Abstract
Genetic alterations of leucine-rich repeat kinase 2 (LRRK2), one of the most important contributors to familial Parkinson’s disease (PD), have been hypothesized to play a role in cancer development due to demographical and preclinical data. Here, we sought to define the prevalence and prognostic significance of LRRK2 somatic mutations across all types of human malignancies by querying the publicly available online genomic database cBioPortal. Ninety-six different studies with 14,041 cases were included in the analysis, and 761/14,041 (5.4%) showed genetic alterations in LRRK2. Among these, 585 (76.9%) were point mutations, indels or fusions, 168 (22.1%) were copy number variations (CNVs), and 8 (1.0%) showed both types of alterations. One case showed the somatic mutation R1441C. A significant difference in terms of overall survival (OS) was noted between cases harboring somatic LRRK2 whole deletions, amplifications, and CNV-unaltered cases (median OS: 20.09, 57.40, and 106.57 months, respectively; p = 0.0008). These results suggest that both LRRK2 amplifications and whole gene deletions could play a role in cancer development, paving the way for future research in terms of potential treatment with LRRK2 small molecule inhibitors for LRRK2-amplified cases.
Keywords: LRRK2, cancer, mutations, CNV, prognostic
1. Introduction
Identification of the PARK8 locus [1] and mutations in the leucine-rich repeat kinase 2 (LRRK2) gene [2,3], located on chromosome 12 (12q12) in familial cases of Parkinson’s disease (PD), more than fifteen years ago is considered a game-changing discovery in our knowledge of this yet incurable neurodegenerative disorder. Today, single LRRK2 mutations represent one of the most frequently known genetic determinants of PD [4].
The LRRK2 gene consists of 51 exons, and it encodes a large protein of 2527 amino acids, retaining two catalytic domains with kinase (MAPKKK domain) and GTPase (ROC, Ras in Complex domain) function and other protein–protein interaction domains (armadillo-like, leucine-rich repeats, WD40) [5]. All PD-associated mutations identified until now are single nucleotide substitutions, the most relevant being G2019S, which accounts for approximately half of LRRK2 mutation in Caucasian populations, underlying approximately 5% of PD cases of autosomal dominant PD and approximately 2% of PD cases with no known family history [6,7,8], and R1441C/G/H. Interestingly, these mutations cluster in the two aforementioned enzymatic domains. The gene encodes for the LRRK2 protein, also known as dardarin, which is widely expressed in different tissues, namely the brain, heart, kidney, and lungs [9], but also in peripheral blood mononuclear cells (PBMCs), including lymphocytes and monocytes [10].
The physiological role of LRRK2 is still not completely understood, despite the enormous amount of research conducted on this protein in the last years. LRRK2 has been hypothesized to have a role in several fundamental cellular processes, primarily autophagy, endocytosis, and mitochondrial and cytoskeletal function [11,12,13,14]. Notably, disruptions in all these processes have been implicated in the neurodegeneration leading to PD [15,16,17,18]. Furthermore, as predictable from the expression of the protein in PBMCs, LRRK2 alterations have also been implicated in dysfunction of immune pathways [19]. Interestingly, genome-wide association studies have linked LRRK2 to at least three chronic inflammatory conditions, namely Crohn’s Disease [20,21,22], leprosy [23], and tuberculosis [24]. Taken together, current evidence strongly suggests a role of LRRK2 misfunction in the pathogenesis of PD, possibly mediated by a role in neuroinflammation [25].
Current evidence in the literature may suggest a negative association between certain neurological diseases like PD or Alzheimer’s disease and cancer [26]. Interestingly, demographical data suggest an increased incidence of cancer in individuals with a germline G2019S LRRK2 mutation, in particular, melanoma [27,28,29,30]; however, findings are not consistent across all studies [31,32]. Moreover, in vitro models recently demonstrated that downregulation of LRRK2 suppresses cholangiocarcinoma cell growth [33] and decreases proliferation of papillary thyroid carcinoma [34]; on the other hand, overexpression of LRRK2 activates survival and proliferation signals in melanocytes and melanoma cells [35]. A series of molecular studies have linked the abnormal activity of this protein to autophagic pathways, inflammation, and mitochondrial dysfunction [11,12,13,14,19], but a defined role in cancer pathogenesis needs yet to be elucidated. Several mechanisms for which LRRK2 could display a tumor-suppressor function have been described, most notably p53 phosphorylation and p21 induction [36], JNK activation [37], and RCAN1 phosphorylation [38]. In contrast, LRRK2 has also a potentially oncogenic role in MET signaling activation [39]. Using multigene testing panels, LRRK2 demonstrated a prognostic significance in different types of cancers, including oral squamous cell carcinomas [40], intrahepatic cholangiocarcinoma [33], non-small-cell lung cancer [41,42], and colon cancer [43]. Recently, great efforts to develop small molecule inhibitors of LRRK2 have been made, hoping for a disease-modifying role in the setting of PD [44,45,46,47]. If an oncogenic role of LRRK2 is confirmed, those drugs are of potential use in the field of cancer-targeted therapy as well.
The current study aims to establish the prevalence of LRRK2 point mutations, indels or fusions (hereby MUTs) and copy-number variations (CNVs) in a large cohort of human malignancies, as well to investigate the potential prognostic value of such genetic alterations.
2. Materials and Methods
A curated set of 176 non-redundant studies publicly available in the online cancer genomic database cBioPortal [48,49,50], comprising 46,595 samples from all types of human malignancies, was initially screened, assessing whether LRRK2 was profiled across those publications; 96 studies endured the screening (full list available in Table S1), encompassing 23,796 different samples from all human cancers excluding those of the eye, the peripheral nervous system, and the thymus. Cases without both MUTs and CNVs data were excluded; a total of 14,286 samples from 14,041 patients remained. The dataset was also queried for MUTs alone, comprising 17,454 patients from 94 studies (2 studies excluded for not profiling LRRK2 MUTs), and for CNVs alone, comprising 15,183 patients from 93 studies (3 studies excluded for not profiling LRRK2 CNVs). For survival analysis of the different types of CNVs, (i.e., amplifications and whole gene deletions), the raw data were downloaded from the cBioPortal database and processed with MedCalc Statistical Software version 19.1.3 (MedCalc Software, Ostend, Belgium). Overall survival was calculated using the Kaplan–Meier Estimate; statistical significance was calculated with a Log-rank Test. Statistical significance of co-occurrence of alterations of LRRK2 and other genes was calculated with Fisher Exact test for p-values and with the Benjamini–Hochberg procedure for q-values.
3. Results
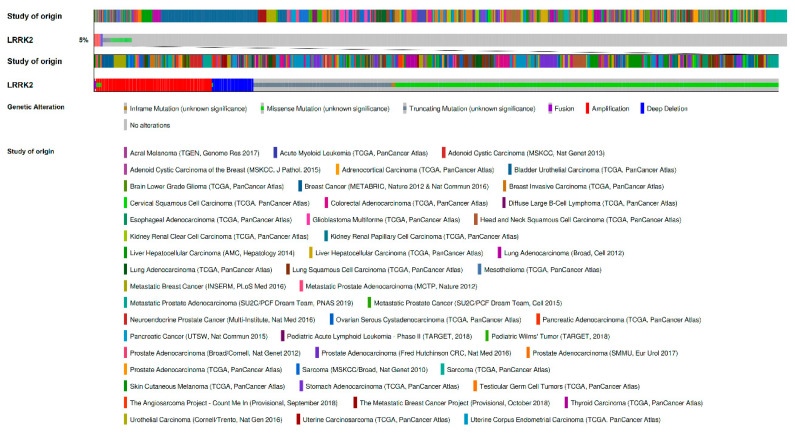
Among 96 non-redundant studies publicly available in the online cancer genomic database cBioPortal [48,49,50], comprising 46,595 samples from all types of human malignancies, the overall observed prevalence of LRRK2 genetic alterations was 761/14041 (5.4%), with MUTs accounting for 585 cases (76.9%; 426 missense, 155 truncating, 3 inframe indels, 1 fusion) and CNVs accounting for 168 cases (22.1%; 123 amplifications and 45 whole gene deletions); eight cases (1.0%) showed both MUTs and CNVs (5 missense and amplification, 1 fusion and amplification, 1 missense and whole gene deletion, 1 inframe indel and amplification), as shown in Figure 1. All these were somatic alterations, and no germinal LRRK2 mutations were present in the dataset.
Figure 1.
Prevalence and distribution of LRRK2 genetic alterations across the analyzed studies. The 5% of the first and second row is expanded to better visualize the data [48,49,50].
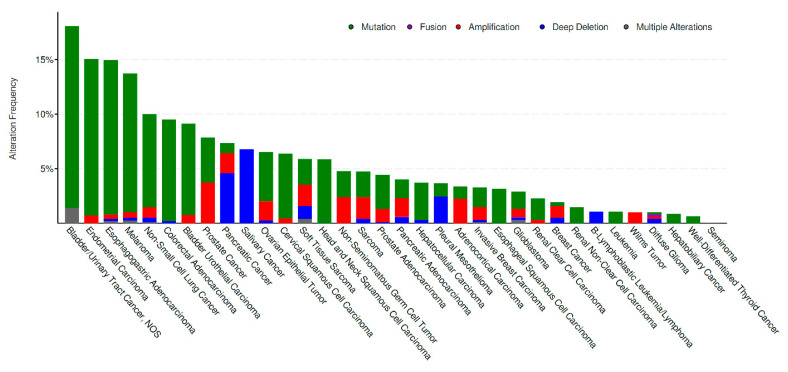
When considering studies with at least 50 patients profiled, alterations across different cancer types ranged from 18.06% (bladder/urinary tract cancer) to 0.0% (seminoma), as shown in Figure 2.
Figure 2.
LRRK2 alteration frequency across cancer types. The types of alterations are color-coded as shown in the legend above [48,49,50].
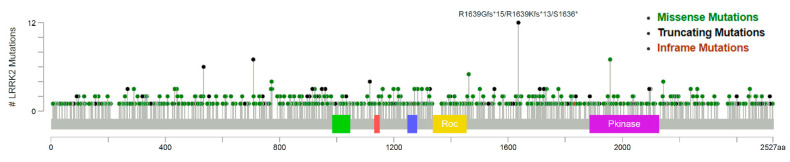
The distribution of missense, truncating, and inframe mutations across the LRRK2 protein is shown in the lollipop plot in Figure 3. Of note, one case of endometrial carcinoma displayed the missense mutation R1441C; one case of stomach adenocarcinoma showed the missense mutation G2019D.
Figure 3.
LRRK2 missense, truncating, and inframe mutations in the studies analyzed. A scattered mutational distribution is evident. The alterations are color-coded as shown in the legend above [48,49,50].
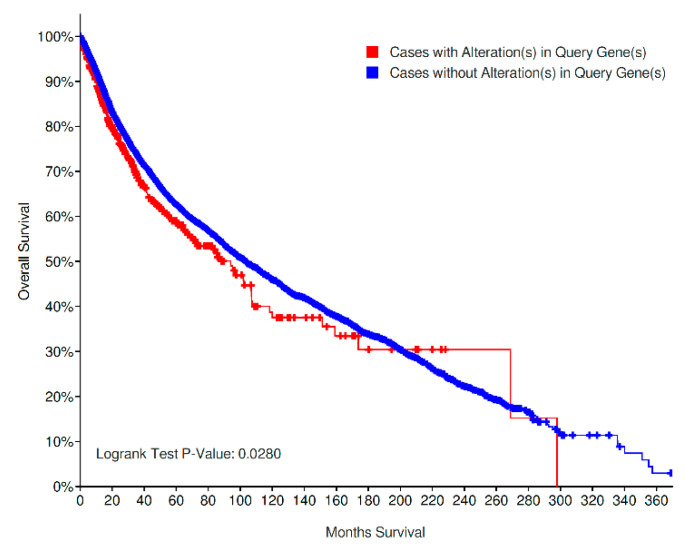
Overall survival (OS) analysis of all cancer types pooled together demonstrated a worse prognosis for cases with LRRK2 alterations (207/613 deceased, median OS: 94.03 months) in comparison to cases with unaltered LRRK2 (4123/11,356 deceased, median OS: 103.26 months) as shown in Figure 4; this result was statistically significant, with p = 0.0280.
Figure 4.
Overall survival of LRRK2 altered (MUT and/or CNV) vs. LRRK2 non-altered cases. A slightly worse prognosis can be observed for altered cases [48,49,50]. MUT: point mutations, indels and fusions; CNV: copy-number variations.
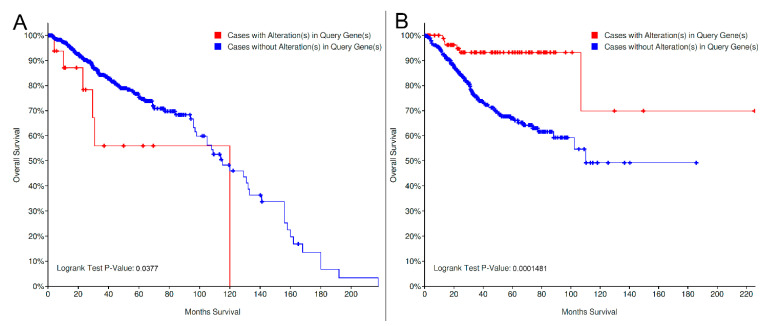
By stratifying for organ sites, a similar result was observed for prostate adenocarcinoma (LRRK2 altered: 6/16 deceased, median OS months: 120, vs. LRRK non-altered: 127/649 deceased, median OS: 115.13 months; p = 0.037); interestingly, an opposite, statistically significant trend was noted for endometrial carcinoma (LRRK2 altered: 6/85 deceased, median OS: NA, vs. LRRK non-altered: 110/479 deceased, median OS: 110.10 months; p = 0.0001481; Figure 5). No other significant results for other organ sites were noted, with trends pointing toward a better/worse prognosis in different settings (Figure S1).
Figure 5.
Overall survival of LRRK2 altered (MUT and/or CNV) vs. LRRK2 non-altered cases in prostatic adenocarcinoma (A) and endometrial carcinoma (B). The prognostic significance of LRRK2 altered cases is negative for prostatic cancer and positive for endometrial cancer [48,49,50]. MUT: point mutations, indels and fusions; CNV: copy-number variations.
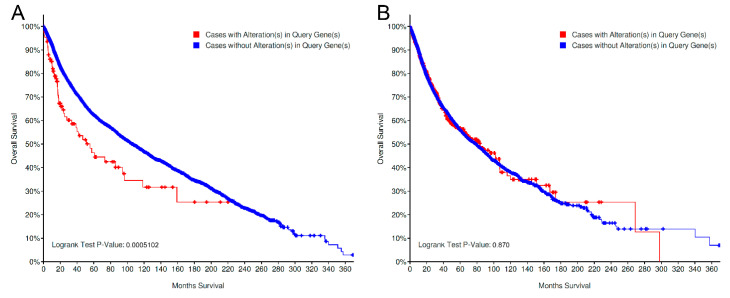
When analyzing MUTs and CNVs alone, a significant (p = 0.0005102) worse survival was noted within the CNV-altered cases (52/111 deceased, median OS: 55.47 months) in contrast to CNV-unaltered ones (4799/13198 deceased, mean OS: 106.57 months); no difference was noted in the cohort analyzed for MUTs only (827/17454; mutated cases: 210/597 deceased, mean OS: 84 months; non-mutated cases: 3711/10945, median OS: 78.8 months; p = 0.870; Figure 6). The majority of CNV-altered cases included in the survival analysis are composed of LRRK2 amplifications (85 amplifications and 26 deep deletions).
Figure 6.
Prognostic significance of LRRK2 CNV-altered vs. CNV-unaltered cases (A) and in MUT-altered vs. MUT-unaltered cases (B). A significant difference is noted for cases analyzed for CNVs, but not for cases profiled for MUTs [48,49,50]. MUT: point mutations, indels and fusions; CNV: copy-number variations.
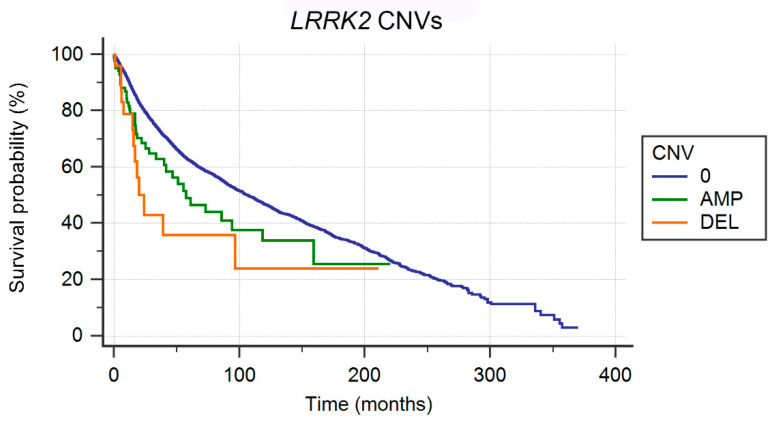
By stratifying for amplifications and deletions, survival analysis shows a significantly (p = 0.0008) worse prognosis in terms of OS for whole gene deleted cases (13/26 deceased, median OS: 20.09 months) in comparison to amplified cases (39/85 deceased, median OS: 57.40 months) and unaltered cases (4799/13,198 deceased, median OS: 106.57 months; Figure 7).
Figure 7.
Survival of cases showing LRRK2 whole gene deletions (DEL, orange curve), amplifications (AMP, green curve), and no copy-number alterations (0, blue curve).
The co-occurrence of CNV alterations of LRRK2 and other genes located at 12q12 is shown in Table 1. The co-occurrence of CNV-alterations of LRRK2 and other genes involved in cancer pathogenesis located at 12q13-12q15 is shown in Table 2.
Table 1.
Co-occurrence of amplifications and whole gene deletions of genes located at 12q12 and LRRK2.
| Gene | Alteration | Altered Group | Unaltered Group | Log Ratio | p-Value | q-Value | Enriched in |
|---|---|---|---|---|---|---|---|
| SLC2A13 | Amp | 99 (57.56%) | 26 (0.17%) | 8.40 | 5.76 × 10−183 | 2.83 × 10−178 | Altered group |
| CNTN1 | Amp | 95 (55.23%) | 32 (0.21%) | 8.04 | 2.10 × 10−170 | 5.16 × 10−166 | Altered group |
| C12ORF40 | Amp | 88 (51.16%) | 27 (0.18%) | 8.18 | 2.46 × 10−158 | 4.02 × 10−154 | Altered group |
| ABCD2 | Amp | 85 (49.42%) | 30 (0.20%) | 7.98 | 3.71 × 10−150 | 4.55 × 10−146 | Altered group |
| KIF21A | Amp | 84 (48.84%) | 35 (0.23%) | 7.74 | 2.61 × 10−145 | 2.56 × 10−141 | Altered group |
| PDZRN4 | Amp | 82 (47.67%) | 35 (0.23%) | 7.70 | 3.80 × 10−141 | 3.11 × 10−137 | Altered group |
| MUC19 | Amp | 74 (43.02%) | 21 (0.14%) | 8.29 | 1.52 × 10−132 | 1.06 × 10−128 | Altered group |
| CPNE8 | Amp | 82 (47.67%) | 132 (0.86%) | 5.79 | 1.09 × 10−110 | 6.65 × 10−107 | Altered group |
| ALG10B | Amp | 76 (44.19%) | 137 (0.90%) | 5.62 | 5.19 × 10−99 | 2.83 × 10−95 | Altered group |
| PPHLN1 | Amp | 63 (36.63%) | 54 (0.35%) | 6.70 | 1.25 × 10−95 | 6.15 × 10−92 | Altered group |
| YAF2 | Amp | 62 (36.05%) | 50 (0.33%) | 6.78 | 4.27 × 10−95 | 1.91 × 10−91 | Altered group |
| GXYLT1 | Amp | 60 (34.88%) | 45 (0.29%) | 6.89 | 4.42 × 10−93 | 1.81 × 10−89 | Altered group |
| ZCRB1 | Amp | 60 (34.88%) | 46 (0.30%) | 6.86 | 1.01 × 10−92 | 3.81 × 10−89 | Altered group |
| PRICKLE1 | Amp | 61 (35.47%) | 53 (0.35%) | 6.68 | 3.28 × 10−92 | 1.15 × 10−88 | Altered group |
| ADAMTS20 | Amp | 59 (34.30%) | 61 (0.40%) | 6.42 | 4.67 × 10−86 | 1.53 × 10−82 | Altered group |
| PUS7L | Amp | 55 (31.98%) | 52 (0.34%) | 6.55 | 1.96 × 10−81 | 6.00 × 10−78 | Altered group |
| IRAK4 | Amp | 54 (31.40%) | 53 (0.35%) | 6.50 | 2.62 × 10−79 | 7.15 × 10−76 | Altered group |
| TWF1 | Amp | 53 (30.81%) | 57 (0.37%) | 6.37 | 2.42 × 10−76 | 5.93 × 10−73 | Altered group |
| TMEM117 | Amp | 54 (31.40%) | 82 (0.54%) | 5.87 | 5.64 × 10−72 | 1.32 × 10−68 | Altered group |
| NELL2 | Amp | 53 (30.81%) | 80 (0.52%) | 5.88 | 1.08 × 10−70 | 2.40 × 10−67 | Altered group |
| CNTN1 | DeepDel | 38 (22.09%) | 15 (0.10%) | 7.81 | 4.09 × 10−64 | 8.73 × 10−61 | Altered group |
| DBX2 | Amp | 44 (25.58%) | 62 (0.41%) | 5.98 | 2.37 × 10−59 | 4.85 × 10−56 | Altered group |
| C12ORF40 | DeepDel | 31 (18.02%) | 5 (0.03%) | 9.11 | 5.82 × 10−57 | 1.14 × 10−53 | Altered group |
| ANO6 | Amp | 44 (25.58%) | 76 (0.50%) | 5.68 | 2.06 × 10−56 | 3.89 × 10−53 | Altered group |
| SLC2A13 | DeepDel | 35 (20.35%) | 20 (0.13%) | 7.28 | 4.60 × 10−56 | 8.35 × 10−53 | Altered group |
| KIF21A | DeepDel | 30 (17.44%) | 6 (0.04%) | 8.79 | 3.24 × 10−54 | 5.67 × 10−51 | Altered group |
| PLEKHA8P1 | Amp | 41 (23.84%) | 66 (0.43%) | 5.79 | 1.86 × 10−53 | 3.14 × 10−50 | Altered group |
| ABCD2 | DeepDel | 29 (16.86%) | 5 (0.03%) | 9.01 | 5.03 × 10−53 | 8.23 × 10−50 | Altered group |
| PDZRN4 | DeepDel | 32 (18.60%) | 19 (0.12%) | 7.22 | 6.05 × 10−51 | 9.57 × 10−48 | Altered group |
| SCAF11 | Amp | 38 (22.09%) | 77 (0.50%) | 5.45 | 1.49 × 10−46 | 2.08 × 10−43 | Altered group |
| ADAMTS20 | DeepDel | 31 (18.02%) | 26 (0.17%) | 6.73 | 1.57 × 10−46 | 2.13 × 10−43 | Altered group |
| GXYLT1 | DeepDel | 28 (16.28%) | 13 (0.09%) | 7.58 | 3.17 × 10−46 | 4.15 × 10−43 | Altered group |
| ARID2 | Amp | 38 (22.09%) | 79 (0.52%) | 5.42 | 3.22 × 10−46 | 4.15 × 10−43 | Altered group |
| CPNE8 | DeepDel | 27 (15.70%) | 10 (0.07%) | 7.91 | 6.85 × 10−46 | 8.62 × 10−43 | Altered group |
| ALG10B | DeepDel | 25 (14.53%) | 10 (0.07%) | 7.79 | 4.00 × 10−42 | 4.90 × 10−39 | Altered group |
| LINC02402 | Amp | 29 (16.86%) | 31 (0.20%) | 6.38 | 1.64 × 10−41 | 1.96 × 10−38 | Altered group |
| YAF2 | DeepDel | 26 (15.12%) | 16 (0.10%) | 7.17 | 3.27 × 10−41 | 3.82 × 10−38 | Altered group |
| ZCRB1 | DeepDel | 25 (14.53%) | 12 (0.08%) | 7.53 | 3.96 × 10−41 | 4.52 × 10−38 | Altered group |
| PPHLN1 | DeepDel | 25 (14.53%) | 14 (0.09%) | 7.31 | 3.17 × 10−40 | 3.53 × 10−37 | Altered group |
| PRICKLE1 | DeepDel | 26 (15.12%) | 19 (0.12%) | 6.93 | 4.66 × 10−40 | 5.08 × 10−37 | Altered group |
| PUS7L | DeepDel | 26 (15.12%) | 22 (0.14%) | 6.71 | 5.09 × 10−39 | 5.43 × 10−36 | Altered group |
| IRAK4 | DeepDel | 25 (14.53%) | 22 (0.14%) | 6.66 | 2.89 × 10−37 | 2.78 × 10−34 | Altered group |
| NELL2 | DeepDel | 24 (13.95%) | 25 (0.16%) | 6.41 | 1.25 × 10−34 | 9.57 × 10−32 | Altered group |
| RACGAP1P | Amp | 27 (15.70%) | 47 (0.31%) | 5.67 | 1.65 × 10−34 | 1.24 × 10−31 | Altered group |
| TWF1 | DeepDel | 22 (12.79%) | 21 (0.14%) | 6.54 | 2.29 × 10−32 | 1.29 × 10−29 | Altered group |
| MUC19 | DeepDel | 17 (9.88%) | 2 (0.01%) | 9.56 | 4.64 × 10−32 | 2.50 × 10−29 | Altered group |
| SCAF11 | DeepDel | 24 (13.95%) | 36 (0.24%) | 5.89 | 6.43 × 10−32 | 3.43 × 10−29 | Altered group |
| TMEM117 | DeepDel | 23 (13.37%) | 30 (0.20%) | 6.09 | 1.21 × 10−31 | 6.27 × 10−29 | Altered group |
| LINC00938 | Amp | 27 (15.70%) | 65 (0.43%) | 5.21 | 1.65 × 10−31 | 8.34 × 10−29 | Altered group |
| DBX2 | DeepDel | 21 (12.21%) | 21 (0.14%) | 6.47 | 1.20 × 10−30 | 5.75 × 10−28 | Altered group |
| ARID2 | DeepDel | 25 (14.53%) | 54 (0.35%) | 5.36 | 3.63 × 10−30 | 1.65 × 10−27 | Altered group |
| PLEKHA8P1 | DeepDel | 21 (12.21%) | 25 (0.16%) | 6.22 | 1.49 × 10−29 | 6.63 × 10−27 | Altered group |
| ANO6 | DeepDel | 21 (12.21%) | 27 (0.18%) | 6.11 | 4.69 × 10−29 | 2.00 × 10−26 | Altered group |
| LINC00938 | DeepDel | 21 (12.21%) | 27 (0.18%) | 6.11 | 4.69 × 10−29 | 2.00 × 10−26 | Altered group |
| RACGAP1P | DeepDel | 20 (11.63%) | 21 (0.14%) | 6.40 | 6.07 × 10−29 | 2.57 × 10−26 | Altered group |
| LINC02402 | DeepDel | 10 (5.81%) | 7 (0.05%) | 6.99 | 4.09 × 10−16 | 3.74 × 10−14 | Altered group |
Table 2.
Genes located in 12q12-12q15 for which CNVs are associated with cancer, and correlation with LRRK2 CNVs.
| Gene | Cytoband | Alteration | Altered Group | Unaltered Group | Log Ratio | p-Value | q-Value | Enriched in |
|---|---|---|---|---|---|---|---|---|
| MDM2 | 12q15 | Amp | 36 (20.93%) | 480 (3.14%) | 2.74 | 5.05 × 10−19 | 7.37 x 10−17 | Altered group |
| MDM2 | 12q15 | DeepDel | 1 (0.58%) | 4 (0.03%) | 4.47 | 0.0544 | 0.0963 | Altered group |
| CDK4 | 12q14.1 | Amp | 26 (15.12%) | 350 (2.29%) | 2.72 | 8.28 × 10−14 | 5.38 × 10−12 | Altered group |
| ERBB3 | 12q13.2 | Amp | 22 (12.79%) | 133 (0.87%) | 3.88 | 2.18 × 10−18 | 2.81 × 10−16 | Altered group |
| ERBB3 | 12q13.2 | DeepDel | 9 (5.23%) | 14 (0.09%) | 5.84 | 1.52 × 10−12 | 7.93 × 10−11 | Altered group |
| KMT2D | 12q13.12 | Amp | 11 (6.40%) | 38 (0.25%) | 4.68 | 4.77 × 10−12 | 2.32 × 10−10 | Altered group |
| KMT2D | 12q13.12 | DeepDel | 7 (4.07%) | 21 (0.14%) | 4.89 | 1.82 × 10−8 | 3.28 × 10−7 | Altered group |
No significant clustering of LRRK2 CNV-altered versus LRRK2 CNV-unaltered cases was observed in terms of neoplasm staging (Figure S2). Sex and age at diagnosis distribution for LRRK2 CNV-altered and LRRK2 CNV-unaltered cases are shown in Figure S3; both results were statistically non-significant (p > 0.05).
4. Discussion
To our knowledge, this is the first study to assess the prevalence, characteristics, and prognostic implications of LRRK2 alterations in a large cohort of human malignancies. A significant proportion of cancers (5%, Figure 1) across the majority of tumor subtypes (Figure 2) harbor MUTs and/or CNVs of this gene involved in the pathogenesis of PD; however, the role of LRRK2 in cancer development remains to be elucidated, and the possible driver or passenger role of LRRK2 alterations in cancer needs yet to be established.
The mutational landscape of LRRK2 in this cohort suggests a passenger role of MUTs, given their scattered distribution and absence of clustering in specific domains such as Pkinase and Roc (Figure 3). It is also worth noting that the two most commonly encountered mutations of LRRK2 in PD (and therefore pathogenic, albeit in a different setting of human disease), i.e., G2019S and R1441C, are seldom encountered (R1441C: n = 1, G2019S: n = 0, with a case showing a somatic mutation at the same codon: G2019D). An explanation could be represented by a hypothetical tumor-suppressor role of LRRK2 in normal cellular development, for which a multitude of different inactivating mutations could result in a decrease of LRRK2 levels and activity; however, this hypothesis is not supported by previously published data [33] and our subsequent analysis.
The negative prognostic implications of LRRK2 alterations in the studies analyzed (Figure 4) seem to suggest, on the other hand, that LRRK2 does not simply represent a bystander during the process of cancer pathogenesis; the conflicting results for prostate adenocarcinoma and endometrial carcinoma also seem to indicate a site-specific biological meaning of such alterations. By querying for MUTs and CNVs alone, the two different Kaplan–Meier plots unravel the negative prognostic role of CNV-altered cases, while the curves for mutated/nonmutated cases almost completely overlap.
By stratifying for the type of copy-number alteration, in comparison to unaltered cases, both whole gene deleted cases (CNV = −2) and amplified cases (CNV = 2) show a worse prognosis, the former being more marked. Notably, LRRK2 amplification has been observed to be of indirect oncogenic potential in papillary renal cell carcinoma and thyroid carcinomas trough MET signaling [39].
Given the nature of the alterations which confer a negative prognostic value to LRRK2, i.e., amplifications and deletions, other genes located within or near the locus 12q12 need to be taken into account when trying to explain a putative role of LRRK2 in cancer development, since duplications and deletions can involve large segments of DNA strands. The list of genes located at 12q12 and their frequency of CNV alterations in LRRK2 CNV-altered and CNV unaltered cases is shown in Table 1; notably, all genes are significantly co-altered along with LRRK2 (percentage of co-alterations range: 5.81–57.56%; percentage of alterations in LRRK2 unaltered cases range: 0.07-0.54%; Log ratio range: 5.21–9.56). By querying the OncoKB database [51,52] for genes located at 12q12, only ARID2 deletion was found to be likely oncogenic, specifically in non-small cell lung cancer [53] and hepatocellular carcinoma [54]; in our analysis, ARID2 whole gene deletion co-occurred in 14.53% of LRRK2 CNV-altered cases.
CNVs of other genes located at 12q12-12q15 are known to be oncogenic, such as KMT2D deletions [55], ERRB3 amplifications [56], CDK4 amplifications [57], and MDM2 amplifications [58]. Correlations between CNV of those genes and LRRK2 in our analysis are presented in Table 2. Notably, amplifications of MDM2 and CDK4 are both associated with liposarcoma [57,58]. Given the oncogenic role of amplification of ERRB3, CDK4, and MDM2, it could be speculated that LRRK2 represents only a passenger of 12q amplification; it must be noted, though, that the major impact on prognosis is observed in LRRK2 whole gene deleted cases, and the concordance between LRRK2 and KMT2D whole gene deletions is 4.07% in our analysis, and a significant proportion of LRRK2-amplified cases lack amplification of MDM2, CDK4, and ERBB3. Besides, MDM2 acts as a down-regulator of p53 by initiating its ubiquitination which leads to proteasomal degradation [59], and LRRK2 has been described to phosphorylate p53 [36], therefore protecting it from MDM2-induced degradation; in this view, the 20.93% of cases harboring both LRRK2 and MDM2 amplifications could not rely on the latter as a driver of tumor progression. The same applies to the 15.12% of CKD4-amplified cases, as LRRK2 induces p21, a known CKD4 inhibitor [60], via p53 phosphorylation [36].
The hypothesized role of an increase in kinase activity of LRRK2 in cancer development, if confirmed via in vitro studies, potentially paves the way for the use of small molecule LRRK2 inhibitors beyond PD: LRRK2-amplificated cases are putative candidates for such treatment. Our analysis indeed shows that LRRK2 amplification, given its negative prognostic significance, could play a role in cancer development and may be clinically actionable. And yet, we also note a dismal prognosis for LRRK2 whole gene deleted cases. In this view, an inhibition of LRRK2 activity could prove to be more harmful than beneficial. However, when considering that both amplifications (gain of function) and whole gene deletions (loss of function) of LRRK2 are associated with a worse prognosis in cancer, it must be acknowledged that the modulation of the kinase activity by LRRK2 small molecule inhibitors in G2019S-mutated PD patients might lead to a normal kinase function rather than a loss of kinase activity. However, given the complex interplay between the different domains of LRRK2, an inhibition of its kinase domain could have pleiotropic effects on the function of the whole protein [61,62]. Further research is needed, both in vitro and in vivo, in order to better characterize the potential anticancer effects of LRRK2 small molecule inhibitors.
Our analysis provides insights on the potential biological balancing role of LRRK2, a fact hinted at by the different biological processes in which this gene is involved. A number of other genes have been described to possess both oncogenic and tumor-suppressor functions [63]. A subset of these genes possesses kinase activity (e.g., MAP2K4, MAP3K4, PRKAR1A, PRKCB), similarly to LRRK2. In this view, any CNV alteration could play a role in the disruption of normal cellular homeostasis which ultimately leads to cancer. Notably, a small molecule LRRK2 inhibitor, LRRK2-IN-1, demonstrated activity in colorectal and pancreatic cancer via direct inhibition of DCLK1 [64]; the clinical utility of such therapeutic agents could also spawn further from LRRK2-overexpressed cases.
5. Conclusions
Herein, we demonstrated that a significant subset of human malignancies shows LRRK2 genetic alterations. The most frequently encountered mutations in PD, G2019S, and R1441C are almost nonexistent in sporadic human malignancies. LRRK2 CNV, both amplifications and whole gene deletions, confer a poorer prognosis in terms of OS in comparison to unaltered cases; the latter cases show the worse prognosis. In contrast, LRRK2 somatic MUTs show no prognostic significance. These data support the fact that both LRRK2 overexpression and complete loss-of-function could play a role in cancer development, and potentially pave the way for future research to investigate the potential treatment of amplified cases with LRRK2 small molecule inhibitors; the dismal prognosis of LRRK2 whole gene deleted cases also needs further research. In contrast, LRRK2 non-activating MUTs are of probable passenger significance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/8/846/s1, Table S1. Studies included in the analysis using cBioPortal. Figure S1. Kaplan–Meier survival estimates of LRRK2-altered vs. LRRK2-wild-type cases in (A) urothelial cancer, (B) esophagogastric cancer, (C) melanoma, (D) non-small cell lung cancer, (E) colorectal cancer, and (F) pancreatic cancer [48,49,50]. Figure S2. Neoplasm staging for LRRK2 CNV-altered and LRRK2 CNV-unaltered cases. No significant clustering can be observed [48,49,50]. Figure S3. Sex (A) and age at diagnosis (B) distribution for LRRK2 CNV-altered and LRRK2 CNV-unaltered cases (A, p > 0.05, Chi-squared test; B, altered group: median 61.2, interquartile range 52.1-65.0; unaltered group, median 61.16, interquartile range 52.0-69.6; p > 0.05, Kruskal–Wallis Test) [48,49,50].
Author Contributions
Conceptualization, G.L. (Gianluca Lopez) and G.L. (Giulia Lazzeri); formal analysis, G.L. (Gianluca Lopez) and A.R.; investigation, G.L. (Gianluca Lopez) and G.L. (Giulia Lazzeri); methodology, A.R., E.G.-R., and V.V.; supervision, G.I., F.M.C., E.G.-R., S.F., V.V., and A.D.F.; validation, G.L. (Gianluca Lopez), A.R., V.V., and A.D.F.; visualization, G.L. (Gianluca Lopez) and A.R.; writing – original draft, G.L. (Gianluca Lopez) and G.L. (Giulia Lazzeri); writing – review & editing, G.L. (Giuseppe Isimbaldi), F.M.C., S.F., V.V., and A.D.F. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by by the Italian Ministry of Health “Ricerca Corrente 2020” program to S.F.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Funayama M., Hasegawa K., Kowa H., Saito M., Tsuji S., Obata F. A New Locus for Parkinson’s Disease (PARK8) Maps to Chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51 doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 2.Paisán-Ruíz C., Jain S., Evans E., Gilks W., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A., Khan N., et al. Cloning of the Gene Containing Mutations That Cause PARK8-linked Parkinson’s Disease. Neuron. 2004:44. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R., Calne D., et al. Mutations in LRRK2 Cause Autosomal-Dominant Parkinsonism With Pleomorphic Pathology. Neuron. 2004:44. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Monfrini E., Di Fonzo A. Leucine-Rich Repeat Kinase (LRRK2) Genetics and Parkinson’s Disease. Adv. Neurobiol. 2017:14. doi: 10.1007/978-3-319-49969-7_1. [DOI] [PubMed] [Google Scholar]
- 5.Rideout H.J., Stefanis L. The Neurobiology of LRRK2 and Its Role in the Pathogenesis of Parkinson’s Disease. Neurochem. Res. 2014:39. doi: 10.1007/s11064-013-1073-5. [DOI] [PubMed] [Google Scholar]
- 6.Gilks W., Abou-Sleiman P., Gandhi S., Jain S., Singleton A., Lees A., Shaw K., Bhatia K., Bonifati V., Quinn N., et al. A Common LRRK2 Mutation in Idiopathic Parkinson’s Disease. Lancet. 2005:365. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 7.Nichols W., Pankratz N., Hernandez D., Paisán-Ruíz C., Jain S., Halter C., Michaels V., Reed T., Rudolph A., Shults C., et al. Genetic Screening for a Single Common LRRK2 Mutation in Familial Parkinson’s Disease. Lancet. 2005:365. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 8.Di Fonzo A., Rohé C., Ferreira J., Chien H., Vacca L., Stocchi F., Guedes L., Fabrizio E., Manfredi M., Vanacore N., et al. A Frequent LRRK2 Gene Mutation Associated With Autosomal Dominant Parkinson’s Disease. Lancet. 2005:365. doi: 10.1016/S0140-6736(05)17829-5. [DOI] [PubMed] [Google Scholar]
- 9.Westerlund M., Belin A., Anvret A., Bickford P., Olson L., Galter D. Developmental Regulation of Leucine-Rich Repeat Kinase 1 and 2 Expression in the Brain and Other Rodent and Human Organs: Implications for Parkinson’s Disease. Neuroscience. 2008:152. doi: 10.1016/j.neuroscience.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 10.Hakimi M., Selvanantham T., Swinton E., Padmore R., Tong Y., Kabbach G., Venderova K., Girardin S., Bulman D., Scherzer C., et al. Parkinson’s Disease-Linked LRRK2 Is Expressed in Circulating and Tissue Immune Cells and Upregulated Following Recognition of Microbial Structures. J. Neural Transm. 2011:118. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takagawa T., Kitani A., Fuss I., Levine B., Brant S.R., Peter I., Tajima M., Nakamura S., Strober W. An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis. Sci. Transl. Med. 2018:10. doi: 10.1126/scitranslmed.aan8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berwick D., Heaton G., Azeggagh S., Harvey K. LRRK2 Biology From Structure to Dysfunction: Research Progresses, but the Themes Remain the Same. Mol. Neurodegener. 2019:14. doi: 10.1186/s13024-019-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alegre-Abarrategui J., Christian H., Lufino M., Mutihac R., Venda L., Ansorge O., Wade-Martins R. LRRK2 Regulates Autophagic Activity and Localizes to Specific Membrane Microdomains in a Novel Human Genomic Reporter Cellular Model. Hum. Mol. Genet. 2009:18. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin N., Jeong H., Kwon J., Heo H., Kwon J., Yun H., Kim C., Han B., Tong Y., Shen J., et al. LRRK2 Regulates Synaptic Vesicle Endocytosis. Exp. Cell Res. 2008:314. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Manzoni C. The LRRK2-macroautophagy Axis and Its Relevance to Parkinson’s Disease. Biochem. Soc. Trans. 2017:45. doi: 10.1042/BST20160265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schapira A., Cooper J., Dexter D., Clark J., Jenner P., Marsden C. Mitochondrial Complex I Deficiency in Parkinson’s Disease. J. Neurochem. 1990:54. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 17.Schapansky J., Khasnavis S., DeAndrade M., Nardozzi J., Falkson S., Boyd J., Sanderson J., Bartels T., Melrose H., LaVoie M. Familial Knockin Mutation of LRRK2 Causes Lysosomal Dysfunction and Accumulation of Endogenous Insoluble α-Synuclein in Neurons. Neurobiol. Dis. 2018:111. doi: 10.1016/j.nbd.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manzoni C., Mamais A., Dihanich S., McGoldrick P., Devine M., Zerle J., Kara E., Taanman J., Healy D., Marti-Masso J., et al. Pathogenic Parkinson’s Disease Mutations Across the Functional Domains of LRRK2 Alter the Autophagic/Lysosomal Response to Starvation. Biochem. Biophys. Res. Commun. 2013:441. doi: 10.1016/j.bbrc.2013.10.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallings R.L., Tansey M.G. LRRK2 regulation of immune-pathways and inflammatory disease. Biochem. Soc. Trans. 2019;47:1581–1595. doi: 10.1042/BST20180463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manzoni C., Denny P., Lovering R.C., Lewis P.A. Computational analysis of the LRRK2 interactome. PeerJ. 2015;3:e778. doi: 10.7717/peerj.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui K.Y., Fernandez-Hernandez H., Hu J., Schaffner A., Pankratz N., Hsu N.Y., Chuang L.S., Carmi S., Villaverde N., Li X., et al. Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med. 2018:10. doi: 10.1126/scitranslmed.aai7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett J., Hansoul S., Nicolae D., Cho J., Duerr R., Rioux J., Brant S., Silverberg M., Taylor K., Barmada M., et al. Genome-wide Association Defines More Than 30 Distinct Susceptibility Loci for Crohn’s Disease. Nat. Genet. 2008:40. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., Huang W., Chen S., Sun L., Liu H., Li Y., Cui Y., Yan X., Yang H., Yang R., et al. Genomewide Association Study of Leprosy. N. Engl. J. Med. 2009:361. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Arat S., Magid-Slav M., Brown J. Meta-analysis of Human Gene Expression in Response to Mycobacterium Tuberculosis Infection Reveals Potential Therapeutic Targets. BMC Syst. Biol. 2018:12. doi: 10.1186/s12918-017-0524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabezudo D., Baekelandt V., Lobbestael E. Multiple-Hit Hypothesis in Parkinson’s Disease: LRRK2 and Inflammation. Front. Neurosci. 2020:14. doi: 10.3389/fnins.2020.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibáñez K., Boullosa C., Tabarés-Seisdedos R., Baudot A., Valencia A. Molecular Evidence for the Inverse Comorbidity Between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-Analyses. PLoS Genet. 2014:10. doi: 10.1371/journal.pgen.1004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders-Pullman R., Barrett M.J., Stanley K.M., Luciano M.S., Shanker V., Severt L., Hunt A., Raymond D., Ozelius L.J., Bressman S.B. LRRK2 G2019S mutations are associated with an increased cancer risk in Parkinson disease. Mov. Disord. 2010;25:2536–2541. doi: 10.1002/mds.23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inzelberg R., Cohen O.S., Aharon-Peretz J., Schlesinger I., Gershoni-Baruch R., Djaldetti R., Nitsan Z., Ephraty L., Tunkel O., Kozlova E., et al. The LRRK2 G2019S mutation is associated with Parkinson disease and concomitant non-skin cancers. Neurology. 2012;78:781–786. doi: 10.1212/WNL.0b013e318249f673. [DOI] [PubMed] [Google Scholar]
- 29.Waro B.J., Aasly J.O. Exploring cancer in LRRK2 mutation carriers and idiopathic Parkinson’s disease. Brain Behav. 2018;8:e00858. doi: 10.1002/brb3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agalliu I., Ortega R.A., Luciano M.S., Mirelman A., Pont-Sunyer C., Brockmann K., Vilas D., Tolosa E., Berg D., Waro B., et al. Cancer outcomes among Parkinson’s disease patients with leucine rich repeat kinase 2 mutations, idiopathic Parkinson’s disease patients, and nonaffected controls. Mov. Disord. 2019;34:1392–1398. doi: 10.1002/mds.27807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allegra R., Tunesi S., Cilia R., Pezzoli G., Goldwurm S. LRRK2-G2019S mutation is not associated with an increased cancer risk: A kin-cohort study. Mov. Disord. 2014;29:1325–1326. doi: 10.1002/mds.25969. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Martinez J., de la Riva P., Rodriguez-Oroz M.C., Mondragon Rezola E., Bergareche A., Gorostidi A., Gago B., Estanga A., Larranaga N., Sarasqueta C., et al. Prevalence of cancer in Parkinson’s disease related to R1441G and G2019S mutations in LRRK2. Mov. Disord. 2014;29:750–755. doi: 10.1002/mds.25778. [DOI] [PubMed] [Google Scholar]
- 33.Gu S., Chen J., Zhou Q., Yan M., He J., Han X., Qiu Y. LRRK2 Is Associated with Recurrence-Free Survival in Intrahepatic Cholangiocarcinoma and Downregulation of LRRK2 Suppresses Tumor Progress In Vitro. Dig. Dis. Sci. 2019 doi: 10.1007/s10620-019-05806-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y., Zhao L., Li J., Zhong L. Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J. Cell Physiol. 2019;234:20980–20991. doi: 10.1002/jcp.28702. [DOI] [PubMed] [Google Scholar]
- 35.Cai W., Liu P., Schwarzschild M., Chen X. Exploring the Role of LRRK2 in Melanomagenesis. In: Thirty Second Annual Symposium on Etiology, Pathogenesis, and Treatment of Parkinson Disease and Other Movement Disorders. Mov. Disord. 2019;34:S1–S19. doi: 10.1002/mds.27654. [DOI] [PubMed] [Google Scholar]
- 36.Ho D.H., Kim H., Kim J., Sim H., Ahn H., Seo H., Chung K.C., Park B.J., Son I., Seol W. Leucine-Rich Repeat Kinase 2 (LRRK2) phosphorylates p53 and induces p21(WAF1/CIP1) expression. Mol. Brain. 2015;8:54. doi: 10.1186/s13041-015-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Z.C., Chen X.J., Zhou Q., Gong X.H., Chen X., Wu W.J. Downregulated LRRK2 gene expression inhibits proliferation and migration while promoting the apoptosis of thyroid cancer cells by inhibiting activation of the JNK signaling pathway. Int. J. Oncol. 2019;55:21–34. doi: 10.3892/ijo.2019.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han K.A., Yoo L., Sung J.Y., Chung S.A., Um J.W., Kim H., Seol W., Chung K.C. Leucine-Rich Repeat Kinase 2 (LRRK2) Stimulates IL-1beta-Mediated Inflammatory Signaling through Phosphorylation of RCAN1. Front Cell Neurosci. 2017;11:125. doi: 10.3389/fncel.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Looyenga B.D., Furge K.A., Dykema K.J., Koeman J., Swiatek P.J., Giordano T.J., West A.B., Resau J.H., Teh B.T., MacKeigan J.P. Chromosomal amplification of leucine-rich repeat kinase-2 (LRRK2) is required for oncogenic MET signaling in papillary renal and thyroid carcinomas. Proc. Natl. Acad. Sci. USA. 2011;108:1439–1444. doi: 10.1073/pnas.1012500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Wang Y., Kong F., Han R., Song W., Chen D., Bu L., Wang S., Yue J., Ma L. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J. Cell Physiol. 2020;235:3056–3068. doi: 10.1002/jcp.29210. [DOI] [PubMed] [Google Scholar]
- 41.Ma Q., Xu Y., Liao H., Cai Y., Xu L., Xiao D., Liu C., Pu W., Zhong X., Guo X. Identification and validation of key genes associated with non-small-cell lung cancer. J. Cell Physiol. 2019;234:22742–22752. doi: 10.1002/jcp.28839. [DOI] [PubMed] [Google Scholar]
- 42.Shi K., Li N., Yang M., Li W. Identification of Key Genes and Pathways in Female Lung Cancer Patients Who Never Smoked by a Bioinformatics Analysis. J. Cancer. 2019;10:51–60. doi: 10.7150/jca.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng Q., Lin K., Chang T., Zou L., Xing P., Shen Y., Zhu Y. Identification of genomic expression differences between right-sided and left-sided colon cancer based on bioinformatics analysis. OncoTargets. 2018;11:609–618. doi: 10.2147/OTT.S154207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatcher J.M., Choi H.G., Alessi D.R., Gray N.S. Small-Molecule Inhibitors of LRRK2. Adv. Neurobiol. 2017;14:241–264. doi: 10.1007/978-3-319-49969-7_13. [DOI] [PubMed] [Google Scholar]
- 45.Ding X., Ren F. Leucine-rich Repeat Kinase 2 Inhibitors: A Patent Review (2014-present) Expert Opin. Ther. Pat. 2020:30. doi: 10.1080/13543776.2020.1729354. [DOI] [PubMed] [Google Scholar]
- 46.Tolosa E., Vila M., Klein C., Rascol O. LRRK2 in Parkinson Disease: Challenges of Clinical Trials. Nat. Rev. Neurol. 2020:16. doi: 10.1038/s41582-019-0301-2. [DOI] [PubMed] [Google Scholar]
- 47.Taymans J.M., Greggio E. LRRK2 Kinase Inhibition as a Therapeutic Strategy for Parkinson’s Disease, Where Do We Stand? Curr. Neuropharmacol. 2016:14. doi: 10.2174/1570159X13666151030102847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.cBioPortal for Cancer Genomics. [(accessed on 26 May 2020)]; Available online: http://www.cbioportal.org/
- 49.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakravarty D., Gao J., Phillips S., Kundra R., Zhang H., Wang J., Rudolph J., Yaeger R., Soumerai T., Nissan M., et al. OncoKB: A Precision Oncology Knowledge Base. Jco Precis. Oncol. 2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.OncoKB. [(accessed on 26 May 2020)]; Available online: https://www.oncokb.org/
- 53.Manceau G., Letouzé E., Guichard C., Didelot A., Cazes A., Corté H., Fabre E., Pallier K., Imbeaud S., Le Pimpec-Barthes F., et al. Recurrent Inactivating Mutations of ARID2 in Non-Small Cell Lung Carcinoma. Int. J. Cancer. 2013:132. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- 54.Zhao H., Wang J., Han Y., Huang Z., Ying J.X.B., Zhao J., Fang Y., Zhou H., Zhou J. ARID2: A New Tumor Suppressor Gene in Hepatocellular Carcinoma. Oncotarget. 2011:2. doi: 10.18632/oncotarget.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J., Dominguez-Sola D., Hussein S., Lee J., Holmes A., Bansal M., Vlasevska S., Mo T., Tang H., Basso K., et al. Disruption of KMT2D Perturbs Germinal Center B Cell Development and Promotes Lymphomagenesis. Nat. Med. 2015:21. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desbois-Mouthon C. The HER3/ErbB3 Receptor: A Promising Target in Cancer Drug Therapy. Gastroenterol. Clin. Et Biol. 2010:34. doi: 10.1016/j.gcb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Dickson M., Tap W., Keohan M., D’Angelo S., Gounder M., Antonescu C., Landa J., LX Q., Rathbone D., Condy M., et al. Phase II Trial of the CDK4 Inhibitor PD0332991 in Patients With Advanced CDK4-amplified Well-Differentiated or Dedifferentiated Liposarcoma. J. Clin. Oncol. 2013:31. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ray-Coquard I., Blay J., Italiano A., Le Cesne A., Penel N., Zhi J., Heil F., Rueger R., Graves B., Ding M., et al. Effect of the MDM2 Antagonist RG7112 on the P53 Pathway in Patients With MDM2-amplified, Well-Differentiated or Dedifferentiated Liposarcoma: An Exploratory Proof-Of-Mechanism Study. Lancet. Oncol. 2012:13. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 59.Zhao K., Yang Y., Zhang G., Wang C., Wang D., Wu M., Mei Y. Regulation of the Mdm2-p53 Pathway by the Ubiquitin E3 Ligase MARCH7. Embo Rep. 2018:19. doi: 10.15252/embr.201744465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He G., Siddik Z., Huang Z., Wang R., Koomen J., Kobayashi R., Khokhar A., Kuang J. Induction of p21 by p53 Following DNA Damage Inhibits Both Cdk4 and Cdk2 Activities. Oncogene. 2005:24. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z., Mobley J., DeLucas L., Kahn R., West A. LRRK2 Autophosphorylation Enhances Its GTPase Activity. FASEB J. 2016:30. doi: 10.1096/fj.15-277095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guaitoli G., Raimondi F., Gilsbach B.K., Gómez-Llorente Y., Deyaert E., Renzi F., Li X., Schaffner A., Jagtap P.K., Boldt K., et al. Structural Model of the Dimeric Parkinson’s Protein LRRK2 Reveals a Compact Architecture Involving Distant Interdomain Contacts. Proc. Natl. Acad. Sci. USA. 2016:113. doi: 10.1073/pnas.1523708113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen L., Shi Q., Wang W. Double agents: Genes with both oncogenic and tumor-suppressor functions. Oncogenesis. 2018:7. doi: 10.1038/s41389-018-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weygant N., Qu D., Berry W.L., May R., Chandrakesan P., Owen D.B., Sureban S.M., Ali N., Janknecht R., Houchen C.W. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer. 2014;13:103. doi: 10.1186/1476-4598-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.