah et al. also analyzed 10 non-synonymous variants identified from 588 Somali and Kenyan individuals using HEK293T/c17 cells [59], in which P86L, P237L, A513V, T793I, V941A, and P1023S exhibited significantly reduced DPD activities. We have characterized 21 DPD allelic variants identified from 1070 Japanese individuals by transient expression in 293FT cells [60]. Among these, 10 (T298M, V313L, V335M, A380V, V434L, V515I, R592W, T768K, H807R, and V826M) showed significantly reduced CLint values relative to wild-type DPD, and the 5-FU metabolic activity of G926V was practically zero. These reports have yielded consistent results for DPYD*2A, which exhibited decreased activity, and for DPYD*5 (I543V) and *6 (V732I), which exhibited activities that were not considerably different from that of wild-type DPD. In contrast, there are variants such as DPYD*9A (C29R) and M166V, whose reported activities differ significantly among previous reports. Ogura et al. and our group found that M166V had a lower activity compared with that of wild-type DPD, while Offer et al. reported a reduction in activity for M166V. The differences in these activities are believed to be due to the differences in assay conditions and cell lines used. Notably, we and Ogura et al. reported DPD variants that were identified almost exclusively in Japanese individuals. Therefore, this raises awareness of the possibility of unidentified rare and relevant ethno-specific variants, which could lead to severe FP-related toxicity.
Table 1.
DPYD variants reported in in vitro analysis.
| dbSNP rsID | PharmVar ID | Location | Nucleotide Change | Amino Acid Substitution | Domain | Expression System | Substrates | Effect | References |
|---|---|---|---|---|---|---|---|---|---|
| rs150036960 | PV00901 | Exon 2 | 46C > G | L16V | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs72549310 | PV01042 | Exon 2 | 61C > T | R21X | I | HEK293T/c17 | 5-FU | No function | [58] |
| rs80081766 | PV01307 | Exon 2 | 62G > A | R21Q | I | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 2 | 74A > G | H25R | I | 293FT | 5-FU | 156% of CLint ratio | [60] |
| rs1801265 | PV00910 | Exon 2 | 85T > C (DPYD*9A) | C29R | I | HEK293T/c17 HEK293 Flp-In |
5-FU Thymine |
Increased function Decreased function |
[54] [55] |
| rs371587702 | PV00962 | Exon 3 | 194C > T | T65M | I | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 4 | 257C > T | P86L | I | HEK293T/c17 | 5-FU | No function | [59] |
| rs143986398 | PV00887 | Exon 4 | 274C > G | P92A | I | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs72549309 | PV01041 | Exon 4 | 295delTCAT (DPYD*7) |
F100fs | I | HEK293T/c17 | 5-FU | No function | [58] |
| rs150385342 | PV00902 | Exon 4 | 313G > A | A105T | I | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 5 | 325T > A | Y109N | I | 293FT | 5-FU | 79% of CLint ratio | [60] |
| rs141462178 | PV00878 | Exon 5 | 343A > G | M115V | I | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs200562975 | PV00927 | Exon 5 | 451A > G | N151D | I | 293FT HEK293T/c17 |
5-FU 5-FU |
107% of CLint ratio Normal function |
[60] [58] |
| rs2297595 | PV0943 | Exon 6 | 496A > G | M166V | I | 293FT HEK293T/c17 HEK293 Flp-In |
5-FU 5-FU Thymine |
77% of CLint ratio Increased function Decreased function |
[60] [58] [55] |
| rs139834141 | PV00871 | Exon 6 | 498G > A | M166I | I | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs371792178 | – | Exon 6 | 524C > T | S175L | II | 293FT | 5-FU | 131% of CLint ratio | [60] |
| rs115232898 | PV00862 | Exon 6 | 557A > G | Y186C | II | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs72549308 | PV01040 | Exon 6 | 601A > C | S201R | II | HEK293T/c17 | 5-FU | No function | [58] |
| rs72549307 | PV01039 | Exon 6 | 632A > G | Y211C | II | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs1801266 | PV00911 | Exon 7 | 703C > T (DPYD*8) |
R235W | II | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs780025995 | PV01299 | Exon 7 | 710C > T | P237L | II | HEK293T/c17 | 5-FU | Decreased function | [59] |
| rs45589337 | PV00984 | Exon 8 | 775A > G | K259E | II | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs777220476 | PV01275 | Exon 9 | 851G > T | G284V | II | HEK293 Flp-In | Thymine | No function | [56] |
| rs146356975 | PV00895 | Exon 9 | 868A > G | K290E | III | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs143878757 | PV00886 | Exon 9 | 893C > T | T298M | III | 293FT | 5-FU | 50% of CLint ratio | [60] |
| rs183105782 | PV00914 | Exon 9 | 910T > C | Y304H | III | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs150437414 | PV00904 | Exon 9 | 929T > C | L310S | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs145112791 | PV00891 | Exon 9 | 934C > T | L312F | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 9 | 937G > T | V313L | III | 293FT | 5-FU | 30% of CLint ratio | [60] |
| rs201018345 | PV00933 | Exon 10 | 967G > A | A323T | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs72549306 | PV01038 | Exon 10 | 1003G > A | V335M | III | 293FT | 5-FU | 47% of CLint ratio | [60] |
| rs72549306 | PV01037 | Exon 10 | 1003G > T (DPYD*11) |
V335L | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs183385770 | PV00915 | Exon 10 | 1024G > A | D342N | III | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs190577302 | PV00919 | Exon 10 | 1054C > G | L352V | III | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs143154602 | PV00882 | Exon 10 | 1057C > T | R353C | III | HEK293T/c17 | 5-FU | No function | [58] |
| – | – | Exon 10 | 1097G > C | G366A | III | 293FT Escherichia coli |
5-FU 5-FU |
71% of CLint ratio 47% of CLint ratio |
[60] [57] |
| rs72549305 | PV01036 | Exon 10 | 1108A > G | I370V | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 11 | 1139C > T | A380V | III | 293FT | 5-FU | 33% of CLint ratio | [60] |
| – | – | Exon 11 | 1150A > G | K384E | III | 293FT | 5-FU | 68% of CLint ratio | [60] |
| rs78060119 | PV01302 | Exon 11 | 1156G > T(DPYD*12) | E386X | III | HEK293T/c17 | 5-FU | No function | [58] |
| rs140602333 | PV00874 | Exon 11 | 1180C > T | R394W | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs143815742 | PV00883 | Exon 11 | 1181G > T | R394L | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs143815742 | PV00884 | Exon 11 | 1181G > A | R394Q | III | HEK293T/c17 | 5-FU | Normal function | [59] |
| – | – | Exon 11 | 1201G > A | G401R | III | HEK293 Flp-In | Thymine | Decreased function | [55] |
| rs61622928 | PV01018 | Exon 11 | 1218G > A | M406I | III | HEK293T/c17 HEK293 Flp-In |
5-FU Thymine |
Normal function Normal function |
[58] [55] |
| rs200064537 | PV00925 | Exon 11 | 1260T > A | N420K | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs764666241 | PV0183 | Exon 11 | 1278G > T | M426I | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs200693895 | PV00931 | Exon 11 | 1280T > C | V427A | III | HEK293 Flp-In | Thymine | Normal function | [56] |
| rs142512579 | PV00880 | Exon 11 | 1294G > A | D432N | III | HEK293T/c17 | 5-FU | Normal function | [58] |
| – | – | Exon 11 | 1300G > C | V434L | III | 293FT | 5-FU | 44% of CLint ratio | [60] |
| rs186169810 | PV00916 | Exon 11 | 1314T > G | F438L | III | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs72975710 | PV01043 | Exon 12 | 1349C > T | A450V | II | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs144395748 | PV00888 | Exon 12 | 1358C > G | P453R | II | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs199549923 | PV00921 | Exon 12 | 1403C > A | T468N | II | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs72549304 | PV01035 | Exon 12 | 1475C > T | S492L | II | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs111858276 | PV00857 | Exon 12 | 1484A > G | D495G | II | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs138391898 | PV00867 | Exon 12 | 1519G > A | V507I | II | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs760663364 | PV01150 | Exon13 | 1538C > T | A513V | II | HEK293T/c17 | 5-FU | Decreased function | [59] |
| rs148994843 | PV00900 | Exon 13 | 1543G > A | V515I | II | 293FT HEK293T/c17 |
5-FU 5-FU |
36% of CLint ratio Normal function |
[60] [58] |
| – | – | Exon 13 | 1567C > T | L523F | II | HEK293T/c17 | 5-FU | Normal function | [59] |
| rs190951787 | PV00920 | Exon 13 | 1577C > G | T526S | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs1180771326 | PV00864 | Exon 13 | 1582A > G | I528V | IV | HEK293T/c17 | 5-FU | Normal function | [59] |
| rs1801158 | PV00907 | Exon 13 | 1601G > A (DPYD*4) | S534N | IV | HEK293T/c17 HEK293 Flp-In |
5-FU Thymine |
Increased function Decreased function |
[54] [55] |
| rs142619737 | – | Exon 13 | 1615G > C | G539R | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs1801159 | PV00908 | Exon 13 | 1627A > G (DPYD*5) | I543V | IV | 293FT HEK293T/c17 HEK293 Flp-In |
5-FU 5-FU Thymine |
102% of CLint ratio Normal function Normal function |
[60] [54] [55] |
| rs55886062 | PV01000 | Exon 13 | 1679T > G (DPYD*13) | I560S | IV | HEK293T/c17 | 5-FU | Decreased function | [54] |
| rs201615754 | PV00937 | Exon 13 | 1682G > T | R561L | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs59086055 | PV01015 | Exon 14 | 1774C > T | R592W | IV | 293FT HEK293T/c17 |
5-FU 5-FU |
2% of CLint ratio No function |
[60] [58] |
| rs138616379 | PV00869 | Exon 14 | 1775G > A | R592Q | IV | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs145773863 | PV00894 | Exon 14 | 1777G > A | G593R | IV | HEK293T/c17 | 5-FU | No function | [58] |
| rs147601618 | PV00898 | Exon 14 | 1796T > C | M599T | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| Rs72549304 | PV01034 | Exon 4 | 1898delC (DPYD*3) |
P633fs | IV | HEK293T/c17 | 5-FU | No function | [58] |
| rs3918289 | PV00982 | Exon 14 | 1905C > T/G | N635K | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs3918290 | PV00983 | Intron 14 | 1905 + 1G > A (DPYD*2A) | Exon 14 skipping | IV | HEK293T/c17 | 5-FU | No function | [54] |
| rs55971861 | PV01003 | Exon 15 | 1906A > C | I636L | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs138545885 | PV00868 | Exon 16 | 1990G > T | A664S | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs137999090 | PV00866 | Exon 16 | 2021G > A | G674D | IV | HEK293T/c17 | 5-FU | No function | [58] |
| – | – | Exon 17 | 2096G > C | R699T | IV | HEK293T/c17 | 5-FU | Normal function | [59] |
| rs145548112 | PV00893 | Exon 17 | 2161G > A | A721T | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs146529561 | PV00896 | Exon 18 | 2186C > T | A729V | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs1801160 | PV00909 | Exon 18 | 2194G > A (DPYD*6) | V732I | IV | 293FT HEK293T/c17 HEK293 Flp-In |
5-FU 5-FU Thymine |
114% of CLint ratio Normal function Decreased function |
[60] [54] [55] |
| rs60511679 | PV01017 | Exon 18 | 2195T > G | V732G | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs112766203 | PV00858 | Exon 18 | 2279C > T | T760I | IV | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs56005131 | PV01004 | Exon 19 | 2303C > A | T768K | IV | 293FT HEK293T/c17 E. coli |
5-FU 5-FU 5-FU |
48% of CLint ratio Normal function 83% of CLint ratio |
[60] [58] [57] |
| rs199634007 | PV00922 | Exon 19 | 2336C > A | T779N | IV | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs547099198 | PV00994 | Exon 19 | 2378C > T | T793I | IV | HEK293T/c17 | 5-FU | Decreased function | [59] |
| – | – | Exon 19 | 2420A > G | H807R | IV | 293FT | 5-FU | 50% of CLint ratio | [60] |
| – | – | Exon 20 | 2476G > A | V826M | IV | 293FT | 5-FU | 35% of CLint ratio | [60] |
| rs200687447 | PV00930 | Exon 20 | 2482G > A | E828K | IV | HEK293T/c17 | 5-FU | Increased function | [58] |
| rs60139309 | PV01016 | Exon 20 | 2582A > G | K861R | V | HEK293T/c17 | 5-FU | Increased function | [58] |
| rs201035051 | PV00934 | Exon 21 | 2623A > C | K875Q | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs55674432 | PV00996 | Exon 21 | 2639G > T | G880V | V | HEK293T/c17 | 5-FU | No function | [58] |
| rs147545709 | PV00897 | Exon 21 | 2656C > T | R886C | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs1801267 | PV00912 | Exon 21 | 2657G > A (DPYD*9B) |
R886H | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs188052243 | PV00918 | Exon 21 | 2678A > G | N893S | V | 293FT HEK293T/c17 |
5-FU 5-FU |
61% of CLint ratio Decreased function |
[60] [58] |
| – | – | Exon 22 | 2777G > T | G926V | V | 293FT | 5-FU | No function | [58] |
| – | – | Exon 22 | 2822T > C | V941A | V | HEK293T/c17 | 5-FU | Decreased function | [59] |
| – | – | Exon 22 | 2843T > C | I948T | V | HEK293 Flp-In | Thymine | Decreased function | [56] |
| rs67376798 | PV01031 | Exon 22 | 2846A > T | D949V | V | HEK293T/c17 HEK293 Flp-In |
5-FU Thymine |
Decreased function Decreased function |
[58] [55] |
| rs141044036 | PV00876 | Exon 22 | 2872A > G | K958E | V | HEK293T/c17 | 5-FU | No function | [58] |
| rs145529148 | PV00892 | Exon 23 | 2915A > G | Q972R | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs72547602 | PV01033 | Exon 23 | 2921A > T | D974V | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs72547601 | PV01032 | Exon 23 | 2933A > G | H978R | V | HEK293T/c17 | 5-FU | No function | [58] |
| rs61757362 | PV01019 | Exon 23 | 2948C > T | T983I | V | HEK293T/c17 | 5-FU | Decreased function | [58] |
| rs202144771 | PV00941 | Exon 23 | 2977C > T | L993F | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs139459586 | PV00870 | Exon 23 | 2978T > G | L993R | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs1801268 | PV00913 | Exon 23 | 2983G > T (DPYD*10) |
V995F | V | HEK293T/c17 | 5-FU | No function | [58] |
| rs140114515 | PV00873 | Exon 23 | 3049G > A | V1017I | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs148799944 | PV00899 | Exon 23 | 3061G > C | V1021L | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs114096998 | PV00860 | Exon 23 | 3067C > A | P1023T | V | HEK293T/c17 | 5-FU | Normal function | [58] |
| rs114096998 | PV00861 | Exon 23 | 3067C > T | P1023S | V | HEK293T/c17 | 5-FU | Decreased function | [59] |
From a biochemical perspective, human DPD is a flavoprotein containing a single flavin mononucleotide (FMN), a single flavin adenine dinucleotide (FAD), and four iron-sulfur (FeS) clusters. Human DPD consists of five major domains [61,62,63,64]. Domain I (residues 27–172) and domain V (residues 1–26, 848–1025) each contain two FeS clusters. FAD- and nicotinamide adenine dinucleotide phosphate (NADPH)-binding sites are located in domain II (residues 173–286, 442–524) and domain III (residues 287–441), respectively. FMN and the substrate both bind to domain IV (residues 525–847). Human DPD form a dimer, in which electrons from NADPH are transferred to the FeS clusters to catalyze the reduction of bound substrates [65]. Domains II and IV are essential for DPD activity in the structural analysis of variants. Amino acid substitutions that have been observed to affect protein conformation adjacent to the FeS clusters have also caused a significant decrease in enzyme activity.
Henricks et al. described a prediction method using an activity score system and divided DPYD alleles into three categories, consisting of fully functional alleles (wild-type; value of 1), reduced activity alleles (c.2846A > T and HapB3; value of 0.5), and nonfunctional alleles (DPYD*2A and *13; value of 0) [66]. Allele values are totaled for a given patient, leading to an individual gene activity score that represents the DPD phenotype of the patient. Moreover, Shrestha et al. developed a DPYD-specific variant classifier (DPYD-Varifier) using machine learning of in vitro functional data from 156 variants [67]. This model exhibited an accuracy of 85% and outperformed other in silico prediction tools, including PROVEAN, SIFT, and Polyphen-2. In the future, it may be possible to easily predict in vivo DPD activity using machine learning by creating compound databases by gathering detailed information from in vitro analyses. Recently, a list of DPYD variants has been added to the Pharmacogene Variation Consortium website (https://www.pharmvar.org/gene/DPYD). It is expected that evidence-based decisions on FP therapeutic regimens and patient-specific dose guidelines could be applied on the basis of an activity score formula, as has been recommended and implemented with other clinically relevant metabolic enzymes.
3. Dihydropyrimidinase (DHP)
DHP, as previously mentioned, catalyzes the hydrolytic ring opening of FUH2 and UH2 and is expressed mainly in the liver and kidneys [15,68]. The human DHP gene (DPYS) consists of 10 exons mapped to chromosome 8q22, and features a 1560 bp open reading frame, corresponding to a 519 amino acid protein [17].
DHP deficiency is an autosomal recessive disease characterized by the accumulation of UH2 and dihydrothymine (TH2) in blood, urine, and cerebrospinal fluid [69]. The clinical phenotype of DHP-deficient patients is highly variable, ranging from asymptomatic to exhibiting symptomatology similar to that of DPD deficiency, including seizures, intellectual disability, growth impairment, and dysmorphic facial features [70,71,72]. To date, 35 genetically confirmed patients with DHP deficiency have been reported [33,73,74,75,76,77]. However, potential asymptomatic deficiencies might be present in a population with a low frequency of DPD deficiencies. In screening 21,200 healthy Japanese infants, Sumi et al. estimated the deficiency frequency to be approximately 1/10,000 [73]. Akai et al. analyzed the DPYS coding regions from 183 Japanese individuals, in which the c.349T > C (p.W117R) and c.1001A > G (p.Q334R) variants were identified with an allelic frequency of 0.27% and 1.09%, respectively [78].
To date, multiple studies have reported on the relationship between DPD deficiency and the risk of developing FP-related toxicity. However, there is an increasing awareness that patients with DHP deficiencies are also prone to the development of severe FP-associated toxicity. One such study identified severe FP-related toxicity in a female breast cancer patient with the DPYS heterozygous mutation c.833G > A (p.G278D) [21]. We previously reported about a patient with severe capecitabine-associated toxicity and DHP deficiency caused by a compound DPYS heterozygous mutation, c.1001A > G (p.Q334R) and c.1393C > T (p.R465X), including a genetic analysis of the patient’s family [79]. Urinary pyrimidine analysis of the patient’s family revealed that the UH2/uracil ratio of heterozygous individuals was similar to that of wild-type individuals. Although heterozygous patients are predominantly asymptomatic, severe toxicity might occur during chemotherapy containing FPs, rendering the need for genetic testing before FP administration [80].
It is noteworthy that a sizable number of DHP-deficient patients have been identified in East Asian populations. Hamajima et al. identified a single frameshift mutation and five DPYS missense variants in six Japanese patients with dihydropyrimidinuria [17]. Nakajima et al. reported two Chinese pediatric patients with DHP deficiency caused by the compound DPYS heterozygous mutation c.1001A > G and c.1443 + 5G > A (exon 8 skipping) [81]. Moreover, Nakajima et al. identified eight variants, including four novel missense mutations and one novel deletion in four DHP-deficient patients [77]. Thus, DPYS polymorphisms could emerge as novel pharmacogenomic markers associated with severe FP-related toxicity in diverse global populations.
Recently, in vitro functional characterization of DHP variants using heterologous expression systems, including E. coli and mammalian cells, has been reported (Table 2). Van Kuilenburg et al. reported that in the case of 14 variants (L7V, M70T, D81G, G278D, R302Q, L337P, T343A, W360R, V364M, S379R, R412M, R465X, R475X, and R490C) expressed in E. coli, the hydrolytic ring opening of radiolabeled UH2 was markedly altered [71,76]. Hamajima et al. and Thomas et al. reported that six variants (L7V, T68R, Q334R, W360R, G435R, and R490C) showed lower activities than wild-type DHP in COS-7 and RKO cells expression systems [17,82]. We have characterized 21 DHP variants and wild-type DHP expressed in 293FT cells using UH2 and FUH2 as substrates [83]. Among these, 13 variants (N16K, T68R, M70T, D81G, G278D, R302Q, L337P, W360R, S379R, G435R, R465X, R475X, and R490C) demonstrated no enzymatic activity, and five variants (W117R, Q334R, T343A, V364M, and R412M) showed significantly lower CLint values than wild-type DHP. Except for L7V, the results of this study corroborated those of other in vitro studies, suggesting that the specific experimental conditions reflected the in vivo activities of the assayed variants. The divergence observed for L7V might be due to differences in assay conditions, substrate concentrations, or expression systems used.
Table 2.
DPYS variants reported in in vitro analysis.
| dbSNP rsID | Location | Nucleotide Change | Amino Acid Substitution | Expression System | Substrates | Effect | References |
|---|---|---|---|---|---|---|---|
| rs199618701 | Exon 1 | 17G > A | R6Q | 293FT | FUH2 | 120% of CLint ratio | [83] |
| rs57732538 | Exon 1 | 19C > G | L7V | 293FT RKO E. coli |
FUH2UH2UH2 | 116% of CLint ratio 65% of wild-type DHP No function |
[83] [82] [76] |
| rs572241599 | Exon 1 | 48C > G | N16K | 293FT | FUH2 | No function | [83] |
| – | Exon 1 | 203C > G | T68R | 293FT COS-7 |
FUH25-bromo-UH2 | No function 1.5% of wild-type DHP |
[83] [17] |
| rs370718225 | Exon 1 | 209T > C | M70T | 293FT E. coli |
FUH2UH2 | No function No function |
[83] [76] |
| – | Exon 1 | 242A > G | D81G | 293FT E. coli |
FUH2UH2 | No function No function |
[83] [76] |
| – | Exon 2 | 349T > C | W117R | 293FT | FUH2 | 44% of CLint ratio | [83] |
| rs36027551 | Exon 3 | 541C > T | R181W | 293FT RKO |
FUH2UH2 | 110% of CLint ratio 99% of wild-type DHP |
[83] [82] |
| rs751371011 | Exon 4 | 750G > A | M250I | HEK293 | UH2 | 2% of wild-type DHP | [77] |
| – | Exon 5 | 833G > A | G278D | 293FT E. coli |
FUH2UH2 | No function No function |
[83] [21] |
| – | Exon 5 | 884A > G | H295R | HEK293 | UH2 | 9.8% of wild-type DHP | [77] |
| rs200913682 | Exon 5 | 905G > A | R302Q | 293FT E. coli |
FUH2UH2 | No function 3.9% of wild-type DHP |
[83] [76] |
| rs121964923 | Exon 6 | 1001A > G | Q334R | 293FT HEK293 COS-7 |
FUH2UH25-bromo-UH2 | 20% of CLint ratio 9.7% of wild-type DHP 2.5% of wild-type DHP |
[83] [77] [17] |
| rs530911437 | Exon 6 | 1010T > C | L337P | 293FT E. coli |
FUH2UH2 | No function No function |
[83] [76] |
| rs201457190 | Exon 6 | 1027A > G | T343A | 293FT E. coli |
FUH2UH2 | 43% of CLint ratio 49% of wild-type DHP |
[83] [76] |
| rs121964924 | Exon 6 | 1078T > C | W360R | 293FT E. coli E. coli COS-7 |
FUH2UH2UH25-bromo-UH2 | No function No function No function 1.2% of wild-type DHP |
[83] [71] [76] [17] |
| rs138282507 | Exon 6 | 1090G > A | V364M | 293FT E. coli |
FUH2UH2 | 8% of CLint ratio No function |
[83] [76] |
| rs201258823 | Exon 7 | 1137C > A | S379R | 293FT E. coli |
FUH2UH2 | No function 0.20–.9% of wild-type DHP |
[83] [76] |
| rs267606774 | Exon 7 | 1235G > T | R412M | 293FT E. coli |
FUH2UH2 | 36% of CLint ratio No function |
[83] [71] |
| – | Exon 8 | 1253C > T | T418I | HEK293 | UH2 | 64% of wild-type DHP | [77] |
| rs267606773 | Exon 8 | 1303G > A | G435R | 293FT COS-7 |
FUH25-bromo-UH2 | No function 5.1% of wild-type DHP |
[83] [17] |
| rs201280871 | Exon 8 | 1393C > T | R465X | 293FT E. coli |
FUH2UH2 | No function No function |
[83] [76] |
| rs61758444 | Exon 8 | 1423C > T | R475X | 293FT E. coli |
FUH2UH2 | No function 0.2–0.9% of wild-type DHP |
[83] [76] |
| rs142574766 | Exon 9 | 1468C > T | R490C | 293FT E. coli COS-7 |
FUH2UH25-bromo-UH2 | No function 0.2–0.9% of wild-type DHP 1.7% of wild-type DHP |
[83] [76] [17] |
| Rs189448963 | Exon 9 | 1469G > A | R490H | HEK293 | UH2 | 0.3% of wild-type DHP | [77] |
Hsieh et al. reported that dimer formation is essential for DHP activity [84]. Within the cell, DHP is known to form a tetramer composed of subunits containing two zinc ions each [85,86,87]. Each DHP subunit consists of two domains, a large (β/α)8-barrel domain that binds the catalytic dimetal center and a small β-sandwich domain [88]. Each subunit also has two dynamic loops, which act as a lid for the substrate-binding pocket. DHP activity is exerted by the interaction of the C-terminus with the dynamic loop of the neighboring subunit [89,90,91]. We have performed immunoblotting assays of native proteins following blue native polyacrylamide gel electrophoresis and showed that oligomer formation is very important for DHP activity [83]. In the reduced or null-activity variants, the ability of DHP to form oligomers was reduced. The five variants G435R, R465X, R475X, R490C, and R490H introduce mutations in the C-terminus or lead to truncation of the C-terminus, thus affecting oligomer formation and resulting in loss of enzymatic activity. In contrast, the substitutions T68R, M70T, D81G, W117R, M250I, G278D, R302Q, Q334R, L337P, T343A, and R412M exist near the active site of the two dynamic loops, which result in conformational changes in the active site that reduce or eliminate activity. Thus, it has been clarified that changes in DHP activity are associated with amino acid substitutions, as well as changes in oligomer formation and the resulting three-dimensional structure. DHP deficiencies are rarely reported in Caucasians but are highly prevalent in Asians. Thus, we consider that these variants could serve as novel pharmacogenomic markers for the prevention of FP-related toxicity, especially in populations that have a low frequency of symptomatic DPD-deficiency cases.
4. β-Ureidopropionase (β-UP)
β-UP catalyzes the irreversible last step, converting FUPA and β-ureidopropionic acid (bUPA) to fluoro-β-alanine and β-alanine, respectively. The human β-UP gene, UPB1, is located on chromosome 22q11, contains 10 exons, and features an 1155 bp open reading frame; the gene encodes a polypeptide containing 384 amino acids [18]. Human β-UP activity has been detected predominantly in the liver and kidney [26,92].
β-UP deficiency is an autosomal recessive disease characterized by the accumulation of bUPA and N-carbamoyl-β-aminoisobutyric acid (NCBA) in urine, blood, and cerebrospinal fluid [93,94]. To date, 33 genetically confirmed patients with β-UP deficiency have been reported [94,95,96,97,98,99,100]. The clinical phenotype of these patients is highly variable but tends to center around neurological problems. Similar to DHP deficiency, β-UP deficiency is often reported in East Asian populations, including Japan and China. Although it has been reported that severe FP-related toxicity is caused by DPD and DHP deficiencies, little is known about the relationship between β-UP deficiency and FP-related toxicity.
There have been several reports of the in vitro analysis of 13 UPB1 variants with amino acid substitutions identified in β-UP-deficient patients (Table 3). Van Kuilenburg et al. and Thomas et al. reported that variant A85E expressed in E. coli and RKO cells was inactive [93,101]. In a separate study, van Kuilenburg et al., using an E. coli expression system, analyzed six β-UP variants (L13S, G235R, R236W, S264R, R326Q, and T359M) that had been previously identified in 16 β-UP-deficient patients, showing a significant reduction or loss of activity in all of them [95]. Nakajima et al. reported that the G31S, E271K, and R326Q variants expressed in HEK293 cells showed profound reductions in activity [97]. Moreover, Nakajima et al. performed native polyacrylamide gel electrophoresis of β-UP expressed in HEK293 cells and showed that octamer formation is necessary for β-UP activity as well as DHP activity. The majority of variants showed a significant reduction in enzymatic activity. However, whether these variants contribute to the development of FP-related toxicity remains unclear.
Table 3.
UPB1 variants identified in β-UP deficient patients.
| db SNP rsID | Location | Nucleotide Change | Amino Acid Substitution | Expression System | Substrates | Effect | References |
|---|---|---|---|---|---|---|---|
| – | Exon 1 | c.38T > C | p.L13S | E. coli | bUPA | 6% of wild-type β-UP | [95] |
| rs200145797 | Exon 1 | c.91G > A | p.G31S | HEK293 | bUPA | 52% of wild-type β-UP | [97] |
| rs121908066 | Exon 2 | c.209G > C | p.R70P | No reports of in vitro study | [98] | ||
| rs34035085 | Exon 2 | c.254C > A | p.A85E |
E. coli RKO |
bUPA bUPA |
No function 2.7% of wild-type β-UP |
[93] [101] |
| – | Exon 6 | c.703G > A | p.G235R | E. coli | bUPA | No function | [95] |
| rs144135211 | Exon 6 | c.706C > T | p.R236W | E. coli | bUPA | No function | [95] |
| rs145766755 | Exon 7 | c.792C > A | p.S264R | E. coli | bUPA | 20% of wild-type β-UP | [95] |
| – | Exon 7 | c.811G > A | p.E271K | HEK293 | bUPA | 0.7% of wild-type β-UP | [97] |
| – | Exon 7 | c.851G > T | p.C284F | No reports of in vitro study | [99] | ||
| rs1375840064 | Exon 7 | c.853G > A | p.A285T | No reports of in vitro study | [99] | ||
| – | Exon 7 | c.857T > C | p.I286T | HEK293 | bUPA | 70% of wild-type β-UP | [97] |
| rs118163237 | Exon 9 | c.977G > A | p.R326Q |
E. coli HEK293 |
bUPA bUPA |
No function 1.3% of wild-type β-UP |
[95] [97] |
| rs369879221 | Exon 10 | c.1076C > T | p.T359M | E. coli | bUPA | No function | [95] |
Fidlerova et al. performed an analysis of the entire UPB1 coding sequence from 113 Czech cancer patients treated using FP-based chemotherapy [102]. Nine UPB1 variants were detected in a subpopulation of patients exhibiting severe toxicity, including a novel mutation affecting the coding sequence. An analysis of the effect of UPB1 variants on FP-related toxicity in the population of all analyzed patients revealed an association between the c.−80C > G (rs2070474) variant and gastrointestinal toxicity. In addition, a strong positive correlation was found between carriers of the homozygous c.−80G variant and the development of severe mucositis. Thomas et al. deduced that the c.−80G variants might alter the potential binding sites of transcription factors, resulting in a statistically non-significant decrease in UPB1 gene expression in patients who are homozygous for the c.−80G allele. This indicates the possibility that UPB1 variants have an additive and relatively minor effect on the development of FP-related toxicity compared with that of the DPYD and DPYS variants.
5. Other Considerations
Genetic variations in TYMS, MTHFR, and miR-27a have also been associated with FP-related toxicity. Clinical and preclinical studies have shown the importance of intracellular levels of TYMS, a target for 5-FU involved in DNA repair and synthesis [103], as a determinant of sensitivity to 5-FU treatment. Its overexpression stemming from polymorphic TYMS variations lead to differing response rates to 5-FU therapy [104]. The three most studied TYMS genetic polymorphisms are the variable numbers of tandem repeat (VNTR) polymorphisms comprising 28 bp sequence repeats (rs34743033), rs2853542C > G, and the 3’-untranslated region polymorphism 1494delTTAAAG (rs34489327). These polymorphisms alter gene expression, mRNA stability, or TYMS expression levels, resulting in the development of treatment resistance and toxicity [105,106,107]. MTHFR plays a role in the metabolism of folate and forms the reduced folate cofactor essential for TYMS inhibition by 5-FU. Two non-synonymous variants, c.677C > T (p.A222V, rs1801133) and c.1298A > C (p.E429A, rs1801131), alter intracellular folate distribution and decrease enzymatic activity [105,107]. The micro RNA miR-27a polymorphism (rs895819A > G) has been associated with FP-related toxicity, more so in DPD-deficient patients, as increased miR-27a expression leads to decreased DPD mRNA expression [108,109,110]. To date, however, studies involving these genetic polymorphisms have yielded inconsistent results, and further assessment is needed to assess their clinical utility and potential use as biomarkers.
6. Conclusions
FPs are degraded by three metabolic enzymes (DPD, DHP, and β-UP), and a reduction or elimination of their activities leads to severe FP-related toxicity. Therefore, predicting enzymatic activity is critical before the administration of FPs, in which in vitro testing has proven to be a useful complementary method to in vivo testing. This review summarized the findings on the functional characterization of DPD, DHP, and β-UP using in vitro analysis. To date, a large number of DPD variants have been analyzed, giving rise to a significant body of evidence regarding the four most commonly identified risk variants in Caucasians (DPYD*2A, DPYD*13, c.1129 − 5923C > G/hapB3, and c.2846A > T) that are associated with an increased risk of 5-FU-related toxicity. Additionally, a system for predicting in vivo DPD activity has been developed on the basis of in vitro analysis results. This has provided further evidence that rare DHP variants might be useful predictive biomarkers of FP-related toxicity in populations with low frequencies of DPD deficiency, as is the case for Asians. Notably, β-UP is not known to be associated with FP-related toxicity, although variants with reduced function have been identified. Currently, studies comprising in vivo and in vitro correlation of frequent DPYD polymorphisms are advancing applicability as well as underlying the importance of including infrequent DPYD, DPYS, and UPB1 variants, as their collective data is insufficient to establish their clinical consequences fully. Additional in vitro and large-scale in vivo studies using standardized methodologies are needed to generate clear evidence for rare variants and verify existing associative studies.
Recently, the underlying mechanisms by which amino acid substitutions alter enzymatic activities by influencing three-dimensional structures have been elucidated; these findings have significant implications toward the interpretation of previously acquired data and how they could be further used to aid clinical decision making for optimal treatments and forewarning the need for alternative chemotherapy regimens. We expect that this report and others related to genetic FP-metabolizing enzyme variants will be useful in the development and further validation of pharmacogenetic testing with the future inclusion of additional biomarkers. In this way, these developments could lead to optimal personalized medicine grounded on genetic polymorphisms.
Author Contributions
Conceptualization, E.H. and M.H.; writing—original draft preparation, E.H.; writing—review and editing, E.H., E.G.R., and M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Japan Agency for Medical Research and Development, AMED (under grant number 20kk0305009h0002).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pan X., Wang C., Wang F., Li P., Hu Z., Shan Y., Zhang J. Development of 5-Fluorouracil derivatives as anticancer agents. Curr. Med. Chem. 2011;18:4538–4556. doi: 10.2174/092986711797287584. [DOI] [PubMed] [Google Scholar]
- 2.Kilic L., Ordu C., Yildiz I., Sen F., Keskin S., Ciftci R., Pilanci K.N. Current adjuvant treatment modalities for gastric cancer: From history to the future. World J. Gastrointest. Oncol. 2016;8:439–449. doi: 10.4251/wjgo.v8.i5.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas A.S., O’Neil B.H., Goldberg R.M. A decade of advances in cytotoxic chemotherapy for metastatic colorectal cancer. Clin. Colorectal. Cancer. 2011;10:238–244. doi: 10.1016/j.clcc.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Yap Y.S., Kwok L.L., Syn N., Chay W.Y., Chia J.W.K., Tham C.K., Wong N.S., Lo S.K., Dent R.A., Tan S., et al. Predictors of Hand-Foot Syndrome and Pyridoxine for Prevention of Capecitabine-Induced Hand-Foot Syndrome: A Randomized Clinical Trial. JAMA Oncol. 2017;3:1538–1545. doi: 10.1001/jamaoncol.2017.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamberti M., Porto S., Zappavigna S., Addeo E., Marra M., Miraglia N., Sannolo N., Vanacore D., Stiuso P., Caraglia M. A mechanistic study on the cardiotoxicity of 5-fluorouracil in vitro and clinical and occupational perspectives. Toxicol. Lett. 2014;227:151–156. doi: 10.1016/j.toxlet.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y.A., Chung H.C., Choi H.J., Rha S.Y., Seong J.S., Jeung H.C. Intermediate dose 5-fluorouracil-induced encephalopathy. Jpn. J. Clin. Oncol. 2006;36:55–59. doi: 10.1093/jjco/hyi214. [DOI] [PubMed] [Google Scholar]
- 7.Twelves C., Wong A., Nowacki M.P., Abt M., Burris H., 3rd, Carrato A., Cassidy J., Cervantes A., Fagerberg J., Georgoulias V., et al. Capecitabine as adjuvant treatment for stage III colon cancer. N. Engl. J. Med. 2005;352:2696–2704. doi: 10.1056/NEJMoa043116. [DOI] [PubMed] [Google Scholar]
- 8.Saltz L.B., Niedzwiecki D., Hollis D., Goldberg R.M., Hantel A., Thomas J.P., Fields A.L., Mayer R.J. Irinotecan fluorouracil plus leucovorin is not superior to fluorouracil plus leucovorin alone as adjuvant treatment for stage III colon cancer: Results of CALGB 89803. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007;25:3456–3461. doi: 10.1200/JCO.2007.11.2144. [DOI] [PubMed] [Google Scholar]
- 9.Lam S.W., Guchelaar H.J., Boven E. The role of pharmacogenetics in capecitabine efficacy and toxicity. Cancer Treat. Rev. 2016;50:9–22. doi: 10.1016/j.ctrv.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Kunicka T., Prochazka P., Krus I., Bendova P., Protivova M., Susova S., Hlavac V., Liska V., Novak P., Schneiderova M., et al. Molecular profile of 5-fluorouracil pathway genes in colorectal carcinoma. BMC Cancer. 2016;16:795. doi: 10.1186/s12885-016-2826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.Y., Shin E., Kim J.W., Lee H.S., Lee D.W., Kim S.H., Lee J.O., Kim Y.J., Kim J.H., Bang S.M., et al. Impact of intratumoral expression levels of fluoropyrimidine-metabolizing enzymes on treatment outcomes of adjuvant S-1 therapy in gastric cancer. PLoS ONE. 2015;10:e0120324. doi: 10.1371/journal.pone.0120324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daher G.C., Harris B.E., Diasio R.B. Metabolism of pyrimidine analogues and their nucleosides. Pharmacol. Ther. 1990;48:189–222. doi: 10.1016/0163-7258(90)90080-L. [DOI] [PubMed] [Google Scholar]
- 13.Heggie G.D., Sommadossi J.P., Cross D.S., Huster W.J., Diasio R.B. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–2206. [PubMed] [Google Scholar]
- 14.Porter D.J., Harrington J.A., Almond M.R., Lowen G.T., Spector T. (R)-5-fluoro-5,6-dihydrouracil: Kinetics of oxidation by dihydropyrimidine dehydrogenase and hydrolysis by dihydropyrimidine aminohydrolase. Biochem. Pharmacol. 1994;48:775–779. doi: 10.1016/0006-2952(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 15.Kikugawa M., Kaneko M., Fujimoto-Sakata S., Maeda M., Kawasaki K., Takagi T., Tamaki N. Purification, characterization and inhibition of dihydropyrimidinase from rat liver. Eur. J. Biochem. 1994;219:393–399. doi: 10.1111/j.1432-1033.1994.tb19951.x. [DOI] [PubMed] [Google Scholar]
- 16.Lu Z.H., Zhang R., Diasio R.B. Purification and characterization of dihydropyrimidine dehydrogenase from human liver. J. Biol. Chem. 1992;267:17102–17109. [PubMed] [Google Scholar]
- 17.Hamajima N., Kouwaki M., Vreken P., Matsuda K., Sumi S., Imaeda M., Ohba S., Kidouchi K., Nonaka M., Sasaki M., et al. Dihydropyrimidinase deficiency: Structural organization, chromosomal localization, and mutation analysis of the human dihydropyrimidinase gene. Am. J. Hum. Genet. 1998;63:717–726. doi: 10.1086/302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vreken P., van Kuilenburg A.B., Hamajima N., Meinsma R., van Lenthe H., Gohlich-Ratmann G., Assmann B.E., Wevers R.A., van Gennip A.H. cDNA cloning, genomic structure and chromosomal localization of the human BUP-1 gene encoding beta-ureidopropionase. Biochim. Biophys. Acta. 1999;1447:251–257. doi: 10.1016/S0167-4781(99)00182-7. [DOI] [PubMed] [Google Scholar]
- 19.Amstutz U., Farese S., Aebi S., Largiader C.R. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: A haplotype assessment. Pharmacogenomics. 2009;10:931–944. doi: 10.2217/pgs.09.28. [DOI] [PubMed] [Google Scholar]
- 20.van Kuilenburg A.B.P., Haasjes J., Richel D.J., Zoetekouw L., Van Lenthe H., De Abreu R.A., Maring J.G., Vreken P., van Gennip A.H. Clinical Implications of Dihydropyrimidine Dehydrogenase (DPD) Deficiency in Patients with Severe 5-Fluorouracil-associated Toxicity: Identification of New Mutations in the DPD Gene. Clin. Cancer Res. 2000;6:4705–4712. [PubMed] [Google Scholar]
- 21.van Kuilenburg A.B., Meinsma R., Zonnenberg B.A., Zoetekouw L., Baas F., Matsuda K., Tamaki N., van Gennip A.H. Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003;9:4363–4367. [PubMed] [Google Scholar]
- 22.Amstutz U., Froehlich T.K., Largiader C.R. Dihydropyrimidine dehydrogenase gene as a major predictor of severe 5-fluorouracil toxicity. Pharmacogenomics. 2011;12:1321–1336. doi: 10.2217/pgs.11.72. [DOI] [PubMed] [Google Scholar]
- 23.Meulendijks D., Cats A., Beijnen J.H., Schellens J.H. Improving safety of fluoropyrimidine chemotherapy by individualizing treatment based on dihydropyrimidine dehydrogenase activity—Ready for clinical practice? Cancer Treat. Rev. 2016;50:23–34. doi: 10.1016/j.ctrv.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Sistonen J., Buchel B., Froehlich T.K., Kummer D., Fontana S., Joerger M., van Kuilenburg A.B., Largiader C.R. Predicting 5-fluorouracil toxicity: DPD genotype and 5,6-dihydrouracil:uracil ratio. Pharmacogenomics. 2014;15:1653–1666. doi: 10.2217/pgs.14.126. [DOI] [PubMed] [Google Scholar]
- 25.Hiratsuka M. In vitro assessment of the allelic variants of cytochrome P450. Drug Metab. Pharmacokinet. 2012;27:68–84. doi: 10.2133/dmpk.DMPK-11-RV-090. [DOI] [PubMed] [Google Scholar]
- 26.van Kuilenburg A.B., van Lenthe H., van Gennip A.H. Activity of pyrimidine degradation enzymes in normal tissues. Nucleosides Nucleotides Nucleic Acids. 2006;25:1211–1214. doi: 10.1080/15257770600894576. [DOI] [PubMed] [Google Scholar]
- 27.Wei X., Elizondo G., Sapone A., McLeod H.L., Raunio H., Fernandez-Salguero P., Gonzalez F.J. Characterization of the human dihydropyrimidine dehydrogenase gene. Genomics. 1998;51:391–400. doi: 10.1006/geno.1998.5379. [DOI] [PubMed] [Google Scholar]
- 28.Bakkeren J.A., De Abreu R.A., Sengers R.C., Gabreels F.J., Maas J.M., Renier W.O. Elevated urine, blood and cerebrospinal fluid levels of uracil and thymine in a child with dihydrothymine dehydrogenase deficiency. Clin. Chim. Acta Int. J. Clin. Chem. 1984;140:247–256. doi: 10.1016/0009-8981(84)90206-7. [DOI] [PubMed] [Google Scholar]
- 29.Berger R., Stoker-de Vries S.A., Wadman S.K., Duran M., Beemer F.A., de Bree P.K., Weits-Binnerts J.J., Penders T.J., van der Woude J.K. Dihydropyrimidine dehydrogenase deficiency leading to thymine-uraciluria. An inborn error of pyrimidine metabolism. Clin. Chim. Acta Int. J. Clin. Chem. 1984;141:227–234. doi: 10.1016/0009-8981(84)90014-7. [DOI] [PubMed] [Google Scholar]
- 30.van Kuilenburg A.B. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur. J. Cancer. 2004;40:939–950. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Al-Sanna’a N.A., Van Kuilenburg A.B., Atrak T.M., Abdul-Jabbar M.A., Van Gennip A.H. Dihydropyrimidine dehydrogenase deficiency presenting at birth. J. Inherit. Metab. Dis. 2005;28:793–796. doi: 10.1007/s10545-005-4218-0. [DOI] [PubMed] [Google Scholar]
- 32.Boisdron-Celle M., Remaud G., Traore S., Poirier A.L., Gamelin L., Morel A., Gamelin E. 5-Fluorouracil-related severe toxicity: A comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett. 2007;249:271–282. doi: 10.1016/j.canlet.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi K., Kidouchi K., Sumi S., Mizokami M., Orito E., Kumada K., Ueda R., Wada Y. Possible prediction of adverse reactions to pyrimidine chemotherapy from urinary pyrimidine levels and a case of asymptomatic adult dihydropyrimidinuria. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1996;2:1937–1941. [PubMed] [Google Scholar]
- 34.Ezzeldin H., Diasio R. Dihydropyrimidine dehydrogenase deficiency, a pharmacogenetic syndrome associated with potentially life-threatening toxicity following 5-fluorouracil administration. Clin. Colorectal. Cancer. 2004;4:181–189. doi: 10.3816/CCC.2004.n.018. [DOI] [PubMed] [Google Scholar]
- 35.Mounier-Boutoille H., Boisdron-Celle M., Cauchin E., Galmiche J.P., Morel A., Gamelin E., Matysiak-Budnik T. Lethal outcome of 5-fluorouracil infusion in a patient with a total DPD deficiency and a double DPYD and UTG1A1 gene mutation. Br. J. Clin. Pharmacol. 2010;70:280–283. doi: 10.1111/j.1365-2125.2010.03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahu A., Ramaswamy A., Ostwal V. Dihydro pyrimidine dehydrogenase deficiency in patients treated with capecitabine based regimens: A tertiary care centre experience. J. Gastrointest. Oncol. 2016;7:380–386. doi: 10.21037/jgo.2016.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sistonen J., Smith C., Fu Y.K., Largiader C.R. A new DPYD genotyping assay for improving the safety of 5-fluorouracil therapy. Clin. Chim. Acta Int. J. Clin. Chem. 2012;414:109–111. doi: 10.1016/j.cca.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Iyer S.N., Singhal R.S., Hegde M.R., Ankala A. Genetic variation in dihydropyrimidine dehydrogenase (DPYD) gene in a healthy adult Indian population. Ann. Hum. Biol. 2015;42:97–100. doi: 10.3109/03014460.2014.942365. [DOI] [PubMed] [Google Scholar]
- 39.Vaudo C.E., Gil B., Galuski K., Zarwan C., Nugent F.W. Early-Onset 5-Fluorouracil Toxicity in a Patient Negative for Dihydropyrimidine Dehydrogenase Mutations: The Clinical Course of Reversal with Uridine Triacetate. Pharmacotherapy. 2016;36:e178–e182. doi: 10.1002/phar.1841. [DOI] [PubMed] [Google Scholar]
- 40.Thomas F., Hennebelle I., Delmas C., Lochon I., Dhelens C., Garnier Tixidre C., Bonadona A., Penel N., Goncalves A., Delord J.P., et al. Genotyping of a family with a novel deleterious DPYD mutation supports the pretherapeutic screening of DPD deficiency with dihydrouracil/uracil ratio. Clin. Pharmacol. Ther. 2016;99:235–242. doi: 10.1002/cpt.210. [DOI] [PubMed] [Google Scholar]
- 41.Henricks L.M., Siemerink E.J.M., Rosing H., Meijer J., Goorden S.M.I., Polstra A.M., Zoetekouw L., Cats A., Schellens J.H.M., van Kuilenburg A.B.P. Capecitabine-based treatment of a patient with a novel DPYD genotype and complete dihydropyrimidine dehydrogenase deficiency. Int. J. Cancer. 2018;142:424–430. doi: 10.1002/ijc.31065. [DOI] [PubMed] [Google Scholar]
- 42.Gross E., Meul C., Raab S., Propping C., Avril S., Aubele M., Gkazepis A., Schuster T., Grebenchtchikov N., Schmitt M., et al. Somatic copy number changes in DPYD are associated with lower risk of recurrence in triple-negative breast cancers. Br. J. Cancer. 2013;109:2347–2355. doi: 10.1038/bjc.2013.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Kuilenburg A.B., Meijer J., Mul A.N., Meinsma R., Schmid V., Dobritzsch D., Hennekam R.C., Mannens M.M., Kiechle M., Etienne-Grimaldi M.C., et al. Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity. Hum. Genet. 2010;128:529–538. doi: 10.1007/s00439-010-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin F., Zhang H., Huang Y., Yang L., Yu F., Liu X., Fu L., Gu F., Ma Y. Effect of dihydropyrimidine dehydrogenase single nucleotide polymorphisms on prognosis of breast cancer patients with chemotherapy. Oncotarget. 2017;8:112060–112075. doi: 10.18632/oncotarget.23033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seck K., Riemer S., Kates R., Ullrich T., Lutz V., Harbeck N., Schmitt M., Kiechle M., Diasio R., Gross E. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005;11:5886–5892. doi: 10.1158/1078-0432.CCR-04-1784. [DOI] [PubMed] [Google Scholar]
- 46.Amstutz U., Henricks L.M., Offer S.M., Barbarino J., Schellens J.H.M., Swen J.J., Klein T.E., McLeod H.L., Caudle K.E., Diasio R.B., et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018;103:210–216. doi: 10.1002/cpt.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lunenburg C., van der Wouden C.H., Nijenhuis M., Crommentuijn-van Rhenen M.H., de Boer-Veger N.J., Buunk A.M., Houwink E.J.F., Mulder H., Rongen G.A., van Schaik R.H.N., et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2020;28:508–517. doi: 10.1038/s41431-019-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin J.G., Cheong H.S., Kim J.Y., Kim L.H., Han C.S., Kim J.O., Kim H.D., Kim Y.H., Chung M.W., Han S.Y., et al. Screening of dihydropyrimidine dehydrogenase genetic variants by direct sequencing in different ethnic groups. J. Korean Med. Sci. 2013;28:1129–1133. doi: 10.3346/jkms.2013.28.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maekawa K., Saeki M., Saito Y., Ozawa S., Kurose K., Kaniwa N., Kawamoto M., Kamatani N., Kato K., Hamaguchi T., et al. Genetic variations and haplotype structures of the DPYD gene encoding dihydropyrimidine dehydrogenase in Japanese and their ethnic differences. J. Hum. Genet. 2007;52:804–819. doi: 10.1007/s10038-007-0186-6. [DOI] [PubMed] [Google Scholar]
- 50.He Y.F., Wei W., Zhang X., Li Y.H., Li S., Wang F.H., Lin X.B., Li Z.M., Zhang D.S., Huang H.Q., et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in Chinese cancer patients. J. Clin. Pharm. Ther. 2008;33:307–314. doi: 10.1111/j.1365-2710.2008.00898.x. [DOI] [PubMed] [Google Scholar]
- 51.Lunenburg C., Henricks L.M., Guchelaar H.J., Swen J.J., Deenen M.J., Schellens J.H.M., Gelderblom H. Prospective DPYD genotyping to reduce the risk of fluoropyrimidine-induced severe toxicity: Ready for prime time. Eur. J. Cancer. 2016;54:40–48. doi: 10.1016/j.ejca.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Takimoto C.H., Lu Z.H., Zhang R., Liang M.D., Larson L.V., Cantilena L.R., Jr., Grem J.L., Allegra C.J., Diasio R.B., Chu E. Severe neurotoxicity following 5-fluorouracil-based chemotherapy in a patient with dihydropyrimidine dehydrogenase deficiency. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1996;2:477–481. [PubMed] [Google Scholar]
- 53.Lu Z., Zhang R., Diasio R.B. Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: Population characteristics, newly identified deficient patients, and clinical implication in 5-fluorouracil chemotherapy. Cancer Res. 1993;53:5433–5438. [PubMed] [Google Scholar]
- 54.Offer S.M., Wegner N.J., Fossum C., Wang K., Diasio R.B. Phenotypic profiling of DPYD variations relevant to 5-fluorouracil sensitivity using real-time cellular analysis and in vitro measurement of enzyme activity. Cancer Res. 2013;73:1958–1968. doi: 10.1158/0008-5472.CAN-12-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuilenburg A., Meijer J., Tanck M.W.T., Dobritzsch D., Zoetekouw L., Dekkers L.L., Roelofsen J., Meinsma R., Wymenga M., Kulik W., et al. Phenotypic and clinical implications of variants in the dihydropyrimidine dehydrogenase gene. Biochim. Biophys. Acta. 2016;1862:754–762. doi: 10.1016/j.bbadis.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 56.van Kuilenburg A.B., Meijer J., Maurer D., Dobritzsch D., Meinsma R., Los M., Knegt L.C., Zoetekouw L., Jansen R.L., Dezentje V., et al. Severe fluoropyrimidine toxicity due to novel and rare DPYD missense mutations, deletion and genomic amplification affecting DPD activity and mRNA splicing. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:721–730. doi: 10.1016/j.bbadis.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Ogura K., Ohnuma T., Minamide Y., Mizuno A., Nishiyama T., Nagashima S., Kanamaru M., Hiratsuka A., Watabe T., Uematsu T. Dihydropyrimidine dehydrogenase activity in 150 healthy Japanese volunteers and identification of novel mutations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005;11:5104–5111. doi: 10.1158/1078-0432.CCR-05-0217. [DOI] [PubMed] [Google Scholar]
- 58.Offer S.M., Fossum C.C., Wegner N.J., Stuflesser A.J., Butterfield G.L., Diasio R.B. Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer Res. 2014;74:2545–2554. doi: 10.1158/0008-5472.CAN-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elraiyah T., Jerde C.R., Shrestha S., Wu R., Nie Q., Giama N.H., Sarangi V., Roberts L.R., Offer S.M., Diasio R.B. Novel Deleterious Dihydropyrimidine Dehydrogenase Variants May Contribute to 5-Fluorouracil Sensitivity in an East African Population. Clin. Pharmacol. Ther. 2017;101:382–390. doi: 10.1002/cpt.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hishinuma E., Narita Y., Saito S., Maekawa M., Akai F., Nakanishi Y., Yasuda J., Nagasaki M., Yamamoto M., Yamaguchi H., et al. Functional Characterization of 21 Allelic Variants of Dihydropyrimidine Dehydrogenase Identified in 1070 Japanese Individuals. Drug Metab. Dispos. Biol. Fate Chem. 2018;46:1083–1090. doi: 10.1124/dmd.118.081737. [DOI] [PubMed] [Google Scholar]
- 61.Schnackerz K.D., Dobritzsch D., Lindqvist Y., Cook P.F. Dihydropyrimidine dehydrogenase: A flavoprotein with four iron-sulfur clusters. Biochim. Biophys. Acta. 2004;1701:61–74. doi: 10.1016/j.bbapap.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Dobritzsch D., Ricagno S., Schneider G., Schnackerz K.D., Lindqvist Y. Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. J. Biol. Chem. 2002;277:13155–13166. doi: 10.1074/jbc.M111877200. [DOI] [PubMed] [Google Scholar]
- 63.Dobritzsch D., Schneider G., Schnackerz K.D., Lindqvist Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001;20:650–660. doi: 10.1093/emboj/20.4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattison L.K., Johnson M.R., Diasio R.B. A comparative analysis of translated dihydropyrimidine dehydrogenase cDNA; conservation of functional domains and relevance to genetic polymorphisms. Pharmacogenetics. 2002;12:133–144. doi: 10.1097/00008571-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 65.Lohkamp B., Voevodskaya N., Lindqvist Y., Dobritzsch D. Insights into the mechanism of dihydropyrimidine dehydrogenase from site-directed mutagenesis targeting the active site loop and redox cofactor coordination. Biochim. Biophys. Acta. 2010;1804:2198–2206. doi: 10.1016/j.bbapap.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Henricks L.M., Lunenburg C.A., Meulendijks D., Gelderblom H., Cats A., Swen J.J., Schellens J.H., Guchelaar H.J. Translating DPYD genotype into DPD phenotype: Using the DPYD gene activity score. Pharmacogenomics. 2015;16:1277–1286. doi: 10.2217/pgs.15.70. [DOI] [PubMed] [Google Scholar]
- 67.Shrestha S., Zhang C., Jerde C.R., Nie Q., Li H., Offer S.M., Diasio R.B. Gene-Specific Variant Classifier (DPYD-Varifier) to Identify Deleterious Alleles of Dihydropyrimidine Dehydrogenase. Clin. Pharmacol. Ther. 2018;104:709–718. doi: 10.1002/cpt.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamajima N., Matsuda K., Sakata S., Tamaki N., Sasaki M., Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/S0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 69.Duran M., Rovers P., de Bree P.K., Schreuder C.H., Beukenhorst H., Dorland L., Berger R. Dihydropyrimidinuria: A new inborn error of pyrimidine metabolism. J. Inherit. Metab. Dis. 1991;14:367–370. doi: 10.1007/BF01811705. [DOI] [PubMed] [Google Scholar]
- 70.Ohba S., Kidouchi K., Sumi S., Imaeda M., Takeda N., Yoshizumi H., Tatematsu A., Kodama K., Yamanaka K., Kobayashi M., et al. Dihydropyrimidinuria: The first case in Japan. Adv. Exp. Med. Biol. 1994;370:383–386. doi: 10.1007/978-1-4615-2584-4_83. [DOI] [PubMed] [Google Scholar]
- 71.van Kuilenburg A.B., Meijer J., Dobritzsch D., Meinsma R., Duran M., Lohkamp B., Zoetekouw L., Abeling N.G., van Tinteren H.L., Bosch A.M. Clinical, biochemical and genetic findings in two siblings with a dihydropyrimidinase deficiency. Mol. Genet. Metab. 2007;91:157–164. doi: 10.1016/j.ymgme.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 72.Yeung C.W., Yau M.M., Ma C.K., Siu T.S., Tam S., Lam C.W. Diagnosis of dihydropyrimidinase deficiency in a Chinese boy with dihydropyrimidinuria. Hong Kong Med. J./Xianggang Yi Xue Za Zhi. 2013;19:272–275. doi: 10.12809/hkmj133598. [DOI] [PubMed] [Google Scholar]
- 73.Sumi S., Imaeda M., Kidouchi K., Ohba S., Hamajima N., Kodama K., Togari H., Wada Y. Population and family studies of dihydropyrimidinuria: Prevalence, inheritance mode, and risk of fluorouracil toxicity. Am. J. Med Genet. 1998;78:336–340. doi: 10.1002/(SICI)1096-8628(19980724)78:4<336::AID-AJMG6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 74.van Gennip A.H., Abeling N.G., Stroomer A.E., van Lenthe H., Bakker H.D. Clinical and biochemical findings in six patients with pyrimidine degradation defects. J. Inherit. Metab. Dis. 1994;17:130–132. doi: 10.1007/BF00735416. [DOI] [PubMed] [Google Scholar]
- 75.van Gennip A.H., de Abreu R.A., van Lenthe H., Bakkeren J., Rotteveel J., Vreken P., van Kuilenburg A.B. Dihydropyrimidinase deficiency: Confirmation of the enzyme defect in dihydropyrimidinuria. J. Inherit. Metab. Dis. 1997;20:339–342. doi: 10.1023/A:1005309423960. [DOI] [PubMed] [Google Scholar]
- 76.van Kuilenburg A.B., Dobritzsch D., Meijer J., Meinsma R., Benoist J.F., Assmann B., Schubert S., Hoffmann G.F., Duran M., de Vries M.C., et al. Dihydropyrimidinase deficiency: Phenotype, genotype and structural consequences in 17 patients. Biochim. Biophys. Acta. 2010;1802:639–648. doi: 10.1016/j.bbadis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 77.Nakajima Y., Meijer J., Dobritzsch D., Ito T., Zhang C., Wang X., Watanabe Y., Tashiro K., Meinsma R., Roelofsen J., et al. Dihydropyrimidinase deficiency in four East Asian patients due to novel and rare DPYS mutations affecting protein structural integrity and catalytic activity. Mol. Genet. Metab. 2017;122:216–222. doi: 10.1016/j.ymgme.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 78.Akai F., Hosono H., Hirasawa N., Hiratsuka M. Novel single nucleotide polymorphisms of the dihydropyrimidinase gene (DPYS) in Japanese individuals. Drug Metab. Pharmacokinet. 2015;30:127–129. doi: 10.1016/j.dmpk.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Hiratsuka M., Yamashita H., Akai F., Hosono H., Hishinuma E., Hirasawa N., Mori T. Genetic polymorphisms of dihydropyrimidinase in a Japanese patient with capecitabine-induced toxicity. PLoS ONE. 2015;10:e0124818. doi: 10.1371/journal.pone.0124818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fidlerova J., Kleiblova P., Bilek M., Kormunda S., Formankova Z., Novotny J., Kleibl Z. Contribution of dihydropyrimidinase gene alterations to the development of serious toxicity in fluoropyrimidine-treated cancer patients. Cancer Chemother. Pharmacol. 2010;65:661–669. doi: 10.1007/s00280-009-1071-0. [DOI] [PubMed] [Google Scholar]
- 81.Nakajima Y., Meijer J., Zhang C., Wang X., Kondo T., Ito T., Dobritzsch D., Van Kuilenburg A.B. Altered Pre-mRNA Splicing Caused by a Novel Intronic Mutation c.1443+5G>A in the Dihydropyrimidinase (DPYS) Gene. Int. J. Mol. Sci. 2016;17:86. doi: 10.3390/ijms17010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomas H.R., Ezzeldin H.H., Guarcello V., Mattison L.K., Fridley B.L., Diasio R.B. Genetic regulation of dihydropyrimidinase and its possible implication in altered uracil catabolism. Pharm. Genom. 2007;17:973–987. doi: 10.1097/FPC.0b013e3282f01788. [DOI] [PubMed] [Google Scholar]
- 83.Hishinuma E., Akai F., Narita Y., Maekawa M., Yamaguchi H., Mano N., Oda A., Hirasawa N., Hiratsuka M. Functional characterization of 21 allelic variants of dihydropyrimidinase. Biochem. Pharmacol. 2017;143:118–128. doi: 10.1016/j.bcp.2017.06.121. [DOI] [PubMed] [Google Scholar]
- 84.Hsieh Y.C., Chen M.C., Hsu C.C., Chan S.I., Yang Y.S., Chen C.J. Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: Metal and structural requirements for post-translational modification and function. J. Biol. Chem. 2013;288:30645–30658. doi: 10.1074/jbc.M113.496778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brooks K.P., Jones E.A., Kim B.D., Sander E.G. Bovine liver dihydropyrimidine amidohydrolase: Purification, properties, and characterization as a zinc metalloenzyme. Arch. Biochem. Biophys. 1983;226:469–483. doi: 10.1016/0003-9861(83)90316-8. [DOI] [PubMed] [Google Scholar]
- 86.Lohkamp B., Andersen B., Piskur J., Dobritzsch D. The crystal structures of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J. Biol. Chem. 2006;281:13762–13776. doi: 10.1074/jbc.M513266200. [DOI] [PubMed] [Google Scholar]
- 87.Tzeng C.T., Huang Y.H., Huang C.Y. Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1: Insights into the molecular basis of formation of a dimer. Biochem. Biophys. Res. Commun. 2016;478:1449–1455. doi: 10.1016/j.bbrc.2016.08.144. [DOI] [PubMed] [Google Scholar]
- 88.Kim G.J., Kim H.S. C-terminal regions of D-hydantoinases are nonessential for catalysis, but affect the oligomeric structure. Biochem. Biophys. Res. Commun. 1998;243:96–100. doi: 10.1006/bbrc.1997.8037. [DOI] [PubMed] [Google Scholar]
- 89.Niu L., Zhang X., Shi Y., Yuan J. Subunit dissociation and stability alteration of D hydantoinase deleted at the terminal amino acid residue. Biotechnol. Lett. 2007;29:303–308. doi: 10.1007/s10529-006-9238-9. [DOI] [PubMed] [Google Scholar]
- 90.Abendroth J., Niefind K., Schomburg D. X-ray structure of a dihydropyrimidinase from Thermus sp. at 1.3 A resolution. J. Mol. Biol. 2002;320:143–156. doi: 10.1016/S0022-2836(02)00422-9. [DOI] [PubMed] [Google Scholar]
- 91.Gojkovic Z., Rislund L., Andersen B., Sandrini M.P., Cook P.F., Schnackerz K.D., Piskur J. Dihydropyrimidine amidohydrolases and dihydroorotases share the same origin and several enzymatic properties. Nucleic Acids Res. 2003;31:1683–1692. doi: 10.1093/nar/gkg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakamoto T., Sakata S.F., Matsuda K., Horikawa Y., Tamaki N. Expression and properties of human liver beta-ureidopropionase. J. Nutr. Sci. Vitaminol. 2001;47:132–138. doi: 10.3177/jnsv.47.132. [DOI] [PubMed] [Google Scholar]
- 93.van Kuilenburg A.B., Meinsma R., Beke E., Assmann B., Ribes A., Lorente I., Busch R., Mayatepek E., Abeling N.G., van Cruchten A., et al. beta-Ureidopropionase deficiency: An inborn error of pyrimidine degradation associated with neurological abnormalities. Hum. Mol. Genet. 2004;13:2793–2801. doi: 10.1093/hmg/ddh303. [DOI] [PubMed] [Google Scholar]
- 94.Akiyama T., Shibata T., Yoshinaga H., Kuhara T., Nakajima Y., Kato T., Maeda Y., Ohse M., Oka M., Kageyama M., et al. A Japanese case of β-ureidopropionase deficiency with dysmorphic features. Brain Dev. 2017;39:58–61. doi: 10.1016/j.braindev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 95.van Kuilenburg A.B., Dobritzsch D., Meijer J., Krumpel M., Selim L.A., Rashed M.S., Assmann B., Meinsma R., Lohkamp B., Ito T., et al. ß-ureidopropionase deficiency: Phenotype, genotype and protein structural consequences in 16 patients. Biochim. Biophys. Acta. 2012;1822:1096–1108. doi: 10.1016/j.bbadis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 96.Lee J.H., van Kuilenburg A.B., Abeling N.G., Vasta V., Hahn S.H. A Korean Case of β-Ureidopropionase Deficiency Presenting with Intractable Seizure, Global Developmental Delay, and Microcephaly. JIMD Rep. 2015;19:117–121. doi: 10.1007/8904_2014_379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakajima Y., Meijer J., Dobritzsch D., Ito T., Meinsma R., Abeling N.G., Roelofsen J., Zoetekouw L., Watanabe Y., Tashiro K., et al. Clinical, biochemical and molecular analysis of 13 Japanese patients with beta-ureidopropionase deficiency demonstrates high prevalence of the c.977G > A (p.R326Q) mutation [corrected] J. Inherit. Metab. Dis. 2014;37:801–812. doi: 10.1007/s10545-014-9682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yaplito-Lee J., Pitt J., Meijer J., Zoetekouw L., Meinsma R., van Kuilenburg A.B. Beta-ureidopropionase deficiency presenting with congenital anomalies of the urogenital and colorectal systems. Mol. Genet. Metab. 2008;93:190–194. doi: 10.1016/j.ymgme.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 99.Fang Y., Cai C., Wang C., Sun B., Zhang X., Fan W., Hu W., Meng Y., Lin S., Zhang C., et al. Clinical and genetic analysis of 7 Chinese patients with beta-ureidopropionase deficiency. Medicine. 2019;98:e14021. doi: 10.1097/MD.0000000000014021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shu J., Lv X., Jiang S., Zhang Y., Zhang C., Meng Y., Situ A., Xu H., Song L. Genetic analysis of the UPB1 gene in two new Chinese families with beta-ureidopropionase deficiency and the carrier frequency of the mutation c.977G>A in Northern China. Child’s Nerv. Syst. Off. J. Int. Soc. Pediatric Neurosurg. 2014;30:2109–2114. doi: 10.1007/s00381-014-2541-1. [DOI] [PubMed] [Google Scholar]
- 101.Thomas H.R., Ezzeldin H.H., Guarcello V., Mattison L.K., Fridley B.L., Diasio R.B. Genetic regulation of beta-ureidopropionase and its possible implication in altered uracil catabolism. Pharm. Genom. 2008;18:25–35. doi: 10.1097/FPC.0b013e3282f2f134. [DOI] [PubMed] [Google Scholar]
- 102.Fidlerova J., Kleiblova P., Kormunda S., Novotny J., Kleibl Z. Contribution of the beta-ureidopropionase (UPB1) gene alterations to the development of fluoropyrimidine-related toxicity. Pharmacol. Rep. 2012;64:1234–1242. doi: 10.1016/S1734-1140(12)70919-2. [DOI] [PubMed] [Google Scholar]
- 103.Matsusaka S., Lenz H.J. Pharmacogenomics of fluorouracil -based chemotherapy toxicity. Expert Opin. Drug Metab. Toxicol. 2015;11:811–821. doi: 10.1517/17425255.2015.1027684. [DOI] [PubMed] [Google Scholar]
- 104.de Bock C.E., Garg M.B., Scott N., Sakoff J.A., Scorgie F.E., Ackland S.P., Lincz L.F. Association of thymidylate synthase enhancer region polymorphisms with thymidylate synthase activity in vivo. Pharm. J. 2011;11:307–314. doi: 10.1038/tpj.2010.43. [DOI] [PubMed] [Google Scholar]
- 105.Amirfallah A., Kocal G.C., Unal O.U., Ellidokuz H., Oztop I., Basbinar Y. DPYD, TYMS and MTHFR Genes Polymorphism Frequencies in a Series of Turkish Colorectal Cancer Patients. J. Pers. Med. 2018;8:45. doi: 10.3390/jpm8040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lima A., Azevedo R., Sousa H., Seabra V., Medeiros R. Current approaches for TYMS polymorphisms and their importance in molecular epidemiology and pharmacogenetics. Pharmacogenomics. 2013;14:1337–1351. doi: 10.2217/pgs.13.118. [DOI] [PubMed] [Google Scholar]
- 107.Loganayagam A., Arenas Hernandez M., Corrigan A., Fairbanks L., Lewis C.M., Harper P., Maisey N., Ross P., Sanderson J.D., Marinaki A.M. Pharmacogenetic variants in the DPYD, TYMS, CDA and MTHFR genes are clinically significant predictors of fluoropyrimidine toxicity. Br. J. Cancer. 2013;108:2505–2515. doi: 10.1038/bjc.2013.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amstutz U., Offer S.M., Sistonen J., Joerger M., Diasio R.B., Largiader C.R. Polymorphisms in MIR27A Associated with Early-Onset Toxicity in Fluoropyrimidine-Based Chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015;21:2038–2044. doi: 10.1158/1078-0432.CCR-14-2817. [DOI] [PubMed] [Google Scholar]
- 109.Meulendijks D., Henricks L.M., Amstutz U., Froehlich T.K., Largiader C.R., Beijnen J.H., de Boer A., Deenen M.J., Cats A., Schellens J.H. Rs895819 in MIR27A improves the predictive value of DPYD variants to identify patients at risk of severe fluoropyrimidine-associated toxicity. Int. J. Cancer. 2016;138:2752–2761. doi: 10.1002/ijc.30014. [DOI] [PubMed] [Google Scholar]
- 110.Offer S.M., Butterfield G.L., Jerde C.R., Fossum C.C., Wegner N.J., Diasio R.B. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol. Cancer Ther. 2014;13:742–751. doi: 10.1158/1535-7163.MCT-13-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]