Abstract
Background
Genomic methods for identifying causative variants for trait loci applicable to a wide range of germplasm are required for plant biologists and breeders to understand the genetic control of trait variation.
Results
We implemented Cas9-targeted sequencing for fine-mapping in apple, a method combining CRISPR-Cas9 targeted cleavage of a region of interest, followed by enrichment and long-read sequencing using the Oxford Nanopore Technology (ONT). We demonstrated the capability of this methodology to specifically cleave and enrich a plant genomic locus spanning 8 kb. The repeated mini-satellite motif located upstream of the Malus × domestica (apple) MYB10 transcription factor gene, causing red fruit colouration when present in a heterozygous state, was our exemplar to demonstrate the efficiency of this method: it contains a genomic region with a long structural variant normally ignored by short-read sequencing technologies
Cleavage specificity of the guide RNAs was demonstrated using polymerase chain reaction products, before using them to specify cleavage of high molecular weight apple DNA. An enriched library was subsequently prepared and sequenced using an ONT MinION flow cell (R.9.4.1). Of the 7,056 ONT reads base-called using both Albacore2 (v2.3.4) and Guppy (v3.2.4), with a median length of 9.78 and 9.89 kb, respectively, 85.35 and 91.38%, aligned to the reference apple genome. Of the aligned reads, 2.98 and 3.04% were on-target with read depths of 180 × and 196 × for Albacore2 and Guppy, respectively, and only five genomic loci were off-target with read depth greater than 25 × , which demonstrated the efficiency of the enrichment method and specificity of the CRISPR-Cas9 cleavage.
Conclusions
We demonstrated that this method can isolate and resolve single-nucleotide and structural variants at the haplotype level in plant genomic regions. The combination of CRISPR-Cas9 target enrichment and ONT sequencing provides a more efficient technology for fine-mapping loci than genome-walking approaches.
Keywords: Causative variant, SNP, Apple, MYB10, Red flesh, Oxford nanopore, QTL cloning
Background
Representatives of most plant species important to primary industries have been fully sequenced following the dramatic reduction of sequencing cost. However, the individuals from which reference genomes have been assembled are often chosen because they are highly homozygous (doubled haploids or inbred lines). Typically, they do not represent the full complement of genes and variability of the germplasm that is used for selective breeding. Indeed, plant breeders often use wild, undomesticated relatives as donors of alleles for pest and disease resistance, as well as novel quality traits. The genomes of these wild relatives are often even further substantially different, in both sequence and structure, from the reference species’ genomes. To address this issue, researchers have focused on cataloguing the most abundant genomic variations present on any genome—single nucleotide polymorphisms (SNPs)—using the reference genomes as guides for aligning (re)sequencing data. SNPs can be readily identified and visualized using SNP arrays or short-read sequencing approaches, such as Whole Genome Sequencing (WGS), or restriction-enzyme-based enrichment approaches, such as Genotyping by Sequencing (GBS) [1] or restriction site associated DNA (RAD) tags [2]. However, a growing body of evidence demonstrates that there are other types of genomic variants [e.g. structural variants (SVs), copy number variations (CNV) and epimutations] that are also directly causative for key traits. These genomic variants can have complex allelic diversity, which is not reliably captured by the typical read length (several hundred bases) of short-read sequencing technologies. Sequence assembly of these areas is hampered by repeats and transposable elements, which is an issue when using short-read sequencing methods.
Not only do the causative variants for key plant breeding trait loci provide ideal markers for efficient selection, but developing an understanding of the nature of the genetic variants may allow similar alleles to be generated by gene-editing methods in species other than the original host, enabling the introduction of valuable new traits into these crops. However, identification of such causative variants has been hampered due to limitations of using SNPs, which do not encode allelic variation in all traits. New methods to identify variants that are applicable to a wide range of germplasm are required. While WGS and alignment to a reference genome approach is possible, this often relies on massive paralleled short-read sequencing, which suffers from low sensitivity (30–70%) and a high false discovery rate (up to 85%) of SVs [3] because they are inferred directly from aberrant short-read alignments based on the identification of an unexpected depth of coverage, or inconsistent orientation or distance between the alignment of paired-end reads [4, 5]. Targeted capture by in-solution hybridization of coding and non-coding regions can be more cost-effective for the identification of variants, especially for very large, repetitive genomes [6, 7]. Capture-based methods, however, can only infer SVs by analysing the depth of coverage of those sequenced regions [8]. Complete de novo haplotype-resolved assembly is considered the best approach for the full characterization of the genetic diversity, however, this method struggles to identify low-copy repeats, also known as segmental duplications [9]. Targeted approaches employing long-read sequencing (LRS) are therefore an attractive alternative. The main advantage of LRS compared with short-read sequencing is that long reads (> 10 kb on average) can be obtained from single DNA molecules in real time without polymerase chain reaction (PCR) amplification. Although long-fragment targeted capture methods have been developed in plants, they are based on in-solution hybridization of predesigned long probes from a reference genome and the enrichment of the targeted regions via post-capture PCR amplification before sequencing [10–13], which generates biases from both the reference genome and the use of PCR. The sequencing of native molecules (both DNA and RNA) based on Oxford Nanopore Technology (ONT) and the preparation of a sequencing library without PCR amplification would be preferable. Such an approach is possible using the CRISPR-Cas9 system.
Clustered regularly interspaced short palindromic repeat (CRISPR) is a bacterial and archaeal defence system against foreign nucleic acids from invading viruses and plasmids, via nucleases, such as Cas9 that target specific sequences [14–16]. The most studied CRISPR system is the type II CRISPR-Cas system from Streptococcus pyogenes, which relies on only a single protein, the nuclease Cas9, and two non-coding RNAs, a 20-bp crRNA and a tracrRNA to target DNA. These two RNAs are fused into an artificial short guide RNA (sgRNA). The combination of the Cas9 protein and the sgRNA produces a Cas9/sgRNA complex, which binds on double-stranded DNA by the match-recognition of the first 17–20 nucleotides of the sgRNA if the target sequence presents a protospacer adjacent motif (PAM). Once bound, both Cas9 cleaves each of the DNA strands independently at three bases upstream of the PAM, generating a blunt and double DNA strand end [17, 18]. In addition to being widely adopted as a tool for genome editing [18–21] and for epigenome remodelling, when dead Cas9 (dCas9) is used [22], CRISPR/Cas9 is also a valuable tool for in vitro DNA cleavage, where it can act as a precise, custom restriction endonuclease, enabling CRISPR methods for targeted sequencing [23].
The use of CRISPR/Cas9 for in vitro DNA cleavage purposes was first reported in 2015 for Cas9-assisted targeting of chromosome segments (CATCH) for cloning of large intact bacterial genomic DNA fragments (up to 100 kb) via Gibson assembly [24], and for transformation-associated recombination (TAR)-cloning in yeast of large human genomic sequences (up to 150 kb) [25]. Optimization of the CATCH protocol was subsequently achieved by performing in-gel cleavage and the separation of the target region from the rest of the genomic DNA by pulse field gel electrophoresis [26, 27]. However, it was the utility of CATCH coupled with targeted ONT sequencing that turned the method into a powerful tool for the isolation, in real time, of very large native nucleotide molecules. This method was first developed to isolate a human breast cancer gene (BRCA1), obtaining sufficient coverage (70 × on average) to characterize variants, albeit only 1% of the reads were on target [28]. Another approach includes the use of dCas9 to ‘pull down’ targeted regions by using streptavidin magnetic beads to attach to the tracrRNA and ligate to ONT adapters [29]. An updated method for effective targeted enrichment was proposed [30], which started with the dephosphorisation of the genomic DNA, followed by targeted CRISPR-Cas9 cleavage, which enabled the specific ligation of ONT adapters to the digested ends. A similar method has been reported for human diagnostic purposes, targeting genes in order to identify variants associated with a range of diseases [31, 32].
Here, we used CRISPR-Cas9 targeted cleavage for the isolation of a region of interest, followed by enrichment and LRS using ONT (Fig. 1). This method does not require sequencing the entire genome and uses techniques and hardware commonly available in a molecular biology laboratory. We validated this method for use in fine-mapping by isolating the causative variant for red fruit coloration in apple (Type 1 red flesh), previously discovered to be a repeated mini-satellite motif located upstream of the MYB10 transcription factor open reading frame [33].
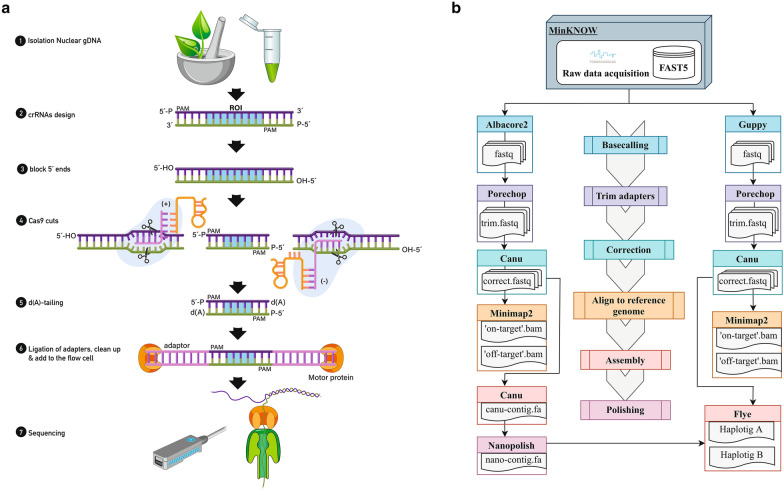
Fig. 1.
a In vitro CRISPR-Cas9 enrichment steps. High molecular weight nuclear DNA is extracted and crRNA probes designed. DNA 5′ ends are dephosphorylated to reduce ligation of sequencing adapters to non-target strands. Cas9 ribonucleoprotein particles (RNPs) bind at each side and cleave the region of interest (ROI). Double strand DNA is cleaved by Cas9 revealing blunt ends with ligatable 5′ phosphates. Cas9 remains bound to the protospacer adjacent motif (PAM)-distal end giving directionality for the strands towards the ROI. All DNA is dA-tailed, preparing the blunt ends for sequencing adapter ligation. ONT adapters are ligated to Cas9 cut sites which are both 3′ dA-tailed and 5′ phosphorylated. Library is cleaned to remove excess adapters using AMPure XP beads. Non-target molecules are not removed from the library. Library is added to the flow cell for sequencing. b Bioinformatics pipeline. Raw FAST5 reads are base called using Albacore2 (v2.3.4, ONT) and Guppy (v3.2.4, ONT) and converted to FASTQ. Reads get adapters trimmed using Porechop (v0.2.3 https://github.com/rrwick/Porechop), corrected using Canu (v1.7) [36] and aligned to the apple reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] using Minimap2 (v2.9) [46] to localize physically the ‘on-target’ and ‘off-target’ enriched regions. A de novo assembly is performed by Canu (v1.7) [36] using Albacore2 (v2.3.4, ONT) corrected reads and polished using Nanopolish (v0.11.1) [37]. Canu_corrected reads and the Canu_nanopolished assembly are used as inputs to run the final assembly performed by Flye (v2.5) [38]
Results
Assessment of guide RNA activity and specificity using an amplicon cleavage test
Four guide RNAs (gRNAs) were designed to test whether CRISPR/Cas9 could specifically excise a 7841 base pair (bp) segment of the Type 1 apple red flesh MYB10 gene locus. Of these, two gRNAs specifically targeted the upstream promotor region and two targeted regions downstream of the gene. The four gRNAs had 100% specificity scores (Table 1) for the production of specific fragment lengths based on the apple reference genome assembly (Malus × domestica ‘Golden Delicious’ double haploid GDDH13v1.1) [34]. Sanger sequencing of PCR products containing the gRNA targets amplified from DNA of the Type 1 red flesh apple accession ABGS0131 was used to assess whether variants of the gRNA target sequences were present.
Table 1.
CRISPR RNA oligonucleotide sequences used for cleavage of genomic DNA in the apple MYB10 locus
| Name | Sequence 5′-3′ | Off-target score (%) | On-target activity score (%) | Chr | Start position | End position |
|---|---|---|---|---|---|---|
| crRNA_RF_1_F | GTCATATCTAAGGACCCGCGTGG | 100 | 76.30 | Chr09 | 35542701 | 35542723 |
| crRNA_RF_2_F | TCTGTACTCCGTCTGTCGGTCGG | 100 | 77.90 | Chr09 | 35542848 | 35542870 |
| crRNA_RF_3_R | AGAAGACTGTCAATCCCGAGTGG | 100 | 79.60 | Chr09 | 35550689 | 35550711 |
| crRNA_RF_4_F | TGTCTGGAAAGTTTCTAACGCGG | 100 | 70.80 | Chr09 | 35551878 | 35551900 |
Cleavage specificity was tested using PCR products (Table 2) from ABGS0131. The cleavage products of the four different gRNAs (Table 1) visualized on a Fragment Analyser (Advanced Analytical Technologies, Inc.) showed the expected fragment sizes after digestion of each amplicon (Fig. 2). There was a SNP present between two of the four gRNAs and the ABGS0131 sequence (the bolded “G” in crRNA_RF_2F: “TCTGTACTCCGTCTGTCGGTCGG” and “T” in crRNA_RF_4F: “TGTCTGGAAAGTTTCTAACGCGG” replaced by “A” in the ABGS0131 sequence). However, since successful digestion of all amplicons was observed regardless of the SNPs, both crRNAs were used for subsequent experiments.
Table 2.
Primer sequences used for PCR amplification of CRISPR/Cas9 target sites of the apple MYB10 locus
| Name | crRNA | Sequence 5′-3′ | Tm | %GC | Start position | End position |
|---|---|---|---|---|---|---|
| Chr9_35542587_F | crRNA_RF_1, crRNA_RF_2 | AACAAGATGATGACGACGTG | 56.2 | 45 | 35542587 | 35542606 |
| Chr9_35542966_R | crRNA_RF_1, crRNA_RF_2 | GATGCACGAACTGATACTGT | 55.6 | 45 | 35542947 | 35542966 |
| Chr9_35550584_F | crRNA_RF_3 | CCCTGTATGCGAAAGACAAT | 55.8 | 45 | 35550584 | 35550603 |
| Chr9_35550962_R | crRNA_RF_3 | AAAAGACCACATGCATGCTG | 57.3 | 45 | 35550943 | 35550962 |
| Chr9_35551563_F | crRNA_RF_4 | TGATTGAATGTCTCCACCA | 53.6 | 42.1 | 35551563 | 35551581 |
| Chr9_35552158_R | crRNA_RF_4 | CACATGTGAGAGAGATTTGC | 54.4 | 45 | 35552158 | 35552177 |
crRNA CRISPR RNA, Tm melting temperature (°C), %GC GC content, start and end positions are in base pairs on apple chromosome 9 (‘Golden Delicious’ double haploid GDDH13v1.1) [34].
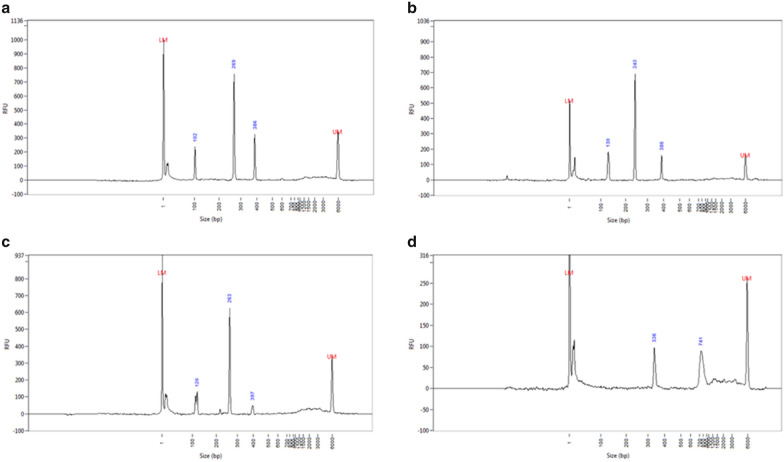
Fig. 2.
Guide RNA Cas9 digestion efficiency in PCR-amplicons linked to the apple MYB10 locus. Fragment analysis evaluating the four gRNAs designed: graphics A to D are for crRNA_RF_1 to 4, respectively. The first two peaks on A–C correspond to both fragments after Cas9 digestion. D shows a single digested fragment because the digestion occurred in the middle of the original PCR amplicon. The longest fragments correspond to undigested original PCR amplicons. Lower marker (LM) set to 1 bp and 6000 bp upper marker (UM) from the high sensitivity NGS fragment analysis kit (1–6000 bp), DNF-474, Advance Analytical
Long-read sequencing of enriched libraries
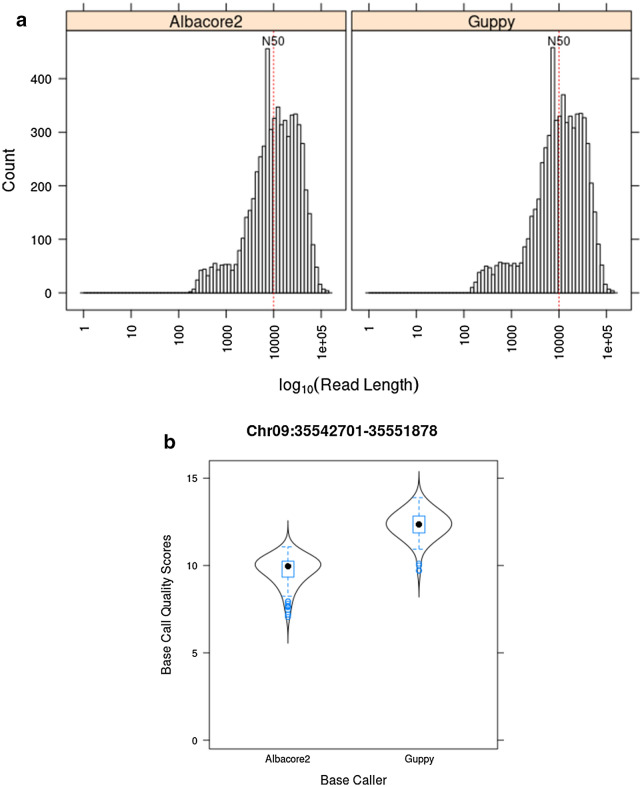
Enriched libraries from ABGS0131 genomic DNA were prepared and sequenced using the ONT Ligation Sequencing Kit (catalog no. SQL-LSK109) and a single ONT MinION flow cell (R.9.4.1). A total of 7056 reads were base-called using Albacore (v2.3.4, ONT) and Guppy (v3.2.4, ONT). The N50 values were similar for both base callers: 30,910 and 30,825 bp for Albacore2 and Guppy, respectively (Table 3). The read length distribution histogram exhibited a high proportion of sequenced reads close to 8 kb in length, corresponding with the targeted locus length of 7841 bp (Fig. 3a). The average read quality distribution ranged from 7.05 to 11.07 and from 9.69 to 13.88, and the median read quality was 9.95 and 12.36 for Albacore2 and Guppy, respectively (Fig. 3b).
Table 3.
Oxford Nanopore Technology sequencing reads base-called using Albacore2 and Guppy
| Base-called reads | Bases | Median read length | N50 | Median read quality | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All | Pass | All | Pass | All | Pass | All | Pass | All | Pass | |
| Albacore2 | 7056 | 6082 | 112578716 | 106589486 | 9783.5 | 11359 | 30416 | 30910 | 9.11 | 9.28 |
| Guppy | 7056 | 6377 | 113972292 | 109479250 | 9891.5 | 11008 | 30635 | 30825 | 11.22 | 11.39 |
Pass reads are those with a mean quality of 7 or more. Software versions: Albacore (v2.3.4, ONT) and Guppy (v3.2.4, ONT).
Fig. 3.
Read length and quality statistics for Oxford Nanopore Technology sequencing. a Read length statistics: log-transformed histogram of read length from base-called reads coming from Albacore (v2.3.4, ONT) and Guppy (v3.2.4, ONT) base callers. Vertical blue line indicates the read N50. b Average quality score distribution of ‘on-target’ reads coming from Albacore2 and Guppy base callers
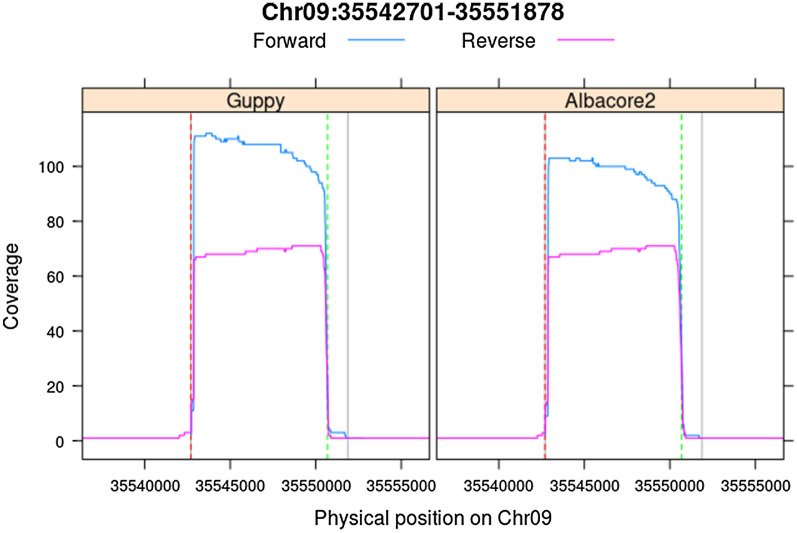
The ONT reads were mapped to the ‘Golden Delicious’ double haploid GDDH13v1.1 apple reference genome [34]. A total of 6022 (85.35%) and 6448 (91.38%) reads were successfully aligned with average mapping qualities of 12.9 and 20.7 using Albacore2 and Guppy, respectively. The percentage of ‘on-target’ reads was 2.98 and 3.04% and the coverage was 180 × and 196 × for Albacore2 and Guppy, respectively (Table 4; Fig. 4). Two of the crRNAs were present in these sequences (crRNA_RF_1F and crRNA_RF_3R), however, the two crRNAs with SNPs (crRNA_RF_2F and crRNA_RF_4F) were not retrieved. In both alignments, reads containing either the R1 (a repeat unit, present in both white- and red-fleshed varieties) or R6 (six repeat units, present in heterozygous state in red-fleshed varieties) promoter alleles of the MYB10 [33] gene could be observed when loaded onto the integrative genome viewer (IGV) [35] without performing any structural variant calling.
Table 4.
Enrichment statistics of Oxford Nanopore Technology sequencing reads against the target apple MYB10 locus
| Aligned reads | On-target reads | On-target coverage | On-target percentage | ||
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| Albacore2 | 6022 | 107 | 73 | 180 | 2.98% |
| Guppy | 6448 | 121 | 75 | 196 | 3.04% |
Total aligned read count against the apple genome (GDDH13v1.1), on-target reads (within 9177 bp on targeted locus), on-target total coverage and percentage.
Fig. 4.
Sequencing coverage plots at the apple MYB10 locus. Physical locations of crRNA_RF_2_F and crRNA_RF_3_R in the targeted locus on apple chromosome 9 (‘Golden Delicious’ double haploid GDDH13v1.1) [34] are shown by the dotted vertical red and green lines respectively. Physical locations of crRNA_RF_1_F and crRNA_RF_4_F are shown by grey vertical lines
Only four genomic regions had ‘off-target’ sequences with a read depth greater than 25 × . These regions, located on chromosomes 0, 14, 15 and 16, had small fragment lengths ranging between 795 and 8496 bp (Table 5). The largest ‘off-target’ region on chromosome 16 was only detected using Guppy base-called reads and spanned two predicted gene models of unknown functions. None of the four crRNA sequences were present in these ‘off-target’ regions (Additional file 1: Table S1 and Additional file 2: Figure S1).
Table 5.
Off-target enrichment statistics of Oxford Nanopore Technology sequencing reads against the apple genome
| Base-caller | Chromosome | Start (bp) | End (bp) | Total size (bp) | Maximum coverage | Annotation |
|---|---|---|---|---|---|---|
| Guppy | Chr00 | 48263504 | 48264621 | 1117 | 28 | within MD00G1202200 |
| Guppy | Chr14a | 30289152 | 30291917 | 2765 | 26 | 2,351 bp upstream of MD14G1221200 |
| Albacore2 | Chr14a | 30289890 | 30291921 | 2031 | 26 | |
| Guppy | Chr15 | 47766093 | 47766888 | 795 | 26 | within MD15G1386400 |
| Guppy | Chr16 | 38910980 | 38919476 | 8496 | 41 | spanning two genes: MD16G1282900 & MD16G1283000 |
Off-targets sites presented had mapping quality equal or higher than 40 × and coverage equal or higher than 25 × .
bp: base pairs
aBoth chromosome 14 regions are the same. The apple reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] was used as reference.
De novo assembly of the Type 1 apple red flesh locus MYB10
A de novo assembly was built using the Albacore2-called reads. The 6105 reads were corrected and assembled using Canu (v1.7) [36] and further improved using Nanopolish (v0.11.1) [37]. The assembly was composed of a total of seven contigs with lengths before polishing of 130,229, 17,146, 14,585, 16,132, 8006, 7927 and, 7197 bp.
Alignment against the apple reference genome showed that five of these contigs had a mapping quality < 6. Two of them had mapped reads with quality ≥ 60. One of these was a contig of 130,229 bp, which mapped partially (120 kb) to chromosome 3, however, this mapping contained many gaps and the maximum coverage was lower than 15 × . Only the 8006 bp contig (hereafter called “canu-contig”) aligned to the target MYB10 locus. The canu-contig contained the complete R6 motif upstream of the MYB10 locus and was flanked by the gRNA target sequences (crRNA_RF_1F and crRNA_RF_3R). After polishing, the canu-contig lost 95 bp in total, which corresponded to the repeat units 3b, 5, 4 and 3a of the R6 motif (Additional file 3: Figure S2). Coverage across the canu-contig had an average and maximum coverage of 128 × and 132 × , respectively (Additional file 4: Figure S3), and only the first 155 bp had a coverage less than 35 × . No other contig mapped to the MYB10 target region, which indicated the 8006 bp of canu-contig was a consensus sequence that had both haplotypes collapsed. Both haplotypes were successfully split by Flye (v2.5) [38] using Canu-corrected reads and the Canu_nanopolished assembly together as input. The two split haplotigs corresponded to both haplotypes of the MYB10 locus in their reverse and complement orientation. The haplotigs obtained were 7915 and 8029 bp long, and contained the full R1 and R6 motif of the MYB10 locus, respectively (Additional file 5: Figure S4) and where identical to the alleles cloned by Espley et al. [33].
Discussion
We have demonstrated the utility of the CRISPR-Cas9 targeted enrichment method followed by Oxford Nanopore Technology LRS for targeted genome sequencing and allele mining in a plant species. Proof-of-concept was demonstrated by retrieving the causative variant for the Type 1 red flesh locus in apple [39], the R6 motif that is located in the promoter region of the MYB10 transcription factor gene and derived from M. × domestica ‘Niedzwetzkyana’. Enrichment was achieved by cleaving previously dephosphorylated nuclear DNA using two Cas9/guide RNA ribonucleoprotein complexes flanking the MYB10 locus and spanning the R6 motif [33]. The cleavage of Cas9 leaves a 5′-phosphate group on DNA ends ensuring that the ligation of ONT sequencing adapters is preferential to the region of interest.
Native DNA strands enriched for the MYB10 locus were sequenced in a single MinION ONT sequencing run without the necessity of performing PCR amplification, allowing the possibility of detecting any modified nucleotides from the raw electrical data. The efficiency of this enrichment protocol was demonstrated by the high percentage of reads mapping to the target region (~ 3% for both base calling software packages) and the high read depth in that region (greater than 180 × for both base calling software packages). The enriched ‘on-target’ region had much higher read depth than off-target regions. The ‘on-target’ region was enriched by just one of the two pairs of crRNAs used in this experiment and further confirmed by examining the sequence content of the contigs constructed by de novo assembly. Regarding the other pair of crRNAs, the low cleavage of crRNA_RF_2_F, was caused by the presence of a SNP that disrupted the target PAM, where the bolded “G” in crRNA_RF_2F: “TCTGTACTCCGTCTGTCGGTCGG” was replaced by “A” in the ABGS0131 sequence. In the case of crRNA_RF_4_F, two factors might have affected its cleavage efficiency. Firstly, the presence of a SNP in the protospacer sequence, which, although being a less severe modification than those occurring at the PAM, may still remove the target locus for ‘off-target’ cleavage. Secondly, the forward orientation of this crRNA that provides directionality for the strands away from the region of interest as Cas9, remains tightly bound after cleavage at the PAM-distal end, blocking the ligation of ONT adapters. With the SNP presence in the protospacer sequence, some strands might have been released from the Cas9′s bond with the result that they were ligated with adapters. ‘Off-target’ reads were distributed randomly across the genome for both base callers (data not shown). None of the four crRNA sequences were found in any ‘off-target’ regions that had a read depth greater than 25 × , indicating that they arose during ligation of ONT adapters to random exposable ends that were not dephosphorylated during the library preparation. One region had a read depth of 41 × and spanned a large fragment (> 8 kb), however this region was not detected when using the Albacore2 (v2.3.4, ONT) base caller software. Remarkably, all sequence reads in the enriched region aligned to the correct chromosome 9 region upstream of the MYB10 gene, however, the reads did not align with the homologous duplicated region on chromosome 17 [40], upstream of MYB110 [41], demonstrating the high fidelity of the technique.
ONT sequencing has been documented as being error-prone, requiring careful error correction, usually through use of high sequencing depth or polishing using more accurate short reads. The read depth obtained by our enrichment method was sufficient to error correct the raw reads by computing k-mer counts in preparation to computing overlaps and producing consensus sequences as performed by the Canu (v1.7) [36] correction step. The first de novo assembly attempt that was performed under standard parameters (see methods) collapsed the two haplotypes of the target region into a single contig. This was expected as most assembly software aims to produce haploid genome assemblies, thus ignoring or collapsing allelic variants together that may be involved in important biological functions. Hence, haplotype phasing becomes a required post-processing assembly step when dealing with diploid or polypoid genomes: a computationally intensive task if allowing for several degrees of divergence within the algorithms in order to maintain haplotype separation and allow for the correct error rate.
Although Cas9-enriched data require much fewer computing resources than that for analysing whole genome sequencing data, we found that Canu took several days to build the first assembly. Therefore, we also used Flye assembler (v2.5) [38], which was able to generate two haplotigs that corresponded with the two haplotypes found at the MYB10 locus on the sequenced Type 1 red flesh accession ABGS0131 in a much shorter run time and without the necessity of enabling the option to retain alternative haplotypes (–keep-haplotypes) included in the latest version of this software.
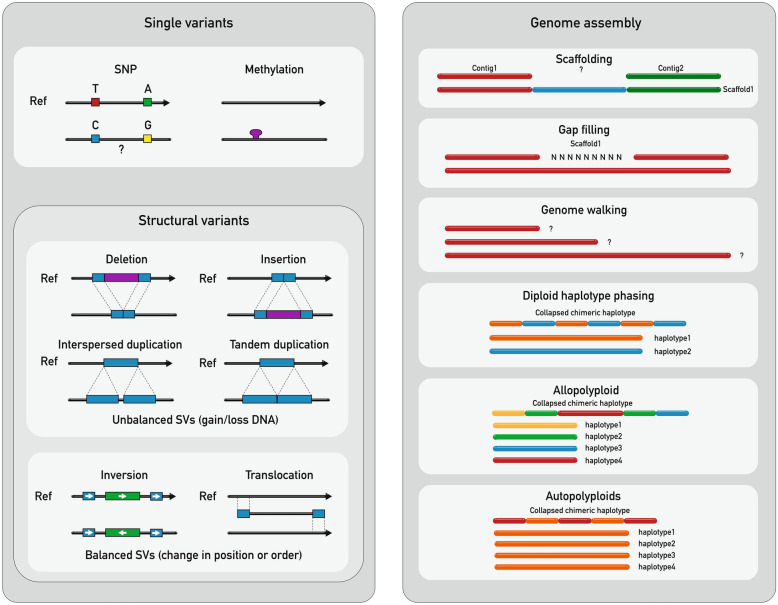
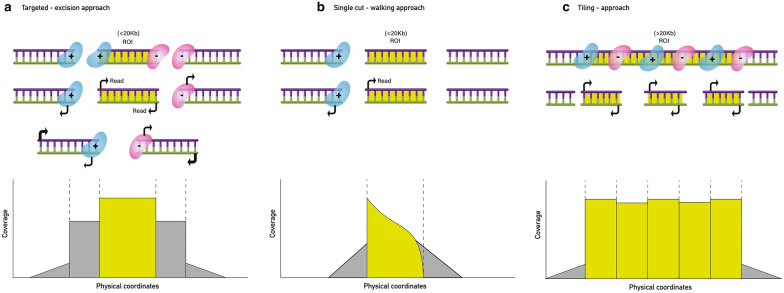
This method has the potential to be widely applied as a tool for identifying single nucleotide variants as a first step in developing trait-associated SNP-based markers for marker-assisted selection. It can also help characterize structural rearrangements, repetitive regions and methylation variants (Fig. 5). The design of crRNA probes to either side of an unknown sequence within a genome scaffold or the design of a single crRNA at one end of a target region (or the end of a scaffold) to read into the unknown could help in the scaffolding or genome walking process by closing sequencing gaps present in most published genome assemblies. A tiling approach could be applied for a deeper characterization of large regions of interest. However, some conserved or syntenic regions, from which crRNA probes could be designed targeting several smaller and overlapping fragments, would need to be known to obtain an even coverage across the region of interest (Fig. 6). In addition, our method could be employed to accurately detect in vitro off-targets mutations induced by Cas-RNP complexes before these are used in vivo for genome-editing purposes.
Fig. 5.
CRISPR-Cas9 targeted enrichment applications. The method can be used to identify single nucleotide genomic and epigenomic variants as well as long structural variants (SVs) (both, balanced and unbalanced SVs) by knowing one or both ends of the region of interest (ROI). It can improve genome assembly contiguity by assisting in the scaffolding and gap filling processes when two ends of the ROI are known or by genome walking approaches when just one side of the ROI is known. Phasing targeted loci of genome assemblies for both diploids and polyploid species is also possible with this technique
Fig. 6.
crRNA design approaches,
adapted from Oxford Nanopore Technology (ONT). a Targeted-excision approach. The crRNAs are designed to either side of the region of interest (ROI). Both ends of the ROI must be known to be able to design the crRNAs. Indicated when the ROI is shorter than 20 kb (specific genes or genomic regions). b Single cut-walking. A single crRNA is designed at one known end of a ROI to walk towards the unknown. Indicated to target a ROI when just one side is known and for genome walking approaches of targeted regions shorter than 20 kb. c Tiling approach. Several crRNAs are designed across a ROI (which can be longer than 20 kb) in 5–10 kb overlapping pieces to get an even coverage along the region. Indicated especially when the length of the input DNA is limited
The major consideration for successfully using this sequence-capture approach is DNA quality.
Our preliminary evaluations found that total genomic DNA isolated using common CTAB extraction methods were unsuitable for CRISPR-Cas9 digestion and enrichment, while high molecular weight nuclear DNA was effective. Although, it involves a few more steps than standard total genomic DNA preparations, it is worth optimizing the protocol to avoid wasting valuable sequence data on organellar DNA. Furthermore, inhibitors affecting DNA digestion and reducing the sequencing of very long reads are removed using this protocol. The other critical aspect for success is efficient DNA digestion by RNP complexes in vitro. It is known that guide RNAs do not always work as efficiently as predicted [42, 43], so we evaluated the activity of RNP complexes to digest upon PCR amplicons. However, an alternative strategy could be to use several RNPs targeting a single region, either within the same digest, or separately, and then pooled for DNA enrichment and sequencing. This approach is likely to be particularly valuable if the CRISPR target sites have not been thoroughly sequenced (e.g. sequencing a locus from many samples), or when there are several haplotypes (e.g. isolating a locus from polyploid species).
Future improvements on our methodology will move into CRISPR-Cas9 multiplexing enrichment. The use of barcodes to index different samples enables a more efficient use of the ONT sequencing flow cells, increasing pore occupancy and therefore expanding the life of pores while sequencing.
We have shown that haplotype phasing is possible with this method due to the high-coverage ONT data. As the base-calling and variant-calling algorithms improve, we also anticipate a broader use of this methodology to phase targeted ONT sequencing reads into parental alleles as well as to determine dosage of specific alleles in polyploid species and diversity within breeding populations.
Conclusion
We demonstrated the efficiency of combining an enriched CRISPR-Cas9 library with ONT sequencing for fine mapping the red flesh locus on the apple physical genome. This method could potentially be used for a broader range of trait and variant types, for example CpG methylation variants. Larger loci may be resolved using very long sequencing reads or by tiling the loci with multiple overlapping cleavage products. In future, several loci may also be multiplexed in one single run and multiplexed individual samples could be used employing barcoded adapters.
Methods
Plant material and DNA isolation
Intact and fully expanded but young leaves (20 g) were collected from a Type 1 red flesh M. × domestica accession (ABGS0131) grown at The New Zealand Institute for Plant and Food Research Limited, Havelock North (New Zealand) [33]. Nuclear genomic DNA was extracted as described at https://www.protocols.io/view/plant-nuclear-genomic-dna-preps-rncd5aw.
Guide RNA design
Functional guide RNA duplexes were built from synthetic CRISPR RNAs (crRNAs; Integrated DNA Technologies (IDT), custom designed) and tracer RNA (tracrRNA, IDT, 1072532) to cut in complementary strands flanking the region of interest. The crRNA sequences (Table 1) were designed using Geneious’ 10.0.9 (https://www.geneious.com) ‘Find CRISPR sites’ tool using the apple reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] as a template for selecting the highest scored on-targets and avoiding off-targets. In addition, a blastn + [44] (command line mode “blastn-short”) search was performed (Additional file 1: Table S1) for the four selected crRNAs against the apple reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] to check the degree of sequence specificity.
From gRNA complexes to Cas9 ribonucleoprotein complexes
All crRNAs were re-suspended and pooled in nuclease-free duplex buffer (IDT, 11010301) to a final concentration of 100 µM. Guide RNA complexes were built by adding equal amounts of pooled crRNAs and tracrRNA in duplex buffer to a final concentration of 10 µM and incubating for 5 min at 95 °C before the reaction was allowed to cool at room temperature for 5 min. Cas9 ribonucleoprotein complexes (RNPs) were assembled by mixing 10 pmol of gRNA duplexes with 10 pmol of Cas9 Nuclease V3 (IDT, 1081058) in 1 × CutSmart buffer (NEB, B7204) to a final volume of 30 µL. The complexes were incubated at room temperature for 30 min, then stored at −20 °C until use.
Cas9 cleavage
The efficiency of Cas9 cleavage was verified by determining the absence of any variant on the crRNA sequences by PCR amplification and Sanger sequencing of cloned amplicons surrounding the region of interest. Primer sequences for amplifying crRNA amplicons can be found in Table 2. PCR reactions were carried out in a 50 µL volume containing 1 × PCR buffer mix (Invitrogen), 100 μM of each dNTP, 1.5 mM MgCl2, 0.5 μM each primer, 0.5 U Platinum™ DNA polymerase (Thermo Fisher Scientific, 10966034) and 20 ng template DNA. Amplifications were carried out on a MasterCycler ProS thermocycler (Eppendorf). The conditions of the touchdown PCR included an initial denaturing at 94 °C for 2 min, then five cycles (94 °C for 55 s, 60 °C for 55 s (decreased 1 °C each cycle), 72 °C for 1 min and 39 s), then 35 cycles (94 °C for 55 s, 55 °C for 55 s and 72 °C for 1 min and 39 s) and a final extension at 72 °C for 10 min. The Cas9 cleavage reactions were performed using 20 ng of amplicon DNA, 2 µL of Cas9-RNP complex specific to that amplicon, 2 µL of 10 × NEB buffer 3 and 12 µL of DNA-free water. Cleaved fragments of the PCR amplicons were separated by electrophoresis using a 5200 Fragment Analyzer (Advanced Analytical Technologies, Inc.).
A total of 5 µg of high molecular weight nuclear genomic DNA re-suspended in 30 µL of 1 × NEB CutSmart buffer (NEB, B7204) was dephosphorylated using 3 µL of Shrimp Alkaline Phosphatase (rSAP: NEB, M0371S) for 30 min at 37 °C, followed by a heat-inactivation step for 5 min at 65 °C and a cooling step to return the sample to room temperature. The dephosphorylated sample was mixed with 10 µL of pre-built pooled Cas9-RNP complexes, 1 µL of 10 mM dATP (Invitrogen, 10297-018) and 1 µL of Taq polymerase (NEB, M0267) and incubated for 30 min at 37 °C for Cas9 cleavage, followed by 5 min at 72 °C for A-tailing.
Oxford Nanopore Technology sequencing library preparation
AMX adapters from ONT Ligation Sequencing Kit (SQL-LSK109) were ligated to the dA-tailed DNA ends using 20 µL of the following adapter mix: 10 µL of NEBNext Quick T4 DNA Ligase (NEB, M2200), 20 µL of Oxford Nanopore Ligation buffer (LNB, ONT, SQL-LSK109) and 3 µL of nuclease-free water. This reaction was incubated for 10 min at room temperature. The ligated sample was purified twice on a magnetic rack using 0.3 × volume of AMPure XP Beads (Becman Coulter, A63881) and 250 µL of long-fragment buffer (ONT, SQL-LSK109). Elution of the ligated sample was performed by adding 13 µL of elution buffer (ONT, SQL-LSK109), incubating for 10 min at room temperature and placing the tube back on the magnetic rack to collect 12 µL of the eluate. The Oxford Nanopore DNA sequencing library was then prepared by adding 25 µL of sequencing buffer (ONT, SQL-LSK109) and 13 µL of loading beads (ONT, SQL-LSK109) to the eluate. Flow cell priming mix was prepared by placing 30 µL of flush tether (ONT, SQL-LSK109) into a tube of flush buffer (ONT, SQL-LSK109).
Oxford Nanopore sequencing
The sample library was run on a 9.4.1 version flow cell using an ONT MinION sequencer using MinKNOW software (ONT). Prior to loading the DNA library, the flow cell was primed by drawing back 230 µL of the buffer from the priming port followed by the addition of 200 µL of the priming mix. The DNA library was then added via the SpotON sample port in a dropwise fashion.
Sequencing data analysis and alignment against the apple reference genome
Nanopore 9.4.1 raw FAST5 reads were base called using Albacore (v2.3.4, ONT) and Guppy (v3.2.4, ONT) and converted to FASTQ format. The quality of base-called sequencing reads was assessed using PycoQC (v2.2.3.3) [45]. Adapters were trimmed from read ends using Porechop (v0.2.3 https://github.com/rrwick/Porechop). Reads were corrected by using Canu’s (v1.7) [36] command ‘canu-correct’ and aligned to the apple reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] using Minimap2 (v2.9) [46]. Alignments were observed using the integrative genome viewer (IGV) [35]. Samtools (v1.9) [47] depth and bedtools (v2.27.1) [48] coverage were used to determine the per-nucleotide coverage. Coverage clustering was performed using the ‘bincov’ function from SURVIVOR (v1.0.7) [49] tool package and plotted using a custom-based Rmarkdown script found in https://github.com/PlantandFoodResearch/ONT_CRISPR-Cas9_enrichment.
De novo assembly
The de novo assembly pipeline followed is shown in Fig. 1. The first draft de novo assembly was performed by the Canu (v1.7) assembler [36] using Albacore2 (v2.3.4, ONT) corrected reads as input. The command used was ‘nanopore-corrected’ with of 200 kb of genome length estimated based on the total length of loci that were aligned against the reference genome (‘Golden Delicious’ double haploid GDDH13v1.1) [34] and the possibility of getting de novo assembled loci. This draft assembly was further improved using Nanopolish (v0.11.1) software [37], which created an index to link raw reads with their signal-level data in the FAST5. Then, raw indexed reads were aligned to the draft assembly producing a consensus polished variant calling file (VCF) that was used to call true positive variants using a minor allele frequency of 10% to get the first Canu_nanopolished assembly. Subsequently, Canu_corrected reads and the Canu_nanopolished assembly were used as inputs to run the final assembly performed by Flye (v2.5) [38]. We evaluated the sequence and structural similarity of these assemblies using dnadiff wrapper for nucmer software [50].
Characterization of haplotypes on enriched targeted loci and off-targets
Both Albacore2 and Guppy corrected reads were aligned by Minimap2 (v2.9) [46] against: (1) the Canu_nanopolished assembly to identify the contig containing the target locus; (2) Chr09 of the apple reference ‘Golden Delicious’ double haploid GDDH13v1.1 genome [34] to characterize the variants and mutations between the red-fleshed accession and ‘Golden Delicious’; and (3) Chr17 from apple reference genome GDDH13v1.1 [34] to ascertain whether the reads aligned to Chr09’s homologous chromosome.
Supplementary information
Additional file 1: Table S1. Location of crRNAs sequences on the apple reference genome. Blastn+ hits of crRNAs in ‘Golden Delicious’ double haploid GDDH13v1.1 [34]. qseq: query sequence name; sseq:subject sequence name; pident: percentage of identical matches; qcovs: query coverage per subject; length: alignment length; mism: mismatches; gap: gap openings; qstart: start alignment in query; qend: end alignment query; sstart: start of alignment in subject; send: end of alignment in subject; sframe: subject frame; evalue: expected value; bitscore: bit score; qseq: aligned part of query sequence. Extended explanation of features at “Fassler CPJ. BLAST(R) help. Bethesda, MD: National Center for Biotechnology Information (US). https://www.ncbi.nlm.nih.gov/books/NBK62051/.”.
Additional file 2: Figure S1. Location of crRNAs sequences in relation to the apple ‘type 1’ red flesh locus and the MYB10 candidate gene. Pink arrows represent the location of crRNA_RF_1_F and crRNA_RF_3_R sequences on R1 (found in both white and red-fleshed apple varieties) and the R6 promoter (showing the repeat units found on the ‘Type 1’ red fleshed allele of the MYB10 gene) of the M. x domestica accession (ABGS0131) studied here, as previously shown in [33]. The vertical arrow indicates the ATG start side of the MYB10 gene.
Additional file 3: Figure S2. Pair-wise alignment between contig sequences generated by Canu assembler (v1.7) [36] and after polishing it by Nanopolish (v0.11.1) [37]. In pink is highlighted the protospacer adjacent motif (PAM) site and the 3 bp upstream of the PAM site of crRNA_RF_1_F where Cas9 performed the cleavage (^).The PAM site of crRNA_RF_3_R and one upstream bp before the cleavage site is highlighted in blue. In yellow is highlighted the sequence of crRNA_RF_2_F is highlighted in yellow. The nucleotide differences between both contigs are highlighted in green. The repeat units present in R1 and R6 promoter allele of MYB10 gene are highlighted in grey tones.
Additional file 4: Figure S3. Per base coverage plot of the alignment of canu-corrected reads against the de novo assembled canu-contig.
Additional file 5: Figure S4. Pair-wise alignment between haplotig A and haplotig B sequences generated by Flye assembler (v2.5) [38]. The protospacer adjacent motif (PAM) is site and the 3 bp upstream of the PAM site of crRNA_RF_1_F where Cas9 performed the cleavage (^) are highlighted in pink. The PAM site of crRNA_RF_3_R and one upstream bp before the cleavage site are highlighted in blue. The sequence of crRNA_RF_2_F is highlighted in yellow. The nucleotide differences between the contigs are highlighted in green. The repeat units present in R1 and R6 promoter allele of MYB10 gene are highlighted in grey tones. The sequences referred in line numbers preceded by (-) or (+) were not included in the de novo assembled contigs shown here.
Acknowledgements
We thank Deepa Bowatte and Giulia Pasqualetto for their help with plant material collection and Sue Gardiner for helpful advice and critical reading of the manuscript.
Authors’ contributions
ELG, NWA, SJT, and DC conceived and designed the work. ELG, CK, EH and MES prepared the sample and constructed libraries. ELG, MWD and SJT designed and performed the analysis. ELG, NWA, MWD, SJT and DC wrote and prepared the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the New Zealand Ministry of Business, Innovation and Employment (MBIE) Strategic Science Investment Fund (SSIF) allocated to Plant & Food Research (‘Discovery Science’ project).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional information files. Sequencing data from the plant sample used in this study are available from SRA, under the BioProject ID PRJNA629607.
The code and documentation used for the analysis of the data can be found in https://github.com/PlantandFoodResearch/ONT_CRISPR-Cas9_enrichment.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena López-Girona, Email: elena.lopez-girona@plantandfood.co.nz.
Marcus W. Davy, Email: Marcus.Davy@plantandfood.co.nz
Nick W. Albert, Email: Nick.Albert@plantandfood.co.nz
Elena Hilario, Email: Elena.Hilario@plantandfood.co.nz.
Maia E. M. Smart, Email: maia@smart.co.nz
Chris Kirk, Email: Chris.Kirk@plantandfood.co.nz.
Susan J. Thomson, Email: Susan.Thomson@plantandfood.co.nz
David Chagné, Email: David.Chagne@plantandfood.co.nz.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13007-020-00661-x.
References
- 1.Rowan BA, Seymour DK, Chae E, Lundberg DS, Weigel D. Methods for genotyping-by-sequencing. In: White S, Cantsilieris S, editors. Genotyping. Methods in molecular biology. New York, NY: Springer New York; 2017. pp. 221–242. [DOI] [PubMed] [Google Scholar]
- 2.Jamann TM, Balint-Kurti PJ, Holland JB. QTL mapping using high-throughput sequencing. In: Alonso J, Stepanova A, editors. Plant functional genomics. Methods in molecular biology. New York, NY: Springer; 2015. pp. 257–285. [DOI] [PubMed] [Google Scholar]
- 3.Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19(6):329–346. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 4.Yao R, et al. Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data. Mol Cytogenet. 2017;10(1):30. doi: 10.1186/s13039-017-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huddleston J, Eichler EE. An incomplete understanding of human genetic variation. Genetics. 2016;202(4):1251–1254. doi: 10.1534/genetics.115.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill CB, et al. Targeted enrichment by solution-based hybrid capture to identify genetic sequence variants in barley. Sci Data. 2019;6(1):12. doi: 10.1038/s41597-019-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronn R, et al. Targeted enrichment strategies for next-generation plant biology. Am J Bot. 2012;99(2):291–311. doi: 10.3732/ajb.1100356. [DOI] [PubMed] [Google Scholar]
- 8.De Coster W, Van Broeckhoven C. Newest methods for detecting structural variations. Trends Biotechnol. 2019;37(9):973–982. doi: 10.1016/j.tibtech.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Chaisson MJP, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10(1):1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bethune K, et al. Long-fragment targeted capture for long-read sequencing of plastomes. Appl Plant Sci. 2019;7(5):e1243–e1243. doi: 10.1002/aps3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andermann T, et al. A guide to carrying out a phylogenomic target sequence capture project. Front Genet. 2020;10:1407. doi: 10.3389/fgene.2019.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giolai M, Paajanen P, Verweij W, Witek K, Jones JDG, Clark MD. Comparative analysis of targeted long read sequencing approaches for characterization of a plant’s immune receptor repertoire. BMC Genomics. 2017;18(1):564. doi: 10.1186/s12864-017-3936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van de Weyer A-L, et al. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell. 2019;178(5):1260–1272.e14. doi: 10.1016/j.cell.2019.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mojica FJM, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 15.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 16.Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014;2(2):59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shola DTN, Yang C, Kewaldar V-S, Kar P, Bustos V. New additions to the CRISPR toolbox: CRISPR-CLONInG and CRISPR-CLIP for donor construction in genome editing. Cris J. 2020;3(2):109–122. doi: 10.1089/crispr.2019.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brezgin S, Kostyusheva A, Kostyushev D, Chulanov V. Dead Cas systems: types, principles, and applications. Int J Mol Sci. 2019;20(23):6041. doi: 10.3390/ijms20236041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loose M. Finding the needle: targeted Nanopore sequencing and CRISPR-Cas9. Cris J. 2018;1(4):265–267. doi: 10.1089/crispr.2018.29028.mlo. [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Zhao X, Gabrieli T, Lou C, Ebenstein Y, Zhu TF. Cas9-assisted targeting of chromosome cloning of large gene clusters. Nat Commun. 2015;6:1–8. doi: 10.1038/ncomms9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee NCO, Larionov V, Kouprina N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015;43(8):e55–e55. doi: 10.1093/nar/gkv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Zhu TF. Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments. Nat Protoc. 2016;11(5):960–975. doi: 10.1038/nprot.2016.055. [DOI] [PubMed] [Google Scholar]
- 27.Bennett-Baker PE, Mueller JL. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 2017;45(19):e165. doi: 10.1093/nar/gkx749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabrieli T, Sharim H, Fridman D, Arbib N, Michaeli Y, Ebenstein Y. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH) Nucleic Acids Res. 2018;46(14):e87–e87. doi: 10.1093/nar/gky411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilpatrick T. Cas9 targeted enrichment for nanopore profiling of methylation at known cancer drivers. 2018. https://nanoporetech.com/resource-centre/cas9-targeted-enrichment-nanopore-profiling-methylation-known-cancer-drivers.
- 30.Gilpatrick T, Lee I, Graham JE, Raimondeau E, Bowen R, Heron A, Downs B, Sukumar S, Sedlazeck FJ, Timp W. Targeted nanopore sequencing with Cas9 for studies of methylation, structural variants and mutations. Nat Biotechnol. 2019;38(4):433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson CM, et al. Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications. Lab Invest. 2020;100(1):135–146. doi: 10.1038/s41374-019-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilpatrick T, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat Biotechnol. 2020;38(4):433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espley RV, et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell Online. 2009;21(1):168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daccord N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Publ Gr. 2017;49(7):1099–1106. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 35.Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77(21):e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only Nanopore sequencing data. Nat Methods. 2015;12:733. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 38.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 39.Chagné D, et al. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics. 2007;8:212. doi: 10.1186/1471-2164-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velasco R, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 41.Chagné D, et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013;161(1):225–239. doi: 10.1104/pp.112.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G, Zhang Y, Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput Struct Biotechnol J. 2020;18:35–44. doi: 10.1016/j.csbj.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naim F, et al. Are the current gRNA ranking prediction algorithms useful for genome editing in plants? PLoS ONE. 2020;15(1):e0227994. doi: 10.1371/journal.pone.0227994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leger A, Leonardi T. pycoQC, interactive quality control for Oxford Nanopore Sequencing. J Open Source Softw. 2019;4:2–5. [Google Scholar]
- 46.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durbin R, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeffares DC, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8(1):14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50(4):488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Location of crRNAs sequences on the apple reference genome. Blastn+ hits of crRNAs in ‘Golden Delicious’ double haploid GDDH13v1.1 [34]. qseq: query sequence name; sseq:subject sequence name; pident: percentage of identical matches; qcovs: query coverage per subject; length: alignment length; mism: mismatches; gap: gap openings; qstart: start alignment in query; qend: end alignment query; sstart: start of alignment in subject; send: end of alignment in subject; sframe: subject frame; evalue: expected value; bitscore: bit score; qseq: aligned part of query sequence. Extended explanation of features at “Fassler CPJ. BLAST(R) help. Bethesda, MD: National Center for Biotechnology Information (US). https://www.ncbi.nlm.nih.gov/books/NBK62051/.”.
Additional file 2: Figure S1. Location of crRNAs sequences in relation to the apple ‘type 1’ red flesh locus and the MYB10 candidate gene. Pink arrows represent the location of crRNA_RF_1_F and crRNA_RF_3_R sequences on R1 (found in both white and red-fleshed apple varieties) and the R6 promoter (showing the repeat units found on the ‘Type 1’ red fleshed allele of the MYB10 gene) of the M. x domestica accession (ABGS0131) studied here, as previously shown in [33]. The vertical arrow indicates the ATG start side of the MYB10 gene.
Additional file 3: Figure S2. Pair-wise alignment between contig sequences generated by Canu assembler (v1.7) [36] and after polishing it by Nanopolish (v0.11.1) [37]. In pink is highlighted the protospacer adjacent motif (PAM) site and the 3 bp upstream of the PAM site of crRNA_RF_1_F where Cas9 performed the cleavage (^).The PAM site of crRNA_RF_3_R and one upstream bp before the cleavage site is highlighted in blue. In yellow is highlighted the sequence of crRNA_RF_2_F is highlighted in yellow. The nucleotide differences between both contigs are highlighted in green. The repeat units present in R1 and R6 promoter allele of MYB10 gene are highlighted in grey tones.
Additional file 4: Figure S3. Per base coverage plot of the alignment of canu-corrected reads against the de novo assembled canu-contig.
Additional file 5: Figure S4. Pair-wise alignment between haplotig A and haplotig B sequences generated by Flye assembler (v2.5) [38]. The protospacer adjacent motif (PAM) is site and the 3 bp upstream of the PAM site of crRNA_RF_1_F where Cas9 performed the cleavage (^) are highlighted in pink. The PAM site of crRNA_RF_3_R and one upstream bp before the cleavage site are highlighted in blue. The sequence of crRNA_RF_2_F is highlighted in yellow. The nucleotide differences between the contigs are highlighted in green. The repeat units present in R1 and R6 promoter allele of MYB10 gene are highlighted in grey tones. The sequences referred in line numbers preceded by (-) or (+) were not included in the de novo assembled contigs shown here.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional information files. Sequencing data from the plant sample used in this study are available from SRA, under the BioProject ID PRJNA629607.
The code and documentation used for the analysis of the data can be found in https://github.com/PlantandFoodResearch/ONT_CRISPR-Cas9_enrichment.