Abstract
Infection by the Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) results in the novel coronavirus disease COVID-19, which has posed a serious threat globally. Infection of SARS-CoV-2 during pregnancy is associated with complications such as preterm labor and premature rupture of membranes, and a proportion of neonates born to infected mothers are also positive for the virus. During pregnancy, the placental barrier protects the fetus from pathogens and ensures healthy development. To predict if the placenta is permissive to SARS-CoV-2, we utilized publicly available single-cell RNA-seq data to identify if the placental cells express the necessary factors required for infection. SARS-CoV-2 binding receptor ACE2 and the S protein priming protease TMPRSS2 are co-expressed by a subset of syncytiotrophoblasts (STB) in the first trimester and extravillous trophoblasts (EVT) in the second trimester human placenta. In addition, the non-canonical receptor BSG/CD147 and other proteases (CTSL, CTSB, and FURIN) are detected in most of the placental cells. Other coronavirus family receptors (ANPEP and DPP4) were also expressed in the first and second trimester placental cells. Additionally, the term placenta of multiple species including humans expressed ACE2, DPP4, and ANPEP along with the viral S protein proteases. The ACE2- and TMPRSS2-positive (ACE2 + TMPRSS2 +) placental subsets expressed mRNA for proteins involved in viral budding and replication. These cells also had the mRNA for proteins that physically interact with SARS-CoV-2 in host cells. Further, we discovered unique signatures of genes in ACE2 + TMPRSS2 + STBs and EVTs. The ACE2 + TMPRSS2 + STBs are highly differentiated cells and express genes involving mitochondrial metabolism and glucose transport. The second trimester ACE2 + TMPRSS2 + EVTs are enriched for markers of endovascular trophoblasts. Both these subtypes abundantly expressed genes in the Toll-like receptor pathway. The second trimester EVTs are also enriched for components of the JAK-STAT pathway that drives inflammation. We carried out a systematic review and identified that in 12% of pregnant women with COVID-19, the placenta was infected with SARS-CoV-2, and the virus was detected in STBs. To conclude, herein we have uncovered the cellular targets for SARS-CoV-2 entry and have shown that these cells can potentially drive viremia in the developing human placenta. Our results provide a basic framework toward understanding the paraphernalia involved in SARS-CoV-2 infections in pregnancy.
Keywords: placenta, trophoblast, SARS-CoV-2, coronaviruses, receptors, single-cell RNA-seq, inflammation, COVID-19
Introduction
Epidemiologic evidence indicates that pregnant women are at higher risk of severe illness and mortality from viral infections such as influenza, Ebola and Lassa fever (Silasi et al., 2015). Certain viral infections during pregnancy can lead to several adverse pregnancy outcomes such as spontaneous abortion, mother-to-child transmission resulting in congenital viral syndromes, still-births and intrauterine fetal deaths (Silasi et al., 2015; Arora et al., 2018). Furthermore, viral infection also predisposes the pregnancy toward preterm birth, which has major long-term health implications for the newborn. Thus, understanding the health risks of viral infections during pregnancy is vital for designing appropriate approaches for its clinical management. The importance of understanding the role of viral infection during pregnancy gains further relevance as we are confronted with newer pandemics, which may affect the pregnant mother and the fetus.
Coronaviruses (CoV) are positive-sense RNA viruses that, upon zoonotic transmission, lead to respiratory disease in humans and some animals. Previous outbreaks of zoonotic coronaviruses, including the severe acute respiratory syndrome (SARS)-CoV and the Middle East respiratory syndrome (MERS)-CoV, have proven to be of great public health concern. Another outbreak of severe acute respiratory syndrome called coronavirus disease-2019 (COVID-19) has been recently reported, which is due to infection by a novel coronavirus termed SARS-CoV-2 (Zhu et al., 2020). The infection has spread rapidly worldwide due to high human-to-human transmission, resulting in a public health emergency of international concern (Bharti et al., 2020; Gupta et al., 2020; Li R. et al., 2020). Presently, there are no specific treatments available for COVID-19, and there is an urgent need to identify the drugs and vaccines targeted against this virus (Prajapat et al., 2020).
Since SARS-CoV-2 has not been detected in humans before, limited information is available about its health effects; negligible information is available for pregnant women. In pregnant women, COVID-19 is associated with severe pregnancy complications such as preterm labor and premature rupture of membranes (Gajbhiye et al., 2020). Furthermore, a proportion of neonates born to mothers with COVID-19 are positive for the virus, suggesting the possibility of vertical transmission through the placental barrier (Gajbhiye et al., 2020; Knight et al., 2020).
The placenta is a highly specialized organ that maintains the equilibrium between immunological and biochemical factors required for fetal development (Deshpande and Balasinor, 2018). It also acts as a barrier for vertical transmission of pathogens (Maltepe and Fisher, 2015; Burton et al., 2016). However, some viruses such as Zika can infect the placental cells via receptors on trophoblasts, leading to fetal malformation and pregnancy complications (Hirsch et al., 2018; Tabata et al., 2018). Interestingly, placental and amniotic fluid infections of SARS-CoV-2 have been reported (Baud et al., 2020; Zamaniyan et al., 2020). For SARS-CoV-2 to be able to infect the placenta, the host cells must harbor the necessary receptors and virus-processing machinery. It has been shown that SARS-CoV-2 binds and infects host cells by utilizing the membrane-bound Angiotensin-Converting Enzyme II (ACE2), which is considered its canonical mode of action (Jagtap et al., 2020; Letko et al., 2020; Shang et al., 2020). In addition, SARS-CoV-2 binds to CD147/Basigin (BSG) on the cell surface that may act as an alternate non-canonical receptor (Wang et al., 2020). Upon receptor binding, the viral-encoded S protein requires cleavage by host proteases for efficient membrane fusion. The main host protease that mediates S protein priming and initiates viral entry is the Type II transmembrane serine protease TMPRSS2 (Hoffmann et al., 2020). The endosomal protease cathepsin L (CTSL) can also enhance viral entry (Hoffmann et al., 2020). Thus, the presence of such receptors and S protein primer proteases in host cells is a key determinant of SARS-CoV-2 infection. Indeed, expression of ACE2 and TMPRSS2 have been detected in lung airway cells and the upper respiratory epithelium, the primary site of SARS-CoV-2 action (Sungnak et al., 2020; Ziegler et al., 2020). Beyond respiratory distress, some patients with SARS-CoV-2 viremia develop multiple organ injuries, and cells of these tissues also express ACE2 and TMPRSS2 (Qi et al., 2020; Seow et al., 2020; Zou et al., 2020).
The binding of enveloped viruses like SARS-CoV-2 to its receptors results in events related to membrane fusion and/or endocytosis followed by establishment of the primary infection. Following its entry and uncoating, coronavirus replication is initiated by translation of its non-structural proteins including the replicases that allow viral RNA synthesis and capping. This course requires a network of host factors to create an optimal environment for facilitating viral entry, gene expression, RNA synthesis and virus release (de Wilde et al., 2018). Further, most enveloped viruses bud at the plasma membrane by recruiting the host endosomal sorting complex required for transport (ESCRT) machinery (Ahmed et al., 2019; Gatta and Carlton, 2019). While the precise host proteins in SARS-CoV-2 entry and replication are not yet understood, its host interactome has been characterized (Gordon et al., 2020). The host proteins that interact with SARS-CoV-2 are involved in endocytosis and replication of viruses (Gordon et al., 2020). Thus, elucidating tissue and cell-type-specific host machinery that not only mediate viral entry but also replication and budding from the host cell is essential to understand the pathogenesis of SARS-CoV-2 infection.
Single-cell RNA sequencing (scRNA-seq) of different tissues has transformed our ability to map the types, subsets and states of cells in healthy and diseased conditions in an unprecedented manner (Sharma et al., 2018; Szabo et al., 2019; Iyer et al., 2020). Recently, scRNA-seq has been applied to expand our understanding of the cellular landscape during viral infection including that of SARS-CoV-2 (Russell et al., 2018; Galinato et al., 2019; Liao et al., 2020). scRNA-seq has also been used in the identification of various tissues and cells that are potential targets of SARS-CoV-2, and these studies have immensely contributed toward expanding our understanding of the molecular characteristics of the host cells that are targets of viral infection (Colaco et al., 2020; Lukassen et al., 2020; Qi et al., 2020; Seow et al., 2020; Singh et al., 2020; Sungnak et al., 2020; Zhang et al., 2020).
To gain an insight into the pathogenesis of SARS-CoV-2 infection during pregnancy, it is essential to identify and characterize the placental cell types that express the viral receptors ACE2 and BSG/CD147, along with the proteases TMPRSS2 and CTSL. Recent studies have reported ACE2-positive cells in early embryonic trophoblasts as well as first trimester human placenta (Colaco et al., 2020; Li M. et al., 2020; Singh et al., 2020). ACE2 protein is also detected in term human placenta1. Studies have also shown that BSG/CD147 is expressed in first trimester human trophoblasts and gestational day 18 mouse placenta (Bharadwaj et al., 2011; Lee et al., 2013). However, information regarding the cells co-expressing various coronavirus receptors and S protein proteases as well as their detailed characteristics in the placenta is unknown. Herein (Ashray et al., 2020), we surveyed the publicly available scRNA-seq data of human placenta for the expression of the SARS-CoV-2 receptors ACE2 and BSG/CD147, along with the S protein proteases TMPRSS2 and CTSL. We also surveyed the placental cells for the expression of DPP4 and ANPEP, which are utilized by MERS-CoV and CoV-229E, respectively. The study also aimed to characterize the ACE2- and TMPRSS2-positive placental cells for their possible roles in viral endocytosis, replication, SARS-CoV-2 interactions and viral budding. The results reveal that placental cells are potential targets for SARS-CoV-2 infection.
Materials and Methods
To identify the population of human placental cells that express ACE2, BSG, TMPRSS2, and CTSL at single-cell resolution, we analyzed scRNA-seq data of first and second trimester human placenta (Liu et al., 2018) [Accession number GSE89497]. This dataset is derived out of 7 first trimester (8 weeks) placentae and 1 second trimester placenta (24 weeks) in which single cells were isolated by enzymatic digestion followed by enrichment of cells using a combination of methods. Extravillous trophoblasts (EVTs) were enriched by Magnetic Activated Cell Sorting (MACS) using anti-HLA-G antibody, cytotrophoblasts (CTBs) were enriched using anti-CDH1 antibody, and syncytiotrophoblasts (STBs) were manually sorted. The HLA-G- and CDH1-negative fraction was designated to be villous stromal cells (STR). To isolate EVTs from the placenta at 24 weeks, the basal plate was dissected from the villi of the placenta, and the single cells were prepared using enzymatic digestion followed by MACS using anti-HLA-G antibody. We deliberately chose this “index sorted” dataset over the unbiased agnostic scRNA-seq datasets since our major focus was to identify the cellular targets of SARS-CoV-2 specifically in the placenta, and such a priori approach allows analysis of homogeneous cell populations and accurate linking of rare transcripts with the index sorted cells.
To understand changes in the expression of ACE2, BSG, TMPRSS2, and CTSL in the second and third trimester placenta, bulk RNA-seq data was analyzed [Accession number GSE124282]. This dataset is derived out of 4 second trimester and term human placentae (Wang et al., 2019). Bulk RNA-seq data (Armstrong et al., 2017) was also analyzed of term placenta from human, cow, dog, armadillo, elephant, opossum, mouse and bonobo samples [Accession number GSE79121] to understand if the expression of the SARS-CoV-2 receptors in the term placenta is evolutionarily conserved. Pseudo-bulk scRNA-seq data of human first trimester decidua (Suryawanshi et al., 2018) was then analyzed to understand the distribution of ACE2, BSG, TMPRSS2, and CTSL in different maternal cell populations.
We profiled the mRNA levels of 27 host proteins involved in human ESCRT for viruses (Ahmed et al., 2019) and mRNA levels of 30 proteins involved in viral replication (de Wilde et al., 2018) in placental cells that co-express ACE2 and TMPRSS2 in first and second trimester (Supplementary Table 1). We also analyzed the transcript profiles of 332 human proteins that physically interact with SARS-CoV-2 in placental cells that co-express ACE2 and TMPRSS2 (Gordon et al., 2020). To guard against viral infection, host cells express a plethora of genes that sense the presence of the virus on the cell surface, in cytosol and in endosomes. This in turn activates the host defense mechanisms to limit or eliminate viral infection and restore tissue homeostasis. We profiled the expression of 487 host viral response genes in placental cells that co-express ACE2 and TMPRSS2 (Supplementary Table 1).
To characterize the trophoblast cells that co-express ACE2 and TMPRSS2 as well as their counterparts that do not express both of these genes; we carried out pseudo-bulk analysis of ACE2- and TMPRSS2-positive (ACE2 + TMPRSS2 +) and ACE2- and TMPRSS2-negative (ACE2–TMPRSS2–) trophoblast cells. Single-cell data for ACE2 + TMPRSS2 + and ACE2–TMPRSS2– cells was independently aggregated and the mean TPM values were computed. The data was filtered for genes whose mean values were ≥0.1 TPM and the ratio of the mean value in ACE2 + TMPRSS2 + cells over ACE2–TMPRSS2– cells was calculated. The genes that had a ratio of ≥1.5 or ≤0.5 and p-value of <0.05 were filtered and the data was deconvoluted for single cells. Gene ontology (GO) analysis was performed using the PANTHER database and over-representation tests were performed using reference genes of PANTHER pathways2.
To determine if SARS-CoV-2 is detected in the placenta, one author (AB) carried out a systematic review with the keywords “placenta and SARS-CoV-2,” “COVID-19 and placenta,” “SARS-CoV-2 and pregnancy,” “coronaviruses” and “pregnancy.” The primary outcome was to determine the number of cases reporting the presence of SARS-CoV-2 in placental tissue; the secondary outcome was to determine the placental cell types positive for SARS-CoV-2. Searches were carried out in PubMed, Google Scholar, MedRxiv, bioRxiv and other preprint databases. Articles reporting primary data in which detection of SARS-CoV-2 was carried out by either RT-PCR and/or by immunohistochemistry or electron microscopy were included. Reviews, blog and newspaper reports were excluded. Data was entered in tabular format and was independently verified by another author (DM).
All the data was processed using R Studio version 3.6.2. Heatmaps were plotted using the heatmap.2 function from the gplots R package and the pheatmap R package. Uniform Manifold Approximation and Projection (UMAP) analysis was performed using the UMAP 0.2.6.0 R package. To identify the cell clusters, 500 genes with high SD and average log2-transformed expression >1 were selected. Next, these 500 genes were given as an input to UMAP for calculating the projections of all cells. Statistical analysis was done using the Welch’s t-test and the graphs were plotted in GraphPad Prism version 8.0.
Results
Trophoblast Cells Express mRNA for SARS-CoV-2 Receptors and Spike Protein Processing Enzymes
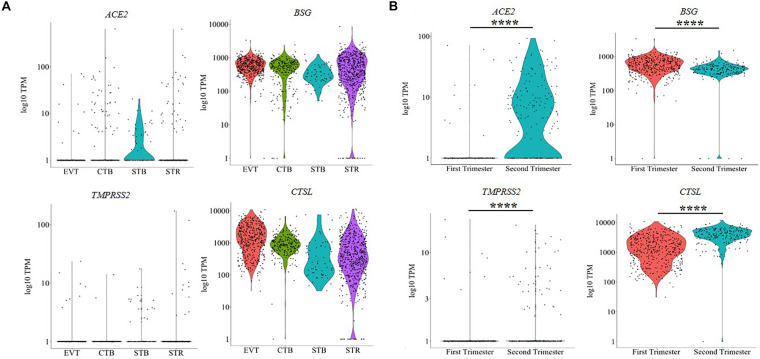
The human placenta is characterized by four distinct cell lineages: extravillous trophoblasts (EVT), cytotrophoblasts (CTB), syncytiotrophoblasts (STB), and villous stromal cells (STR). To understand the distribution of the SARS-CoV-2 receptors in these cell types, we analyzed publicly available single-cell transcriptome data from human placenta. The results revealed that ACE2, BSG, TMPRSS2 and CTSL were expressed in all the cell types of first trimester placenta; however, not every cell of each type expressed these genes (Figure 1A). As evident, STBs represented the highest proportion of ACE2-expressing cells (39%) in first trimester placenta, whereas only 2% of EVTs had ACE2 expression. BSG was abundantly expressed in almost all the cells (96–100%) of EVT, CTB, STB, and STR of the first trimester (Supplementary Table 2). Very few cells of the human placenta expressed TMPRSS2. The highest TMPRSS2 expression was detected in STBs of first trimester placenta (23%), whereas only 1% of CTBs had TMPRSS2 expression (Supplementary Table 2). CTSL was expressed in nearly all STBs, CTBs, EVTs and STRs of first trimester placenta (Figure 1A). The numbers of cells that express these genes individually are given in Supplementary Table 2.
FIGURE 1.
Trophoblast cells express mRNA for SARS-CoV-2 receptors and spike protein processing enzymes. (A) mRNA level of SARS-CoV-2 receptors (ACE2 and BSG) and spike protein primer enzymes (TMPRSS2 and CTSL) in different cell types of first trimester human placenta [EVT (n = 440), CTB (n = 248), STB (n = 64), STR (n = 615)]. (B) Comparison of the mRNA levels of ACE2, BSG, TMPRSS2, and CTSL in EVT of first (n = 440 cells) and second (n = 200 cells) trimester human placenta. Each dot represents data of a single cell, the Y axis represents log10 Transcripts Per Million (TPM). Bar within the cluster denotes mean. Horizontal black bars denote significantly different values (**** indicates p-value ≤ 0.0001). Data was extracted from single-cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497]. EVT, extravillous trophoblast; CTB, cytotrophoblast; STB, syncytiotrophoblast; STR, villous stromal cell.
We then compared the expression of these genes in EVTs of the first trimester and second trimester human placenta (Figure 1B). 2% of first trimester EVTs and 62% of second trimester EVTs expressed ACE2. Similarly, the numbers of EVTs expressing TMPRSS2 also increased in the second trimester as compared to the first trimester (2 vs. 19%) (Supplementary Table 2). In both cases, the increase was statistically significant (p-value ≤ 0.001). Nearly all EVT cells in the first and the second trimesters expressed BSG and CTSL (Supplementary Table 2). BSG expression was significantly reduced in second trimester EVTs as compared to the first trimester (p-value ≤ 0.001); the expression of CTSL was significantly higher in second trimester EVTs as compared to first trimester EVTs (p-value ≤ 0.001) (Figure 1B).
We also studied the expression of other SARS-CoV receptors DPP4 and ANPEP, which are utilized by MERS-CoV and CoV-229E, respectively. ANPEP was detected in the EVTs, CTBs and STRs but not in the STBs of the first trimester placenta. DPP4, on the other hand, was expressed by all CTBs, STBs, EVTs, and STRs (Supplementary Figure 1A). As compared to first trimester EVTs, the levels of ANPEP significantly decreased (p-value ≤ 0.001), while those of DPP4 significantly increased (p-value ≤ 0.001) in the second trimester EVTs (Supplementary Figure 1B).
Bulk transcriptome analysis revealed that, in comparison to the second trimester, the levels of ACE2, BSG, and DPP4 were reduced in the term placenta; however, a statistically significant reduction was observed only in the case of BSG (p-value ≤ 0.05). Although the levels of TMPRSS2 and ANPEP were higher in the term placenta as compared to second trimester placenta, this increase was not statistically significant. CTSL levels were similar in the second trimester and term placentae (Supplementary Figure 2A). In the absence of publicly available index sorted scRNA-seq data of term placenta, we cannot comment on the cell types that express these genes.
The expression of coronavirus receptors and spike protein proteases was compared in term placenta of different species (Supplementary Figure 2B). ACE2 transcripts were detected in the placenta of most mammals except bonobo. BSG mRNA was expressed in term placenta of all the species, albeit at varying levels. TMPRSS2 mRNA was expressed in the placenta of most species except dog, armadillo, and elephant. CTSL mRNA was detected in human, cow, dog, mouse, and bonobo placentae. ANPEP was only expressed in human, armadillo and mouse placentae, and DPP4 was expressed in placenta of all species except mouse.
To determine if endometrial cells express these genes, we analyzed pseudo-bulk data of the first trimester feto-maternal interface (Supplementary Figure 3). Expression of ACE2 was detected in smooth muscle cells, and a low abundance of ACE2 transcripts was also detected in decidual stromal cells and fibroblasts, vascular endothelial cells and NK cells. TMPRSS2 transcripts were detected in endometrial epithelial cells and lymphatic endothelial cells. BSG and CTSL were detected in all the maternal cells of the first trimester feto-maternal interface.
Co-expression of mRNA of SARS-CoV-2 Receptors and Spike Protein Processing Enzymes in Human Placental Cells
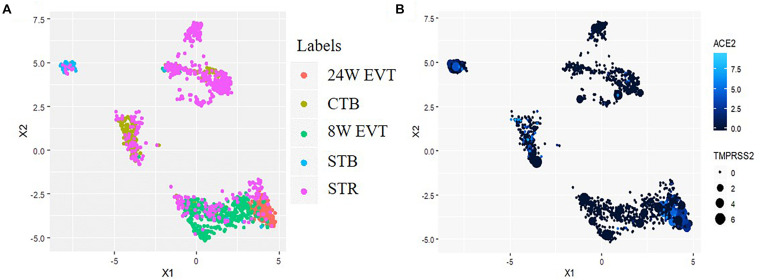
Uniform Manifold Approximation and Projection revealed distinct clusters of CTBs and STBs, and EVTs in the first trimester placenta; the second trimester EVTs also formed an independent cluster (Figure 2A). Some STR cells clustered independently while others clustered with the different trophoblast cell types, suggesting that the STRs are not a pure population. This finding was expected since the STRs were the post-enrichment leftover fractions of CTBs, STBs and EVTs. The data on this population was hence excluded in further analysis. SARS-CoV-2 infection in host cells requires coordinated expression of the entry receptor ACE2 and S protein primer TMPRSS2. UMAP analysis revealed that a subset of CTBs, STBs and EVTs co-expressed both ACE2 and TMPRSS2 (Figure 2B).
FIGURE 2.
Uniform Manifold Approximation and Projection (UMAP) plot illustrating cell clusters and ACE2 & TMPRSS2 expressing cells in the human placenta. Data was extracted from single-cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497]. (A) Clusters consist of three trophoblast lineages (EVT, extravillous trophoblast; CTB, cytotrophoblast; STB, syncytiotrophoblast) and the villous stromal cells (STR). (B) The points colored blue are the cells co-expressing ACE2 and TMPRSS2. In each blue point, the intensity of the color represents ACE2 expression and the size of the point represents the extent of TMPRSS2 expression.
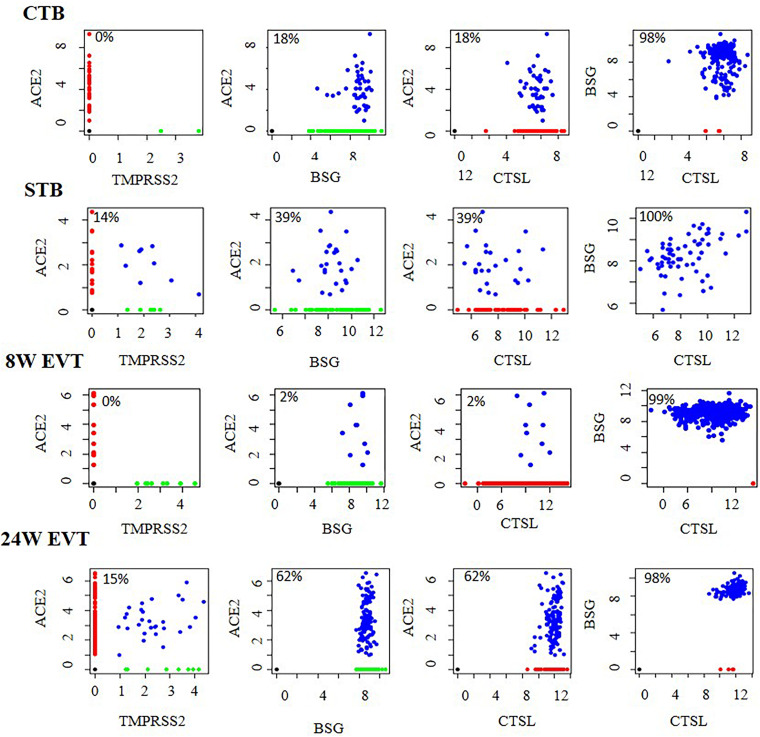
We next evaluated CTBs, STBs and EVTs co-expressing ACE2, TMPRSS2, BSG, and CTSL in different combinations (Figure 3). The results revealed that a subset of STBs (14%) in the first trimester placenta co-expressed ACE2 and TMPRSS2 (Supplementary Table 3). No other cell types in first trimester placenta expressed this receptor and S protein primer protease pair, although there were cells expressing ACE2 (Figure 3). However, 15% of EVTs in the second trimester placenta co-expressed ACE2 and TMPRSS2 (Supplementary Table 3).
FIGURE 3.
Co-expression of mRNA of SARS-CoV-2 receptors and spike protein processing enzymes in human placental cells. Co-expression of ACE2 and TMPRSS2, ACE2 and BSG, ACE2 and CTSL, and BSG and CTSL in STB (n = 64), CTB (n = 248), first trimester EVT (8W EVT, n = 440) and second trimester EVT (24W EVT, n = 200). Each dot represents data of a single cell. Values in each box are the percentage of co-expressing cells. Co-expressing cells are blue; single-positive cells are red and green. X and Y axes represent log2 Transcripts Per Million (TPM) values for that gene. Data was extracted from single cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497]. EVT, extravillous trophoblast; CTB, cytotrophoblast; STB, syncytiotrophoblast.
In the first trimester placenta, all the ACE2-positive trophoblast subtypes co-expressed BSG and CTSL, and all the BSG-positive cells co-expressed CTSL. All the ACE2-positive second trimester EVTs co-expressed BSG and CTSL, and all the BSG-positive cells co-expressed CTSL (Figure 3). The absolute numbers and percentages of the co-expressing cells are given in Supplementary Table 3.
ACE2 + TMPRSS2 + First Trimester Syncytiotrophoblast Cells Are Highly Differentiated and Express the Machinery for Viral Endocytosis, Replication and Budding
Since only STBs co-expressed ACE2 and TMPRSS2 in the first trimester, we carried out an in-depth characterization of these cells. 14% of the total STB population of the first trimester placenta expressed both ACE2 and TMPRSS2, 52% did not express either, and the rest of the cells expressed either ACE2 or TMPRSS2 (Supplementary Table 3). We compared the expression profiles of classical STB genes between the ACE2 and TMPRSS2 co-expressing cells and ACE2- and TMPRSS2-negative STBs. We observed that both cell types abundantly expressed the transcripts for human chorionic gonadotropin beta 5 (CGB5) and somatomammotropin [placental lactogen (CSH1)], as well as steroid hormone biosynthesis enzymes (HSD17B1 and CYP19A1). Further, both these subsets of STBs abundantly expressed the other putative SARS-CoV-2 S protein primers FURIN and Cathepsin B (CTSB) (Supplementary Figure 4).
We next characterized the transcriptome differences between the ACE2 + TMPRSS2 + versus the ACE2–TMPRSS2– STB cells. Pseudo-bulk analysis identified 817 genes (including ACE2 and TMPRSS2) between these two cell types (Supplementary Table 4). Of these, 444 were over represented while the others were under represented in the ACE2 + TMPRSS2 + cells as compared to ACE2–TMPRSS2– cells. These genes were heterogeneously expressed in the ACE2–TMPRSS2– STBs, while most ACE2 + TMPRSS2 + cells uniformly expressed these genes (Figure 4A). The biological processes enriched by these genes included regulation of G1/S cell-cycle checkpoints, actin polymerization/depolymerization, regulation of mitochondrial membrane permeability and electron-transport-coupled ATP synthesis, monosaccharide transport and unfolded protein response (Figure 4B). Most of the ACE2 + TMPRSS2 + cells significantly overexpressed the transcription factor OVOL1 (a terminal STB differentiation marker), and the glucose transport regulators GPC3 and SLC2A9 (p-value ≤ 0.05). The expression of ACTN1 (an actin binding protein) was significantly downregulated in ACE2 + TMPRSS2 + versus ACE2–TMPRSS2– STBs (p-value ≤ 0.05) (Figure 4C).
FIGURE 4.
ACE2 + TMPRSS2 + first trimester syncytiotrophoblast cells are terminally differentiated and express the machinery for viral endocytosis, replication and budding (A) Distribution of 817 differentially expressed genes in ACE2- and TMPRSS2-positive (n = 9) (ACE2 + TMPRSS2 +) and ACE2- and TMPRSS2-negative (ACE2– TMPRSS2–) cells (n = 33) of the first trimester syncytiotrophoblast. Rows represent genes and columns represent individual cells, presented on a relative color scale. (B) Biological processes enriched in the differentially expressed genes of the first trimester syncytiotrophoblast. The Y axis indicates the enriched biological processes and X axis is the -log10 of the raw p-values. (C) mRNA levels of OVOL1, GPC3, SLC2A9, and ACTN1 in ACE2–TMPRSS2– and ACE2 + TMPRSS2 + cells. Each dot represents data of a single cell, the Y axis represents Transcripts Per Million (TPM). Bars denote mean ± SD. Horizontal black bars denote significantly different values (* indicates p-value ≤ 0.05, ** indicates p-value ≤ 0.001). (D) Heatmap showing the expression of genes involved in endosomal sorting complexes required for transport (ESCRT), replication and host genes involved in SARS-CoV-2 interaction. In all heat maps, each row depicts a gene and each column depicts a single ACE2- and TMPRSS2-positive (n = 9) (ACE2 + TMPRSS2 +) cell in the first trimester syncytiotrophoblast. The data is presented on a relative color scale in which the minimum and maximum values in each row are used to convert values to colors. Data was extracted from single cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497].
We next analyzed the mRNA levels of 27 genes involved in human ESCRT of viruses and 30 host genes involved in SARS-CoV replication in ACE2- and TMPRSS2-positive STB cells. All the ACE2 + TMPRSS2 + STBs uniformly expressed most of these genes (Figure 4A); however, the other cells showed heterogeneous expression across the different subtypes (Supplementary Figure 5). We also analyzed the mRNA levels of 332 host proteins that are known to interact with SARS-CoV-2 and found that there was minimal heterogeneity in expression of these genes in the first trimester STBs (Figure 4D) as compared to other cell types (Supplementary Figure 5).
Second Trimester ACE2 + TMPRSS2 + Cells Are Invasive Extravillous Trophoblasts and Express Markers of Endovascular Trophoblasts
Amongst the second trimester EVTs, 15% of cells were ACE2 + TMPRSS2 + while 33% did not express either of the transcripts (Supplementary Table 3). Two populations of second trimester EVTs are reported and characterized by the expression of TAC3. Type 1 EVTs are TAC3-high and express genes involved in migration and invasion; type 2 EVTs are TAC3-low cells that express genes involved in cell proliferation (Liu et al., 2018). In addition to TAC3, the type 1 EVTs also express JAM2, SERPENIN1 and PRG2 (Liu et al., 2018). We observed that the levels of TAC3 were marginally but not significantly higher in ACE2 + TMPRSS2 + EVTs (Supplementary Figure 6A), and the mRNA levels of other genes were identical in ACE2 + TMPRSS2 + EVTs as compared to cells not expressing either of the two genes (ACE2– TMPRSS2–) (Supplementary Figure 6A). Principal component analysis did not reveal major differences in the transcriptome of the ACE2 + TMPRSS2 + and ACE2–TMPRSS2– cells (Supplementary Figure 6B). We compared the expression profiles of classical EVT genes between the ACE2 + TMPRSS2 + and ACE2–TMPRSS2– cells and observed that both the cell types abundantly expressed the transcripts for HLA-G and ITGB1 (Supplementary Figure 6C). Both these subsets of EVTs also abundantly expressed other SARS-CoV-2 S protein primer proteins FURIN and CTSB of which the levels of FURIN were significantly higher (p-value ≤ 0.05) in ACE2 + TMPRSS2 + EVTs as compared to ACE2–TMPRSS2– EVTs (Supplementary Figure 6C).
To characterize if there are any specific classes of genes differentially abundant between the ACE2 + TMPRSS2 + versus the ACE2–TMPRSS2– EVT cells, pseudo-bulk analysis was carried out. There were 983 differentially abundant genes (including ACE2 and TMPRSS2) between these two cell types (Supplementary Table 5) of which 931 were overrepresented and 52 were underrepresented in the ACE2 + TMPRSS2 + cells as compared to ACE2–TMPRSS2– cells. Further, these genes were heterogeneously expressed in the ACE2–TMPRSS2– EVTs while most ACE2 + TMPRSS2 + cells uniformly expressed these genes (Figure 5A). Most of these differentially abundant genes enriched several GO biological processes such as viral entry, release and intracellular transport. The other enriched GO biological processes were nucleic acid replication, epithelial morphogenesis and cell migration (Figure 5B).
FIGURE 5.
Second trimester ACE2 + TMPRSS2 + cells are invasive extravillous trophoblasts and express markers of endovascular trophoblasts. (A) Distribution of 983 differentially expressed genes in ACE2- and TMPRSS2-positive (n = 29) (ACE2 + TMPRSS2 +) and ACE2- and TMPRSS2-negative (n = 66) (ACE2–TMPRSS2–) cells of the second trimester extravillous trophoblast. Rows represent genes and columns represent individual cells, presented on a relative color scale. (B) Biological processes enriched in the ACE2 + TMPRSS2 + cells (n = 29) of the second trimester extravillous trophoblast. The Y axis indicates the enriched biological processes and X axis is the enrichment score. (C) Comparison of mRNA level of CDH5, VCAM, CCR1, CD59, OVOL2, ICAM, and AKT1 in ACE2–TMPRSS2- (n = 66) and ACE2 + TMPRSS2 + (n = 29) cells. Each dot represents data of a single cell, the Y axis represents Transcripts Per Million (TPM). Bars denote mean ± SD. Horizontal black bars denote significantly different values (* indicates p-value ≤ 0.05, ** indicates p-value ≤ 0.001, **** indicates p-value < 0.0001). (D) Heatmap showing the expression of genes involved in endosomal sorting complexes required for transport (ESCRT), replication and host genes involved in SARS-CoV-2 interaction. In all heat maps, each row depicts a gene and each column depicts a single ACE2 + TMPRSS2 + cell of the second trimester extravillous trophoblast. The data is presented on a relative color scale in which the minimum and maximum values in each row are used to convert values to colors. Data was extracted from single cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497].
The ACE2 + TMPRSS2 + cells significantly overexpressed the markers of endovascular trophoblasts CDH5, VCAM, CCR1 and CD59 (p-value ≤ 0.05) (Figure 5C). These cells also significantly overexpressed OVOL2, the marker of terminally differentiated EVTs, and the invasion-related marker AKT1 (p-value ≤ 0.05) (Figure 5C). ICAM and GJA5 are markers for EVTs in anchoring cell columns, and their levels were identical in the ACE2 + TMPRSS2 + and the ACE2–TMPRSS2– EVT cells.
Analysis of the mRNA levels of 27 genes involved in human ESCRT of viruses and 30 host genes involved in SARS-CoV replication in ACE2- and TMPRSS2-positive cells at single-cell resolution revealed that all the ACE2 + TMPRSS2 + EVTs uniformly expressed most of these genes (Figure 5D), while the first trimester EVTs that had no ACE2 + TMPRSS2 + cells had a very heterogeneous expression of these genes (Supplementary Figure 5). We also analyzed the mRNA levels of 332 host proteins that interact with SARS-CoV-2 and observed that almost all these genes were expressed in most ACE2 + TMPRSS2 + second trimester EVTs (Figure 5D), while the first trimester EVTs had heterogeneous expression (Supplementary Figure 5).
Unique Signatures of Genes Involved in Viral Response in First Trimester Syncytiotrophoblast Cells and Second Trimester Extravillous Trophoblasts
We studied the baseline expression of 487 genes involved in viral response in both first trimester STBs and second trimester EVTs (Supplementary Table 1). Only a subset of viral response genes were expressed in ACE2 + TMPRSS2 + EVTs and STBs (Figure 6A). The heatmaps showed minimal heterogeneity across cells but high variability in expression across genes involved in the viral response. To characterize these genes in EVT and STB cells, genes were clustered by a hierarchical clustering method using the “hclust” function available in R. Genes were grouped based on the most optimum threshold, which resulted in four different gene clusters (Figure 6B). The average of the gene expression in Clusters 1 and 2 were identical in both EVTs and STBs. However, average expression of genes in Clusters 3 and 4 was more abundant in EVTs as compared to STBs (Figure 6C).
FIGURE 6.
Viral response genes in first trimester syncytiotrophoblasts and second trimester extravillous trophoblasts. (A) Heatmap showing the expression of genes involved in viral response in ACE2- and TMPRSS2-positive (ACE2 + TMPRSS2 +) cells of first trimester STB (n = 9) and second trimester EVT (n = 29). Each row is a gene and each column represents data for a single ACE2 and TMPRSS2 co-expressing cell. (B) Cluster dendrogram of first trimester STB and second trimester EVT cells. The horizontal axis of the dendrogram represents the distance or dissimilarity between clusters. The vertical axis represents the clusters. (C) Mean of four clusters of upregulated genes in first trimester STB and second trimester EVT cells. Y axis represents cluster mean expression and X axis represents different cell types. (D) Pathways enriched in the in four clusters of first trimester STBs and second trimester EVTs, with their enrichment score and p-values. EVT, extravillous trophoblast; STB, syncytiotrophoblast. Data was extracted from single cell RNA-seq of human placenta (Liu et al., 2018) [Accession number GSE89497].
Next, GO analysis was performed using the PANTHER database for all four gene clusters. For each cluster, an over-representation test was performed using reference genes of PANTHER pathways, and a pathway with the highest fold-enrichment value was selected as the enriched pathway for a given gene cluster. GO classification of these clusters revealed that most of the genes in Cluster 1 had a role in the Toll-like receptor (TLR) signaling response and the genes in Cluster 2 had a role in apoptosis. Additionally, Cluster 3, which included genes of the JAK-STAT pathway, and Cluster 4, which included genes for axon guidance mediated by semaphorins, were enriched in EVTs as compared to STBs (p-value ≤ 0.05) (Figure 6D).
SARS-CoV-2 Infects the Human Placenta and Is Localized in Syncytiotrophoblast Cells
To determine if SARS-CoV-2 can infect human placenta, a systematic review was carried out (Supplementary Table 6). Seventeen studies that reported analysis of SARS-CoV-2 in placental tissue from 93 pregnant women with COVID-19 were identified. In these studies, SARS-CoV-2 was detected by reverse transcriptase PCR (RT-PCR), immunohistochemistry or electron microscopy. Of the 93 placentae, 5 were second trimester placentae and 88 were term/preterm placentae. In all, 12% of placentae were reported to be positive for SARS-CoV-2. Second trimester placentae from all 5 women with COVID-19 were positive for SARS-CoV-2 while term/preterm placenta from ∼7% of women with COVID-19 had SARS-CoV-2 positivity. Immunohistochemistry of 1 second trimester and 2 term placentae revealed the presence of SARS-CoV-2 protein in STBs. Viral particles were also identified in STBs of second trimester and preterm (28 weeks) placenta by electron microscopy (Supplementary Table 6).
Discussion
Herein, we utilized scRNA-seq to identify and characterize the potential cellular targets of SARS-CoV-2 infection in human placenta. To review the data presented: (1) The mRNA for coronavirus receptors (ACE2, BSG, DPP4, and ANPEP) and the S protein proteases (TMPRSS2 and CTSL) are expressed in the first trimester to term placenta, (2) The SARS-CoV-2 binding receptor ACE2 and the S protein priming protease TMPRSS2 are co-expressed by a subset of syncytiotrophoblasts (STB) in the first trimester and extravillous trophoblasts (EVT) in the second trimester human placenta, (3) These ACE2 + TMPRSS2 + subsets are highly differentiated STBs and endovascular EVTs, (4) The ACE2 + TMPRSS2 + placental subsets readily express mRNA for proteins involved in ESCRT of viruses and replication, in addition to transcripts for proteins that are known to interact with SARS-CoV-2 structural and non-structural proteins, and (5) The STBs and EVTs differentially express genes involved in the host response to viral infection.
Using a scRNA-seq dataset (Vento-Tormo et al., 2018), ACE2-positive CTBs and STBs are reported in the first trimester human placenta (Li M. et al., 2020; Singh et al., 2020). Corroborating these findings using a different dataset (Liu et al., 2018), we show the presence of ACE2 in CTBs and STBs of first trimester placenta. However, along with STBs and CTBs, we also identified ACE2 expression in EVTs of the first and second trimester placenta, which has not been reported earlier. In addition to ACE2-expressing STBs and EVTs, our study revealed that BSG/CD147, the alternate receptor for SARS-CoV-2 (Wang et al., 2020), is expressed by almost all the placental cells. We also detected abundant expression of DPP4 (the receptor for MERS-CoV) and ANPEP (the receptor for CoV-229E) in the cells of the placenta. Like ACE2, DPP4 was detected in all the cell types of the first trimester placenta and also in EVTs of the second trimester; very few STBs expressed ANPEP. Similar observations are made using different datasets of scRNA-seq of first trimester human placenta (Pique-Regi et al., 2020; Singh et al., 2020). In addition to the first and second trimester placenta, ACE2, TMPRSS2, BSG, ANPEP, and DPP4 transcripts are also detected in human term placenta by bulk RNA-seq. However, we cannot comment on the cell types that express these genes due to the lack of publicly available scRNA-seq datasets of term human placenta. Interestingly, these genes are also expressed in the term placenta of different species including mice, cows, dogs, armadillos, elephants, opossums and bonobos. Thus, we propose that multiple cell types in the placental tissue could be targets of different coronaviruses throughout gestation.
While ACE2 is the primary receptor for SARS-CoV-2 entry, the S protein of SARS-CoV-2 undergoes cleavage by a cell surface protease, TMPRSS2 (Hoffmann et al., 2020). Whether ACE2 and TMPRSS2 are required on the same cell to activate SARS-CoV-2 S protein to invade ACE2 single-positive cells is a matter of investigation. However, as active S protein has a finite lifetime (Shulla et al., 2011), its processing at the plasma membrane will make it most effective for viral entry. Thus, we assumed that for SARS-CoV-2, the ACE2 and TMPRSS2 co-expressing cells would have the highest infectivity. Our analysis revealed that a proportion of STBs (14%) in the first trimester and a subset of EVTs (15%) in second trimester human placenta co-express ACE2 and TMPRSS2. Contradicting this proposition, Pique-Regi et al. (2020) reported that co-expression of ACE2 and TMPRSS2 is negligible in the trophoblasts of human first, second and third trimester placenta. Differences in the methods of tissue sampling, cell isolation and inefficiencies in detection of low-abundance transcripts in scRNA-seq can underestimate the actual frequencies of ACE2 + cells in a given tissue. Indeed, the dataset used in this study is exclusively of MACS enriched trophoblast preparations while the datasets used by Pique-Regi et al. (2019, 2020) are a mixed population of cells from the feto-maternal interface. It is known that fractionated cell preparations allow better identification of low abundance transcripts in rare cell populations (Nguyen et al., 2018). These factors could be the possible reasons that our analysis could identify more numbers of ACE2 and TMPRSS2 co-expressing cell types in the human placenta. While the numbers of placental cells co-expressing ACE2 and TMPRSS2 may appear insignificant considering the total placental volume, it must be borne in mind that only 3–6% of lung airway epithelial cell subtypes (the primary site of SARS-CoV-2 action) co-express both ACE2 and TMPRSS2 (Ziegler et al., 2020).
Beyond the canonical ACE2 and TMPRSS2 based entry, SARS-CoV-2 also utilizes BSG/CD147 as the non-canonical mode of entry (Wang et al., 2020). Presently, the mechanism by which BSG/CD147 mediates viral entry in host cells is unknown. In other cells, BSG/CD147 promotes entry of viruses by endocytosis (Pushkarsky et al., 2001). It is possible that the same mechanism may be operative in the case of SARS-CoV-2. In this context, it is interesting that all the BSG/CD147-positive cells abundantly co-expressed the endosomal protease CTSL. Further, we observed that almost all the ACE2 + STBs and EVTs co-expressed BSG/CD147, suggesting that more than one mechanism may operate for viral entry in these cells of the human placenta. Beyond TMPRSS2, studies have identified that SARS-CoV-2 may have a FURIN cleavage site, leading to a broader set of host proteases that could mediate S protein priming (Coutard et al., 2020). The ACE2 + TMPRSS2 + STBs and EVTs abundantly express FURIN as well as another endosomal protease, CTSB. Together our data conclusively show that multiple cells of human placenta are targets for SARS-CoV-2 binding and entry with S protein priming by both canonical and non-canonical pathways.
We next aimed to characterize the placental cells that are potential targets for SARS-CoV-2 infection. As the ACE2-mediated viral entry is a well-established mechanism, we focused only on characterizing the STBs of the first trimester and EVTs of the second trimester placenta that co-express both ACE2 and TMPRSS2 in a proportion of cells while others are devoid of these transcripts. In the developing placenta, trophoblast stem cells differentiate into cytotrophoblasts, which undergo further differentiation to form the non-self-renewing cytotrophoblasts, extravillous trophoblasts and syncytiotrophoblasts (Turco and Moffett, 2019; Hemberger et al., 2020). The syncytiotrophoblasts covering the villi are major hormone secreting cells and function as a protective immunological barrier (Maltepe and Fisher, 2015; Gupta et al., 2016; Liu et al., 2018; Vento-Tormo et al., 2018; Turco and Moffett, 2019). We observed that STBs that co-express both ACE2 and TMPRSS2 also express the mRNA for the peptide hormones and enzymes for steroid hormone biosynthesis. However, their levels are not significantly different from their ACE2–TMPRSS2– counterparts, suggesting that both these cell types retain the basic functions of STBs. However, pseudo-bulk analysis revealed that the ACE2 + TMPRSS2 + cells are enriched for genes involved in cell cycle checkpoints, actin filament remodeling, mitochondrial functions, hexose transport and type I interferon signaling. Indeed, the terminally differentiated STBs have replicative senescence and require extensive cytoskeletal remodeling for syncytialization; the mitochondria of STBs play a key role in progesterone synthesis by providing cholesterol (Martinez et al., 2015). Additionally, these cells are enriched in OVOL1, the transcription factor required for STB specification, as well as proteins involved in glucose transport across the feto-maternal barrier, a key function of well differentiated STBs (Jansson and Ylve, 2002; Renaud et al., 2015; Vento-Tormo et al., 2018; Turco and Moffett, 2019). These results imply that the ACE2 + TMPRSS2 + cells are a subset of highly differentiated STBs and these cells are potential targets for viral entry. Indeed, SARS-CoV-2 mRNA, protein and virions are detected in STBs of second trimester and term/preterm placenta from a woman with COVID-19 (Hosier et al., 2020). Additionally, increased syncytiotrophoblastic knots are observed in placenta from pregnant women with COVID-19 (Chen et al., 2020), which is suggestive of injury to the STBs in the placenta.
We next probed the second trimester EVTs, 15% of which co-express ACE2 and TMPRSS2. The EVTs differentiate from cytotrophoblast stem cells and populate the tips of the placental villi to form the anchoring villi, thus defining the boundary between mother and fetus. The EVTs are central to placentation as they invade into the maternal decidua and are involved in remodeling of maternal spiral arteries, veins and lymphatic ducts (Sharma et al., 2016; Pollheimer et al., 2018). We observed that the ACE2 + TMPRSS2 + cells abundantly express the classical EVT marker ITGB1 and also HLA-G that induces tolerogenic immune responses leading to acceptance of the semi-allogeneic fetus. Two kinds of EVTs are reported in the second trimester human placenta: the proliferative EVTs in the cell columns and the invasive endovascular or interstitial EVTs (Pollheimer et al., 2018; Turco and Moffett, 2019), and both of these have a unique transcript signature (Liu et al., 2018). Herein, we observed that while the levels of TAC3 and other molecules associated with columnar versus invasive EVTs are not significantly different between ACE2 + TMPRSS2 + and ACE2–TMPRSS2– second trimester EVTs, the double-positive cells overexpressed key invasive EVT markers such as OVOL2, GJA5, ICAM and AKT1 (Sharma et al., 2016; Bai et al., 2018; Liu et al., 2018; Jeyarajah et al., 2020), suggesting that these cells are invasive trophoblasts. We further observed that many of the ACE2 + TMPRSS2 + cells were enriched for genes having a role in cell migration. The EVTs can either invade the decidua (designated as interstitial EVTs) or remodel the spiral arteries (designated as endovascular EVTs). While both these EVTs are invasive in nature, they have differential expression of certain marker genes. For example, endovascular EVTs overexpress the CDH5 and VCAM, they also have higher expression of CCR1 and CD59 (Bulla et al., 2005; Cartwright and Balarajah, 2005; Liu et al., 2018; Ueda et al., 2019; Sato, 2020). Intriguingly, the ACE2 + TMPRSS2 + cells also overexpressed several of the key endovascular EVT markers including CDH5, CCD5, CD59, and VCAM, indicating that the ACE2 + TMPRSS2 + population of second trimester EVTs are potentially endovascular trophoblasts and are targets of SARS-CoV-2 infection. Indeed, pseudo-bulk analysis of the ACE2 + TMPRSS2 + cells and ACE2–TMPRSS2– cells revealed significant enrichment of genes with GO terms involving regulation of viral release from host cells. Most of the ACE2 + TMPRSS2 + endovascular EVTs abundantly expressed most genes whose protein products in the host are known to be involved in human endocytosis and budding of viruses and replication. Together, this data shows that SARS-CoV-2 may affect the invading EVTs at the feto-maternal interface in the second trimester and can result in damaged vasculature. In this context, it is important to note that the maternal endothelial cells in the decidua also express ACE2 and BSG, making the maternal endothelium another entry point of SARS-CoV-2 infection at the feto-maternal interface. Any impairment in functions of these cells can cause placental damage and vertical transmission of the virus. Indeed, increased intervillous and subchorionic fibrin deposition and fetal thrombotic vasculopathy with zones of avascular fibrotic villi are observed in placenta of women infected with coronaviruses including SARS CoV-2 (Ng et al., 2006; Baud et al., 2020; Hosier et al., 2020; Mulvey et al., 2020). Together, these results indicate that the integrity of endovascular trophoblasts and the endothelial compartment of the feto-maternal interface may be compromised in women with SARS-CoV-2 infection.
Once the virus binds to its receptors on host cells and gains entry, it utilizes a plethora of host genes for its replication. Post replication, most enveloped viruses complete their life-cycle by forming vesicles that bud from the plasma membrane via the cellular ESCRT (endosomal sorting complexes required for transport) machinery. Interestingly, we observed that the ACE2 + TMPRSS2 + STBs and EVTs were enriched for the key genes that encode for proteins involved in ESCRT and viral replication. Using affinity-purification mass spectrometry, 332 human proteins that interact with SARS-CoV-2 have been identified, and many of these play a role in ESCRT and viral replication (Gordon et al., 2020). We observed that ACE2 + TMPRSS2 + STBs and EVTs abundantly expressed most of these genes. Thus SARS-CoV-2 may hijack proteins in the EVTs and STBs thereby interfering with normal placental functions. In this context, it is important to highlight that a significant proportion of the human SARS-CoV-2 interacting proteins also interact with proteins of other viruses including Zika and Hepatitis C virus, which replicate in the trophoblast cells (Giugliano et al., 2015; Tabata et al., 2016, 2018; Gordon et al., 2020). Together our data strongly implies that first trimester STBs and second trimester EVTs are not just targets for SARS-CoV-2 entry, but also the virus may be potentially pathogenic to these cells.
A proportion of SARS-CoV-2 proteins target the components of innate immune signaling pathways, including NF-kappa-B (Gordon et al., 2020). We decided to probe this in detail by profiling the STBs and EVTs for 487 genes whose protein products are involved in viral response in host cells. We observe that only a proportion of these genes are expressed in most EVTs and STBs. Based on their expression levels in the host cells, they could be classified in four clusters, and interestingly, the first trimester STBs and the second trimester EVTs expressed genes in the TLR signaling pathway, the primary response to viral infection. Previous studies in SARS-CoV have identified involvement of TLR pathways in protection against viral response (Dosch et al., 2009; Totura et al., 2015). However, we found that the genes in the JAK-STAT pathway were overexpressed in the EVTs but not in the STBs. However, this is not surprising as the JAK-STAT pathway is required for physiological functions of EVTs, namely invasion (Fitzgerald et al., 2010; Suman et al., 2013; Sharma et al., 2016; Godbole et al., 2017). The JAK-STAT pathway is also central for mounting a host response to viral infection, and treatment with interferon gamma induces the expression of interferon-stimulated genes in EVT cells (Verma et al., 2020). Thus, EVTs are not only the entry sites for SARS-CoV-2 infection, but they also possess the cellular machinery to mount an inflammatory response toward an infection. With regard to coronaviruses (including SARS-CoV-2), an overexuberant inflammatory response is observed even at lower viral titres, which contributes to the viral pathogenicity in the lung (Liao et al., 2020). Further, the ACE2 receptors are induced by interferon signaling in the lung (Ziegler et al., 2020), thereby amplifying the infectious cycle in host tissues. Whether or not a similar mechanism is operative in placental cells is under investigation, but the heightened baseline expression of the JAK-STAT pathway genes in the EVTs itself could readily lead to placental inflammation that may be detrimental to pregnancy. Infiltration of leukocytes and chorioamnionitis is observed in placenta from women with coronavirus infection (Ng et al., 2006; Baud et al., 2020). Since inflammation of the feto-maternal interface causes preterm births (Silasi et al., 2015; Surve et al., 2016), it is plausible that the increased incidence of preterm delivery in women with COVID-19 could be linked to this process.
Beyond preterm births, the demonstration that the ACE2 + TMPRSS2 + subpopulation of EVTs consists of invasive endovascular trophoblasts is clinically relevant in conditions like preeclampsia. The invasion of the trophoblast cells and remodeling of the spiral arteries deep into the myometrium is essential for normal fetal growth and development (Norwitz, 2006; Soares et al., 2015). If the arteries are not sufficiently remodeled, there is disordered perfusion of blood and an inadequate supply of nutrients and oxygen, resulting in fetal growth restriction, stillbirth, preeclampsia, placental abruption and preterm labor (Brosens et al., 2019). Since SARS-CoV-2 and other coronaviruses may target the endovascular trophoblasts, it is plausible that the infection could lead to other adverse pregnancy outcomes. Indeed, higher incidence of preeclampsia, preterm labor, fetal distress and premature rupture of membranes are reported in pregnant women infected with SARS-CoV-2 in the third trimester (Gajbhiye et al., 2020). Also, high rates of miscarriages, preterm birth and premature rupture of membranes have been reported for other human coronavirus infections (Alfaraj et al., 2019; Mullins et al., 2020). These observations imply that SARS-CoV-2 infection is detrimental to pregnancy due the possible infection of placental cells such as EVTs.
To determine whether SARS-CoV-2 can infect the placental cells, we carried out a systematic review to identify studies that report presence or absence of the virus in placenta of women with COVID-19. The results revealed that ∼12% (11/93) of placentae obtained from mothers with COVID-19 had detectable levels of SARS-CoV-2 RNA (Supplementary Table 6). Viral RNA, non-structural proteins and intact virions are detected in STBs of the second trimester and preterm/term placenta. This is definitive evidence of placental infection by SARS-CoV-2. However, this may not be an accurate estimate of the frequency of placental infection as most studies included women at term with unknown viral loads and duration of infection. Nevertheless, the fact that some studies have also detected the virus in the amniotic fluid and fetal membranes (Baud et al., 2020; Zamaniyan et al., 2020) as well as IgM in fetal blood (Zeng et al., 2020) implies that the placenta does get infected and the virus can cross the transplacental barrier to infect the fetus. In a systematic review and primary data from a large cohort of pregnant women, mother-to-child transmission of the virus is observed in 5–8% of cases (Gajbhiye et al., 2020; Knight et al., 2020). Thus, it appears likely that the placenta is not just permissive to viral entry but can be a site of active viremia that can lead to the breakthrough of SARS-CoV-2 infection from mother to fetus.
To summarize, this is the first in-depth survey to identify the cellular basis of SARS-CoV-2 infection in the human placenta. However, this data is limited by the constraints of scRNA-seq which includes host and environmental factors that may affect the expression of receptors and proteases. Experimental variations like sites of tissue collection, cell isolation techniques and statistical cut-offs may lead to inclusion or exclusion of specific cell types. Furthermore, our observations need to be corroborated for protein expression. Nevertheless, our results provide a basic framework in understanding of the paraphernalia involved in SARS-CoV-2 infections in pregnancy. It will be essential to determine how SARS-CoV-2 infection alters the temporal dynamics of host responses at the single-cell resolution in the placenta. We believe that this work will aid in developing rational strategies for management of COVID-19 and other coronavirus infections in pregnancy.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
NA, AB, and PC analyzed and interpreted the data and prepared the figures. SC and AM were involved in data analysis and preparing the manuscript. KC prepared the figures and edited the manuscript. DM conceived the idea and planned this study. DM and MJ spearheaded the study and were involved in data interpretation. All authors were involved in manuscript writing and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Mr. Domdatt Singh for help in editing the manuscript. The study makes use of the datasets bearing the accession numbers GSE89497, GSE124282, and GSE79121. We express our gratitude to the authors for making these datasets publically available. KC is thankful to Nehru-Fulbright Scholarship. AM is thankful to University Grants Commission (UGC) Government of India for research fellowship. NA and AB are thankful to DST-SERB for project assistantship. The manuscript bears the NIRRH ID RA/908/05-2020. The pre-print of this manuscript is available at https://www.preprints.org/manuscript/202005.0195/v1.
Funding. DM lab was funded by grants from Indian Council of Medical Research (ICMR), Government of India and IMPRINT IIC grant (IMP/2019/000115/HT) from Department of Science and Technology, Government of India. MJ was supported by Ramanujan Fellowship (SB/S2/RJN-049/2018) awarded by SERB, DST, Government of India. SC was supported by funding from Department of Health Research, Government of India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00783/full#supplementary-material
References
- Ahmed I., Akram Z., Iqbal H. M. N., Munn A. L. (2019). The regulation of endosomal sorting complex required for transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview. Int. J. Biol. Macromol. 127 1–11. 10.1016/j.ijbiomac.2019.01.015 [DOI] [PubMed] [Google Scholar]
- Alfaraj S. H., Al-Tawfiq J. A., Memish Z. A. (2019). Middle East respiratory syndrome coronavirus (MERS-CoV) infection during pregnancy: report of two cases & review of the literature. J. Microbiol. Immunol. Infect. 52 501–503. 10.1016/j.jmii.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D. L., McGowen M. R., Weckle A., Pantham P., Caravas J., Agnew D., et al. (2017). The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 57 71–78. 10.1016/j.placenta.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora N., Sadovsky Y., Dermody T. S., Coyne C. B. (2018). Microbial vertical transmission during human pregnancy. Cell 21 561–567. 10.1016/j.chom.2017.04.007.Microbial [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashray N., Bhide A., Chakarborty P., Colaco S., Mishra A., Chhabria K., et al. (2020). Single-cell RNA-seq identifies cell subsets in human placenta that highly expresses factors to drive pathogenesis of SARS-CoV-2. Preprints 2020050195. 10.20944/preprints202005.0195.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R., Kusama K., Nakamura K., Sakurai T., Kimura K., Ideta A., et al. (2018). Down-regulation of transcription factor OVOL2 contributes to epithelial-mesenchymal transition in a noninvasive type of trophoblast implantation to the maternal endometrium. FASEB J. 32 3371–3384. 10.1096/fj.201701131RR [DOI] [PubMed] [Google Scholar]
- Baud D., Greub G., Frave G., Gengler C., Jaton K., Dubruc E., et al. (2020). Second-Trimester Miscarriage in a Pregnant Woman With SARS-CoV-2 Infection. JAMA J. Am. Med. Assoc. 323 2198–2200. 10.1001/jama.2020.7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj M. S., Strawn W. B., Groban L., Yamaleyeva L. M., Chappell M. C., Horta C., et al. (2011). Angiotensin-converting enzyme 2 deficiency is associated with impaired gestational weight gain and fetal growth restriction. Hypertension 58 852–858. 10.1161/HYPERTENSIONAHA.111.179358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti S., Narad P., Chugh P., Choudhury A., Bhatnagar S., Sengupta A. (2020). Multi-parametric disease dynamics study and analysis of the COVID-19 epidemic and implementation of population-wide intrusions: the Indian perspective. medRxiv [Preprint]. 10.1101/2020.06.02.20120360 [DOI] [Google Scholar]
- Brosens I., Puttemans P., Benagiano G. (2019). Placental bed research: I. The placental bed: from spiral arteries remodeling to the great obstetrical syndromes. Am. J. Obstet. Gynecol. 221 437–456. 10.1016/j.ajog.2019.05.044 [DOI] [PubMed] [Google Scholar]
- Bulla R., Villa A., Bossi F., Cassetti A., Radillo O., Spessotto P., et al. (2005). VE-cadherin is a critical molecule for trophoblast-endothelial cell interaction in decidual spiral arteries. Exp. Cell Res. 303 101–113. 10.1016/j.yexcr.2004.09.015 [DOI] [PubMed] [Google Scholar]
- Burton G. J., Fowden A. L., Thornburg K. L. (2016). Placental origins of chronic disease. Physiol. Rev. 96 1509–1565. 10.1152/physrev.00029.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright J. E., Balarajah G. (2005). Trophoblast interactions with endothelial cells are increased by interleukin-1β and tumour necrosis factor α and involve vascular cell adhesion molecule-1 and α4β1. Exp. Cell Res. 304 328–336. 10.1016/j.yexcr.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Chen S., Huang B., Luo D. J., Li X., Yang F., Zhao Y., et al. (2020). [Pregnant women with new coronavirus infection: a clinical characteristics and placental pathological analysis of three cases]. Chinese J. Pathol. 49:E005. 10.3760/cma.j.cn112151-20200225-00138 [DOI] [PubMed] [Google Scholar]
- Colaco S., Chhabria K., Singh N., Bhide A., Singh D., Singh A., et al. (2020). Expression of SARS-CoV-2 Receptor ACE2 and the Spike Protein Processing Enzymes in Developing Human Embryos. Available online at: http://arxiv.org/abs/2004.04935 (accessed May 7, 2020). [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N. G., Decroly E. (2020). The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176:104742. 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A. H., Snijder E. J., Kikkert M., van Hemert M. J. (2018). Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 49 1–42. 10.1007/82_2017_25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S. S., Balasinor N. H. (2018). Placental defects: an epigenetic perspective. Reprod. Sci. 25 1143–1160. 10.1177/1933719118766265 [DOI] [PubMed] [Google Scholar]
- Dosch S. F., Mahajan S. D., Collins A. R. (2009). SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-κB pathway in human monocyte macrophages in vitro. Virus Res. 142 19–27. 10.1016/j.virusres.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J. S., Germeyer A., Huppertz B., Jeschke U., Knöfler M., Moser G., et al. (2010). Governing the invasive trophoblast: current aspects on intra- and extracellular regulation. Am. J. Reprod. Immunol. 63 492–505. 10.1111/j.1600-0897.2010.00824.x [DOI] [PubMed] [Google Scholar]
- Gajbhiye R., Modi D., Mahale S. (2020). Pregnancy outcomes, newborn complications and maternal-fetal transmission of SARS-CoV-2 in women with COVID-19: a systematic review. medRxiv [Preprint]. 10.1101/2020.04.11.20062356 [DOI] [Google Scholar]
- Galinato M., Shimoda K., Aguiar A., Hennig F., Boffelli D., McVoy M. A., et al. (2019). Single-cell transcriptome analysis of CD34+stem cell-derived myeloid cells infected with human cytomegalovirus. Front. Microbiol. 10:577. 10.3389/fmicb.2019.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta A. T., Carlton J. G. (2019). The ESCRT-machinery: closing holes and expanding roles. Curr. Opin. Cell Biol. 59 121–132. 10.1016/j.ceb.2019.04.005 [DOI] [PubMed] [Google Scholar]
- Giugliano S., Petroff M. G., Warren B. D., Jasti S., Linscheid C., Ward A., et al. (2015). Hepatitis C virus sensing by human trophoblasts induces innate immune responses and recruitment of maternal NK cells: potential implications for limiting vertical transmission. J. Immunol. 195 3737–3747. 10.4049/jimmunol.1500409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbole G., Suman P., Malik A., Galvankar M., Joshi N., Fazleabas A., et al. (2017). Decrease in expression of HOXA10 in the decidua after embryo implantation promotes trophoblast invasion. Endocrinology 158 2618–2633. 10.1210/en.2017-00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Praharaj I., Bhatnagar T., Thangaraj J. W. V., Giri S., Chauhan H., et al. (2020). Severe acute respiratory illness surveillance for coronavirus disease 2019, India, 2020. Indian J. Med. Res. 151 236–240. 10.4103/ijmr.IJMR_1035_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Malhotra S. S., Malik A., Verma S., Chaudhary P. (2016). Cell signaling pathways involved during invasion and syncytialization of trophoblast cells. Am. J. Reprod. Immunol. 75 361–371. 10.1111/aji.12436 [DOI] [PubMed] [Google Scholar]
- Hemberger M., Hanna C. W., Dean W. (2020). Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 21 27–43. 10.1038/s41576-019-0169-4 [DOI] [PubMed] [Google Scholar]
- Hirsch A. J., Roberts V. H. J., Grigsby P. L., Haese N., Schabel M. C., Wang X., et al. (2018). Zika virus infection in pregnant rhesus macaques causes placental dysfunction and immunopathology. Nat. Commun. 9 1–15. 10.1038/s41467-017-02499-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 271.e8–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosier H., Farhadian S. F., Morotti R. A., Deshmukh U., Lu-Culligan A., Campbell K. H., et al. (2020). SARS-CoV-2 infection of the placenta. J. Clin. Invest. (in press). 10.1172/JCI139569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A., Gupta K., Sharma S., Hari K., Lee Y. F., Ramalingam N. N., et al. (2020). Integrative analysis and machine learning based characterization of single circulating tumor cells. J. Clin. Med. 9:1206. 10.3390/jcm9041206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap D., Selvaa Kumar C., Mahale S., Patel V. (2020). Modelling and docking of Indian SARS-CoV-2 spike protein 1 with ACE2: implications for co-morbidity and therapeutic intervention. arXiv [Preprint]. Available online at: http://arxiv.org/abs/2004.06361 (accessed June 12, 2020). [Google Scholar]
- Jansson T., Ylve K. (2002). Glucose transport and system a activity in syncytiotrophoblast microvillous and basal plasma membranes in intrauterine growth. Placenta 23 392–399. 10.1053/plac.2002.0826 [DOI] [PubMed] [Google Scholar]
- Jeyarajah M. J., Jaju Bhattad G., Hillier D. M., Renaud S. J. (2020). The transcription factor OVOL2 represses ID2 and drives differentiation of trophoblast stem cells and placental development in mice. Cells 9:840. 10.3390/cells9040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M., Bunch K., Vousden N., Morris E., Simpson N., Gale C., et al. (2020). Characteristics and outcomes of pregnant women admitted to hospital with confirmed SARS-CoV-2 infection in UK: national population based cohort study. BMJ 369:m2107. 10.1136/bmj.m2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. L., Lam M. P. Y., Lam K. K. W., Leung C. O. N., Pang R. T. K., Chu I. K., et al. (2013). Identification of CD147 (basigin) as a mediator of trophoblast functions. Hum. Reprod. 28 2920–2929. 10.1093/humrep/det355 [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5 562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen L., Zhang J., Xiong C., Li X. (2020). The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One 15:e0230295. 10.1371/journal.pone.0230295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., et al. (2020). Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 368 489–493. 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. (2020). The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv [Preprint]. 10.1101/2020.02.23.20026690 [DOI] [Google Scholar]
- Liu Y., Fan X., Wang R., Lu X., Dang Y. L., Wang H. H., et al. (2018). Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28 819–832. 10.1038/s41422-018-0066-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R. L., Trefzer T., Kahn N. C., Schneider M. A., Muley T., et al. (2020). SARS-CoV-2 receptor ACE2 and TMPRSS2 are predominantly expressed in a transient secretory cell type in subsegmental bronchial branches. bioRxiv [preprint]. 10.1101/2020.03.13.991455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltepe E., Fisher S. J. (2015). Placenta: the forgotten organ. Annu. Rev. Cell Dev. Biol. 31 523–552. 10.1146/annurev-cellbio-100814-125620 [DOI] [PubMed] [Google Scholar]
- Martinez F., Olvera-Sanchez S., Esparza-Perusquia M., Gomez-Chang E., Flores-Herrera O. (2015). Multiple functions of syncytiotrophoblast mitochondria. Steroids 103 11–22. 10.1016/j.steroids.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Mullins E., Evans D., Viner R. M., O’Brien P., Morris E. (2020). Coronavirus in pregnancy and delivery: rapid review. Ultrasound Obstet. Gynecol. 55 586–592. 10.1002/uog.22014 [DOI] [PubMed] [Google Scholar]
- Mulvey J. J., Magro C. M., Ma L. X., Nuovo G. J., Baergen R. N. (2020). Analysis of complement deposition and viral RNA in placentas of COVID-19 patients. Ann. Diagn. Pathol. 46:151529. 10.1016/j.anndiagpath.2020.151529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. F., Wong S. F., Lam A., Mak Y. F., Yao H., Lee K. C., et al. (2006). The placentas of patients with severe acute respiratory syndrome: a pathophysiological evaluation. Pathology 38 210–218. 10.1080/00313020600696280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q. H., Pervolarakis N., Nee K., Kessenbrock K. (2018). Experimental considerations for single-cell RNA sequencing approaches. Front. Cell Dev. Biol. 6:108. 10.3389/fcell.2018.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz E. R. (2006). Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod. Biomed. Online 13 591–599. 10.1016/S1472-6483(10)60649-9 [DOI] [PubMed] [Google Scholar]
- Pique-Regi R., Romero R., Tarca A. L., Luca F., Xu Y., Alazizi A., et al. (2020). Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? BioRxiv [Preprint]. 10.1101/2020.05.18.101485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R., Romero R., Tarca A. L., Sendler E. D., Xu Y., Garcia-Flores V., et al. (2019). Single cell transcriptional signatures of the human placenta in term and preterm parturition. eLife 8:e52004. 10.7554/eLife.52004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollheimer J., Vondra S., Baltayeva J., Beristain A. G., Knöfler M. (2018). Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front. Immunol. 9:2597. 10.3389/fimmu.2018.02597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., et al. (2020). Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 52 56–65. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., et al. (2001). CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. U.S.A. 98 6360–6365. 10.1073/pnas.111583198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Zhou Y., Hua J., Zhang L., Bian J., Liu B., et al. (2020). The scRNA-seq expression profiling of the receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to COVID-19 infection. BioRxiv [Preprint]. 10.1101/2020.04.16.045690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S. J., Chakraborty D., Mason C. W., Karim Rumi M. A., Vivian J. L., Soares M. J. (2015). OVO-like 1 regulates progenitor cell fate in human trophoblast development. Proc. Natl. Acad. Sci. U.S.A. 112 E6175–E6184. 10.1073/pnas.1507397112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Trapnell C., Bloom J. D. (2018). Extreme heterogeneity of influenza virus infection in single cells. eLife 7 1–26. 10.7554/eLife.32303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. (2020). Endovascular trophoblast and spiral artery remodeling. Mol. Cell. Endocrinol. 503:110699. 10.1016/j.mce.2019.110699 [DOI] [PubMed] [Google Scholar]
- Seow J. J. W., Pai R., Mishra A., Shepherdson E., Lim T. K. H., Goh B. K. P., et al. (2020). scRNA-seq reveals ACE2 and TMPRSS2 expression in TROP2+ Liver Progenitor Cells: implications in COVID-19 associated Liver Dysfunction. BioRxiv [Preprint]. 10.1101/2020.03.23.002832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature 581 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Cao E. Y., Kumar V., Zhang X., Leong H. S., Wong A. M. L., et al. (2018). Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy. Nat. Commun. 9:4931. 10.1038/s41467-018-07261-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Godbole G., Modi D. (2016). Decidual control of trophoblast invasion. Am. J. Reprod. Immunol. 75 341–350. 10.1111/aji.12466 [DOI] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. (2011). A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85 873–882. 10.1128/jvi.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silasi M., Cardenas I., Kwon J. Y., Racicot K., Aldo P., Mor G. (2015). Viral infections during pregnancy. Am. J. Reprod. Immunol. 73 199–213. 10.1111/aji.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Bansal V., Feschotte C., Feschotte C. (2020). A single-cell RNA expression map of human coronavirus entry factors. bioRxiv [Preprint]. 10.1101/2020.05.08.084806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M. J., Chakraborty D., Kubota K., Renaud S. J. (2015). Adaptive mechanisms controlling uterine spiral artery remodeling during the establishment of pregnancy. Int. J. Dev. Biol. 58 247–259. 10.1387/ijdb.140083ms [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suman P., Malhotra S. S., Gupta S. K. (2013). LIF-STAT signaling and trophoblast biology. JAK-STAT 2:e25155. 10.4161/jkst.25155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., et al. (2020). SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26 681–687. 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surve M. V., Anil A., Kamath K. G., Bhutda S., Sthanam L. K., Pradhan A., et al. (2016). Membrane vesicles of group B Streptococcus disrupt feto-maternal barrier leading to preterm birth. PLoS Pathog. 12:e1005816. 10.1371/journal.ppat.1005816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H., Morozov P., Straus A., Sahasrabudhe N., Max K. E. A., Garzia A., et al. (2018). A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4 1–12. 10.1126/sciadv.aau4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P. A., Levitin H. M., Miron M., Snyder M. E., Senda T., Yuan J., et al. (2019). Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 10:4706. 10.1038/s41467-019-12464-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Petitt M., Puerta-Guardo H., Michlmayr D., Harris E., Pereira L. (2018). Zika virus replicates in proliferating cells in explants from first-trimester human placentas, potential sites for dissemination of infection. J. Infect. Dis. 217 1202–1213. 10.1093/infdis/jix552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Petitt M., Puerta-Guardo H., Michlmayr D., Wang C., Fang-Hoover J., et al. (2016). Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 20 155–166. 10.1016/j.chom.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A. L., Whitmore A., Agnihothram S., Schäfer A., Katze M. G., Heise M. T., et al. (2015). Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 6:e 00638-15. 10.1128/mBio.00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco M. Y., Moffett A. (2019). Development of the human placenta. Development 146 1–14. 10.1242/dev.163428 [DOI] [PubMed] [Google Scholar]
- Ueda M., Sato Y., Horie A., Tani H., Miyazaki Y., Okunomiya A., et al. (2019). Endovascular trophoblast expresses CD59 to evade complement-dependent cytotoxicity. Mol. Cell. Endocrinol. 490 57–67. 10.1016/j.mce.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R. A., Turco M. Y., Vento-Tormo M., Meyer K. B., et al. (2018). Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563 347–353. 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Kang A. K., Pal R., Gupta S. K. (2020). BST2 regulates interferon gamma-dependent decrease in invasion of HTR-8/SVneo cells via STAT1 and AKT signaling pathways and expression of E-cadherin. Cell Adhes. Migr. 14 24–41. 10.1080/19336918.2019.1710024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang P., Parobchak N., Treff N., Tao X., Wang J., et al. (2019). Integrated RNA-seq and ChIP-seq analysis reveals a feed-forward loop regulating H3K9ac and key labor drivers in human placenta. Placenta 76 40–50. 10.1016/j.placenta.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., et al. (2020). SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv [Preprint]. 10.1101/2020.03.14.988345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamaniyan M., Ebadi A., Aghajanpoor Mir S., Rahmani Z., Haghshenas M., Azizi S. (2020). Preterm delivery in pregnant woman with critical COVID -19 pneumonia and vertical transmission. Prenat. Diagn. [Epub ahead of print]. 10.1002/pd.5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Xu C., Fan J., Tang Y., Deng Q., Zhang W., et al. (2020). Antibodies in Infants Born to Mothers with COVID-19 Pneumonia. JAMA J. Am. Med. Assoc. 323 1848–1849. 10.1001/jama.2020.4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z. Z., et al. (2020). Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut (in press). 10.1136/gutjnl-2020-320953 [DOI] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C., Allon S. J., Nyquist S. K., Mbano I., Miao V. N., Cao Y., et al. (2020). SARS-CoV-2 Receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. SSRN Electron. J. 181 1016.e19–1035.e19. 10.2139/ssrn.3555145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14 185–192. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.