Abstract
Cryptosporidium ryanae is one of the most common species for cryptosporidiosis in cattle. However, little is known of the genetic characteristics of C. ryanae due to the lack of subtyping tools. In the present study, the 60-kDa glycoprotein (gp60) gene of C. ryanae was identified in whole genome sequence data and analyzed for sequence characteristics using bioinformatics tools. The protein it encodes had some of the typical characteristics of GP60 proteins, with a signal peptide, a furin cleavage site, and a glycosylphosphatidylinositol anchor at the C terminus of the protein, and numerous O-glycosylation sites. The gene sequence was used in the development of a subtyping tool, which was used in characterizing C. ryanae from 110 specimens from dairy cattle, 2 from beef cattle, 6 from yaks, and 4 from water buffaloes in China. Altogether, 17 subtypes from 8 subtype families were recognized, namely XXIa to XXIh. Possible host adaption was identified within this species, reflected by the unique occurrence of XXIa, XXIc, and XXIh in dairy cattle, yaks, and water buffaloes, respectively. Some geographical differences were detected in the distribution of subtype families in dairy cattle; specimens from southern China showed higher genetic diversity than from northern China, and the XXIa subtype family was only seen in dairy cattle in southern and eastern China. The gp60-based subtyping tool should be useful in molecular epidemiological studies of the transmission of C. ryanae.
Keywords: Cryptosporidium ryanae, 60-kDa glycoprotein, subtyping tool, host adaptation, geographical differences
1. Introduction
Cryptosporidium spp. are important pathogens for moderate-to-severe diarrhea in humans and various animals, with a worldwide distribution [1]. Up to date, there are over 40 established species, four of which are commonly seen in cattle, including Cryptosporidium parvum, C. bovis, C. ryanae and C. andersoni [2,3]. Among them, C. parvum is almost exclusively seen in pre-weaned calves, C. bovis and C. ryanae mostly in post-weaned calves, and C. andersoni in juvenile and adult cattle [4]. These four species differ significantly in infection patterns, pathogenicity, and human infectivity.
Sequence analysis of the 60-kDa glycoprotein (gp60) gene has been widely used in subtyping C. parvum and other intestinal Cryptosporidium species or genotypes. The application of gp60 subtyping tools has contributed greatly to our understanding of the genetic diversity, transmission dynamics, as well as host adaption in human-pathogenic Cryptosporidium spp. [5,6,7,8,9,10,11]. There is also a subtyping tool for C. andersoni based on sequence analysis of several genetic loci with simple tandem repeats [12]. Subtyping tools, however, are not available for C. ryanae and C. bovis, the two bovine-specific intestinal Cryptosporidium spp. with reduced pathogenicity.
In the present study, the gp60 gene of C. ryanae was identified in whole-genome sequence data, and used in the development of a subtyping tool for the characterization of isolates from dairy cattle, beef cattle, yaks, and water buffaloes.
2. Materials and Methods
2.1. Specimens
DNA extracts from 353 C. ryanae-positive specimens from dairy cattle, beef cattle, yaks, and water buffaloes on 17 farms in China were used for subtype analysis in the present study (additional files: Table S1). They were from previous and ongoing studies of molecular epidemiology of cryptosporidiosis in bovine animals [13,14,15,16,17,18]. They were confirmed to be positive for C. ryanae by PCR and sequence analysis of an ~830-bp fragment of the small subunit (SSU) rRNA gene [19].
2.2. Identification of the gp60 Gene of C. ryanae
The whole genome data of one C. ryanae isolate (45,019) from a dairy calf in Guangdong, China were used to identify the nucleotide sequence of the gp60 gene [20]. The gp60 gene of C. ryanae was identified by blastn analysis of the genome assembly with the gp60 (cgd6_1080) gene sequence of C. parvum. The coding region of the gene was predicted from the identified contig sequence using FGENESH (http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind). The amino acid sequence predicted was analyzed by a blastp search of the NCBI database to confirm the identification of the gp60 gene.
2.3. Subtyping of C. ryanae
Based on the gp60 sequence of C. ryanae, several sets of primers were designed for nested PCR analysis of the gene (additional files: Table S2). The primer set F1F2 was used in the initial PCR analysis of specimens. Those specimens negative in the PCR analysis were re-analyzed using primer sets F3F4, F3F2, and F5F6. The PCR reaction contained 1 μL of DNA (for the primary PCR) or 2 μL of the primary PCR product (for the secondary PCR), 1× Fermentas Mix (Thermo Scientific, Waltham, MA, USA), and 250 nM primary PCR primers or 500 nM secondary PCR primers in a 50-μL reaction. The amplification was conducted on a T100 PCR thermocycler (Bio-Rad, Hercules, CA, USA) using the following program: A pre-denaturation at 94 °C for 5 min; 35 cycles of 94 °C for 45 s, 55 °C for 45 s, and 68 °C for 1 min; and a final extension at 68 °C for 10 min. The secondary PCR products were visualized under UV after 1.5% (W/V) agarose gel electrophoresis.
2.4. Sequence Analysis
Positive secondary PCR products were sequenced in both directions by Sangon Biotech (Shanghai, China). The sequences generated were assembled, edited, and aligned using ChromasPro V1.5 (http://technelysium.com.au/wp/chromaspro/), BioEdit V7.05 (http://www.mbio.ncsu.edu/bioedit/bioedit), and MUSCLE implemented in MEGA V6.0 (https://www.megasoftware.net/), respectively. Repeat sequences were identified using Tandem Repeat Finder (http://www.tandem.bu.edu/trf/trf). The structure (signal peptide and glycosylphosphatidylinositol (GPI) anchor), N-glycosylated sites, O-glycosylated sites, and furin proteolytic cleavage sites were predicted using the PSORT II (http://psort.hgc.jp/form2.html), NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/), YinOYang 1.2 (http://www.cbs.dtu.dk/services/YinOYang/), and ProP 1.0 (http://www.cbs.dtu.dk/services/ProP/) server, respectively. To assess the genetic relationship of C. ryanae subtype families, a maximum likelihood tree was constructed using MEGA V6.0 (http://www.megasoftware.net/). The general time-reversible model was used in substitution rate calculations and 1,000 replicates were used in bootstrap analysis. To identify potential recombination among subtype families, DnaSP V5.10 (http://www.ub.es/dnasp/) was used in the calculation of recombination events. Representative nucleotide sequences from the 17 subtypes (four, two, one, one, four, one, three, and one subtype(s) in the XXIa, XXIb, XXIc, XXId, XXIe, XXIf, XXIg, and XXIh subtype families, respectively) obtained in this study were deposited in GenBank under the accession number MT588090-MT588106.
2.5. Statistical Analysis
Differences in the distribution of C. ryanae XXIa and other subtype families in dairy cattle were analyzed using the χ2 test in SPSS V17.0 (IBM, New York, NY, USA). They were considered significant if p < 0.01.
3. Results
3.1. Features of the gp60 Gene of C. ryanae
The gp60 gene was identified in contig_13 from the whole-genome sequencing of C. ryanae using blastn analysis. Gene prediction using the combination of FGENSH and blastp revealed that the gene was 1548 bp in length and encoded 515 amino acids. It shared significant sequence similarities at the 5′ and 3′ ends with the gp60 gene of C. parvum (AF022929), C. hominis (FJ839883), and C. ubiquitum [10] at the amino acid level, although the overall sequence similarity was only 19.1–22.4% between C. ryanae and the other three species. Despite the much larger size, the structure of the GP60 protein of C. ryanae had some of the classic features of GP60 proteins, including the presence of a signal peptide with a cleavage site between Ser 19 and Ala 20, a furin cleavage site (RSRR) between the GP40 and GP15 fragments of the protein, and a glycosylphosphatidylinositol (GPI) anchor at the C terminus of the protein, one potential N-glycosylation site, and 115 O-glycosylation sites. However, unlike C. parvum and C. hominis, TCA/TCG/TCT trinucleotide repeats were absent in the 5′ region of the gp60 gene of C. ryanae, while GGT trinucleotide repeats encoding a polyglycine tract were recognized by using Tandem Repeat Finder (Figure 1).
Figure 1.
Deduced amino acid sequence of the gp60 gene of Cryptosporidium ryanae (45,019) compared with sequences of C. parvum (AF022929), C. hominis (FJ839883), and C. ubiquitum [10]. Potential N-linked glycosylation sites are indicated in boldface and italic type, and predicted O-linked glycosylation sites are indicated in boldface and underlined type. The first 19 amino acids for the signal peptide and the last 17 amino acids for the GPI anchor are highlighted in green and blue, respectively. The classic furin proteolytic cleavage site sequence RSRR is highlighted in red. Dashes denote amino acid deletions.
3.2. Sequence Polymorphisms in the gp60 Gene of C. ryanae
Among the 353 C. ryanae-positive specimens in this study, 146 specimens yielded an expected band. For Farm Hezhou, 13 of 37 PCR-positive specimens were randomly selected for sequencing, which generated identical sequences. As a result, PCR products from the remaining 24 specimens were not sequenced. Therefore, 122 C. ryanae sequences were used in sequence analysis, including 110 from dairy cattle, 2 from beef cattle, 6 from yaks, and 4 from water buffaloes (Table 1). The nucleotide sequences from 38 specimens were identical to the reference sequence (45,019) from the whole-genome sequencing, while the remaining sequences showed nucleotide differences of 2.1–32.6% (Table 2). The nucleotide differences among divergent sequences were found in several regions with length polymorphism mostly in the form of copy number variations of the trinucleotide repeat. Altogether, 17 subtypes were found among the 122 gp60 sequences obtained.
Table 1.
Cryptosporidium ryanae subtypes identified in bovine animals in China at the gp60 locus.
| Host | Farms | No. of Specimen Analyzed | Subtype Family (No. of Specimen) |
|---|---|---|---|
| Dairy cattle | Harbin | 17 | XXIb (17) |
| Shijiazhuang-1 | 4 | XXIg (4) | |
| Xinghua | 19 | XXIa (7), XXIe (12) | |
| Fengxian-1 | 2 | XXIa (2) | |
| Fengxian-2 | 3 | XXIa (3) | |
| Jinshan | 2 | XXIf (2) | |
| Dali | 9 | XXIa (1), XXId (1), XXIg (7) | |
| Hezhou | 13 | XXIa (13) | |
| Guangzhou-1 | 7 | XXIa (2), XXIb (2), XXIg (3) | |
| Guangzhou-2 | 1 | XXIa (1) | |
| Qingyuan-1 | 2 | XXIe (2) | |
| Qingyuan-2 | 18 | XXIa (18) | |
| Foshan | 5 | XXIa (1), XXIf (4) | |
| Zhaoqing | 8 | XXIa (8) | |
| Beef cattle | Shijiazhuang-2 | 2 | XXIb (1), XXIg (1) |
| Yaks | Guoluo | 6 | XXIc (4), XXIf (2) |
| Water buffalo | Yueyang | 4 | XXId (2), XXIh (2) |
| Total | 122 | XXIa (56), XXIb (20), XXIc (4), XXId (3), XXIe (14), XXIf (8), XXIg (15), XXIh (2) |
Table 2.
Nucleotide sequence similarity among Cryptosporidium ryanae subtype families at the gp60 locus.
| XXIa | XXIb | XXIc | XXId | XXIe | XXIf | XXIg | XXIh | |
|---|---|---|---|---|---|---|---|---|
| XXIa | - | |||||||
| XXIb | 95.5 | - | ||||||
| XXIc | 94.2 | 97.9 | - | |||||
| XXId | 92.0 | 92.3 | 91.7 | - | ||||
| XXIe | 87.7 | 87.0 | 87.1 | 89.5 | - | |||
| XXIf | 83.6 | 83.1 | 83.5 | 87.1 | 93.3 | - | ||
| XXIg | 87.0 | 86.3 | 86.8 | 89.6 | 97.1 | 94.1 | - | |
| XXIh | 67.8 | 67.9 | 67.4 | 70.6 | 71.0 | 71.8 | 70.0 | - |
3.3. Subtype Families and Subtypes of C. ryanae
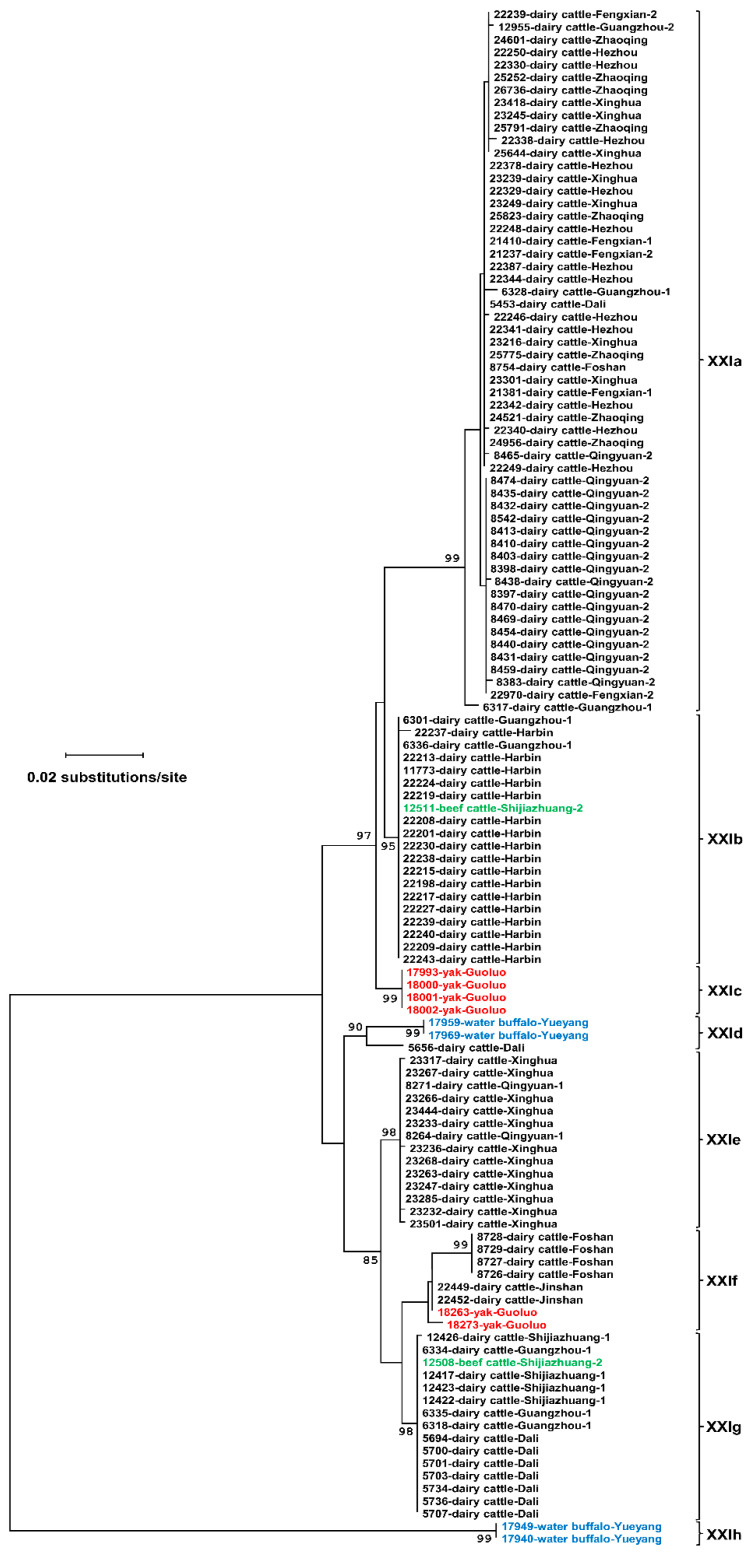
In phylogenetic analysis of the nucleotide sequences generated, the 17 sequence types formed 8 clusters (Figure 2). They were named as the XXIa, XXIb, XXIc, XXId, XXIe, XXIf, XXIg, and XXIh subtype families following the established nomenclature of gp60 subtype families for Cryptosporidium spp. [21]. The gp60 nucleotide sequences of C. ryanae subtype families differed from each other in length by at most 99 bp. Subtype family XXIh formed a highly divergent cluster from the dominant cluster of the remaining seven subtype families (Figure 2). In the dominant cluster, XXIa differed from the remaining six subtype families at both the 5′ and 3′ ends. In contrast, XXIb and XXIc had divergent sequences at the 5′ end, while XXIf and XXIg had divergent sequences at the 3′ end. The sequence differences among subtype families ranged from 2.1% to 32.6% at the nucleotide level (Table 2). DnaSP analysis of the sequences identified six potential recombination events among the eight subtype families (XXIa–XXIh), and five recombination events among the seven more related subtype families (XXIa–XXIg).
Figure 2.
Phylogeny among eight Cryptosporidium ryanae subtype families based on a maximum likelihood analysis of the partial gp60 gene. Sequences from dairy cattle, beef cattle, yaks, and water buffaloes are marked in black, green, red, and blue, respectively. Bootstrap values (>75) are indicated on branches. Scale bar indicates 0.02 nucleotide substitutions per site.
At the amino acid level, most of the sequence differences among the eight subtype families (XXIa–XXIg) occurred at the N terminus of the partial GP60 sequences (Figure 3). Therefore, all the subtype families had a furin cleavage site. Among the eight subtype families, XXIh had more divergent GP60 sequences, with the furin cleavage site having the motif RTRR instead of the RSRR in other subtype families. In addition, it had five to nine fewer O-glycosylation sites than other subtype families in the GP40 region.
Figure 3.
Deduced amino acid sequences of the partial gp60 gene of eight subtype families (XXIa to XXIh) in Cryptosporidium ryanae. Predicted O-linked glycosylation sites are indicated in boldface and underlined type. The classic furin proteolytic cleavage site sequence RSRR/RTRR is shown in red. Dashes denote amino acid deletions.
3.4. Distribution of C. ryanae Subtypes by Host, Farm, and Geographic Region
Among the eight subtype families of C. ryanae, namely XXIa (56), XXIb (20), XXIc (4), XXId (3), XXIe (14), XXIf (8), XXIg (15), and XXIh (2), six were detected in dairy cattle, including XXIa (56), XXIb (19), XXId (1), XXIe (14), XXIf (6), and XXIg (14). XXIa was the predominant subtype family, accounting for about half of the C. ryanae specimens from these animals. The distribution of XXIa was significantly different from that of XXIb (χ2 = 27.695, p = 0.000), XXId (χ2 = 71.628, p = 0.000), XXIe (χ2 = 36.960, p = 0.000), XXIf (χ2 = 56.145, p = 0.000), and XXIg (χ2 = 36.960, p = 0.000). In contrast, XXIb (1) and XXIg (1), and XXIc (4) and XXIf (2) were detected in small numbers in beef cattle and yaks, respectively. In water buffaloes, only XXId (2) and XXIh (2) were identified in the few PCR-positive specimens (Table 1).
Each farm had one to three subtype families. As shown in Table 1, 10 farms had one subtype family, five farms had two, and two farms had three. On Farm Harbin, all 17 sequences obtained belonged to subtype family XXIb. Similar results were obtained from Farm Hezhou (XXIa) and Qingyuan-2 (XXIa). However, although only two specimens were subtyped on Shijiazhuang-2, they belonged to two subtype families (XXIb and XXIg) (Table 1).
Among the six subtype families (XXIa, XXIb, XXId, XXIe, XXIf, and XXIg) in dairy cattle, five were seen in southern China, including XXIa (44), XXIb (2), XXId (1), XXIe (2), XXIf (4), and XXIg (10). In contrast, three subtype families were found in eastern China, including XXIa (12), XXIe (12), and XXIf (2). However, only two subtype families were detected in northern China, including XXIb (17) and XXIg (4) (Table 3).
Table 3.
Distribution of Cryptosporidium ryanae subtype families at the gp60 locus in dairy cattle by geographical origins.
| Geographical Division | Province | No. of Specimen Subtyped | Subtype Family (No. of Specimen) |
|---|---|---|---|
| Northern China | Heilongjiang | 17 | XXIb (17) |
| Hebei | 4 | XXIg (4) | |
| Eastern China | Jiangsu | 19 | XXIa (7), XXIe (12) |
| Shanghai | 7 | XXIa (5), XXIf (2) | |
| Southern China | Guangxi | 13 | XXIa (13) |
| Guangdong | 41 | XXIa (30), XXIb (2), XXIe (2),XXIf (4), XXIg (3) | |
| Yunnan | 9 | XXIa (1), XXId (1), XXIg (7) | |
| Total | 110 | XXIa (56), XXIb (19), XXId (1),XXIe (14), XXIf (6), XXIg (14) |
4. Discussion
Although numerous studies have reported the wide distribution of C. ryanae in cattle, little is known about the genetic diversity and transmission of this species with the absence of subtyping tools [4]. In the present study, we identified the gp60 gene of C. ryanae from the recently published whole-genome sequence data [20]. Using the sequence data from the gene, a subtyping tool was established to assess the genetic diversity of C. ryanae. The application of this subtyping tool in characterizing C. ryanae-positive specimens from dairy cattle, beef cattle, yaks, and water buffaloes has identified possible differences in the distribution of C. ryanae subtypes among hosts and geographic areas.
Several major sequence differences are present in the gp60 gene between C. ryanae and other Cryptosporidium species. The gene in C. ryanae is ~1548 bp in length, much larger than the ~981–1035 bp in C. parvum and C. hominis [22]. It has low similarity to that of C. parvum, C. hominis, and C. ubiquitum at both the nucleotide and amino acid levels, especially in the gp40 region. This sequence difference is probably responsible for the failure of PCR to amply the gp60 locus in C. ryanae using the universal primers designed based on the sequences of C. parvum and C. hominis [21]. Unlike the gp60 gene of C. parvum and other closely related species/genotypes, the TCA/TCG/TCT repeats, which encode a polyserine tract and are used in differentiating subtypes within subtype families, are absent in the gp60 gene of C. ryanae, C. ubiquitum, and C. felis [5,10,23]. In C. ryanae, there is a polyglycine tract encoded by GTT repeats in this region of the gp60 gene. Otherwise, the GP60 protein of C. ryanae has some of the typical features of GP60 proteins of Cryptosporidium spp., including a signal peptide at the N terminus, a furin cleavage site between GP40 and GP15, and a glycosylphosphatidylinositol anchor at the C terminus, and over 100 O-glycosylation sites mostly in the GP40 region [22,24].
Based on the gp60 gene identified, a subtyping tool was developed for C. ryanae for the first time. Sequence analysis of 122 PCR-positive specimens led to the identification of 17 subtypes in 8 subtype families. The high genetic diversity is expected, as C. ryanae has a high infection rate in cattle and a patent period much longer than C. parvum [25,26,27]. This can potentially facilitate the occurrence of genetic recombination among isolates, illustrated in the present study by the identification of genetic recombination events in the overall sequence data and the presence of mosaic sequence patterns among some of the C. ryanae subtype families. It is expected that with the use of the newly developed subtyping tool in other areas, other subtype families will be identified. The presence of high sequence heterogeneity in the gp60 gene of C. ryanae, nevertheless, has made PCR amplification of the gene difficult, which together with the large amplicon could be responsible for the poor amplification efficiency of the current subtyping tool.
The subtyping results obtained in the study suggest a possible occurrence of host adaptation within C. ryanae. Among the eight subtype families, XXIa is the dominant subtype family in dairy cattle but has thus far not been detected in beef cattle, yaks, and water buffaloes. The numbers of specimens analyzed for the latter, however, are small. Nevertheless, XXIc and XXIh are unique subtype families that have been found only in yaks and water buffaloes in the study, respectively. Previous studies of the SSU rRNA sequences showed the existence of host-adapted C. ryanae genotypes in yaks and water buffaloes [16,17,18,28], which is in agreement with the observation of host-adapted subtype families in the present study. In other Cryptosporidium spp., host-adapted gp60 subtype families have been identified in C. parvum, C. hominis, C. tyzzeri, and C. ubiquitum [3].
There appears to be some geographic differences in the distribution of C. ryanae subtype families in dairy cattle. As shown in Table 3, the genetic diversity of C. ryanae in southern China is higher than in northern China, reflected by the presence of three or more subtype families in Guangdong and Yunnan but only one subtype family in Heilongjiang and Hebei. In addition, while XXIa is the most common subtype family in dairy cattle, it is absent from northern China. Instead, XXIb appears to be more common there. In agreement with this, there are also some differences in the prevalence and distribution of Cryptosporidium spp. in dairy cattle between northern and southern China. In Heilongjiang, the prevalence of Cryptosporidium spp. ranged from 5.3% to 47.7% in dairy cattle, with C. andersoni being the predominant species, while in dairy cattle in Guangdong, the prevalence of Cryptosporidium spp. ranged from 4.4% to 23.7%, with C. bovis as the major species [14,29,30,31,32]. Previously, geographic differences have been reported in the distribution of subtypes of C. hominis, C. parvum, and Cryptosporidium chipmunk genotype I in humans, reflecting differences in the transmission dynamics of those pathogens [8,33].
The genetic diversity in C. ryanae could have been underestimated by the current subtyping tool. For some farms, the PCR amplification efficiency at the gp60 locus was low. For example, on Farm Zhaoqing, only 3 of 66 C. ryanae specimens from the second sampling generated the expected products in gp60 PCR, while on Farm Yangjiang, all 37 C. ryanae specimens analyzed were negative at the gp60 locus. Therefore, there could exist sequence polymorphism in the primer regions in some C. ryanae subtypes, which led to the PCR failures [21].
5. Conclusions
The gp60 gene of C. ryanae was identified and a subtyping tool was developed based on the nucleotide sequence. The use of the tool in the analysis of fecal specimens revealed high genetic diversity in C. ryanae, and the data obtained also provided evidence for the likely occurrence of host adaptation and geographical differences in the distribution of C. ryanae subtypes. Further studies are needed to verify these observations and to acquire more whole-genome sequence data from other C. ryanae isolates from diverse hosts and geographical areas. This can lead to a comprehensive understanding of the genetic diversity and transmission dynamics of C. ryanae.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/8/1107/s1, Table S1: Cryptosporidium ryanae specimens used in this study and their gp60 amplification efficiency; Table S2: Primers for nested PCR analysis of the gp60 gene in Cryptosporidium ryanae.
Author Contributions
Y.F. and L.X. conceived and designed the experiments; X.Y., N.H. and W.J. performed the experiments; X.W., N.L., Y.G. and M.K. provided technical assistance; X.Y., Y.F. and L.X. analyzed the data; X.Y., Y.F. and L.X. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (U1901208, 31820103014), 111 Project (D20008), and Innovation Team Project of Guangdong Universities (2019KCXTD001).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kotloff K.L. The burden and etiology of diarrheal illness in developing countries. Pediatr. Clin. 2017;64:799–814. doi: 10.1016/j.pcl.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Holubová N., Zikmundová V., Limpouchová Z., Sak B., Konečný R., Hlásková L., Rajský D., Kopacz Z., McEvoy J., Kváč M. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur. J. Protistol. 2019;69:70–87. doi: 10.1016/j.ejop.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Santin M. Cryptosporidium and Giardia in ruminants. Vet. Clin. Food Anim. Pract. 2020;36:223–238. doi: 10.1016/j.cvfa.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Rojas-Lopez L., Elwin K., Chalmers R.M., Enemark H.L., Beser J., Troell K. Development of a gp60-subtyping method for Cryptosporidium felis. Parasit. Vectors. 2020;13:39. doi: 10.1186/s13071-020-3906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan W., Alderisio K., Roellig D.M., Elwin K., Chalmers R.M., Yang F., Wang Y., Feng Y., Xiao L. Subtype analysis of zoonotic pathogen Cryptosporidium skunk genotype. Infect. Genet. Evol. 2017;55:20–25. doi: 10.1016/j.meegid.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stensvold C.R., Elwin K., Winiecka-Krusnell J., Chalmers R.M., Xiao L., Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J. Clin. Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y., Cebelinski E., Matusevich C., Alderisio K.A., Lebbad M., McEvoy J., Roellig D.M., Yang C., Feng Y., Xiao L. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stensvold C.R., Beser J., Axén C., Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J. Clin. Microbiol. 2014;52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santin M., Fayer R., Kvac M., Ryan U., et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N., Xiao L., Cama V.A., Ortega Y., Gilman R.H., Guo M., Feng Y. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg. Infect. Dis. 2013;19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y., Yang W., Ryan U., Zhang L., Kvác M., Koudela B., Modry D., Li N., Fayer R., Xiao L. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J. Clin. Microbiol. 2011;49:34–41. doi: 10.1128/JCM.01329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N., Wang R., Cai M., Jiang W., Feng Y., Xiao L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int. J. Parasitol. 2019;49:569–577. doi: 10.1016/j.ijpara.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y., Gong X., Zhu K., Li N., Yu Z., Guo Y., Weng Y., Kváč M., Feng Y., Xiao L. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit. Vectors. 2019;12:41. doi: 10.1186/s13071-019-3310-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai M., Guo Y., Pan B., Li N., Wang X., Tang C., Feng Y., Xiao L. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet. Parasitol. 2017;241:14–19. doi: 10.1016/j.vetpar.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Li P., Cai J., Cai M., Wu W., Li C., Lei M., Xu H., Feng L., Ma J., Feng Y., et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 2016;215:58–62. doi: 10.1016/j.vetpar.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Ma J., Li P., Zhao X., Xu H., Wu W., Wang Y., Guo Y., Wang L., Feng Y., Xiao L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015;207:220–227. doi: 10.1016/j.vetpar.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Ma J., Cai J., Ma J., Feng Y., Xiao L. Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet. Parasitol. 2014;202:113–118. doi: 10.1016/j.vetpar.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Xiao L., Morgan U.M., Limor J., Escalante A., Arrowood M., Shulaw W., Thompson R.C., Fayer R., Lal A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999;65:3386–3391. doi: 10.1128/AEM.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z., Li N., Guo Y., Feng Y., Xiao L. Comparative genomic analysis of three intestinal species reveals reductions in secreted pathogenesis determinants in bovine-specific and non-pathogenic Cryptosporidium species. Microb. Genom. 2020 doi: 10.1099/mgen.0.000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y., Lal A.A., Li N., Xiao L. Subtypes of Cryptosporidium spp. in mice and other small mammals. Exp. Parasitol. 2011;127:238–242. doi: 10.1016/j.exppara.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 22.O’Connor R.M., Thorpe C.M., Cevallos A.M., Ward H.D. Expression of the highly polymorphic Cryptosporidium parvum Cpgp40/15 gene in genotype I and II isolates. Mol. Biochem. Parasitol. 2002;119:203–215. doi: 10.1016/S0166-6851(01)00416-9. [DOI] [PubMed] [Google Scholar]
- 23.Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leav B.A., Mackay M.R., Anyanwu A., O’Connor R.M., Cevallos A.M., Kindra G., Rollins N.C., Bennish M.L., Nelson R.G., Ward H.D. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 2002;70:3881–3890. doi: 10.1128/IAI.70.7.3881-3890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Åberg M., Emanuelson U., Troell K., Björkman C. A single-cohort study of Cryptosporidium bovis and Cryptosporidium ryanae in dairy cattle from birth to calving. Vet. Parasitol. Reg. Stud. Rep. 2020;20:100400. doi: 10.1016/j.vprsr.2020.100400. [DOI] [PubMed] [Google Scholar]
- 26.Santín M., Trout J.M., Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet. Parasitol. 2008;155:15–23. doi: 10.1016/j.vetpar.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Fayer R., Santín M., Trout J.M. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus) Vet. Parasitol. 2008;156:191–198. doi: 10.1016/j.vetpar.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y., Karna S.R., Dearen T.K., Singh D.K., Adhikari L.N., Shrestha A., Xiao L. Common occurrence of a unique Cryptosporidium ryanae variant in zebu cattle and water buffaloes in the buffer zone of the Chitwan National Park, Nepal. Vet. Parasitol. 2012;185:309–314. doi: 10.1016/j.vetpar.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 29.Liu A., Wang R., Li Y., Zhang L., Shu J., Zhang W., Feng Y., Xiao L., Ling H. Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol. Res. 2009;105:797–802. doi: 10.1007/s00436-009-1457-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Wang R., Yang F., Zhang L., Cao J., Zhang X., Ling H., Liu A., Shen Y. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China’s Heilongjiang Province. PLoS ONE. 2013;8:e54857. doi: 10.1371/journal.pone.0054857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W., Wang R., Zhang W., Liu A., Cao J., Shen Y., Yang F., Zhang L. MLST subtypes and population genetic structure of Cryptosporidium andersoni from dairy cattle and beef cattle in northeastern China’s Heilongjiang Province. PLoS ONE. 2014;9:e102006. doi: 10.1371/journal.pone.0102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang N., Wu Y., Sun M., Chang Y., Lin X., Yu L., Hu S., Zhang X., Zheng S., Cui Z., et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology. 2019;146:28–32. doi: 10.1017/S0031182018001129. [DOI] [PubMed] [Google Scholar]
- 33.Xiao L., Cama V. Cryptosporidium and Cryptosporidiosis. In: Ortega Y., Sterling C., editors. Food Microbiology and Food Safety. 2nd ed. Springer; Cham, Germany: 2018. pp. 73–117. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.