Abstract
Objective: Herpes simplex viruses (HSVs) are widely spread throughout the world, causing infections from oral, and genital mucous membrane ulcerations to severe viral encephalitis. Glycoprotein B (gB) was the first HSV envelope glycoprotein identified to induce cell fusion. This glycoprotein initiates viral entry and thereby determines the infectivity of HSV, as well as oncolytic HSV (oHSV). Clarifying its molecular characterization and enlarging its motif reservoir will help to engineer oHSV and in cancer treatment applications. Only in recent years has the importance of gB been acknowledged in HSV infection and oHSV engineering. Although gB-modified oHSVs have been developed, the detailed molecular biology of gB needs to be illustrated more clearly in order to construct more effective oHSVs.
Method: Here, we performed a systematic comparative sequence analysis of gBs from the 9 HSV-1 and 2 HSV-2 strains, including HSV-1-LXMW, which was isolated by our lab. Online software was implemented to predict gB secondary structure and motifs. Based on extensive literature reviews, a functional analysis of the predicted motifs was performed.
Results: Here, we reported the DNA and predicted amino acid sequences of our recently isolated HSV-1-LXMW and found that the strain was evolutionarily close to HSV-1 strains F, H129, and SC16 based on gB analysis. The 22 novel motifs of HSV gB were identified for the first time. An amino acid sequence alignment of the 11 HSV strains showed that the gB motifs are conserved among HSV strains, suggesting that they are functional in vivo. Additionally, we found that certain amino acids within the 13 motifs out of the 22 were reported to be functional in vivo. Furthermore, the gB mutants and gB-engineered oHSVs were also summarized.
Conclusion: Our identification of the 22 novel motifs shed light on HSV gB biology and provide new options for gB engineering to improve the efficiency and safety of oHSVs.
Keywords: oncolytic herpes simplex virus (oHSV), glycoprotein B (gB), motif, domain, cancer
Introduction
Cancer is a major threat to human health. Cancer stem cells play an important role in cancer initiation, progression, and drug resistance (1–5). Oncolytic viruses (OVs) are spontaneously occurring or genetically modified viruses that preferentially infect and kill cancer cells, including cancer stem cells (6–8). An oncolytic adenovirus with an E1B deletion, H101, or Oncorine®, was approved to treat cancer in China in 2005, which is the first clinical OV in the world (9). Recently, OVs, including the measles virus (MV), Newcastle disease virus (NDV), herpes simplex virus (HSV), reovirus, adenoviruses, and vesicular stomatitis virus (VSV), provide promising approaches in cancer treatment (10). Engineered HSV is considered a promising agent because of a series of unrivaled merits, including its opulent genome (150 kbp) and exhaustive cognition of viral genes (11). In 2015, the U.S. Food and Drug Administration (FDA) approved T-vec, an oncolytic HSV that was engineered with deletions of genes γ34.5 and US12 (encoding ICP47) and subsequent insertion of the gene encoding human granulocyte macrophage colony-stimulating factor (GM-CSF), to treat melanoma (12). Oncolytic HSVs (oHSVs) have also been made by engineering envelope glycoproteins (13).
HSV entry is a perplexing process, requiring the coordination of glycoprotein ligands gD, gB, gH, and gL alongside cellular receptors. These glycoproteins interact with cellular receptors and lead to virus–cell fusion (14). The cascade is initiated by HSV to heparan sulfate proteoglycans (HSPG) at the cell surface. While HSV-1 gC has a critical role in HSPG attachment during HSV-1 entry, HSV-2 gB is the key glycoprotein for HSV-2 attachment to HSPG (15, 16). gD attaches to one of its own receptors, herpesvirus entry mediator (HVEM), nectin-1, or 3-O-sulfated heparan sulfate (3-OS HS) (17–19), activating gH/gL and changing the conformation of gB (17). The attachment of gH/gL to its own receptors may also be involved in this process (17, 20). gB inserts itself into the cell membrane and is considered as a viral fusogen (21, 22). Virus–cell fusion can only take place when gB interacts with one of the cell's receptors. There are three HSV-1 gB receptors: the myelin-associated glycoprotein (MAG), the paired immunoglobulin-like type 2 receptor α (PILRA), and the myosin heavy chain 9 (MYH9). However, HSV-2 gB-specific receptors remain unknown.
gB is a conservative glycoprotein (23). HSV-1 gB is composed of four parts: a secretory signal, an extracellular region, a transmembrane region, and a cytoplasmic region (24). Furthermore, gB contains five domains: domain I contains residues Ile154 to Val363; domain II is composed of residues Tyr142 to Asn153 and Cys364 to Thr459; domain III is composed of residues Pro117 to Pro133, Ser 500 to Thr572, and Arg661 to Thr669; domain IV encompasses residues Ala111 to Cys116 and Cys573 to Ser 660; domain V ranges from residues Phe670 to Ala725 (23). Four functional regions of gB were identified alongside the domains: (i) the first region consists of domain I, residues Glu697 to Ala725, and contains two internal fusion loops (FLs); (ii) the second region, which is generated by Ile391 to Gly410, Pro454 to Lys475, and a part of domain II, may play a direct role in gH/gL binding; (iii) the third region includes residues within domains III and IV that bind to a cellular receptor; and (iv) the last region contains Met1 to Trp12 (25, 26).
However, the details of gB structure and function are still not fully understood (27). The efficiency of oHSV may be improved upon in the future. (22). Here, we sequenced the DNA encoded the gB of our recently isolated new strain HSV-1-LXMW; compared the gB amino acid sequence with 10 other gBs through phylogenetic analysis, amino acid sequence alignment, and motif predictions; and found 22 new motifs in HSV gB. We further correlated the newly identified motifs with engineered gB reported in HSVs. Our studies may enrich the knowledge on gB structure, and gB-engineered oHSV may contribute greatly to cancer treatment.
Materials and Methods
HSV Genomic DNA Sequencing Analysis
A new HSV strain, named HSV-1 strain LXMW, was obtained previously (28). Briefly, a 45-year-old male patient with acute oral herpes was admitted for HSV-1 isolation. The herpes lesion was punctured with a sterile syringe, the liquid was dipped onto sterile cotton swab, and the swab was placed in a virus collection tube [Youkang Keye Biotechnology (Beijing) Co., Ltd., Cat. No. MT0301] and shipped at 4°C to the laboratory. After successful isolation and identification, genomic HSV DNA sequencing was described in a previous publication (28). The high-quality genomic DNA (500 ng) was submitted to the Beijing Genomics Institute (BGI, http://www.genomics.cn). The UL27 DNA sequence was presented in Supplementary Materials. Afterwards, the UL27 DNA sequence was translated into gB amino acids using the DNAMAN software (http://www.shinegene.org.cn/q2.html). The translation result was also shown in Supplementary Materials. In addition, we applied the Burrows-Wheeler Aligner (BWA) software (https://github.com/lh3/bwa) to perform the alignment. The genomic sequences of 10 other HSV strains were obtained from NCBI Reference Database (Table 1).
Table 1.
The information of HSV strains studied in this article.
| HSV Strain | Gene Bank ID | Tax-ID | Sub-Date | UL27 DNA sequence | University, Country |
|---|---|---|---|---|---|
| HSV-1 strain LXMW |
53088/55802 | Yangtze University, Jingzhou, China | |||
| HSV-1 strain 17 |
JN555585.1 | 10299 | 2011-08-02 | 53059/55795 | RC University, Glasgow, UK |
| HSV-1 strain H129 |
GU734772.1 | 744249 | 2010-2-9 | 53007/56030 | Princeton University, USA |
| HSV-1 strain RH2 |
AB618031.1 | 946522 | 2011-2-28 | 51607/54321 | Osaka University, Japan |
| HSV-1 isolate SC16 |
KX946970.1 | 10309 | 2016-10-30 | 53707/56443 | Severo Ochoa, Spain |
| HSV-1 strain Patton Isolate GFP-Us11 |
MF959544.1 | 10308 | 2017-10-11 | 53071/56093 | NYU, New York, USA |
| HSV-1 strain F |
GU734771.1 | 10304 | 2010-2-9 | 52974/55996 | Princeton University, USA |
| HSV-1 strain KOS |
JQ673480.1 | 10306 | 2012-2-14 | 53022/55736 | University of Kansas, USA |
| HSV-1 strain MP-R15 |
EF177455 | 10298 | 2007-10-09 | 1/2700 | Goteborg University, Sweden |
| HSV-2 strain HG52 |
JN561323.2 | 10315 | 2011-08-05 | 53402/56152 | University of Glasgow, UK |
| HSV-2 strain G |
KU310668.1 | 10314 | 2015-12-16 | 1/2706 | Einstein College, USA |
Phylogenetic Analysis of gB Amino Acids From HSV-1 and HSV-2
The software MEGA7 (http://www.megasoftware.net) was implemented for the phylogenetic analysis of gB amino acid sequences from 11 HSV strains. Based on the General Time Reversible model with “complete deletion,” the Maximum Likelihood method option was applied to infer evolutionary history. The evolutionary history of the taxa analyzed was represented in a bootstrap consensus tree. The percentage of trees, in which the associated taxa were clustered together, is displayed next to the branches. The cladogram with the highest log likelihood is displayed. The details of the method were described previously (28).
Alignment of gB Amino Acid Sequences for 11 HSV Strains
gB amino acids were previously translated from the UL27 DNA sequence using the DNAMAN software (http://www.shinegene.org.cn/q2.html). The 10 other amino acid sequences utilized for the analysis were translated from the above DNA sequences. To determine the conservation of gB amino acid sequences among different HSV strains, the online software EMBL-EBI (https://www.ebi.ac.uk) was implemented to perform the alignment of gB amino acid sequences. The stringent conditions were set as default by the online program. Aberrant amino acids were marked in red.
Prediction of gB Secondary Structure and Motifs
Secondary structure and motifs are the basis for the formation of functional domains and usually play a crucial role in protein function. The online software UCL-CS Bioinformatics (http://bioinf.cs.ucl.ac.uk/introduction/) was deployed to predict the secondary structure of gB. Motifs were predicted through the online software MEME (http://meme-suite.org/). The stringent conditions were set as default by the online program. To show the results briefly and conveniently, one HSV-1 strain (HSV-1 strain KOS), one HSV-2 strain (HSV-2 strain G), and our new HSV-strain LXMW were selected.
Results
HSV-1-LXMW Is Evolutionarily Close to HSV-1 Strains F, H129, and SC16 Based on UL27 DNA Sequence Analysis
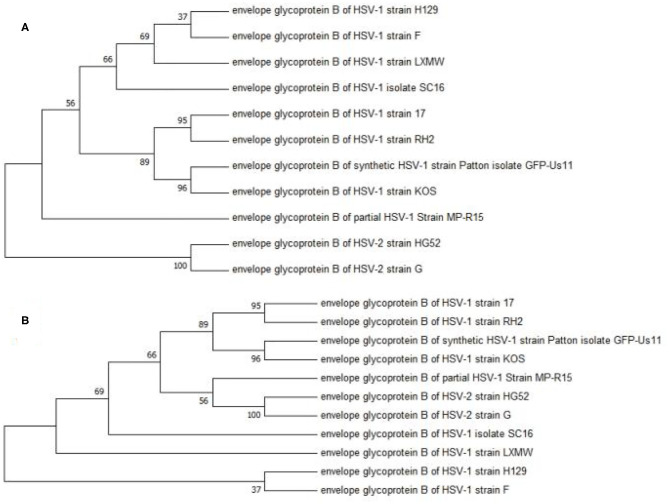
A phylogenetic analysis was performed to analyze the evolutionary relationship among the 11 HSV-1 strains. The UL27 DNA sequence of HSV-1-LXMW, together with 11 HSV strains (Table 1), was translated into amino acid sequences and was then analyzed using the MEGA7 software. The results further supported that HSV-1-LXMW belongs to an HSV-1 strain, but not to an HSV-2 strain. Interestingly, the clades corresponding to HSV-1-LXMW and HSV-1 strains F and H129 were grouped together into one main branch in the phylogenetic trees, indicating a closer evolutionary link to each other likely due to their origin from a common ancestor (Figure 1). However, HSV-1-LXMW is grouped in a different branch from the HSV-1 strain KOS (Figure 1). Ultimately, the analysis suggests that the UL27 gene of HSV-1-LXMW is evolutionarily close to HSV-1 strains F, H129, and SC16.
Figure 1.
Phylogenetic analysis of the gBs of 11 HSV strains. This phylogenetic tree was generated by the neighbor-joining (NJ) method using MEGA7. The percentage of trees in which the associated taxa were clustered together is displayed next to the branches. The cladogram with the highest log likelihood is displayed. (A) The evolutionary tree is drawn to scale, with branch lengths measured in number of substitutions per site. (B) The bootstrap consensus tree is taken to represent the evolutionary history of the taxa analyzed. Our result showed an average distance of about 69% among the strains.
Prediction of the Secondary Structure, Motifs, and Functional Domains of gB
The Prediction of gB Secondary Structure
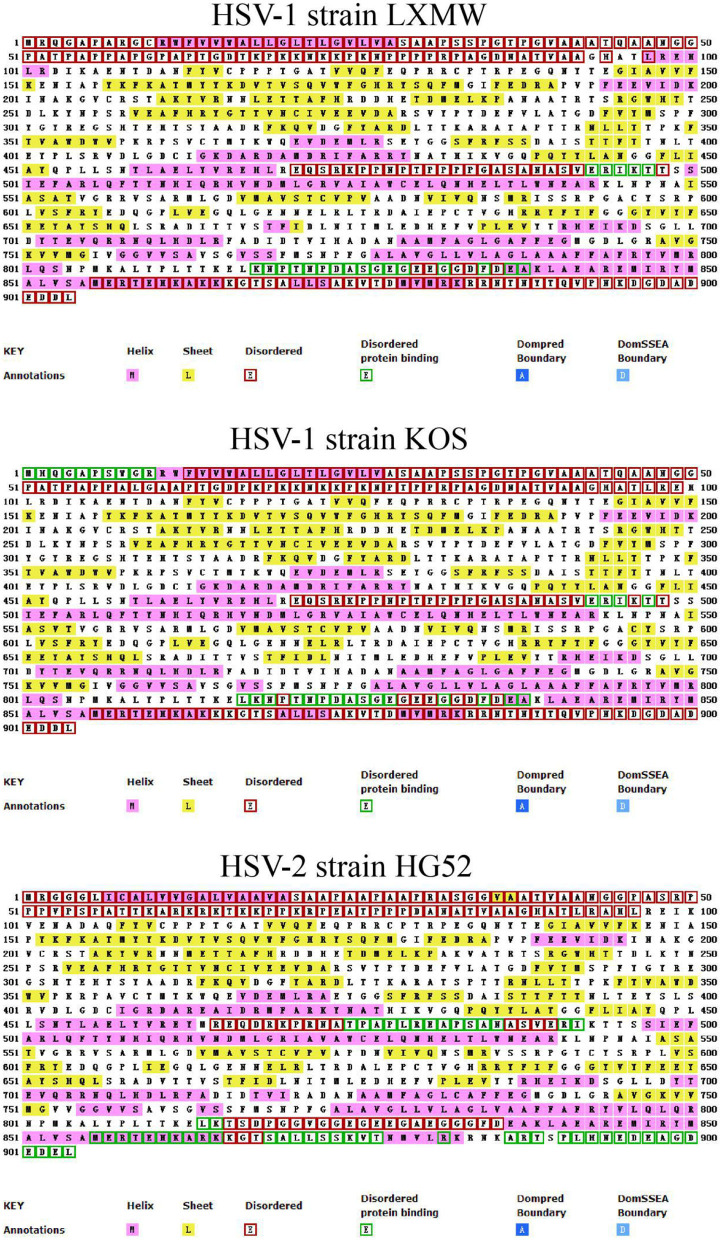
In order to analyze the secondary structure of gB and present it simply, we selected HSV-1 strains LXMW and KOS, and HSV-2 strain HG52, and predicted their secondary structure using the online software UCL-CS Bioinformatics (http://bioinf.cs.ucl.ac.uk/introduction/). As shown in Figure 2, the secondary structural forms of gB are various and abundant and include the helix, sheet, disordered, disordered protein binding, dompred boundary, and DomSSEA boundary formations. To be specific, the disordered protein binding sites in HSV-1 strain LXMW are in residues Glu493-Thr497 and residues Lys818-Ala838. For HSV-1 strain KOS, this site is located at residues Met1-Arg10, residues Glu493-Thr497, and residues Leu817-Ala838. However, for HSV-2 strain HG52, the site is located at residues Thr475-Ile492, Leu814-Lys815, Met856-Thr877, Arg882, and Ala887-Leu904.
Figure 2.
The predicted gB secondary structures of HSV-1 strains LXMW and KOS as well as HSV-2 strain HG52.
The Prediction of 22 Novel gB Motifs From 11 HSV Strains
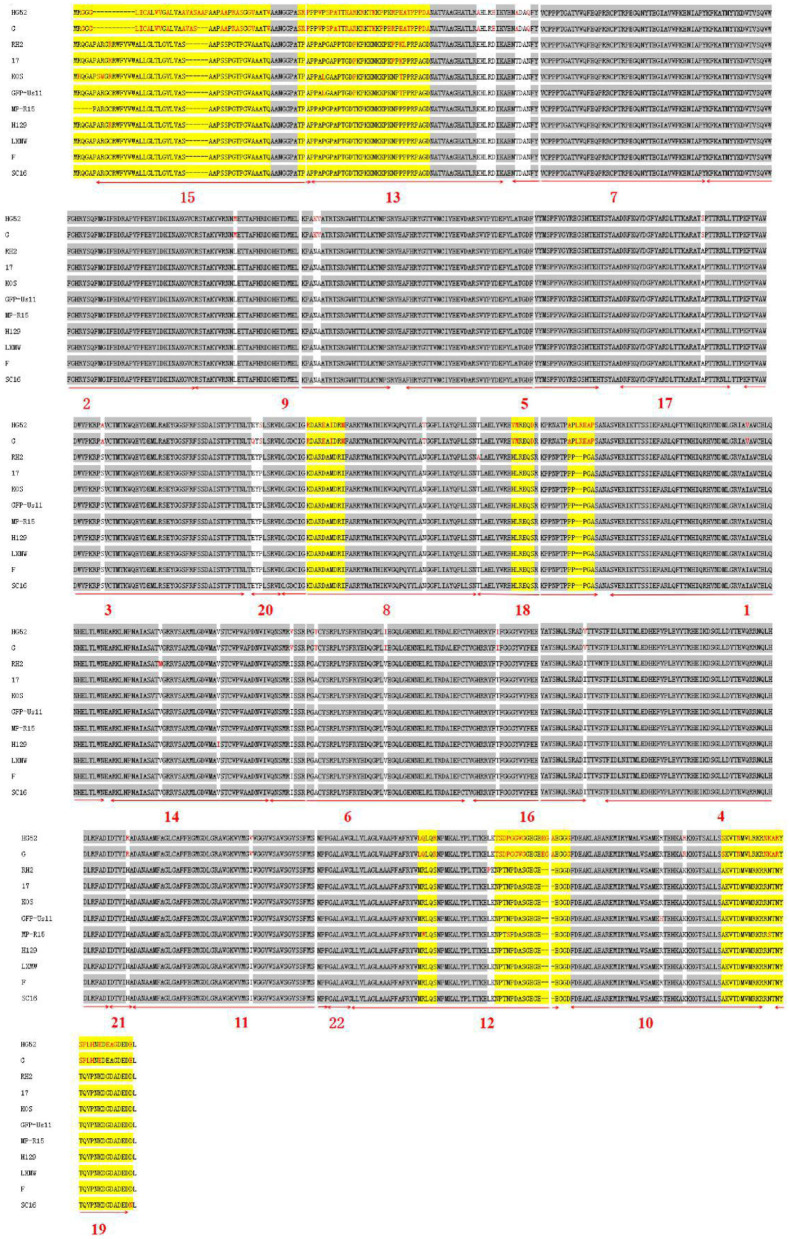
To predict the gB motifs, the online software MEME was used (http://meme-suite.org/). The motif amino acid sequences were marked in Figure 3A and the newly predicted new motifs were named 1–22 in Figure 3B. The result revealed that HSV-2 gB lacked motif 15, located in Pro6 to Ala55. All HSV-1 and HSV-2 gBs share 21 motifs except motif 15 (Figures 3, 5).
Figure 3.
(A) The predicted consensus sequences of the 22 motifs from the gBs of 11 HSV strains. (B) The location of the 22 new gB motifs (1–21, 23) predicted in (A).
gB Motifs Are Conserved Among 11 HSV Strains
To further analyze the conservatism of gB, the gB amino acid sequences of the 11 HSVs were aligned (Figure 4). The alignment showed that there are 28 conserved and 8 variable regions in gB. Moreover, among the gBs of HSV-1 strains, there were fewer mutations among the gBs of HSV-1 strains. However, there were more variations between HSV-1 and HSV-2. In the gB domain of HSV-1 strain H129, two different amino acids (residues Arg10 and Ile570) were found from HSV-1 strain LXMW (residues Cys10, Val570, and Asp903). In the gB region of HSV-1 strain SC16, there was one different amino acid (residue Asn903) compared to HSV-1 strain LXMW. Both the phylogenic analysis and alignment corroborated the conclusion that gB amino acids of LXMW are highly similar to HSV-1 strains F, H129, and SC16.
Figure 4.
The gB amino acid sequence alignment of 11 HSV strains.
For the 11 HSVs analyzed, the 28 conserved sequences among all the gB amino acids are marked by gray shade section, while the 8 variable amino acids are marked by yellow shade section. The different amino acids are marked in red. The 22 motifs predicted by the online software are marked by numerals 1–22 and underlined in bold red.
The Potential Function of the 13 Newly Identified Motifs for gB of HSV
It is well acknowledged that gB is a glycoprotein needed for HSV infection, which can form different dimers to mediate virus–cell fusion and induce the production of neutralizing antibodies (13). To understand the potential function of the identified gB motifs, we performed an extensive literature search on the topic of HSV gB. We summarized the gB mutants and gB-engineered oHSVs (Table 2 and Figure 5). Based on the following pieces of literatures, we, for the first time, systematically compared the relationship of the engineered sites in gB with the conserved regions, functional domains, and motifs, and suggested probable functions for 13 motifs of the total 22. However, we were not able to find any report on any of the other 9 motifs in published studies. Indeed, it is a double-edged sword to engineer HSV gBs. Some mutations and modifications will enhance its functionality while other manipulations will severely undermine it. A summary of these modifications is detailed below.
Table 2.
The function of the identified motifs of gB of HSV.
| Virus | Name | Mutations in gB | Conserved/variable region in gB | Motif in gB | Motif potential function | oHSV effect | Additional modification | References |
|---|---|---|---|---|---|---|---|---|
| HSV-1 strain KOS |
Y179K, W174R A261D | Y179K, W174R A261D | Conserved region 4 | 2 | Motif 2 may be necessary for virus–cell fusion. | Loss of fusion | None | (29) |
| HSV-1 (strain not mentioned) |
T665i5, V667i5I671i5, L673i5 E187i5 | MFKHT inserted after T665, MFKHV inserted after aa667, DLLNI inserted after aa671, NSLNI inserted after aa673, DSLNK inserted after aa187 | Conserved region 22 | 2 and 4 | Motif 2 is important for the conformation of the fusion loops. | Fusion deficient | None | (30) |
| HSV-1 strain KOS |
I671A, H681A, F683A | I671A, H681A, F683A | Conserved region 22 | 4 | Motif 4 may be required for virus–cell fusion possibly via interaction of the arm with coil regions of gB. | Decreased fusion | gH:lacked | (31) |
| HSV-1 strain F |
T887A or truncated at residue 886. | T887A or truncated at residue 886. | Variable region 8 | None | Reduced virus–cell fusion | gH:lacked | (32) | |
| HSV-1 strain KOS |
Y889A LL871AA | Disruption of YTQV motif (aa 889 to 892) and LL motif (aa 871 to 872) | Conserved region 28 and variable region 8 | 19 and 10 | Motif 19 and 10 are involved in cell–cell fusion. | Disruption of YTQV and LL decrease and facilitate virus–cell fusion, respectively | None | (33) |
| HSV-1 strain KOS |
gBΔ28, gBΔ36, A874P | gBΔ28, gBΔ36, A874P | Conserved region 28, variable region 8 | 10 and 19 | Motifs 10 and 19 may participate in virus-induced fusion activity. | Increased cell fusion | None | (34) |
| HSV-1 strain F |
syn3 | R857H | Conserved region 26 | 10 | Motif 10 may promote cell–cell fusion and this mutant effectively reduced primary and metastatic breast tumors in immunocompetent mice. | Effective in the treatment | gK: Ala-to-Val at position 40 | (35) |
| HSV-1 strain KOS |
Truncated at 863 | Lacking the last 41 amino acids of the cytoplasmic domain | Conserved regions 27, 28, variable region 8 | 10 and 19 | Deletion of the carboxy-terminal 41 amino acids had no influence on the function of gB. | No influence | None | (36) |
| HSV-1 strain F |
R-903 | Engineered the scFv to HER2 between AA 43 and 44 | Variable region 1 | 15 | Motif 15 may be either flexible or non-essential and oHSVs were capable to target Her2+ cancer cells | No influence | None | (22) |
| HSV-1 | R-313, R-315, R-317, R-319 | The GCN4 peptide inserted between aa 43 and 44, 81 and 82, 76 and 77, and 95 and 96 | Conserved region 6 | 15 and 13 | Motif 15 may be either flexible or non-essential and gB retargeted oHSV could cultivate in non-cancer cells. | No influence | gD: scFv-HER2 replaced aa 6 to 38 | (37) |
| HSV-1 strain KOS |
D285N/A549T | D285N/A549T | Conserved region 6 and 15 | 5 and 14 | Motif 5 may regulate the interaction of gB with gH/gL, motif 14 may be essential for the conformational change of gB and the double mutant was effective in an orthotopic mouse model of primary human glioma. | Accelerate cells entry; facilitate cell fusion | gD: R222N/F223I gD: An scFv replaced aa 2–24 | (38, 39) |
| HSV-1 strain KOS |
S668N | S668N | Conserved region 21 | None | gH/ gB formation | gH: Residues N753K and A778V in the H3 domain | (40) | |
| HSV-1 strain F |
T53A, T53/480A | T53A, T53/480A | Variable region 2, conserved region 13, | 15 and 17 | Motifs 15 and 17 may be required for the binding of gB to PILRα and virus–cell fusion. | Decreased virus–cell fusion and virus entry | (41) | |
| HSV-1 strain S (a syncytial mutant from HSV-1strain F) |
T787C | T787C of the cytoplasmic domain of mature gB, results in L to P substitution | Conserved region 24 | 12 | Motif 12 may contribute to gB binding to heparan sulfate. | Decreased gB/heparan sulfate binding | None | (42) |
| HSV-1 strain KOS |
gBpK− | Residues 68 to 76 (KPKKNKKPK) were deleted | Variable region 2 | 13 | Motif 13 may be the heparan sulfate binding element of gB. | gC: gC coding sequence (UL44) deleted | (43) | |
| HSV-1 strain 17 | K76L K79L | Replace the aa 75PKPPKP80 with 75PLPPLP80 | Variable region 8 | 13 | Motif 13 may be involved in the interaction between gB and HLA-DR heterodimers. | Destroyed gB/HLA-DR heterodimers | None | (44) |
| HSV-1 strain KOS |
C633S or C596S/C633S | Replace the aa 633C or both 633C and 596C with S | Conserved region 19, 24 | 6 | Motif 6 may translocate gB to the cell surface. | Unable to translocate to the cell surface | None | (45) |
| HSV-2 strain G |
N133Q | N133Q | Conserved region 4 | 7 | Motif 7 may be required for gB trafficking from the ER to the Golgi. | Not transported from the ER to the Golgi | None | (46) |
| HSV-1 strain KOS |
gBΔ(730–747), gBΔ(748–773), gBΔ(730–739), gBΔ(735–744), gBΔ(740–749), gBΔ(750–759), gBΔ(765–773) | gBΔ(730–747), gBΔ(748–773), gBΔ(730–739), gBΔ(735–744), gBΔ(740–749), gBΔ(750–759), gBΔ(765–773) | Conserved region 23,24 | 11 | Motif 11 may be required for the interaction between gB and the cell membrane. | Cannot arrive at the cell surface | None | (47) |
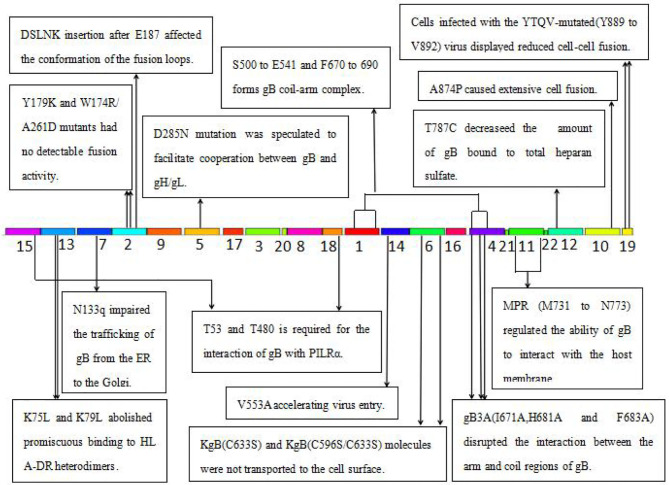
Figure 5.
The function of the identified 22 motifs of gB of HSV. MPR, membrane-proximal region; PILR, the paired immunoglobulin-like type 2 receptor; HLA-DR, human leukocytic antigen DR; ER, endoplasmic reticulum.
Several key motifs of gB play important roles in promoting virus–cell fusion. Sometimes, mutations in some motifs of gB could attenuate virus–cell fusion. Hannah BP et al. found that Y179K and W174R/A261D mutants hardly executed fusion activity (29). Residues Tyr179 and Trp174 are located in motif 2, suggesting that motif 2 is necessary for virus–cell fusion. Silverman JL et al. generated five insertion mutants (T665i5, V667i5, L673i5, I671i5, and E187i5) that caused a fusion-deficient phenotype. They speculated that the mutated region in E187i5 (in motif 2) was involved in the destruction of the fusion loops (30), suggesting that motif 2 is important for the conformation of the fusion loops. Connolly et al. found that three mutations (I671A, H681A, and F683A located in motif 4) in the arm region decreased fusion but did not reduce surface expression (31), suggesting that motif 4 is possibly required for virus–cell fusion via interaction of the arm with gB coil regions. Wisner TW et al. found that the T887A (not in any of the 22 motifs) mutation or truncation at Asn886 markedly reduced virus–cell fusion (32). Beitia et al. discovered that the destruction of YTQV (residues Tyr889 to Val892) decreased cell–cell fusion, but the damage of LL (residues Leu871 to Leu872) facilitated cell–cell fusion. They subsequently hypothesized that the LL mutations returned gB to the cell surface (33). This study suggested that motif 19 (Tyr889) and 10 (Leu871 to Leu872) are involved in cell–cell fusion.
However, there are also certain mutations in some motifs of gB-improved virus–cell fusion. Foster TP et al. concluded that the deletion of 28–36 amino acids in the carboxyl-terminal, or A874P mutation, increased virus–cell fusion, and the carboxyl-terminal 36 amino acids participated in membrane fusion (34). Since carboxyl-terminal 36 amino acids are distributed in motifs 10 and 19, these two motifs were also suggested to participate in virus–cell fusion. Israyelyan et al. demonstrated that fusogenic OncdSyn viruses, which relied on a combination of mutations R857H of gB (syn3) and gK (syn1), were effective in the treatment of breast tumors (35). Huff V et al. found that deletion of the carboxyl-terminal 41 amino acids had no influence on the function of gB (36). Petrovic B et al. inserted single-chain variable fragment (scFv) between residues 43 and 44 of gB, generating a mutant HSV R-903 that retargeted to HER2-positive cancer cells. They found that R-903 did not require activation by other glycoproteins (22). One year later, they inserted the GCN4 peptide into gB to construct a gB retargeted virus that could be cultivated in non-cancer cells (37). Residues 43 and 44 are located in motif 15, suggesting that motif 15 is either flexible or non-essential.
Some motifs are required for the attachment to gH/gL, PILRα, heparan sulfate, and HLA-DR. When gD receptors are nonexistent, gB double mutations D285N (in motif 5) and A549T (in motif 14) could allow HSV to enter into cells processed by facilitating interaction between gB and gH/gL and virus–cell fusion, respectively (38). In 2013, Uchida et al. established an oHSV that retargeted HSV to EGFR by combining the gD and gB mutations. This retargeting system was effective in the treatment of glioma (39). The D285N mutation is located in motif 5 and A549T mutation is located in motif 14. These results suggest that motif 5 may regulate gB interaction with gH/gL and motif 14 may be essential for gB conformational change (34). Uchida H et al. also showed that the lack of traditional gD receptors could also be offset by the combination of gH:KV and gB:S668N (not in any of the 22 motifs) (40). Arii et al. identified that gB-T53A (in motif 15) and gB-T53/480A (in motif 17) mutations distinctly decreased virus–cell fusion and virus entry into cells expressing PILRα (41), suggesting that motifs 15 and 17 are required for the binding of gB to PILRα as well as virus–cell fusion. Diakidi-Kosta et al. revealed that the T787C mutation decreased the amount of gB bound to total heparan sulfate (42). Residue Gly787 is located in motif 12, suggesting that motif 12 contributes to gB and heparan sulfate binding. Laquerre et al. concluded that the binding of gB to heparan sulfate required residues Lys68 to Lys76 (in motif 13) (43), implying that motif 13 is the heparan sulfate binding element of gB. Sievers et al. found that a combination of K76L and K79L (in motif 13) mutations destroyed the interaction between gB and HLA-DR heterodimers (44), suggesting that motif 13 is involved in the formation of gB and HLA-DR heterodimers.
Some motifs are involved in gB translocation and trafficking. Laquerre S et al. constructed KgB (C633S in motif 6) and KgB (C596S/C633S) mutant viruses that resulted in gB being unable to translocate to the cell surface. The mutant gB was not mature and therefore HSV could not infect cells (45). Residue Cys633 is located in motif 6, suggesting that motif 6 is required for gB translocating to the cell surface. Luo et al. demonstrated that the N133Q (Cys133 in motif 7) mutant was not transported from the ER to the Golgi, and therefore gB lost its function (46), suggesting that motif 7 is required for gB trafficking from the ER to the Golgi. Shelly et al. concluded that membrane-proximal region (Ala730 to Asn773 in motif 11) deletion mutants cannot arrive at the cell surface (47), suggesting that motif 11 is required for the gB trafficking to the cell membrane.
Taken together, key motifs can be modified in the aspects as follows to improve the efficiency of oHSV. First, the reasonable modification of motifs 2, 10, and 19 can increase virus–cell fusion. Second, to facilitate the translocation and trafficking of gB, modifications of motifs 6, 7, and 11 should be cautious. Third, motif 15 is flexible for exogenous gene fragment insertion, such as scFv-HER2, GCN4 peptide, etc. Furthermore, key motifs of gB determining antibody and complement neutralization could be explored and modified to evade immune response and improve efficacy of oHSV.
Discussion
The development of more effective oHSVs depends on a deeper understanding of HSV gB structure and function. Here, we performed a systematic comparative sequence analysis of gBs from 9 HSV-1 and 2 HSV-2 strains, including our recently isolated new strain HSV-1-LXMW. Phylogenetic analysis found that HSV-1-LXMW gB is evolutionarily close to HSV-1 strains F, H129, and SC16. For the first time, we identified 22 novel motifs in HSV gB. Amino acid sequence alignment showed that the 22 motifs are conserved among HSV gBs. Through literature analysis, we found that 13 out of the 22 motifs are biologically functional. We also identified 28 conserved regions and 8 variable regions of HSV gB. Our findings may have significant implications on HSV gB biology and the construction of gB-engineered oHSVs with improved the efficacy and safety.
The phylogenetic analysis revealed that the gB amino acid sequence in HSV-1-LXMW was close to those in HSV-1 strains F, H129, and SC16 based on UL27 gene analysis, while our earlier study demonstrated that ICP27 of our new strain is close to those in HSV-1 strains Patton and H129 (27). This study complements previous work demonstrating that our strain may be evolutionarily closer to strain H129. Therefore, any further manipulation done to LXMW strain's gB could benefit from the previously successful engineering strategies of gB from strain H129. The different genes may show some evidence of the phylogenetic relationships of different HSV-1 strains. When more genes are included in the analysis, more accurate phylogenetic relationships of different HSV-1 strains may be obtained.
We identified 22 motifs of HSV gB. Our investigations into previous literature revealed no reports on these motifs yet. Since the motifs were conserved among different HSVs, they are likely functional. Indeed, 13 of the 22 motifs are reported to be functional, which validated our newly identified motifs. However, the 13 motifs reported in the literature and the 9 unreported motifs must be further studied to concretely determine their functions, such as induction of neutralizing antibody responses. These fascinating novel functional motifs may be closely related to gB function and HSV infection, which could attract more and more attention in the future.
The alignment and comparison of different HSV gBs revealed conserved regions of gB that overlapped with some of the predicted motifs. This finding supports that the motifs are conserved and are more likely to be functional. Our analyses not only validated the function of the newly identified motifs in our new strain by using the previous reports but also provided the reference for future oHSV motif engineering in the future. Many of the motifs will be candidates as the sites of modification for generating new oHSVs. The previous functional studies may delete or mutate one or more amino acid(s) of gB randomly, while future functional studies on gB may delete the whole motif based on our work. For example, motif 19 (Tyr889) or motif 10 (Leu871–Leu872), instead of the point mutations, may be deleted to test motif function in virus–cell fusion in future studies (33).
Manipulating envelope glycoproteins of HSVs for oncolytic virotherapy would achieve strict targeting of tumor cells as well as complete de-targeting from healthy tissues to allow maximum oncolytic efficacy and minimum adverse effects. Ideally, such engineering should promote systemic administration of oHSVs, enabling direct targeting of not only the primary tumors but also distant metastatic cancer cells. Some studies showed that intravenous administration of oHSVs produced a significant reduction of primary tumors and was effective in treating metastatic tumors (48–52). Further preclinical and clinical trials are indispensable for the intravenous administration of oHSVs. In addition, although replication of oHSVs in cancer cells is a major determinant of therapeutic efficacy, activating the immune system to kill cancer cells is an important consideration. Targeting tumor specific antigens with modified gB, or with the other modified envelope glycoproteins, does not theoretically address the issue of tumor heterogeneity, which is considered a common cause of treatment failure for targeted therapy. Therefore, the development of multi-targets and combination therapy targeting different tumor cell subgroups is the topic of the research field in the future.
One of the most critical requirements for oHSV is tumor specificity. Although cancer specificity of many oHSVs has been described, the replication and killing specificity of oHSVs still have room to improve. Engineered gB can facilitate specific virus entry, trigger extensive cell fusion, enable oHSV cultivation in the non-cancer cells, and retarget oHSV to cancer cells in combination with gD (26, 31, 34, 43, 44). Although some strategies of manipulating envelope glycoproteins of HSV have been tested successfully in preclinical experiments (22, 37, 53), in oncolytic virotherapy, it remains to be determined whether the strategy of manipulating envelope glycoproteins of HSV is appropriate for clinical translation. Future studies will likely focus on the engineering of gB to ameliorate the efficacy and safety of oHSVs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
FS: contributed to this paper with the design. FS, VX, X-QL, Y-YW, YZ, and J-TC: literature search. FS and W-QC: drafting. FS, XW, YX, and X-CP: revision. H-WX and XW: editing and final approval. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was partly supported by grants from the National Natural Science Foundation of China (81872412 to H-WX, 31700736 to XW, and 81602303 to YX), China Scholarship Council (201908420102), Hubei Medical Youth Tip-Top Talent (XW), Leading Talent Program of Yangtze Talent Project (XW), the College Students Innovative Entrepreneurial Training Programin Yangtze University (2018184 and 2019372), and Yangtze University Fellowship to graduate student FS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01386/full#supplementary-material
References
- 1.Xin HW, Hari DM, Mullinax JE, Ambe CM, Koizumi T, Ray S, et al. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. (2012) 30:591–8. 10.1002/stem.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xin HW, Ambe CM, Ray S, Kim BK, Koizumi T, Wiegand GW, et al. WNT and the cancer niche: paracrine interactions with gastrointestinal cancer cells undergoing asymmetric cell division. J Cancer. (2013) 4:447–57. 10.7150/jca.6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong-Wu Xin, Ambe CM, Miller TC, Chen JQ, Wiegand GW, Anderson AJ, et al. Liver label retaining cancer cells are relatively resistant to the reported anti-cancer stem cell drug metformin. J Cancer. (2016) 7:1142–51. 10.7150/jca.10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong-Wu Xin, Ambe CM, Hari DM, Wiegand GW, Miller TC, Chen JQ, et al. Label-retaining liver cancer cells are relatively resistant to sorafenib. Gut. (2013) 62:1777–86. 10.1136/gutjnl-2012-303261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hari D, Xin HW, Jaiswal K, Wiegand G, Kim BK, Ambe C, et al. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem Cells Dev. (2011) 20:1649–58. 10.1089/scd.2010.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell SJ, Peng KW. Viruses as anticancer drugs. Trends Pharmacol Sci. (2007) 28:326–33. 10.1016/j.tips.2007.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Wang XW, Peng XC, Xiang Y, Song SB, Wang YY, et al. CRISPR/Cas9 genome editing technology significantly accelerated herpes simplex virus research. Cancer Gene Ther. (2018) 25:93–105. 10.1038/s41417-018-0016-3 [DOI] [PubMed] [Google Scholar]
- 8.Wu ZJ, Tang FR, Ma ZW, Peng XC, Xiang Y, Zhang Y, et al. Oncolytic viruses for tumor precision imaging and radiotherapy. Hum Gene Ther. (2018) 29:204–22. 10.1089/hum.2017.189 [DOI] [PubMed] [Google Scholar]
- 9.Garber K. China approves world's first oncolytic virus therapy for cancertreatment. J Natl Cancer Inst. (2006) 98:298–300. 10.1093/jnci/djj111 [DOI] [PubMed] [Google Scholar]
- 10.Wong HH, Lemoine NR, Wang Y. Oncolytic viruses for cancer therapy: overcoming the obstacles. Viruses. (2010) 2:78–106. 10.3390/v2010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campadelli-Fiume G, de Giovanni C, Gatta V, Nanni P, Lollini PL, Menotti L. Rethinking herpes simplex virus: the way to oncolytic agents. Rev Med Virol. (2011) 21:213–26. 10.1002/rmv.691 [DOI] [PubMed] [Google Scholar]
- 12.Pol J, Kroemer G, Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. Oncoimmunology. (2015) 5:E1115641. 10.1080/2162402X.2015.1115641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XQ, Xin HY, Lyu YN, Ma ZW, Peng XC, Xiang Y, et al. Oncolytic herpes simplex virus tumor targeting and neutralization escape by engineering viral envelope glycoproteins. Drug Deliv. (2018) 25:1950–62. 10.1080/10717544.2018.1534895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr Opin Virol. (2012) 2:28–36. 10.1016/j.coviro.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 15.Herold BC, WuDunn D, Soltys N, Spear PG. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. (1991) 65:1090–8. 10.1128/JVI.65.3.1090-1098.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheshenko N, Herold BC. Glycoprotein B plays a predominant role in mediating herpes simplex virus type 2 attachment and is required for entry and cell-to-cell spread. J Gen Virol. (2002) 83(Pt 9):2247–55. 10.1099/0022-1317-83-9-2247 [DOI] [PubMed] [Google Scholar]
- 17.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. (1996) 87:427–36. 10.1016/S0092-8674(00)81363-X [DOI] [PubMed] [Google Scholar]
- 18.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. (1998) 280:1618–20. 10.1126/science.280.5369.1618 [DOI] [PubMed] [Google Scholar]
- 19.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. (1999) 99:13–22. 10.1016/S0092-8674(00)80058-6 [DOI] [PubMed] [Google Scholar]
- 20.Karasneh GA, Shukla D. Herpes simplex virus infects most cell types in vitro: clues to its success. Virol J. (2011) 8:481. 10.1186/1743-422X-8-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol. (2010) 84:12292–9. 10.1128/JVI.01700-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Q, Kopp SJ, Byskosh NC, Connolly SA, Longnecker R. Insertion of a ligand to HER2 in gB retargets HSV tropism and obviates the need for activation of the other entry glycoproteins. PLoS Pathog. (2017) 13:e1006352. 10.1371/journal.ppat.1006352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. (2006) 313:217–20. 10.1126/science.1126548 [DOI] [PubMed] [Google Scholar]
- 24.Chowdary TK, Heldwein EE. Syncytial phenotype of C-terminally truncated herpes simplex virustype 1 gB is associated with diminished membrane interactions. J Virol. (2010) 84:4923–35. 10.1128/JVI.00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, et al. Antigenic and mutational analyses of herpes simplex virusglycoprotein B reveal four functional regions. J Virol. (2007) 81:3827–41. 10.1128/JVI.02710-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, et al. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. (2010) 84:3825–34. 10.1128/JVI.02687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Q, Kopp SJ, Byskosh NC, Connolly SA, Longnecker R. Natural selection of glycoprotein B mutations that rescue the small-plaque phenotype of a fusion-impaired herpes simplex virus mutant. MBio. (2018) 9:e01948–18. 10.1128/mBio.01948-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YY, Lyu YN, Xin HY, Cheng JT, Liu XQ, Wang XW, et al. Identification of putative UL54 (ICP27) transcription regulatory sequences binding to oct-1, V-MYB, PAX-6 and hairy in herpes simplex viruses. J Cancer. (2019) 10:430–40. 10.7150/jca.29787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. (2007) 81:4858–65. 10.1128/JVI.02755-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silverman JL, Sharma S, Cairns TM, Heldwein EE. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J Virol. (2010) 84:2001–12. 10.1128/JVI.01791-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly SA, Longnecker R. Residues within the c-terminal arm of the herpes simplex virus 1 glycoprotein b ectodomain contribute to its refolding during the fusion step of virus entry. J Virol. (2012) 86:6386–93. 10.1128/JVI.00104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisner TW, Wright CC, Kato A, Kawaguchi Y, Mou F, Baines JD, et al. Herpesvirus gb-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral us3 kinase. J Virol. (2009) 83:3115–26. 10.1128/JVI.01462-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beitia Ortiz de Zarate I, Cantero-Aguilar L, Longo M, Berlioz-Torrent C, Rozenberg F. Contribution of endocytic motifs in the cytoplasmic tail of herpes simplex virus type 1 glycoprotein b to virus replication and cell-cell fusion. J Virol. (2007) 81:13889–903. 10.1128/JVI.01231-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster TP, Melancon JM, Kousoulas KG. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology. (2001) 287:18–29. 10.1006/viro.2001.1004 [DOI] [PubMed] [Google Scholar]
- 35.Israyelyan A, Chouljenko VN, Baghian A, David AT, Kearney MT, Kousoulas KG. Herpes simplex virus type-1(hsv-1) oncolytic and highly fusogenic mutants carrying the NV1020 genomic deletion effectively inhibit primary and metastatic tumors in mice. Virol J. (2008) 5:68. 10.1186/1743-422x-5-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huff V, Cai W, Glorioso JC, Levine M. The carboxy-terminal 41 amino acids of herpes simplex virus type 1 glycoprotein B are not essential for production of infectious virus particles. J Virol. (1988) 62:4403–6. 10.1128/JVI.62.11.4403-4406.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrovic B, Leoni V, Gatta V, Zaghini A, Vannini A, Campadelli-Fiume G. Dual ligand insertion in gB and gD of oncolytic herpes simplex viruses for retargeting to a producer vero cell line and to cancer cells. J Virol. (2018) 92:e02122–17. 10.1128/JVI.02122-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, Cohen JB, et al. A double mutation in glycoprotein gb compensates for ineffective Gd-dependent initiation of herpes simplex virus type 1 infection. J Virol. (2010) 84:12200–9. 10.1128/Jvi.01633-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida H, Marzulli M, Nakano K, Goins WF, Chan J, Hong CS, et al. Effective treatment of an orthotopic xenograft model of human glioblastoma using an Egfr-retargeted oncolytic herpes simplex virus. Mol Ther. (2013) 21:561–9. 10.1038/Mt.2012.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchida H, Chan J, Shrivastava I, Reinhart B, Grandi P, Glorioso JC, et al. Novel mutations in GB and GH circumvent the requirement for known GD receptors in herpes simplex virus 1 entry and cell-to-cell spread. J Virol. (2013) 87:1430–42. 10.1128/Jvi.02804-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arii J, Wang J, Morimoto T, Suenaga T, Akashi H, Arase H, et al. A single-amino-acid substitution in herpes simplex virus 1 envelope glycoprotein B at a site required for binding to the paired immunoglobulin-like type 2 receptor alpha (Pilralpha) abrogates pilralpha-dependent viral entry and reduces pathogenesis. J Virol. (2010) 84:10773–83. 10.1128/Jvi.01166-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diakidi-Kosta A, Michailidou G, Kontogounis G, Sivropoulou A, Arsenakis M. A single amino acid substitution in the cytoplasmic tail of the glycoprotein b of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. (2003) 93:99–108. 10.1016/S0168-1702(03)00070-4 [DOI] [PubMed] [Google Scholar]
- 43.Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins b and c, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. (1998) 72:6119–30. 10.1128/Jvi.72.7.6119-6130.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers E, Neumann J, Raftery M, Schonrich G, Eis-Hübinger Am, Koch N. Glycoprotein B from strain 17 of herpes simplex virus type i contains an invariant chain homologous sequence that binds to MHC class ii molecules. Immunology. (2002) 107:129–35. 10.1046/J.1365-2567.2002.01472.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laquerre S, Anderson DB, Argnani R, Glorioso JC. Herpes simplex virus type 1 glycoprotein b requires a cysteine residue at position 633 for folding, processing, and incorporation into mature infectious virus particles. J Virol. (1998) 72:4940–9. 10.1128/Jvi.72.6.4940-4949.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo S, Hu K, He S, Wang P, Zhang M, Huang X, et al. Contribution of N-linked glycans on HSV-2 gB to cell-cell fusion and viral entry. Virology. (2015) 483:72–82. 10.1016/j.virol.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 47.Shelly SS, Cairns TM, Whitbeck JC, Lou H, Krummenacher C, Cohen GH, et al. The membrane-proximal region (MPR) of herpes simplex virus GB regulates association of the fusion loops with lipid membranes. MBIO. (2012) 3:E00429–12. 10.1128/mBio.00429-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braidwood L, Learmonth K, Graham A, Conner J. Potent efficacy signals from systemically administered oncolytic herpes simplex virus (HSV1716) in hepatocellular carcinoma xenograft models. J Hepatocell Carcinoma. (2014) 1:149–61. 10.2147/JHC.S71019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomura N, Kasuya H, Shikano T, Shirota T, Misawa M, Sugimoto H, et al. Considerations for intravascular administration of oncolytic herpes virus for the treatment of multiple liver metastases. Cancer Chemother Pharmacol. (2009) 63:321–30. 10.1007/s00280-008-0742-6 [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Hu P, Rabkin SD, Liu R. Oncolytic herpes simplex virus treatment of metastatic breast cancer. Int J Oncol. (2012) 40:757–63. 10.3892/ijo.2011.1266 [DOI] [PubMed] [Google Scholar]
- 51.Walker JR, McGeagh KG, Sundaresan P, Jorgensen TJ, Rabkin SD, Martuza RL. Local and systemic therapy of human prostate adenocarcinoma with the conditionally replicating herpes simplex virus vector G207. Hum Gene Ther. (1999) 10:2237–43. 10.1089/10430349950017211 [DOI] [PubMed] [Google Scholar]
- 52.Conner J, Braidwood L, Brown SM. A strategy for systemic delivery of the oncolytic herpes virus HSV1716: redirected tropism by antibody-binding sites incorporated on the virion surface as a glycoprotein D fusion protein. Gene Ther. (2008) 15:1579–92. 10.1038/gt.2008.121 [DOI] [PubMed] [Google Scholar]
- 53.Leoni V, Gatta V, Casiraghi C, Nicosia A, Petrovic B, Campadelli-Fiume G. A strategy for cultivation of retargeted oncolytic herpes simplex viruses in non-cancer cells. J Virol. (2017) 91:e00067–17. 10.1128/JVI.00067-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.