Abstract
Competition for scarce water resources and the continued effects of global warming exacerbate current constraints on potato crop production. While plants’ response to drought in above-ground tissues has been well documented, the regulatory cascades and subsequent nutritive changes in developing tubers have been largely unexplored. Using the commercial Canadian cultivar “Vigor”, plants were subjected to a gradual drought treatment under high tunnels causing a 4 °C increase in the canopy temperature. Tubers were sampled for RNAseq and metabolite analysis. Approximately 2600 genes and 3898 transcripts were differentially expressed by at least 4-fold in drought-stressed potato tubers, with 75% and 69% being down-regulated, respectively. A further 229 small RNAs were implicated in gene regulation during drought. Expression of several small RNA clusters negatively correlated with expression of their six target patatin genes, suggesting involvement in the regulation of storage proteins during drought. The comparison of protein homologues between Solanum tuberosum L. and Arabidopsis thaliana L. indicated that down-regulated genes were associated with phenylpropanoid and carotenoid biosynthesis. As is indicative of reduced flow through the phenylpropanoid pathway, phenylalanine accumulated in drought-stressed tubers. This suggests that there may be nutritive implications to drought stress occurring during the potato tuber bulking phase in sensitive cultivars.
Keywords: crop genetics, Solanum tuberosum, abiotic stress, phenylpropanoids, essential amino acid, transcriptome, small RNA, comparative genomics, nutrition
1. Introduction
Potatoes are the fourth most consumed food crop worldwide and are an efficient source of energy, vitamins and minerals in the human diet [1]. High consumption rates and moderate concentrations of dietary antioxidants have led potatoes to be the third-largest source of total phenolics in the American diet [2]. Diets rich in phenolics have been implicated in the prevention of an array of degenerative diseases and concentrations of these compounds vary greatly based on the cultivar, highlighting the potential for the targeted breeding of potato (S. tuberosum L.) to enhance global human health [3]. In potato tubers, the primary polyphenol is chlorogenic acid with the remaining components comprised of carotenoids, anthocyanins, and flavonoids. While the phenolic content of tubers is largely genotype-dependent, the phenolic profiles are driven by the environmental conditions present during growth, tuber bulking, and throughout storage. In general, the production and accumulation of these compounds are favored at lower temperatures, with potatoes grown in warm, dry regions producing lower amounts of phenolics [4]. Environmental parameters could be manipulated to manage the concentrations of desired phytonutrients [4,5,6].
Substrate entry into the general phenylpropanoid pathway in eudicots is driven by phenylalanine ammonia-lyase (PAL), an enzyme that regulates the deamination of phenylalanine to yield the cinnamic acid from which monolignols, flavonoids, and anthocyanins are produced [7]. PAL activity responds to a variety of developmental and environmental cues, with transcriptional regulation occurring by way of MYB, LIM, and KNOX transcription factors [8]. Furthermore, independent MYB transcription factors play a prominent role in the regulation of anthocyanin and flavonoid biosynthesis genes such as flavonol synthase (FLS), flavanone 3-hydroxylase (F3H) and flavonoid 3′–hydroxylase (F3′H), while expression of genes such as dihydroflavonol reductase (DFR) require MYB transcriptional complexes. MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) have notable functions in the regulation of phenylpropanoid biosynthesis in eudicots through the targeting of MYBs, the most notable being miR858, miR828 and TAS4 [7,9]. Causal miRNA and the target MYB transcription factors have been previously identified in potato leaves under drought [10], however, the regulatory cascades present in potato tubers are still unknown.
In addition to phenolics, concentrations of available essential amino acids affect the nutritional value of potato tubers. Up to 50% of the amino acids in tubers are aspartic acid and glutamic acid, with the remaining portion made up of leucine, valine, alanine, lysine, and arginine with the total protein nutritional value of a potato being comparable to an egg white [11]. Genotypes for improving protein quality have been identified among non-traditional potato cultivars [12]. Essential amino acids function both as substrates for secondary metabolism and as a source of energy [13]. As a result, concentrations fluctuate in response to environmental stressors due to concurrent protein degradation and de novo synthesis. The transcriptional regulation of amino acid biosynthesis is highly complex and their function during stress response is still unclear [13].
Drought stress is one of the primary concerns for potato production given the projected increases in aridity. Potato is adapted to temperate climates with optimal tuber growth occurring at temperatures between 15 and 20 °C. Temperatures above this range, coupled with periodic drought, have resulted in reduced yields and increased incidences of tuber physiological defects [14]. Symptoms of drought in potato include reduced leaf size, increased chlorophyll content, reduced stomatal conductance, and wilting. However, rooting depth and plant recovery have been shown as the best indicators of plant susceptibility to drought [15,16]. Through comparisons between genotypes with differing tolerance to drought, novel potato drought-responsive genes and transcript markers for drought tolerance in potato leaves have been identified [17,18]. Gene responses in developing potato tubers to drought conditions are not well-documented outside of targeted metabolic pathways [3].
This study aimed at identifying drought-associated changes in developing potato tubers (i.e., tuber bulking phase) and their impacts on nutritional quality. The assessment of transcriptional changes in genes with metabolic functions and quantification of amino acid concentrations aims to guide production and harvest practices when optimizing the nutritional value of the crop. Analysis of small RNAs seeks to identify components of the drought regulatory cascade in potato tubers which, to our knowledge, has yet to be explored.
2. Materials and Methods
2.1. Experimental Design and Plant Growth Conditions
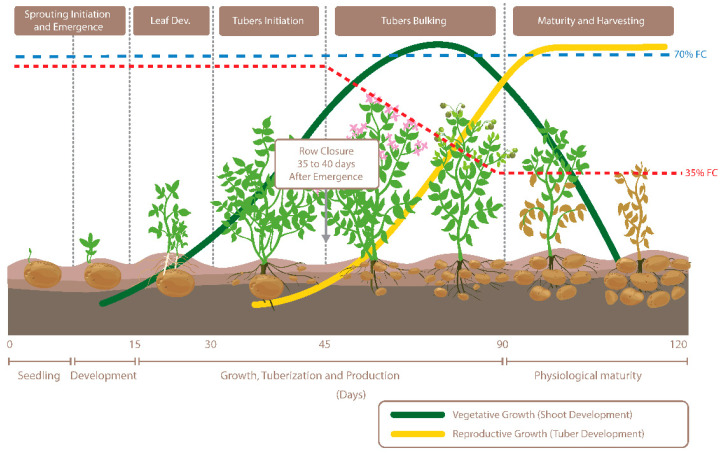
This study was conducted during the summer of 2017 at the Agriculture and Agri-Food Canada’s Canada-Saskatchewan Irrigation Diversification Centre in Outlook (51°29′ N, 107°03′ W, 541 m), Saskatchewan. The cultivar “Vigor”, a cross between “Agria” and “Wischip” from the Agriculture and Agri-Food Canada Lethbridge Research and Development Centre, was evaluated for its performance under soil moisture stress during the tuber bulking phase. Prominent characteristics of the cultivar are its yellow-fleshed tubers and pigmented (red-violet) flowers. Plants were grown under optimum soil moisture conditions at 70% field capacity (FC) and restricted soil moisture conditions at 35% FC under two high tunnels using drip irrigation. Treatments were imposed at the start of the tuber bulking phase for gradual exposure to drought stress, thereby mimicking natural field conditions (Figure 1). High tunnels were opened from all sides but covered with plastic film on top to mimic the open field condition while preventing rainfall (Supplementary Figure S1A). Plots were laid out in a randomized complete block design containing four replicates with guard rows on either side. Each plot consisted of 12 hills. The two end hills were considered as guard hills for yield estimation purposes. Seed pieces were spaced 1 m between-rows and 20 cm within-rows and were planted on 30 May 2017. The crop was raised using standard management practices (i.e., fertility, irrigation, pest control, etc.). Pre-plant basal fertilizer included urea (46-0-0), mono-ammonium phosphate (11-52-0), and potash (0-0-60). Two applications of ammonium sulphate (21.5-0-0-4) were given at 4 and 7 weeks after planting. Soil moisture was monitored using Watermark sensors (Supplementary Figure S1B). Plots were harvested on 2 October 2017 and graded according to commercial-grade standards.
Figure 1.
Visualization of potato growth stages and in-season soil moisture trends when soil moisture was maintained at 35% and 70% field capacity (FC).
2.2. Physiological Measurements
Physiological measurements were taken at the end of the tuber bulking phase (90th day after planting) with readings recorded consistently between 11:30 a.m. and 12:30 p.m. Canopy temperature was assessed with an infra-red thermal imaging camera (FLIR T530, FLIR Systems. Wilsonville, OR, USA), and leaflet chlorophyll content (CCI) was recorded using a chlorophyll content meter (CCM-200- Apogee Instruments, Logan, UT, USA). Quantum yield of dark-adapted leaflets (Fv/Fm) was measured using the portable fluorometer FluorPen FP 100 (PSI, DRASO Czech Republic). Detachable clips were used to dark-adapt the leaflets for 20 min, and Fv/Fm was measured on the adaxial surfaces of the top 3rd and 4th leaflet of each sampled plant (three plants per replication).
2.3. Amino Acid Profiling and Abscisic Acid Content
In both high tunnels from each replication, a tuber was collected from the middle of the plot on 13 September 2017 (106 DAP). The tubers were washed in running water, followed by distilled water, cut into cubes while evading the skin and immediately frozen in liquid nitrogen. Samples were stored at −80 °C until further use.
Amino acids were extracted from 10 mg of ground freeze-dried tissue following Inaba et al. (1994) [19] with some modifications. Briefly, 1 mL of 80% (v/v) ethanol solution (40 °C) was added to each sample, shaken for 30 min at 40 °C and the supernatant was recovered by centrifugation (4000 rpm for 10 min) at 4 °C. The pellets were re-extracted under the same conditions with an additional 500 µL of 80% (v/v) ethanol (40 °C). The supernatants were combined and stored at −20 °C until further use. Amino acids were derivatized following Waters AccQTag Reagent Kit (Waters, Milford, MA, USA; [20]). Briefly, a 10 µL aliquot of sample was mixed with 70 µL borate buffer and 20 µL AccQFluor reagent which was reconstituted in acetonitrile. AccQFluor reagent was reconstituted as follows: 1 mL of AccQFluor reagent diluent was transferred to a vial containing AccQFluor reagent powder and vortexed for 10 s before heating at 55 °C for a maximum of 10 min or until dissolved. The derivatized mixture was transferred to an autosampler vial and incubated at 55 °C for 10 min. High-performance liquid chromatography (HPLC) was conducted, as described in Waters AccQTag’s chemistry package instruction manual, with samples separated on a Waters amino acid column −3.9 × 150 mm and quantified at an excitation wavelength of 285 nm and an emission wavelength of 320 nm using a 2475 scanning fluorescence detector (Waters, Milford, MA, USA). The column was set at 37 °C with a 5 µL injection volume. Waters AccQTag buffer (100 mL AccQTag Buffer concentrate +1000 mL deionized water), acetonitrile, and deionized water were used as mobile phases A, B, and C, respectively.
Abscisic acid content was determined following Yan et al., (2016) [21]. Samples were centrifuged to remove debris, and the pellet was washed twice. The supernatant was evaporated in a SpeedVac, and reconstituted in 1 mL of 1% (v/v) acetic acid. Abscisic acid (ABA) was purified by solid-phase extraction using Oasis HLB, MCX, and WAX cartridge columns (Waters, Milford, MA, USA). The solvent was removed under vacuum and subjected to LC-ESI-MS/MS analysis (Agilent 6410 TripleQuad LC/MS system). An LC (Agilent 1200 series) equipped with a 50 × 2.1 mm, 1.8-μm Zorbax SB-Phenyl column (Agilent Technologies, Santa Clara, CA, USA) was used with a binary solvent system comprised of 0.01% (v/v) acetic acid in water (solvent A) and 0.05% (v/v) acetic acid in acetonitrile (solvent B). Separations were performed using a gradient of increasing acetonitrile content at a flow rate of 0.2 mL min−1. The gradient was increased linearly from 3% B to 50% B over 15 min. The retention time of ABA was 14 min.
2.4. Transcriptome and Small RNA Sequencing
Total RNA was extracted from 100 mg of tuber tissue partitioned from the sample taken for metabolite analysis using the RNeasy plant mini kit (Qiagen, Hilden, Germany). RNA quality and concentration were verified using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). TruSeq RNA and small RNA sequencing libraries were constructed following the standard preparation guide (Illumina, San Diego, CA, USA). All eight RNA samples (four replicates of each treatment) were multiplexed in a lane of a flow cell and paired-end sequencing (125 cycles) was performed using an Illumina HiSeq 2500. Similarly, for small RNA sequencing, all 8 samples were multiplexed in a lane of a flow cell and single-end sequencing was carried out on Illumina HiSeq 2500.
2.5. RNA and Small RNA Read Mapping and Analysis
Before read mapping and expression quantification, all RNA reads were filtered using Trimmomatic (version 0.36; [22]) by (i) removing adapter sequences, (ii) trimming leading and trailing low-quality sequences, (iii) removing sequences when the average quality per base dropped below 15 within a 4-base wide sliding window and (iv) keeping only those pairs where both reads were longer than 75 bp. Clean reads were aligned to the potato reference genome (SolTub_3.0, EnsemblPlants) with STAR (v2.5.2b) and isoform expression was quantified with the RSEM (v1.3.3) algorithm [23]. The expected read counts generated by the RSEM algorithm were rounded off and fed into DESeq2.
The quality of small RNA sequencing reads was assessed using the FASTQC program (v0.11.8; [24]). Reads were quality-filtered and adapter-trimmed using cutadapt (v2.8; [25]). The alignment of filtered reads to the potato reference genome (SolTub_3.0, EnsemblPlants) and annotation and quantification of small RNAs was carried out using ShortStack (v3.8.5; [26]). psRNATarget [27] was used to predict the miRNA and small RNA target genes.
2.6. Differential RNA Expression Analysis
Raw read counts obtained from RNAseq were normalized and assessed for differential expression using the Statistical Software “R” version 3.6.0 and the package DESeq2 [28,29]. Log2 fold change threshold of 2 and a 5% false discovery rate (FDR) were used as cut-off values for continuing to the annotation step. R scripts are available on Bioconductor (https://bioconductor.org/) from the package developers and were adapted for the data presented in this paper. The same technique was repeated for the discovery of differentially expressed small RNA, with target gene identification done using psRNATarget [27]. Gene annotations for S. tuberosum and A. thaliana were obtained from the Ensembl Plants database (http://plants.ensembl.org). Arabidopsis homologs with >50% identity to the original potato gene were input into the online DAVID Bioinformatics Resources version 6.8 (https://david.ncifcrf.gov/) for functional annotation clustering and KEGG pathway mapping analyses.
3. Results
3.1. Physiological Response
A high degree of variability existed within the four control and four treatment plots for the agronomic and physiological traits measured during this study (Table 1). Both crop yield and tuber number per plot were not found to be significantly different from one another, although differences in plot averages were observed. Calculated values from spectral measurements, such as CCI and Fv/Fm, showed no significant differences. However, canopy temperatures measured in plots maintained at 35% FC were 3.9 °C higher than control plots with soil moisture maintained at 70% FC (Table 1). ABA concentrations in the well-watered control and water-deficit conditions were 43.9 and 55.4 ng gDW−1, respectively. Variation was high among the control plants, resulting in no significant differences between treatments.
Table 1.
Agronomic and physiological traits averaged for drought (35% field capacity) and control (70% field capacity) plots with standard error of the mean in parentheses. Bold numbers indicate significance (p < 0.05) between treatments.
| Treatment | Yield (g) | No. of Tubers | CCI | Fv/Fm | Canopy Temp (°C) |
|---|---|---|---|---|---|
| 35% FC | 1381.7 (248.1) | 12.8 (0.8) | 19.5 (0.2) | 0.24 (0.05) | 28.0 (0.3) |
| 70% FC | 1737.3 (198.1) | 18.1 (3.5) | 15.4 (1.6) | 0.38 (0.03) | 24.1 (0.9) |
3.2. Tuber Amino Acid Fluctuations in Response to Soil Moisture Deficit
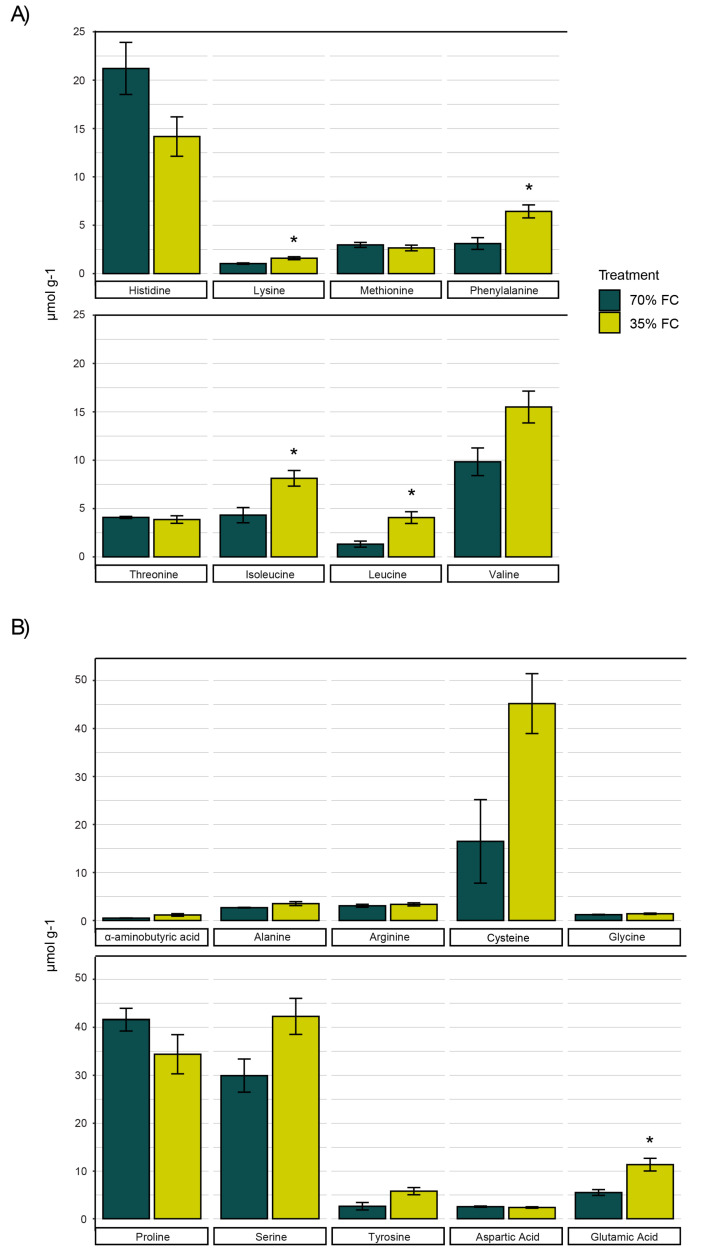
Of the eight essential amino acids, lysine, phenylalanine, isoleucine, and leucine were found to be more abundant in drought-stressed tubers. The largest differences were observed in the concentrations of leucine, phenylalanine, and isoleucine which increased by 3×, 2×, and 1.9×, respectively (Figure 2A). Quantities of branched-chain amino acids, a group that includes leucine, isoleucine, and valine, were therefore significantly higher under drought treatment. Histidine and valine were the most abundant essential amino acids in developing potato tubers (Figure 2A). The majority of non-essential amino acids had similar concentrations in developing tubers regardless of the treatment. Only glutamic acid showed a marked increase of 5.75 µmol g−1 under reduced soil moisture conditions. Concentrations of cysteine, proline, and serine were highest among all amino acids measured (Figure 2B).
Figure 2.
Concentrations of essential (A) and non-essential (B) amino acids in potato tubers (n = 4) sampled during the tuber bulking phase and the associated SEM (p < 0.05) when subjected to 70% and 35% field capacity. * significance (p < 0.05) between treatments.
3.3. Differential Gene Expression and Regulatory Cascades in Developing Tubers Under Drought Stress
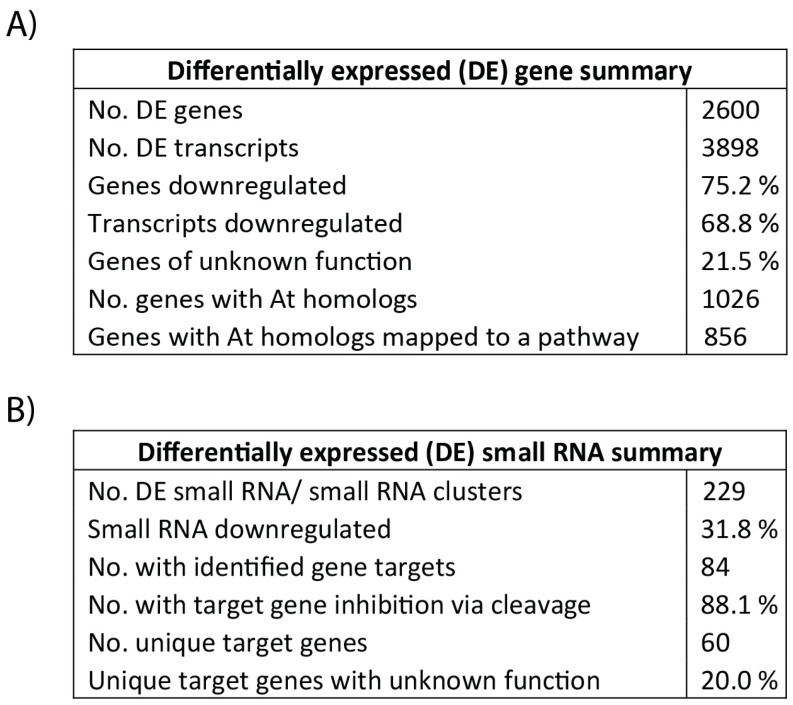
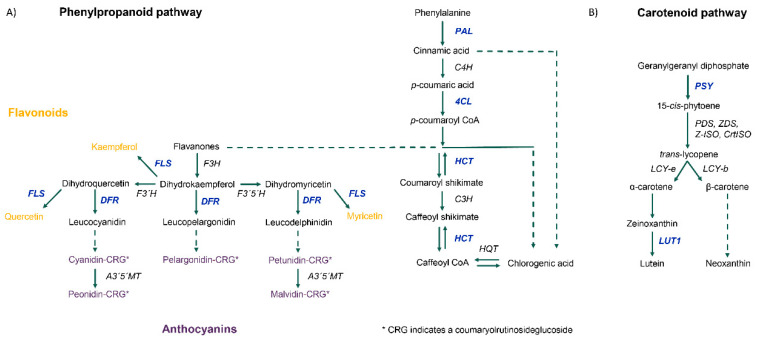
Tubers subjected to soil moisture deficit showed both differential gene and transcript expression, with 75.2% and 68.8% being down-regulated, respectively. One-fifth of genes with differential expression were of unknown function. A full summary of the observed changes is listed in Figure 3A with a list of all differentially expressed genes provided in Supplementary Table S1. Down-regulated genes include those with functions in ABA, auxin and ethylene signaling as well as in auxin, carotenoid, and phenylpropanoid biosynthesis. Up-regulated genes have roles in amino acid biosynthesis, function as molecular chaperones and are involved in ubiquitin-driven proteolysis. Gene names, functional annotation, and the corresponding Arabidopsis homologs used for pathway mapping can be found in Table 2. Focusing on the regulation of the phenylpropanoid pathway, fifteen annotated MYB transcription factors were down-regulated by more than 4-fold under low-soil moisture conditions. Up-regulated transcription factors include two MYB transcription factors and one LIM transcription factor. No other MYB, KNOX or LIM transcription factors were above the cut-off values of 5% FDR and a log-fold change greater than two. Genes with key functions in the phenylpropanoid and carotenoid pathways that were down-regulated in potato tubers at 35% FC are highlighted in Figure 4. Raw read counts for gene expression analysis can be found in Supplementary Table S2.
Figure 3.
Summaries of differentially expressed genes (A) and small RNA (B) in potato tubers at 35% FC using a threshold of 4-fold difference in expression and a 5% FDR. Gene homologs in A. thaliana were considered if identity was greater than 50%. Functional annotation clustering to KEGG pathways was based on Arabidopsis gene IDs using the DAVID Bioinformatics online resource 6.8 (https://david.ncifcrf.gov/).
Table 2.
List of differentially expressed genes in drought-stressed potato tubers, the corresponding homologs in A. thaliana, and the pathway in which they participate. Pathway mapping was done based on the Arabidopsis gene names and similarities between the original S. tuberosum gene and its homolog are expressed as the percentage of identical base pairs in the gene sequences (% ID).
| Gene Regulation | Pathway | Gene Name | Description | Log2 Fold Change | At Homologs | Descriptor | % ID |
|---|---|---|---|---|---|---|---|
| Down- | ABA signaling | PGSC0003DMG400002100 | Abscisic acid receptor PYR1 | −2.07 | AT4G17,870 | PYR1 | 72.2 |
| regulated | AT5G46,790 | PYL1 | 61.0 | ||||
| Auxin | PGSC0003DMG400001589 | Amino acid transporter | −5.30 | AT2G21050 | LAX2 | 86.3 | |
| biosynthesis | PGSC0003DMG400024978 | Indole-3-acetic acid-amido | −5.06 | AT2G14960 | GH3.1 | 77.3 | |
| and signaling | synthetase GH3.3 | AT2G23170 | GH3.3 | 74.1 | |||
| AT4G37390 | GH3.2 | 73.3 | |||||
| AT1G59500 | GH3.4 | 69.8 | |||||
| PGSC0003DMG400024997 | Indole-3-acetic acid-amido synthetase GH3.6 | −2.17 | AT5G54510 | GH3.6 | 70.8 | ||
| PGSC0003DMG400014707 | Flavin monooxygenase | −3.42 | AT4G28720 | YUC8 | 68.3 | ||
| AT5G43890 | YUC5 | 67.2 | |||||
| PGSC0003DMG400026087 | Flavin monooxygenase | −3.09 | AT5G11320 | YUC4 | 57.4 | ||
| AT4G32540 | YUC | 54.3 | |||||
| PGSC0003DMG400003773 | SAUR family protein | −8.34 | AT1G75580 | SAUR51 | 72.2 | ||
| AT1G19830 | SAUR54 | 61.5 | |||||
| PGSC0003DMG400001667 | SAUR family protein | −7.40 | AT4G38860 | SAUR16 | 64.8 | ||
| AT4G34760 | SAUR50 | 64.5 | |||||
| AT2G21220 | SAUR12 | 63.5 | |||||
| AT2G16580 | SAUR8 | 63.0 | |||||
| PGSC0003DMG400001614 | SAUR family protein | −3.75 | AT4G34760 | SAUR50 | 75.7 | ||
| AT4G38860 | SAUR16 | 73.3 | |||||
| AT2G16580 | SAUR8 | 71.3 | |||||
| AT2G21220 | SAUR12 | 71.1 | |||||
| PGSC0003DMG400001668 | SAUR family protein | −3.71 | AT4G38860 | SAUR16 | 77.1 | ||
| AT4G34760 | SAUR50 | 76.6 | |||||
| AT2G21220 | SAUR12 | 75.0 | |||||
| AT2G16580 | SAUR8 | 70.4 | |||||
| PGSC0003DMG400001655 | SAUR family protein | −2.98 | AT4G34750 | SAUR49 | 54.0 | ||
| PGSC0003DMG400022233 | SAUR family protein ARG7 | −2.93 | AT3G12830 | SAUR72 | 64.4 | ||
| AT1G16510 | SAUR41 | 55.1 | |||||
| PGSC0003DMG400001615 | SAUR family protein | −2.06 | AT4G34760 | SAUR50 | 73.8 | ||
| AT4G38860 | SAUR16 | 71.4 | |||||
| AT2G21220 | SAUR12 | 69.2 | |||||
| AT2G16580 | SAUR8 | 68.5 | |||||
| Carotenoid biosynthesis | PGSC0003DMG400028180 | Cytochrome P450-type monooxygenase 97C11 | −2.07 | AT3G53130 | LUT1 | 77.2 | |
| PGSC0003DMG400024063 | Phytoene synthase 1, chloroplastic | −5.07 | AT5G17230 | PSY | 64.3 | ||
| Ethylene signaling | PGSC0003DMG400014204 | Transcription factor TSRF1 | −3.57 | AT3G23240 | ERF1 | 51.4 | |
| Phenylpropanoid | PGSC0003DMG400003605 | Dihydroflavonol 4-reductase | −5.19 | AT5G42800 | DFR | 59.2 | |
| biosynthesis | PGSC0003DMG400014093 | Flavonol synthase | −2.19 | AT5G08640 | FLS1 | 62.5 | |
| AT5G63590 | FLS3 | 50.3 | |||||
| PGSC0003DMG400014152 | Hydroxycinnamoyl transferase | −2.00 | AT5G48930 | HCT | 77.8 | ||
| PGSC0003DMG400023458 | Phenylalanine ammonia- | −4.68 | AT3G10340 | PAL4 | 79.9 | ||
| lyase | AT5G04230 | PAL3 | 73.2 | ||||
| PGSC0003DMG400014223 | 4-coumarate--CoA ligase 2 | −2.30 | AT3G21240 | 4CL2 | 68.5 | ||
| AT1G51680 | 4CL1 | 67.9 | |||||
| AT3G21230 | 4CL4 | 58.9 | |||||
| PGSC0003DMG400028929 | 4-coumarate--CoA ligase 2 | −2.00 | AT3G21240 | 4CL2 | 69.2 | ||
| AT1G51680 | 4CL1 | 68.8 | |||||
| AT3G21230 | 4CL4 | 59.8 | |||||
| Up-regulated | Amino acid biosynthesis | PGSC0003DMG400034102 | Acetolactate synthase | 2.20 | AT3G48560 | CSR1 | 76.9 |
| Protein folding | PGSC0003DMG400008223 | Heat shock factor protein HSF30 | 4.44 | AT2G26150 | HSFA2 | 51.0 | |
| PGSC0003DMG400003219 | Small heat shock protein, chloroplastic | 4.11 | AT4G27670 | Heat shock protein 21 | 53.7 | ||
| PGSC0003DMG400030341 | Small heat shock protein-Class I 17.6kD | 3.99 | AT2G29500 | HSP17.6B | 77.8 | ||
| PGSC0003DMG400024707 | Small heat shock protein | 2.90 | AT1G09080 | Heat shock protein 70 | 75.1 | ||
| PGSC0003DMG402028907 | Small heat shock protein 90 | 2.72 | AT5G52640 | Heat shock protein 90 | 52.0 | ||
| PGSC0003DMG400030426 | Small heat shock protein-Class I 17.6kD | 2.50 | AT2G29500 | HSP17.6B | 74.5 | ||
| Proteolysis | PGSC0003DMG400006185 | Skp1 1 | 2.56 | AT1G75950 | SKP1 | 74.4 | |
| PGSC0003DMG400006184 | Skp1 | 2.20 | AT1G75950 | SKP1 | 75.0 |
Figure 4.
Diagrams depicting key enzymes in the phenylpropanoid (A) and carotenoid (B) biosynthetic pathways. Genes that are significantly down-regulated beyond cut-off values of 5% FDR and a log-fold change greater than 2 are written in blue.
The isolated small RNA were grouped into 87,213 clusters with an additional 10,209 unassigned sequences. Of these, 103 clusters and 126 unassigned sequences were differentially expressed. Additional summary statistics are listed in Figure 3B. Differentially expressed small RNA clusters with identified gene targets are listed in Supplementary Table S3. None of the small RNA clusters with target genes listed in Table 2 showed differential expression between the two treatments. Expression of target MYB transcription factor genes was also not correlated to small RNA cluster expression (Supplementary Table S4). Interestingly, the expression of small RNA clusters primarily targeting patatin genes was significantly up-regulated and negatively correlated to target gene expression (r = −0.61). These clusters and their targets can be found in Table 3.
Table 3.
List of differentially expressed small RNA clusters in drought-stressed potato tubers that negatively correlate to target transcript expression. Target alignments, gene ID, expression and descriptions are included.
| Small RNA Cluster | Log2 Fold Change | Target Alignment | Target Gene | Log2-Fold Change | Protein Description |
|---|---|---|---|---|---|
| Cluster 34023 | 5.03 | AGCUCAUUAAUCUCUUCGAUA | PGSC0003DMG400009921 | −6.24 | Cysteine protease 14 |
| Cluster 23921 | 4.68 | AGGGUUCAAGAAAAUGCAUUA | PGSC0003DMG400029247 | −4.75 | Patatin group O |
| Cluster 15144 | 4.62 | AGGGUUCAAGAAAAUGCAUUA | |||
| Cluster 41775 | 4.49 | ACCUCAGGGUUCAAGAAAAUG | |||
| Cluster 83189 | 5.49 | AGGCACUGGCACUACUUCAGA | PGSC0003DMG400017091 | −4.25 | Patatin-01; Probable lipolytic |
| Cluster 83175 | 4.98 | AGCCAGUAAUAUUCACCAAGU | acyl hydrolase | ||
| Cluster 83174 | 3.45 | AGGCACUGGCACUACUUCAGA | |||
| Cluster 7920 | 4.95 | GGCAGCAAGUUCUUACAUGAC | PGSC0003DMG400008749 | −4.06 | Patatin-05; Probable lipolytic |
| Cluster 68384 | 3.01 | AUCAUUCCGGGUAUCAUUCUC | acyl hydrolase | ||
| Cluster 83190 | 2.87 | UUCCGGGUAUCAUUCUCGAAU | |||
| Cluster 83166 | 2.66 | UCCGGGUAUCAUUCUCGAAU | |||
| Cluster 68380 | 5.49 | AGGCACUGGCACUAAUUCAGA | PGSC0003DMG400014104 | −4.47 | Patatin-2-Kuras 4; Probable |
| Cluster 83164 | 5.49 | AGGCAGCUAAAUGGGGUCCUC | lipolytic acyl hydrolase | ||
| Cluster 20497 | 5.38 | CUGUUGGUGAUCCGGCGUUA | |||
| Cluster 68397 | 5.36 | GUUGCUACUGUUGGUGAUCCG | |||
| Cluster 83182 | 4.97 | GGCACUACUUCAGAGUUUGAU | PGSC0003DMG401017090 | −4.91 | Patatin-3-Kuras 1 |
4. Discussion
Optimal potato tuber growth occurs around 20 °C and plants are susceptible to losses in productivity under hot, arid conditions. Such conditions are expected to increase in the coming decade, therefore functional indicators of plant stress and the cascading effects on the developing tubers were evaluated. In this study, the Canadian potato cultivar “Vigor” was gradually exposed to increasing water deficit to a level of 35% FC beginning at the start of the tuber bulking phase (Figure 1). As seen previously in the literature, fluorescent measurements were not distinguishable between treatments [15] and thus were not dependable indicators of drought stress in potato plants (Table 1). At the plot level, there were no significant differences in yield or tuber number between the treatments (Table 1), however, discrepancies could become more prominent in commercial field production. Canopy temperature was considerably elevated in the drought treatments and has evidence supporting its use for drought stress assessments [30]. Average concentrations of ABA trended upwards in potato tubers exposed to water deficit compared to well-watered controls, suggesting drought responses had been initiated. Gene expression data further corroborated that drought signaling pathways had been activated as there was marked down-regulation of an ABA receptor PYR1, down-regulation of a series of small auxin up-regulated RNA (SAUR) genes involved in cell expansion and organ elongation in response to the environment [31] and the up-regulation of heat shock factor proteins [32] (Table 2). Notably, outside of common changes to the regulation of heat shock proteins, several genes previously identified as differentially expressed in severely drought-stressed potato leaves [32] were inversely regulated in the mildly stressed potato tubers collected in this study. These include the WRKY transcription factor (PGSC0003DMG400001434) and the developmental gene UPA16 (PGSC0003DMG400031742) [32] which had 2-fold and 78-fold increases in expression compared to well-watered control tubers. Lists of genes implicated in drought stress response in potato leaf tissue [33] and potato stolons [34] have been compiled and here we provide those in developing tubers (Table 2; Supplementary Table S1).
Metabolic effects of drought on parameters such as free amino acids, soluble protein, and phenolics were assessed. The gradual drought stress to which the potato tubers were exposed resulted in no significant differences in total free amino acid concentrations, although treatment averages appeared to be divergent with 154.1 and 207.2 µmol g−1 in the control and drought treatments, respectively. These differences could be associated with the up-regulation of genes involved in proteolysis (Table 2). Elevated concentrations of proline have been shown to indicate stress in potato leaves [35], however, similar concentrations were observed in tubers irrespective of treatment. The largest changes occurred in the amino acid profile, where concentrations of branched-chain amino acids leucine and isoleucine increased (Figure 2). This indicated a greater proportion of dietary essential amino acids. Increases in branched-chain amino acids are likely attributable to the up-regulation of acetolactate synthase (Table 2), which is the first enzyme in the branched-chain amino acid synthesis pathway [36].
A major fraction (up to 40%) of the soluble protein in potato tubers consists of glycoproteins, known as patatins, that act both as storage proteins and show activity as non-specific lipid acyl hydrolases (LAH) with potential roles in plant defense against biotic stressors [37,38]. In the case of abiotic stress, it was observed that five patatin genes were down-regulated by at least 16-fold with the regulation of gene expression likely occurring via an increased presence of small RNA (Table 3). A possible consequence is reduced protein content in the resulting potato tubers.
As one of the major sources of plant phenolics in the human diet, potatoes have been targeted in breeding for greater total phenolics and antioxidant capacity [3,39]. Phenolic content is known to show a high degree of environmental plasticity with cooler temperatures during the growing period and storage, attributed to higher average accumulation [4,5]. Under drought conditions, expressions of key enzymes required for the biosynthesis of anthocyanins (DFR), flavonoids (FLS) and chlorogenic acid (HCT) were drastically reduced (Figure 4A). Initial flow into the phenylpropanoid pathway through PAL was also reduced, leading to the accumulation of phenylalanine observed in Figure 2. Similar results have been previously observed in the literature [5]. Key enzymes of the carotenoid pathway were also downregulated (Figure 4B). Environmental conditions leading to the repression of phenolic biosynthesis could minimize gains achieved in breeding programs. Regulation of the phenylpropanoid pathway can occur via MYB transcription factors [8], fifteen of which were significantly suppressed under drought (Supplementary Table S1). Unlike previous findings in potato leaves [10], expressions of small RNA and their target MYB transcription factors were not correlated in the drought-stressed tubers. There was therefore no evidence to suggest that small RNA played a role in regulating the phenylpropanoid or carotenoid pathways under drought conditions in potato tubers (Supplementary Table S4).
5. Conclusions
Potato is among one of the most important food crops, yet maintaining plant productivity in this drought-sensitive crop has become a challenge. From a nutrition perspective, decreasing soil water availability during tuber filling, as a function of a warming climate or as a production practice to induce senescence for an earlier harvest, may lead to a reduction in tuber quality. While mild drought increases the proportion of essential amino acids, potential losses in protein and phenolic content would outweigh the benefit. While MYB transcription factors may be targeted to reduce effects on the phenylpropanoid pathway, identification of small RNA as the regulator of patatin gene expression suggests it may be difficult to maintain patatin expression in drought-susceptible cultivars using current breeding techniques.
Acknowledgments
The authors would like to thank Ken Achtymichuk and Greg Larson (AAFC Canada-Saskatchewan Irrigation Diversification Centre in Outlook, SK) for plot maintenance and Branimir Gjetvaj for assistance with physiological measurements.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/8/864/s1; Figure S1: Experimental conditions; Table S1: DE genes; Table S2: Raw read counts; Table S3: DE small RNA; Table S4: small RNA, targets expression.
Author Contributions
R.S. designed the research; J.W., R.S., R.E., B.B. performed field study; S.K., L.D.R., R.S., M.K. analyzed transcriptome data; S.K. performed bioinformatics; S.P., R.S. performed the amino acid analysis; L.D.R., R.S., S.K. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
R.S., J.W., and B.B. received funding support from Agriculture and Agri-Food Canada.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.King J.C., Slavin J.L. White potatoes, human health, and dietary guidance. Adv. Nutr. 2013;4:393S–401S. doi: 10.3945/an.112.003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun O.K., Kim D.O., Smith N., Schroeder D., Han J.T., Lee C.Y. Daily consumption of phenolics and total antioxidant capacity from fruit and vegetables in the American diet. J. Sci. Food Agric. 2005;85:1715–1724. doi: 10.1002/jsfa.2176. [DOI] [Google Scholar]
- 3.Andre C.M., Oufir M., Guignard C., Hoffmann L., Hausman J.F., Evers D., Larondelle Y. Antioxidant profiling of native Andean potato tubers (Solanum tuberosum L.) reveals cultivars with high levels of β-carotene, α-tocopherol, chlorogenic acid, and petanin. J. Agric. Food Chem. 2007;55:10839–10849. doi: 10.1021/jf0726583. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal S., Jansky S. Effect of production site and storage on antioxidant levels in specialty potato (Solanum tuberosum L.) tubers. J. Sci. Food Agric. 2008;88:2087–2092. doi: 10.1002/jsfa.3318. [DOI] [Google Scholar]
- 5.Payyavula R.S., Navarre D.A., Kuhl J.C., Pantoja A., Pillai S.S. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol. 2012;12:39. doi: 10.1186/1471-2229-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezekiel R., Singh N., Sharma S., Kaur A. Beneficial phytochemicals in potato—A review. Food Res. 2013;50:487–496. doi: 10.1016/j.foodres.2011.04.025. [DOI] [Google Scholar]
- 7.Deng Y., Lu S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017;36:257–290. doi: 10.1080/07352689.2017.1402852. [DOI] [Google Scholar]
- 8.Zhang X., Liu C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant. 2015;8:17–27. doi: 10.1016/j.molp.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen C., Zeng Z., Liu Z., Xia R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hort Res. 2018;5:63–76. doi: 10.1038/s41438-018-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N., Yang J., Wang Z., Wen Y., Wang J., He W., Liu B., Si H., Wang D. Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE. 2014;9:e95489. doi: 10.1371/journal.pone.0095489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pęksa A., Kita A., Kułakowska K., Aniołowska M., Hamouz K., Nemś A. The quality of protein of coloured fleshed potatoes. Food Chem. 2013;141:2960–2966. doi: 10.1016/j.foodchem.2013.05.125. [DOI] [PubMed] [Google Scholar]
- 12.Bártová V., Bárta J., Brabcová A., Zdráhal Z., Horáčková V. Amino acid composition and nutritional value of four cultivated South American potato species. J. Food Compos. Anal. 2015;40:78–85. doi: 10.1016/j.jfca.2014.12.006. [DOI] [Google Scholar]
- 13.Galili G., Amir R., Fernie A.R. The regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 2016;67:153–178. doi: 10.1146/annurev-arplant-043015-112213. [DOI] [PubMed] [Google Scholar]
- 14.Rykaczewska K. The effect of high temperature occurring in subsequent stages of plant development on potato yield and tuber physiological defects. Am. Potato J. 2015;92:339. doi: 10.1007/s12230-015-9436-x. [DOI] [Google Scholar]
- 15.Boguszewska-Mańkowska D., Pieczyński M., Wyrzykowska A., Kalaji H.M., Sieczko L., Szweykowska-Kulińska Z., Zagdańska B. Divergent strategies displayed by potato (Solanum tuberosum L.) cultivars to cope with soil drought. J. Agron. Crop Sci. 2018;204:13–30. doi: 10.1111/jac.12245. [DOI] [Google Scholar]
- 16.Wishart J., George T.S., Brown L.K., White P.J., Ramsay G., Jones H., Gregory P.J. Field phenotyping of potato to assess root and shoot characteristics associated with drought tolerance. Plant Soil. 2014;378:351–363. doi: 10.1007/s11104-014-2029-5. [DOI] [Google Scholar]
- 17.Pieczynski M., Wyrzykowska A., Milanowska K., Boguszewska-Mankowska D., Zagdanska B., Karlowski W., Jarmolowski A., Szweykowska-Kulinska Z. Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 2018;16:603–614. doi: 10.1111/pbi.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprenger H., Erban A., Seddig S., Rudack K., Thalhammer A., Le M.Q., Walther D., Zuther E., Köhl K.I., Kopka J., et al. Metabolite and transcript markers for the prediction of potato drought tolerance. Plant Biotechnol. J. 2018;16:939–950. doi: 10.1111/pbi.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaba K., Fujiwara T., Hayashi H., Chino M., Komeda Y., Naito S. Isolation of an Arabidopsis thaliana mutant, mto1 that overaccumulates soluble methionine (Temporal and spatial patterns of soluble methionine accumulation) Plant Physiol. 1994;104:881–887. doi: 10.1104/pp.104.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen S.A., Michaud D.P. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 21.Yan D., Easwaran V., Chau V., Okamoto M., Ierullo M., Kimura M., Endo A., Yano R., Pasha A., Gong Y., et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016;7:13179. doi: 10.1038/ncomms13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinform. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrew S. FastQC: A quality control tool for high throughput sequence data. [(accessed on 5 September 2018)];2015 Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 25.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 26.Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J. Improved placement of multi-mapping small RNAs. G3 (Bethesda) 2016;6:2103–2111. doi: 10.1534/g3.116.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X., Zhuang Z., Zhao X. psRNATarget: A plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15–28. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prashar A., Yildiz J., McNicol J.W., Bryan G.J., Jones H.G. Infra-red thermography for high throughput field phenotyping in Solanum tuberosum. PLoS ONE. 2013;8:e65816. doi: 10.1371/journal.pone.0065816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren H., Gray W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant. 2015;8:1153–1164. doi: 10.1016/j.molp.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprenger H., Kurowsky C., Horn R., Erban A., Seddig S., Rudack K., Fischer A., Walther D., Zuther E., Köhl K., et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016;39:2370–2389. doi: 10.1111/pce.12780. [DOI] [PubMed] [Google Scholar]
- 33.Obidiegwu J.E., Bryan G.J., Jones H.G., Prashar A. Coping with drought: Stress and adaptive responses in potato and perspectives for improvement. Front. Plant Sci. 2015;6:1–23. doi: 10.3389/fpls.2015.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong L., Zhang H., Gan X., Zhang L., Chen Y., Nie F., Shi L., Li M., Guo Z., Zhang G., et al. Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water-stimulus conditions. PLoS ONE. 2015 doi: 10.1371/journal.pone.0128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasquez-Robinet C., Mane S.P., Ulanov A.V., Watkinson J.I., Stromberg V.K., De Koeyer D., Schafleitner R., Willmot D.B., Bonierbale M., Bohnert H.J., et al. Physiological and molecular adaptations to drought in Andean potato genotypes. J. Exp. Bot. 2008;59:2109–2123. doi: 10.1093/jxb/ern073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dezfulian M., Foreman C., Jalili E., Pal M., Dhaliwal R., Roberto D., Imre K., Kohalmi S., Crosby W. Acetolactate synthase regulatory subunits play divergent and overlapping roles in branched-chain amino acid synthesis and Arabidopsis development. BMC Plant Biol. 2017;17:71. doi: 10.1186/s12870-017-1022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhondt S., Geoffroy P., Stelmach B.A., Legrand M., Heitz T. Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 2000;23:431–440. doi: 10.1046/j.1365-313x.2000.00802.x. [DOI] [PubMed] [Google Scholar]
- 38.Shewry P.R. Tuber storage proteins. Ann. Bot. 2003;91:755–769. doi: 10.1093/aob/mcg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes L.F., Miller J.C., Cisneros-Zevallos L. Antioxidant capacity, anthocyanins and total phenolics in purple-and red-fleshed potato (Solanum tuberosum L.) genotypes. Am. Potato. J. 2005;82:271. doi: 10.1007/BF02871956. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.