Abstract
Seed germination is one of the most important stages during plant life cycle, and DOG1 (Delay of germination1) plays a pivotal regulatory role in seed dormancy and germination. In this study, we have identified the DOG1-Like (DOG1L) family in soybean (Glycine max), a staple oil crop worldwide, and investigated their chromosomal distribution, structure and expression patterns. The results showed that the GmDOG1L family is composed of 40 members, which can be divided into six subgroups, according to their evolutionary relationship with other known DOG1-Like genes. These GmDOG1Ls are distributed on 18 of 20 chromosomes in the soybean genome and the number of exons for all the 40 GmDOG1Ls varied greatly. Members of the different subgroups possess a similar motif structure composition. qRT-PCR assay showed that the expression patterns of different GmDOG1Ls were significantly altered in various tissues, and some GmDOG1Ls expressed primarily in soybean seeds. Gibberellic acid (GA) remarkably inhibited the expression of most of GmDOG1Ls, whereas Abscisic acid (ABA) inhibited some of the GmDOG1Ls expression while promoting others. It is speculated that some GmDOG1Ls regulate seed dormancy and germination by directly or indirectly relating to ABA and GA pathways, with complex interaction networks. This study provides an important theoretical basis for further investigation about the regulatory roles of GmDOG1L family on soybean seed germination.
Keywords: soybean, seed dormancy, seed germination, phytohormone, GmDOG1L family
1. Introduction
Soybean is an important legume plant, and is now widely grown in many countries as a staple oil crop [1]. To meet the growing demand for soybean as food, oil and other resources, it is imperative to increase soybean production, and maize-soybean relay-intercropping is a model to increase soybean yield [2]. Seed germination is an important stage during the plant life cycle, which contributes to the spread and distribution of wild species and enhance the quality and yield of cultivated crops [3]. Most angiosperms begin a new stage of growth and development after seed dormancy, which is a very useful survival mechanism protecting plants from adverse environments [4,5]. Successful germination of seeds in the field is essential for the stable high yield [6]. Therefore, the intensive and extensive investigation of the regulatory mechanisms of seed dormancy and germination has an important scientific significance.
DOG1 is a key gene in the control of seed dormancy and germination, which was first discovered through quantitative trait locus (QTL) analysis in Arabidopsis [7]. Following studies revealed that the amount of DOG1 protein regulates the seed dormancy level and germination capacity [8,9]. DOG1 starts to accumulate during seed maturation, and its protein structure is altered during the post-ripening stage. It has been reported that DOG1 express stably after maturation, and DOG1 protein abundance in freshly harvested seeds is important for seed dormancy release [8]. Interestingly, several studies have demonstrated that DOG1 is highly conserved in diverse plant species, including Lactuca sativa, Brassica rapa, Hordeum vulgare, Triticum aestivum and Oryza sativa [10,11,12,13].
Phytohormone abscisic acid (ABA) and DOG1 protein are essential regulators of seed dormancy and germination. It is believed that DOG1 is associated with ABA signaling pathway via clade A of type 2C protein phosphatases [14]. ABA is essential for DOG1 to function, and DOG1 indirectly promotes the transcription of ABA biosynthesis gene NCED [8]. In addition, AHG1 (ABA-HYPERSENSITIVE GERMINATION 1) and AHG3, other important components in ABA signaling pathway also act downstream of DOG1 and are critical to DOG1 function [14]. This indicated that DOG1 controls seed germination by inhibiting the action of specific PP2Cs, which are the negative regulators of ABA signaling pathways.
The successful germination of soybean seeds is one of the preconditions required for a high yield. According to the previous studies, soybean is rich in oil and protein, which leads to the decrease of soybean germination rate, affecting the final yield [3]. Meanwhile, soybean seed germination is deficient, if exposed to diverse environmental stresses, such as salinity, drought or flooding stress, which further results in a serious decrease in final yield [15,16]. The studies of seed germination and dormancy are mainly focused on ABA and GA, and the molecular mechanisms of DOG1 are extensively documented in Arabidopsis [7]. However, the studies of DOG1 mediating the seed germination and dormancy are largely unknown especially in soybean cultivar, because of the shortage of the genetic information of DOG1L gene family in this species.
In this study, the characteristics and functions of GmDOG1L family are analyzed by several bioinformatics methods. The chromosome distribution, gene structure, gene expression patterns in different tissues and the responsiveness of phytohormone treatment of GmDOG1L family have been thoroughly investigated. We aimed for this study to help us in understanding the characteristics of soybean DOG1 gene and its relationship with phytohormones.
2. Results
2.1. Identification of GmDOG1Ls Genes
The soybean-specific Hidden Markov Model for the DOG1 domain was used to identify GmDOG1-Likes (GmDOG1Ls), upon which a total of 40 non-redundant GmDOG1-Like candidates were identified in the soybean genome. Various data about these 40 GmDOG1Ls, including gene name, chromosome location, the number of exons and length of the coding sequences (CDS) were collected. As such, we named them GmDOG1-Like-1 (GmDOG1-L1) to GmDOG1-Like-40 (GmDOG1-L40) (Table 1). By using the NCBI CDD website, we found that the 40 GmDOG1L genes contained the conservative structural domain of DOG1(PF14144), which suggest that our analysis is reliable. Interestingly, among the 40 GmDOG1Ls identified, several members also contained the b-ZIP domain, presumably because of the three domains (PD870616, PD004114 and PD388003) conferring the characterization of DOG1, and relatively, PD004114 is also presented in the b-ZIP domain containing transcription factors described elsewhere [7].
Table 1.
Characterization of the GmDOGL1s family in soybean.
| Gene Name | Locus ID | CDS (bp) | Chromosome Number | Number of Exons | Length (aa) | pI | MW (Da) |
|---|---|---|---|---|---|---|---|
| GmDOG1-L1 | Glyma.01G084200 | 1461 | 1 | 12 | 486 | 7.32 | 54309.28 |
| GmDOG1-L2 | Glyma.02G097900 | 1404 | 2 | 11 | 467 | 7.32 | 51853.46 |
| GmDOG1-L3 | Glyma.02G176800 | 1455 | 2 | 11 | 484 | 7.77 | 53927.08 |
| GmDOG1-L4 | Glyma.03G127600 | 1383 | 3 | 11 | 460 | 7 | 50928.00 |
| GmDOG1-L5 | Glyma.03G128200 | 873 | 3 | 7 | 290 | 8.82 | 32365.88 |
| GmDOG1-L6 | Glyma.03G142400 | 1476 | 3 | 11 | 491 | 8.22 | 55200.29 |
| GmDOG1-L7 | Glyma.04G151800 | 285 | 4 | 2 | 94 | 5.04 | 10511.23 |
| GmDOG1-L8 | Glyma.04G254800 | 1089 | 4 | 8 | 362 | 8.69 | 40929.00 |
| GmDOG1-L9 | Glyma.05G113200 | 579 | 5 | 2 | 192 | 4.97 | 22591.38 |
| GmDOG1-L10 | Glyma.05G182500 | 1113 | 5 | 8 | 370 | 7.11 | 42096.79 |
| GmDOG1-L11 | Glyma.05G195200 | 858 | 5 | 1 | 285 | 5.28 | 32592.96 |
| GmDOG1-L12 | Glyma.06G090900 | 495 | 6 | 2 | 164 | 9.97 | 18889.84 |
| GmDOG1-L13 | Glyma.06G107300 | 1068 | 6 | 8 | 355 | 6.21 | 39984.68 |
| GmDOG1-L14 | Glyma.07G151600 | 702 | 7 | 1 | 233 | 5.32 | 26409.83 |
| GmDOG1-L15 | Glyma.08G140100 | 1140 | 8 | 8 | 379 | 7.78 | 43105.04 |
| GmDOG1-L16 | Glyma.10G092100 | 1554 | 10 | 10 | 517 | 6.61 | 57921.5 |
| GmDOG1-L17 | Glyma.10G194800 | 696 | 10 | 4 | 231 | 7 | 26599.48 |
| GmDOG1-L18 | Glyma.10G276100 | 1371 | 10 | 11 | 456 | 6.08 | 50993.85 |
| GmDOG1-L19 | Glyma.10G296200 | 999 | 10 | 8 | 332 | 8.94 | 37141.79 |
| GmDOG1-L20 | Glyma.11G183700 | 1482 | 11 | 12 | 493 | 5.87 | 54385.80 |
| GmDOG1-L21 | Glyma.11G236300 | 1095 | 11 | 8 | 364 | 6.28 | 41178.84 |
| GmDOG1-L22 | Glyma.12G088700 | 1506 | 12 | 12 | 501 | 6.61 | 55253.16 |
| GmDOG1-L23 | Glyma.12G184500 | 1467 | 12 | 12 | 488 | 6.75 | 54118.97 |
| GmDOG1-L24 | Glyma.13G085100 | 1113 | 13 | 8 | 370 | 7.13 | 41915.73 |
| GmDOG1-L25 | Glyma.13G193700 | 1410 | 13 | 11 | 469 | 5.98 | 51989.98 |
| GmDOG1-L26 | Glyma.13G228600 | 765 | 13 | 1 | 254 | 8.70 | 28971.62 |
| GmDOG1-L27 | Glyma.13G269500 | 807 | 13 | 2 | 268 | 5.86 | 30487.66 |
| GmDOG1-L28 | Glyma.13G316900 | 1473 | 13 | 12 | 490 | 6.96 | 54630.43 |
| GmDOG1-L29 | Glyma.14G167000 | 1113 | 14 | 8 | 370 | 8.26 | 41888.77 |
| GmDOG1-L30 | Glyma.15G083900 | 873 | 15 | 1 | 290 | 9.37 | 32967.42 |
| GmDOG1-L31 | Glyma.15G182700 | 888 | 15 | 2 | 295 | 6.71 | 33829.21 |
| GmDOG1-L32 | Glyma.15G232000 | 1494 | 15 | 11 | 497 | 6.1 | 55125.35 |
| GmDOG1-L33 | Glyma.17G154000 | 636 | 17 | 3 | 211 | 5.95 | 24440.01 |
| GmDOG1-L34 | Glyma.18G020900 | 1089 | 18 | 8 | 362 | 7.78 | 41052.71 |
| GmDOG1-L35 | Glyma.18G202900 | 699 | 18 | 1 | 232 | 5.72 | 26078.56 |
| GmDOG1-L36 | Glyma.19G130200 | 1380 | 19 | 11 | 459 | 8.53 | 50717.82 |
| GmDOG1-L37 | Glyma.19G145300 | 1476 | 19 | 11 | 491 | 8.71 | 55358.4 |
| GmDOG1-L38 | Glyma.20G113600 | 1368 | 20 | 11 | 455 | 5.98 | 50671.73 |
| GmDOG1-L39 | Glyma.20G195100 | 708 | 20 | 4 | 235 | 8.2 | 26896.70 |
| GmDOG1-L40 | Glyma.20G246400 | 1335 | 20 | 11 | 444 | 5.91 | 49505.41 |
The length of the coding sequences of the 40 GmDOG1Ls genes varied significantly, the longest one being GmDOG1-L16 with 1554 bp, while the shortest one is GmDOG1-L7 with only 285 bp. Accordingly, GmDOG1-L16 had 517 amino acid residues, which was found to be the largest protein in the GmDOG1L family, while the smallest one was GmDOG1-L7 with 94 amino acid residues. Furthermore, their predicted isoelectric points (pI) varied from 4.97 (GmDOG1-L9) to 9.97 (GmDOG1-L12) (Table 1).
2.2. GmDOG1L Genes Structure and Phylogenetic Analysis
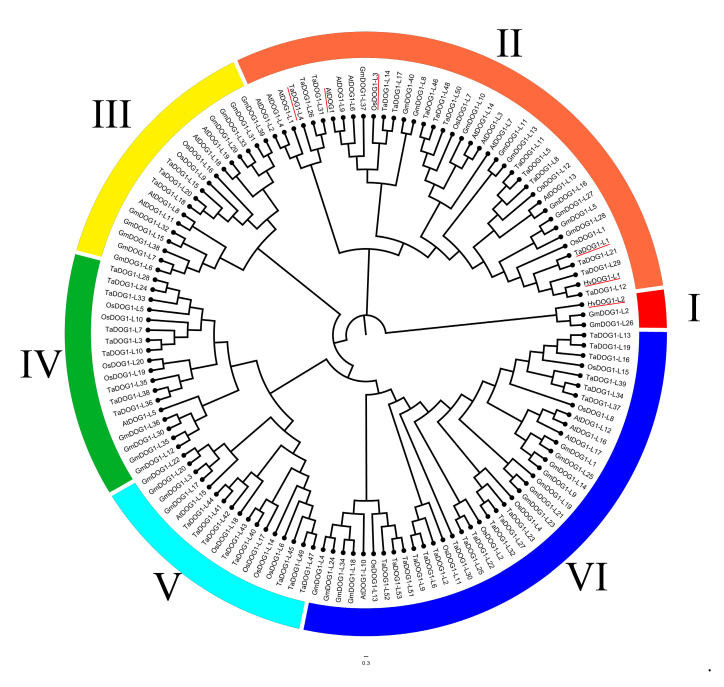
In this study, we used the HMM method to search the DOG1L family in soybean, and as a result, a total of 40 GmDOG1-like genes were found. In addition, by using the same method, we found 20 DOG1-like genes in Arabidopsis thaliana, 20 DOG1-like genes in Oryza sativa and 53 DOG1-like genes in Triticum aestivum; the detailed gene names and gene IDs are shown in Table S1. A total of 135 DOG1Ls from these five species were clustered into six subgroups (I–VI), including 40 GmDOG1-Like, 20 AtDOG1-Like, 53 TaDOG1-Like (Triticum aestivum), 20 OsDOG1-Like (Oryza sativa) and two HvDOG1-Like (Hordeum vulgare) (Figure 1). Among them, AtDOG1 (AT5G45830), AtDOG1-Like-1 (AT4G18660), AtDOG1-Like-2 (AT4G18680), AtDOG1-Like-3 (AT4G18690) and AtDOG1-Like-4 (AT4G18650) were the first five discovered DOG1 genes [7], followed by HvDOG1-L1, HvDOG1-L2, TaDOG1-L1, TaDOG1-L4 and OsDOG1-L3 for which a seed dormancy function has already been confirmed [13]. We found that GmDOG1L was included in all six subgroups. The six DOG1-like genes that have been proven to regulate seed dormancy were concentrated in the subgroups I and II. In addition, we found that HvDOG1-2, GmDOG1-L2 and GmDOG1-L26 were in the subgroup I alone. Meanwhile, GmDOG1-L8, GmDOG1-L37 and GmDOG1-L40 were found to be very close to AtDOG1 in phylogenetic relationship, and GmDOG1-L37 had the closest phylogenetic relationship with AtDOG1. From the phylogenetic tree analysis, we speculated that GmDOG1-L8, GmDOG1-L37 and GmDOG1-L40 might be the potential DOG1 genes in soybean.
Figure 1.
Phylogenetic analysis of DOG1Ls from Glycine max, Arabidopsis, Hordeum vulgare, Triticum aestivum and Oryza sativa. DOG1-like genes for which a seed dormancy function has been confirmed are red underlined. Clustal W software was used to align 135 DOG1Ls. A maximum likelihood phylogenetic tree was constructed by MEGA7.0 software with the Poisson model and 1000 bootstrap replications. The group I–VI use different color difference between them.
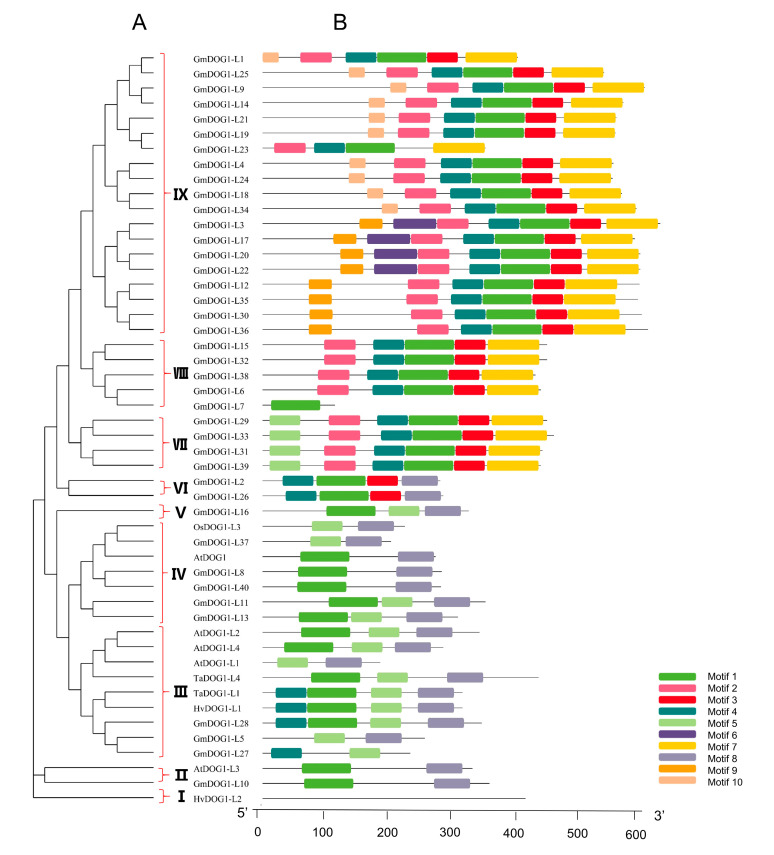
To narrow down the scope of the study, phylogenetic trees were constructed by selecting only GmDOG1Ls, five AtDOG1Ls genes and five other DOG1-like genes that have been proven to regulate seed dormancy [13] (Figure S1). Ten conserved DOG1Ls protein domains were characterized through MEME software analysis. As a result, we found that the motif composition of DOG1-Like in the same group had adequate consistency in phylogenetic trees (Figure 2). For example, in the subgroup IV, AtDOG1 consisted only of motif 1 and motif 8, with GmDOG1-L8, GmDOG1-L40 having the same composition. GmDOG1-L10, GmDOG1-L13 and GmDOG1-L11 all contain two motifs of AtDOG1. AtDOG1-L2, AtDOG1-L3 and AtDOG1-L4 contain motif 1 and motif 8. AtDOG1-L1 consisted of motif 5 and motif 8. These observations probably imply that motif 8 might be the most important component of DOG1. In addition, we found that TaDOG1-L1, TaDOG1-L4, HvDOG1-L1 and OsDOG1-L3 also contained motif 8. On the other hand, 37 of 40 contained both motif 1, whereas GmDOG1-L5, GmDOG1-L27 and GmDOG1-L37 were devoid of motif 1. While GmDOG1-L7 consisted of motif 1 alone. Interestingly, we found no any consistent motif between HvDOG1-L2 and other known DOG1-Like. It is to be noted that motif 8 was found in most of known DOG1 sequences, implying that motif 8 are the conserved structural domains shared by DOG1-like proteins and might play an important role. In summary, from the motif analysis, we speculate GmDOG1-L8, GmDOG1-L10, GmDOG1-L13, GmDOG1-L11 and GmDOG1-L40 as potential DOG1 genes in soybeans. All of them had motifs contained in AtDOG1, and their motifs were similar to the motifs of three other DOG1-Like genes that had been identified to regulate seed dormancy.
Figure 2.
Phylogenetic analysis and conserved motifs of DOG1Ls. (A) The phylogenetic relationship of DOG1Ls. A maximum likelihood phylogenetic tree was constructed by MEGA7.0 software with the Poisson model and 1000 bootstrap replications. (B) Conserved motif arrangements of DOG1Ls. Ten conserved motifs labeled with different colors were found in the DOG1Ls sequences using the MEME program.
The introns and exons of different DOG1Ls genes were varied as shown in the Figure S2. The smallest number of exons was only one, and the largest number was as many as 12. We found that AtDOG1 contained three exons. AtDOG1-L1, AtDOG1-L2, AtDOG1-L3, TaDOG1-L1, HvDOG1-L1 and OsDOG1-L3 had only one exon, but AtDOG1-L4 had two exons. In addition, the exons of HvDOG1-L2 and TaDOG1-L4 were temporarily unavailable. The distribution of exons and introns of DOG1Ls family was found to be complex, and even members of DOG1Ls that were grouped together in the evolutionary analysis had inconsistent exon composition.
In addition, we performed bioinformatics analysis on the DNA sequences of 40 GmDOG1-Like gene promoters, and the analysis results are shown in Figure S3. We analyzed the main role in cis element containing ABRE (cis acting element involved in the abscisic acid responsiveness), the TATC-box (cis acting element involved in gibberellin responsiveness), AuxRR-core (cis acting regulatory element involved in auxin responsiveness) and TGA-box (part of an auxin-responsive element), respectively. We found that GmDOG1-L5, GmDOG1-L10, GmDOG1-L11, GmDOG1-L37 and GmDOG1-L40 contained ABRE elements. GmDOG1-L13 and GmDOG1-L27 contained TGA elements with GmDOG1-L28 containing ABRE and AuxRR elements. Interestingly, GmDOG1-L37 contains three ABRE elements. This result suggested that these GmDOG1Ls might be involved in the regulation of phytohormone response in soybean.
2.3. Chromosomal Location and Gene Duplication
Forty GmDOG1L genes were distributed on 18 chromosomes of the soybean genome, except chromosomes 9 and 16 (Figure S4). Chromosome 1, 7, 8, 14 and 17 contained only one GmDOG1L gene. Chromosome 2, 4, 6, 11, 12, 18 and 19 each contained two GmDOG1L genes, whereas chromosome 3, 5, 15 and 20 each contained three GmDOG1L genes. Chromosome 10 contained four GmDOG1L genes, and chromosome 13 contained five GmDOG1L genes.
According to a previous research, the soybean genome experiences one whole genome triplication (WGT) event and two whole genome duplication (WGD) events with legume WGD and Glycine WGD, as well as about 75% of the genes in soybean, have multiple paralogs [17]. Among the paralog genes, 50% displayed expression of a sub-functionalization that may cause phenotypic variation [18,19]. In addition, dispersed duplicates generally arise by the transposition of DNA or RNA, which might play an important role in creating new genes and changing gene function [20,21]. Finally, we found that 24 of the 40 GmDOG1Ls were distributed in the duplication regions by using the MCScanX program, suggesting that these genes were generated by large-scale duplication events (Figure S5 and Table S2). Furthermore, gene family expansion might be caused by a tandem duplication event, generating consecutive copies of genes in the genome [22,23]. But no tandem duplications were detected in the GmDOG1L gene family.
We then calculated the nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) of these duplicated gene pairs (Table S3). In this study, only the Ka/Ks ratio of GmDOG1-L2 and GmDOG1-L1, GmDOG1-L16 & GmDOG1-L3 and GmDOG1-L32 and GmDOG1-L25 was greater than 1, which is considered to be subject of positive selection, the remaining Ka/Ks ratios were found to be less than 1, which is considered as a purification selection [24]. In addition, the Ks value of GmDOG1-L2 and GmDOG1-L1, GmDOG1-L10 and GmDOG1-L34, GmDOG1-L10 and GmDOG1-L21, GmDOG1-L15 and GmDOG1-L34, GmDOG1-L16 and GmDOG1-L3 and GmDOG1-L32 and GmDOG1-L25 were found to be lesser than 1.3 and greater than 0.3, suggesting that their divergence time was after legume WGD event and before the Glycine WGD. The Ks value of other duplicated gene pairs were less than 0.3, which indicates that their divergence time was after the Glycine whole genome duplication (WGD) event [17,25].
2.4. Expression Profiles of GmDOG1L Genes in Various Tissues of Soybean
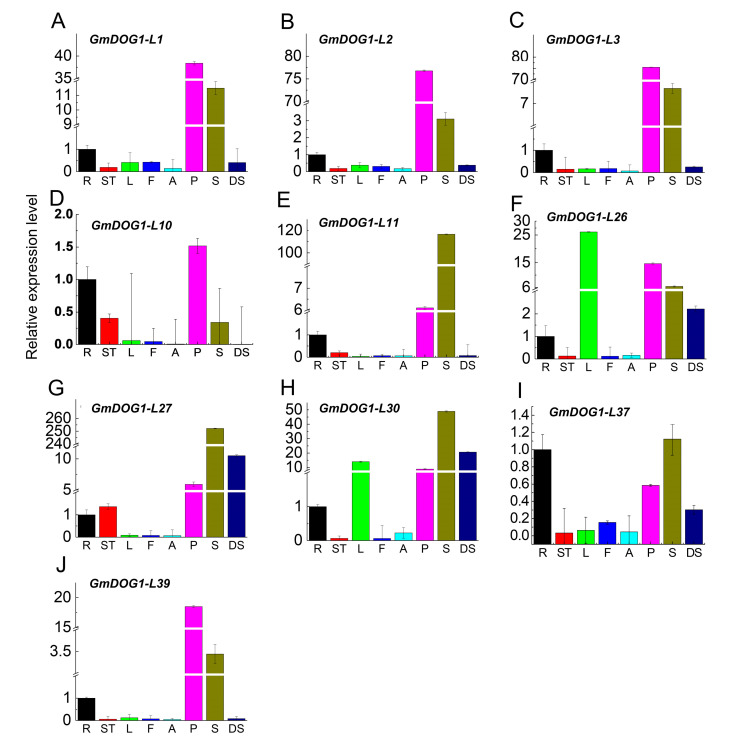
In order to understand the roles of GmDOG1Ls in the growth and development of soybean, we selected 10 GmDOG1L genes from different subgroups for tissue specific expression analysis. First, we selected the GmDOG1-L37 gene from the subgroup II, which was found closest to AtDOG1 in phylogenetic analysis among the 40 members of GmDOG1Ls, and GmDOG1-L37 contained three ABRE elements. Secondly, we selected the GmDOG1-L11 gene from the subgroup II, as GmDOG1-L11 contained AtDOG1’s motif composition along with containing ABRE elements. Third, we chose GmDOG1-L27 from the subgroup II, as it was close to HvDOG1-L1 and TaDOG1-L1, and GmDOG1-L27 also contained TGA elements. Finally, GmDOG1-L10 was also employed for its closest evolutionary relationship to AtDOG1-L3 from the subgroup II, which had the similar motifs composition of AtDOG1, and GmDOG1-L10 contained ABRE elements. In addition, considering that soybean genome experiences one whole genome triplication (WGT) event and two whole genome duplication (WGD) events, functional differentiation might have occurred in some GmDOG1Ls. In order to explore whether GmDOG1Ls had any functional differentiation, we chose GmDOG1-L2 and GmDOG1-L26 from subgroup I, GmDOG1-L39 from subgroup III, GmDOG1-L30 from subgroup IV, GmDOG1-L3 from subgroup V and GmDOG1-L1 from subgroup VI. These 10 GmDOG1L genes were selected from different subgroups as representatives to study whether there were any differences in expression patterns of GmDOG1Ls in different various tissues.
The qRT-PCR assay was employed to investigate the expression patterns of these GmDOG1Ls in several soybean tissues, including root, stem, leaf, flower, apical meristems, pod, developing seed and dry seed. The results showed that all the genes were expressed in eight tissues with different levels (Figure 3). Interestingly, the expression of GmDOG1-L1, GmDOG1-L2, GmDOG1-L3 and GmDOG1-L39 was found to be highest in the pods, followed by dry seeds. GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GmDOG1-L37 were most highly expressed in dry seeds. Furthermore, expression of GmDOG1-L26 was unexpected with high expression level in leaves, followed by pods and dry seeds. In addition, in the phylogenetic analysis, GmDOG1-L26 and GmDOG1-L2 were in subgroup I, but there were significant differences in gene expression in different soybean tissues. Finally, GmDOG1-L26, GmDOG1-L27 and GmDOG1-L30 genes were found to be highly expressed in developing seeds.
Figure 3.
Expression profiles of GmDOG1Ls in different tissues (A–J). Expression levels of 10 selected GmDOG1Ls were examined by qRT-PCR analysis. The housekeeping gene GmTubulin was used as an endogenous reference gene. Roots (R), stems (ST), leaves (L), flowers (F), apical meristems (A), pods (P), dry seeds (S) and developing seeds (DS) were sampled for qRT-PCR analysis. Error bars represent standard errors.
These 10 GmDOG1L genes were all expressed relatively high in seeds, which is consistent with the previous studies reporting that DOG1 is a key gene regulating seed dormancy and germination. In general, GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GmDOG1-L37 were indeed primarily expressed in dry seeds indicating that these genes might mainly function in seed biology. In summary, from tissue specific expression analysis, we believed that GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GmDOG1-L37 might be the potential GmDOG1 genes.
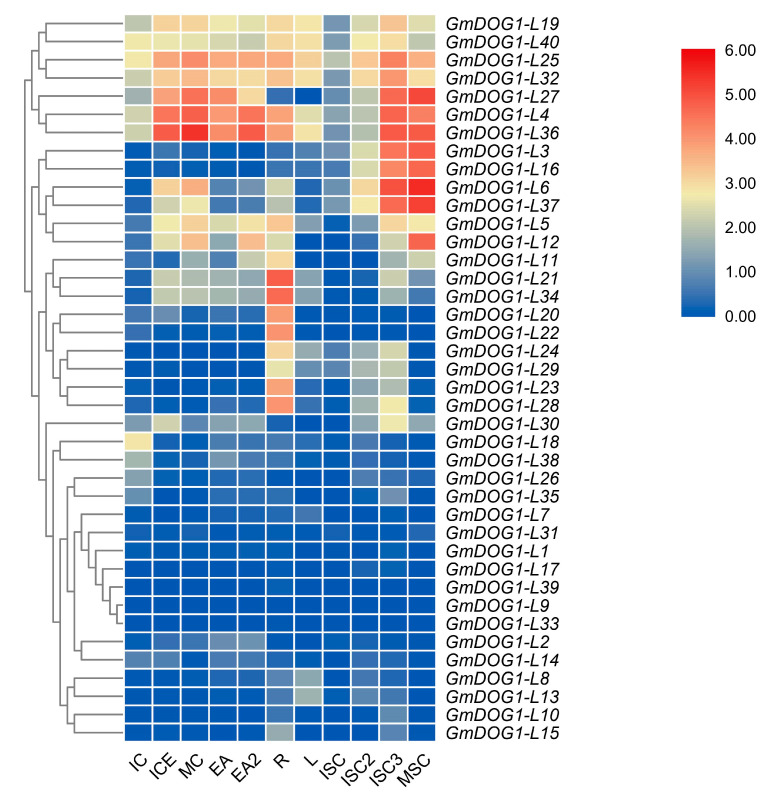
Since some specific primers of GmDOG1Ls members cannot be designed, we obtained transcriptome data of soybean from NCBI GEO DataSets, which contained various organs and different tissues (GEO accession: GSE123655). We extracted 40 GmDOG1Ls expression data from the transcriptome data and used TBtools software to draw the clustering heatmap (Figure 4). We found that GmDOG1-L4, GmDOG1-L19, GmDOG1-L25, GmDOG1-L27, GmDOG1-L32 and GmDOG1-L40 were highly expressed in all tissues, implying that these genes might be important throughout the soybean life cycle. In addition, the transcription expression of GmDOG1-L4, GmDOG1-L5, GmDOG1-L10, GmDOG1-L19, GmDOG1-L25, GmDOG1-L32 and GmDOG1-L40 increased from ISC (immature seed coat-Stage: 5–6 mg), ISC2 (immature seed coat-Stage: 25–50 mg) to ISC3 (immature seed coat-Stage: 200–300 mg), but decreased at the MSC (mature seed coat) stage. Meanwhile, transcription expression of GmDOG1-L3, GmDOG1-L6, GmDOG1-L12, GmDOG1-L16, GmDOG1-L27, GmDOG1-L36 and GmDOG1-L37 gradually increased from ISC, ISC2 and ISC3 to MSC. They gradually increased their expression during seed development period, thus, we speculated that these genes might be the potential GmDOG1 genes.
Figure 4.
Hierarchical clustering of GmDOG1Ls transcriptome expression profiles including 11 samples at different tissue and developmental stages. Immature cotyledon (IC), immature cotyledon without embryo axis (ICE), mature cotyledon (MC), embryo axis-Stage: 300–400 mg (EA), embryo axis-Stage: 200–300 mg (EA2), roots (R), leaves (L), immature seed coat-Stage: 5–6 mg (ISC), immature seed coat-Stage: 25–50 mg (ISC2), immature seed coat-Stage: 200–300 mg (ISC3) and mature seed coat (MSC) were sampled for RNA-seq analysis.
2.5. Expression Analysis of GmDOG1L under Phytohormones Treatment
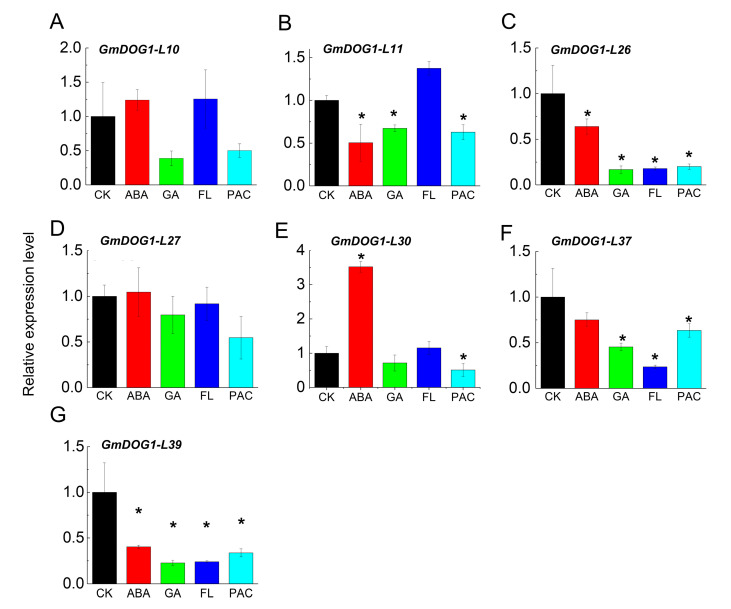
To further investigate the relationship between GmDOG1Ls and phytohormones, seven GmDOG1L genes were selected from different groups for qRT-PCR assay. Compared with control (CK), the expression of GmDOG1-L10, GmDOG1-L27 and GmDOG1-L30 were up-regulated, while the transcription level of GmDOG1-L11, GmDOG1-L26, GmDOG1-L37 and GmDOG1-L39 were down-regulated with distinct levels, after exogenous ABA treatment (Figure 5). Interestingly, the expression of all the GmDOG1L genes was down-regulated under GA treatment, suggesting that the expression of these GmDOG1Ls was inhibited by GA. In addition, the expression of GmDOG1-L10, GmDOG1-L11 and GmDOG1-L30 were up-regulated under FL (fluridone) treatment, while the expression of GmDOG1-L26, GmDOG1-L27, GmDOG1-L37 and GmDOG1-L39 were down-regulated. Intriguingly, the expression of all GmDOG1L genes were down-regulated under PAC (paclobutrazol) treatment.
Figure 5.
Expression profiles of GmDOG1Ls in soybean seeds after phytohormone treatment (A–G). Phytohormones include Abscisic Acid (ABA), Gibberellin (GA), FL and PAC, they all have a concentration of 5 μmol/L. Expression levels of seven selected GmDOG1Ls were examined by qRT-PCR analysis. The housekeeping GmTubulin was used as an endogenous reference gene. The asterisk (*) indicates a significant difference at P < 0.05 by Student’s t-test analysis.
3. Discussions
DOG1s is a key gene regulating seed dormancy and germination [7]. Compared with wild type, dog1 mutant seeds germinated much faster, while the germination was delayed in overexpressed AtDOG1 seeds [7]. In this study, we used the HMM approach to look out for DOG1-like genes in soybeans, and finally found 40 GmDOG1Ls. We performed phylogenetic analysis with 40 GmDOG1-like genes, 20 AtDOG1-Like genes, 20 OsDOG1-like genes, 53 TaDOG1-Like genes and two HvDOG1-like genes. From the phylogenetic analysis, GmDOG1-L37 with AtDOG1 were the closest, followed by GmDOG1-L8 and GmDOG1-L40. Moreover, GmDOG1-L37 was found in between AtDOG1 and OsDOG1-L3, which is one of the genes proven to regulate seed germination. The 135 DOG1-like genes were divided into six subgroups, and we found that AtDOG1 and 5 DOG1-like genes that have been proven to regulate seed dormancy were clustered in the subgroup I and II, which might indicate that these two subgroups played a special role in the phylogenetic analysis. In addition, among the remaining four subgroups, no DOG1-like gene that has been confirmed to regulate seed dormancy was found, which might imply that the DOG1-like genes of these four subgroups are not potential DOG1s and might be those that have lost the ability to regulate seed dormancy.
In Figure 2, the motifs of DOG1Ls had obvious characters in the same subgroup, whereas in Figure S2, the exons of DOG1Ls had no obvious characters. Previous investigations have shown that gene duplication is an important approach to generate new genes, which is actually a protection mechanism for plants to adapt to changing environment [26,27]. In this study, we found that most of the GmDOG1Ls were distributed in duplication blocks, indicating that WGD or segmental duplications plays an important role in the extension of GmDOG1L family [28]. We speculate that the increase in the number of GmDOG1L gene family might have been primarily caused by gene duplication, especially the WGD.
We chose 10 GmDOG1Ls that were relatively highly expressed in dry seeds (Figure 3). Among the 10 genes, GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GmDOG1-L37 showed the highest expression in dry seeds, thus, we speculated them as the potential DOG1 genes in soybean. After exploring the genes selected further, we found that the expression patterns of the genes in the same subgroup showed similar patterns. For example, GmDOG1-L11 and GmDOG1-L27 genes from the group II were highly expressed in dry seeds. GmDOG1-L26 and GmDOG1-L30 had similar motif compositions, and the expression patterns were also similar, in parallel to their high expression in leaves. These tissue-specific expression results suggest that some of the 40 GmDOG1L family members were not potential GmDOG1 genes, thus showing different biological functions. In addition, by analyzing the transcriptome data, we found that the expression of GmDOG1-L3, GmDOG1-L6, GmDOG1-L12, GmDOG1-L16, GmDOG1-L27, GmDOG1-L32, GmDOG1-L36 and GmDOG1-L37 genes were sustained during seed development. Therefore, we speculate that they might be the potential DOG1 genes in soybean.
Phytohormones ABA and GA play a leading role in regulating seed dormancy and germination, and they antagonistically mediate diverse plant developmental processes including seed dormancy and germination [29,30]. For example, when seeds are treated with ABA, dormancy is enhanced and germination is inhibited [6]. Contrarily, when seeds are treated with GA, seed dormancy is inhibited and seed germination is promoted [6]. Analysis of qRT-PCR of GmDOG1Ls under phytohormone treatment showed that the precise correlation between GmDOG1Ls expression and ABA was not detected as ABA inhibits some of the GmDOG1Ls expression while promoting others. Similarly, the correlation between FL and GmDOG1Ls was observed to be the same. In addition, experimental results showed that GA and PAC down-regulated the expression of GmDOG1L genes. The expression of these seven genes had no obvious characters under phytohormone treatment. From this, we speculated that some members of the 40 GmDOG1L may not belong to the GmDOG1 family, and they might not have the function of regulating seed dormancy and germination, thus, their response to phytohormones is inconsistent with the results obtained.
4. Conclusions
In this study, we performed a comprehensive analysis of GmDOG1L gene family, providing a perspective for the evolution of this family. Using DOG1 HMM, we identified 40 GmDOG1Ls from soybean genome. These GmDOG1Ls were distributed on 18 chromosomes of soybean and were divided into five subgroups according to their evolutionary relationship. We found that GmDOG1Ls’ motifs are similar. Gene duplication analysis indicated that the WGD or segmental duplications might lead to the expansion of GmDOG1L family. Expression profile analysis showed that GmDOG1-L11, GmDOG1-L27, GmDOG1-L30 and GmDOG1-L37 had the highest expression in seeds compared to other GmDOG1Ls. The precise correlation between GmDOG1L genes expression and ABA or FL was not detected, but the expression of GmDOG1Ls was inhibited under GA and PAC treatments. More importantly, we identified some potentially useful GmDOG1L genes through this study, such as GmDOG1-L11, GmDOG1-L37 and GmDOG1-L40. Not only were they found closer to identified DOG1-like in phylogenetic tree analysis, but their motif compositions were also similar to identified DOG1-like compositions, and their expression in seeds was the highest in qRT-PCR assay. Among them, we believe that GmDOG1-L37 is the most likely gene to become potential GmDOG1. It was not only found closest to AtDOG1 in phylogenetic analysis, but also had the highest expression in seeds in qRT-PCR analysis. Transcriptome data also confirmed that its expression continues to increase during seed development and their promoter also contained three ABRE elements. Therefore, we believe that GmDOG1-L37 might have the greatest potential to become GmDOG1 gene in soybean. These results provide a basis for further understanding the molecular functions of GmDOG1L family and the specific mechanisms of GmDOG1L family in regulating seed dormancy and germination.
5. Materials and Methods
5.1. Identification of GmDOG1Ls
The soybean genome and protein sequences were downloaded from Phytozome12 website (http://www.phytozome.net/). The Hidden Markov Model (HMM) of conservative structure domain of DOG1 (PF14144) was downloaded from the PFAM database (http://pfam.xfam.org/) [13,31,32]. Predicted GmDOG1Ls were scanned with HMMER 3.1 software using the HMM of conserved domain of DOG1 [33,34]. We then used those protein sequences (E-value < 0.001) to construct a new HMM model of soybean using HMMER 3.1 software. This new soybean-specific HMM was used to identify GmDOG1Ls (E-value < 0.001). In order to ensure the accuracy of the results, 40 GmDOG1Ls proteins sequence were submitted on the NCBI website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for further verification [35].
5.2. Chromosomal Distribution, Gene Structure and Phylogenetic Analysis
The MapGene2Chrom (http://mg2c.iask.in/mg2c_v2.0/) website was used to draw a visual distribution map of GmDOG1Ls on the chromosomes [36]. The exon and intron distribution patterns of GmDOG1Ls were analyzed by Gene Structure Display Server (GSDS2.0: http://gsds.cbi.pku.edu.cn/) [37]. Conserved motifs of GmDOG1Ls were predicted by MEME 5.0.5 software (search parameters: Maximum Number of Motifs:10; Motif E-value Threshold: no limit; Minimum Motif Width: 6; Maximum Motif Width: 50; Minimum Sites per Motif: 2; Maximum Sites per Motif: 50) [38]. Multiple sequence alignments of GmDOG1Ls were analyzed by ClustalW [39]. Phylogenetic analysis was conducted with MEGA 7 software [40]. We used maximum likelihood method to infer the evolutionary relationship of GmDOG1Ls [41]. The bootstrap consensus tree was repeated 1000 times [42]. Figtree 3.1 software was used to analyze and garnish the phylogenetic tree. The theoretical isoelectric point (pI) and molecular weight (MW) of GmDOG1Ls were predicted through ExPASy proteomics server (https://web.expasy.org/protparam/) [43].
5.3. Synteny Analysis of GmDOG1Ls
Duplication events for GmDOG1Ls was performed using the Multiple Collinearity Scan toolkit X (MCScanX) program with the default parameters [44]. According to the results, KaKs-Calculator software was used to calculate the non-synonymous replacement rate (Ka) and synonymous replacement rate (Ks) of duplication genes [45]. We used the Circos tool to draw the positions of genes and segmental duplicated regions on the soybean chromosomes [46,47].
5.4. Gene Expression Analysis
As previously described, we performed the total RNA preparation and first-strand cDNA synthesis as well as a qRT-PCR assay [48]. Total RNA was treated with DNase I, and then 2 μg total RNA was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (200 units per reaction; Promega Corporation), according to the manufacturer’s protocol. The soybean housekeeping gene GmTubulin was used as endogenous reference gene, and each reactions were repeated three times [3].
The qRT-PCR reaction system was 10 μL, which included: 0.4 μL forward primer and reverse primer, 3.6 μL DNase-free ddH2O, 1 μL cDNA and 5 μL Vazyme™ AceQ qPCR SYBR Green Mastermix. The qRT-PCR reaction procedure was set as follows: 94 °C for 2 min 30 s, and then 40 cycles of 94 °C for 10 s and 60 °C for 32 s. Each experiment values represent three biological replicates. The qRT-PCR performed using Vazyme™ AceQ qPCR SYBR Green Master mix on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA) [3]. The expression level of GmDOG1Ls were calculated by the comparative CT method [49]. Online primers were designed using NCBI primer design tool site (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). The detailed information of the primers is shown in Table S4.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/8/937/s1, Figure S1: Phylogenetic analysis of DOG1Ls from soybean, Arabidopsis, Hordeum vulgare, Triticum aestivum and Oryza sativa, Figure S2: Phylogenetic analysis and exon-intron organization of DOG1Ls, Figure S3: Cis-acting element of GmDOG1Ls, Figure S4: Distribution of GmDOG1Ls on soybean chromosomes. Forty GmDOG1L genes were located in 18 chromosomes. The chromosome 9 and chromosome16 has no GmDOG1L genes, Figure S5: The syntenic relationships among GmDOG1Ls, Table S1: Gene Name and Gene ID of DOG1-Like in four species, Table S2: Information about the duplicated regions of GmDOG1Ls, Table S3: Duplication events of GmDOG1Ls, Table S4: Primers sequence used in this study.
Author Contributions
K.S. and Y.Y. designed the research. Y.Y. and C.Z. performed most of the data analysis and experiments. U.C. performed part of the experiments. L.Y., C.L., T.P., X.W., J.D., J.L., F.Y., T.Y. and W.Y. provided valuable suggestions for this study. K.S., W.L. and Y.Y. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the grants from the National Key Research and Development Program of China (2016YFD0300209) and the National Natural Science Foundation of China (31872804, 31701064). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Lee G.-A., Crawford G.W., Liu L., Sasaki Y., Chen X. Archaeological Soybean (Glycine max) in East Asia: Does Size Matter? PLoS ONE. 2011;6:e26720. doi: 10.1371/journal.pone.0026720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng Y., Chen F., Shuai H., Luo X., Ding J., Tang S., Xu S., Liu J., Liu W., Du J., et al. Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Sci. Rep. 2016;6:22073. doi: 10.1038/srep22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou W., Chen F., Zhao S., Yang C., Meng Y., Shuai H., Luo X., Dai Y., Yin H., Du J., et al. DA-6 promotes germination and seedling establishment from aged soybean seeds by mediating fatty acid metabolism and glycometabolism. J. Exp. Bot. 2018;70:101–114. doi: 10.1093/jxb/ery247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng Y., Shuai H., Luo X., Chen F., Zhou W., Yang W., Shu K. Karrikins: Regulators Involved in Phytohormone Signaling Networks during Seed Germination and Seedling Development. Front. Plant Sci. 2017;7:e0157. doi: 10.3389/fpls.2016.02021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou W., Chen F., Luo X., Dai Y., Yang Y., Zheng C., Yang W., Shu K. A matter of life and death: Molecular, physiological, and environmental regulation of seed longevity. Plant Cell Environ. 2019;43:293–302. doi: 10.1111/pce.13666. [DOI] [PubMed] [Google Scholar]
- 6.Shu K., Zhou W., Chen F., Luo X., Yang W. Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2018;9:416. doi: 10.3389/fpls.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentsink L., Jowett J., Hanhart C.J., Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakabayashi K., Bartsch M., Xiang Y., Miatton E., Pellengahr S., Yano R., Seo M., Soppe W.J.J. The Time Required for Dormancy Release in Arabidopsis Is Determined by Delay of Germination1 Protein Levels in Freshly Harvested Seeds. Plant Cell. 2012;24:2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Footitt S., Huang Z., Clay H.A., Mead A., Finch-Savage W.E. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 2013;74:1003–1015. doi: 10.1111/tpj.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huo H., Wei S., Bradford K.J. Delay of Germination1(DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA. 2016;113:e2199–e2206. doi: 10.1073/pnas.1600558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graeber K., Linkies A., Müller K., Wunchova A., Rott A., Leubner-Metzger G. Cross-Species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol. Biol. 2009;73:67–87. doi: 10.1007/s11103-009-9583-x. [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T., et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA. 2010;107:5792–5797. doi: 10.1073/pnas.0911965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashikawa I., Abe F., Nakamura S. DOG1-like genes in cereals: Investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013;208:1–9. doi: 10.1016/j.plantsci.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Née G., Kramer K., Nakabayashi K., Yuan B., Xiang Y., Miatton E., Finkemeier I., Soppe W.J. Delay of Germination1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017;8:72. doi: 10.1038/s41467-017-00113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu K., Qi Y., Chen F., Meng Y., Luo X., Shuai H., Zhou W., Ding J., Du J., Liu J., et al. Salt Stress Represses Soybean Seed Germination by Negatively Regulating GA Biosynthesis While Positively Mediating ABA Biosynthesis. Front. Plant Sci. 2017;8:1372. doi: 10.3389/fpls.2017.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F., Zhou W., Yin H., Luo X., Chen W., Liu X., Wang X., Meng Y., Feng L., Qin Y., et al. Shading of the mother plant during seed development promotes subsequent seed germination in soybean. J. Exp. Bot. 2020;71:2072–2084. doi: 10.1093/jxb/erz553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmutz J., Cannon S.B., Schlueter J.A., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 18.Buggs R.J.A., Elliott N.M., Zhang L., Koh J., Viccini L.F., Soltis U.E., Soltis P.S. Tissue-Specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 2010;186:175–183. doi: 10.1111/j.1469-8137.2010.03205.x. [DOI] [PubMed] [Google Scholar]
- 19.Roulin A., Auer P.L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R.W., Jackson S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2012;73:143–153. doi: 10.1111/tpj.12026. [DOI] [PubMed] [Google Scholar]
- 20.Woodhouse M.R., Pedersen B., Freeling M. Transposed Genes in Arabidopsis Are Often Associated with Flanking Repeats. PLoS Genet. 2010;6:e1000949. doi: 10.1371/journal.pgen.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Wang X., Tang H., Tan X., Ficklin S.P., Feltus F.A., Paterson A.H. Modes of Gene Duplication Contribute Differently to Genetic Novelty and Redundancy, but Show Parallels across Divergent Angiosperms. PLoS ONE. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeling M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 24.Juretic N., Hoen D.R., Huynh M.L., Harrison P.M., Bureau T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh V.K., Jain M. Genome-Wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 2015;6:6. doi: 10.3389/fpls.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh A., Islam T. Genome-Wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016;16:87. doi: 10.1186/s12870-016-0773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Park M., Yeom S.-I., Kim Y.-M., Lee J.M., Lee H.-A., Seo E., Choi J., Cheong K., Kim K.-T., et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014;46:270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 28.Chen F., Yang Y., Luo X., Zhou W., Dai Y., Zheng C., Weiguo L., Yang W., Shu K. Genome-Wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 2019;19:269. doi: 10.1186/s12870-019-1861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu K., Liu X.-D., Xie Q., He Z. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant. 2016;9:34–45. doi: 10.1016/j.molp.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Shu K., Zhou W., Yang W. Apetala 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 2017;217:977–983. doi: 10.1111/nph.14880. [DOI] [PubMed] [Google Scholar]
- 31.Sarris P.F., Cevik V., Dagdas G., Jones J.D.G., Krasileva K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016;14:8. doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang T., Peng Y., Rao Y., Li S., Zeng L. Genome-Wide Identification and Expression Analysis of Sugar Transporter (ST) Gene Family in Longan (Dimocarpus longan L.) Plants. 2020;9:342. doi: 10.3390/plants9030342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2013;42:d222–d230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Suppl. 2):w29–w37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016;45:d200–d203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiangtao C., Yingzhen K., Qian W., Yuhe S., Daping G., Jing L., Guanshan L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hered. Beijing. 2015;37:91. doi: 10.16288/j.yczz.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey T.L., Bodén M., Buske F.A., Frith M.C., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Suppl. 2):w202–w208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 43.Li L., Zheng T., Zhuo X., Li S., Qiu L., Wang J., Cheng T., Zhang Q. Genome-Wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume. PeerJ. 2019;7:e7499. doi: 10.7717/peerj.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Tang H., DeBarry J., Tan X., Li J., Wang X., Lee T.-H., Jin H., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Li J., Zhao X.-Q., Wang J., Wong G.K.-S., Yu J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J.M., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Shi S., Zhou Y., Zhou Y., Yang J., Tang X.-Q. Genome-Wide identification and characterization of GRAS transcription factors in sacred lotus (Nelumbo nucifera) PeerJ. 2016;4:e2388. doi: 10.7717/peerj.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu K., Chen Q., Wu Y., Liu R., Zhang H., Wang P., Li Y., Wang S., Tang S., Liu C., et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016;85:348–361. doi: 10.1111/tpj.13109. [DOI] [PubMed] [Google Scholar]
- 49.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.