ABSTRACT
Background
Around 25% of oral cavity squamous cell carcinoma (OCSCC) are not controlled by the standard of care, but there is currently no validated biomarker to identify those patients. Our objective was to determine a robust biomarker for severe OCSCC, using a biology-driven strategy.
Patients and methods
Tumor and juxtatumor secretome were analyzed in a prospective discovery cohort of 37 OCSCC treated by primary surgery. Independent biomarker validation was performed by RTqPCR in a retrospective cohort of 145 patients with similar clinical features. An 18-gene signature (18 G) predictive of the response to PD-1 blockade was evaluated in the same cohort.
Results
Among 29 deregulated molecules identified in a secretome analysis, including chemokines, cytokines, growth factors, and molecules related to tumor growth and tissue remodeling, only soluble MMP2 was a prognostic biomarker. In our validation cohort, high levels of MMP2 and CD276, and low levels of CXCL10 and STAT1 mRNA were associated with poor prognosis in univariate analysis (Kaplan-Meier). MMP2 (p = .001) and extra-nodal extension (ENE) (p = .006) were independent biomarkers of disease-specific survival (DSS) in multivariate analysis and defined prognostic groups with 5-year DSS ranging from 36% (MMP2highENE+) to 88% (MMP2lowENE-). The expression of 18 G was similar in the different prognostic groups, suggesting comparable responsiveness to anti-PD-1.
Conclusion
High levels of MMP2 were an independent and validated prognostic biomarker, surpassing other molecules of a large panel of the tumor and immune-related processes, which may be used to select poor prognosis patients for intensified neoadjuvant or adjuvant regimens.
KEYWORDS: Biomarker, Metalloproteinase, Prognosis, Secretome, Squamous cell carcinoma, Head and Neck, Oral cavity
Introduction
Oral cavity squamous cell carcinoma (OCSSC) patients treated by primary surgery undergo post-operative surveillance, adjuvant radiotherapy, or chemo-radiotherapy, according to clinical and histopathological parameters that include disease stage, nodal involvement, extranodal extension (ENE), perineural invasion (PNI), lymphovascular invasion (LVI), and resection margin status.1 Despite those numerous clinical decision parameters, around 25% of OCSCC will present an unpredictable early and/or severe recurrence.2–4 Even the local failures that are eligible for the best treatment option, that is salvage surgery,5–7 have a poor prognosis with a median overall survival ranging from 20 to 30 months.4,8 Here, we classified the patient as severe if they had a disease-specific survival (DSS) of less than 36 months and/or a disease-free survival (DFS) of less than 12 months and could not achieve a second remission (unsuccessful salvage procedures and/or permanent palliative treatment). Accurately identifying those high-risk patients would allow proposing them an intensified and risk-adjusted therapy, such as neoadjuvant chemotherapy or immunotherapy. Neoadjuvant chemotherapy has failed to show benefit in head and neck squamous cell carcinoma (HNSCC), possibly because trials were made in unselected Stage III/IV HNSCC population.9,10 Immunotherapy is a new treatment modality, and its interest as neoadjuvant treatment is currently being evaluated.11–13
Numerous prognostic markers have been proposed for OCSCC, but none of them has shown independent validation, and translation to clinical practice.14 For example, among immune-associated biomarkers, high T cell infiltration in tumors has been associated with good,15,16 and high macrophage infiltration to poor prognosis.16 In this study, we used a biology-driven exploratory strategy using a panel of soluble molecules relevant to multiple cancer pathways, such as tumor growth, angiogenesis, tissue remodeling, and the spontaneous immune response to cancer, in order to identify a robust predictive biomarker for early severe recurrence and disease-related death in primary OCSCC after treatment by the standard of care. We found MMP2 as fulfilling those criteria, and when combined to ENE, providing a simple and efficient patient stratification scheme.
To address the question of the best (neo)adjuvant treatment option in high-risk patients, we measured the expression of an 18-gene signature (18 G) predictive of response to PD-1 blockade. This signature was established on a large cohort of patients treated by pembrolizumab for head and neck cancers (n = 107), melanoma (n = 89) and other cancers (n = 119).17 The fact that this signature was established by merging the data from 22 different types of cancers and limited to advanced and recurrent cancers might not reflect the clinical setting of the present study. However, PDL1 and interferon-gamma response genes (STAT1, CXCL9, IDO1, HLADR, HLADQ) were part of this 18 G and were identified as predictive of response to neoadjuvant pembrolizumab in a window-of-opportunity trial including untreated head and neck cancer patients .13 Here, we found no difference in expression of 18 G in the different prognostic groups, which led us to propose intensified treatment schemes oriented by the combination of our MMP2-ENE prognostic biomarker and treatment-specific predictive biomarkers.
Results
Human primary tumor secretome analysis identified 29 deregulated molecules
To identify candidate biomarkers, we chose an unbiased approach applied to human primary tumors, in order to ensure physiopathological relevance. We used a tumor explant-culture system to analyze the soluble microenvironment in a prospective discovery cohort of 37 OCSCC patients treated by primary surgery (Table S1 for patients’ characteristics and Table S2 for information on the treatment of recurrences). Fresh standardized tumor and juxtatumor (non-involved) specimens were cultured for 24 h at 37°C, and we measured a panel of 49 soluble molecules. We identified 25 molecules increased, and 4 decreased, in the tumor tissue (Figure 1, Table S3). The T cell-attracting chemokine CXCL9,18 the metalloproteinases (MMP) MMP1, MMP2, and MMP9, plasminogen activator inhibitor (PAI-1), and resistin were among the molecules most increased in tumors. SCF, multiple cytokines (IL-1b, TNFa, IL-15), growth factors (GM-CSF, VEGF) and several other chemokines (MDC, TARC) were also increased in the tumor, as compared to juxta-tumor samples. The monocyte attracting chemokines MCP-1, MCP-2, and MCP-319,20 were increased in juxtatumors (Figure 1). The cytokines IL-9, TNFb, TSLP, IL-21 were never detected (Figure 1). This provided a global, unbiased protein level profiling of the OCSCC tumor secretome.
Figure 1.

Tumor secretome analysis identified 29 deregulated molecules.
High levels of soluble MMP2 were associated with poor prognosis
Among the 29 deregulated secretome molecules, analyzed as candidate biomarkers, MMP2 was the only molecule expressed at significant higher levels among severe patients as compared to non-severe (p = .007) (Table S4). ROC curve defined 29.3 ng/ml as the optimal cutoff for soluble MMP2, with a sensitivity of 100% and a specificity of 71.4% to identify severe cases (Figure 2a). MMP2high tumors were associated with reduced DSS (p = .001), overall survival (OS) (p = .012) and DFS (p = .003) (Figure 2b). MMP1, MMP9, IL-8, G-CSF, and GRO were expressed over the detection limit of the assay in more than 50% of the tumor samples (Table S3), a fact that may have limited the potential to detect the prognostic value of those molecules in the secretome.
Figure 2.
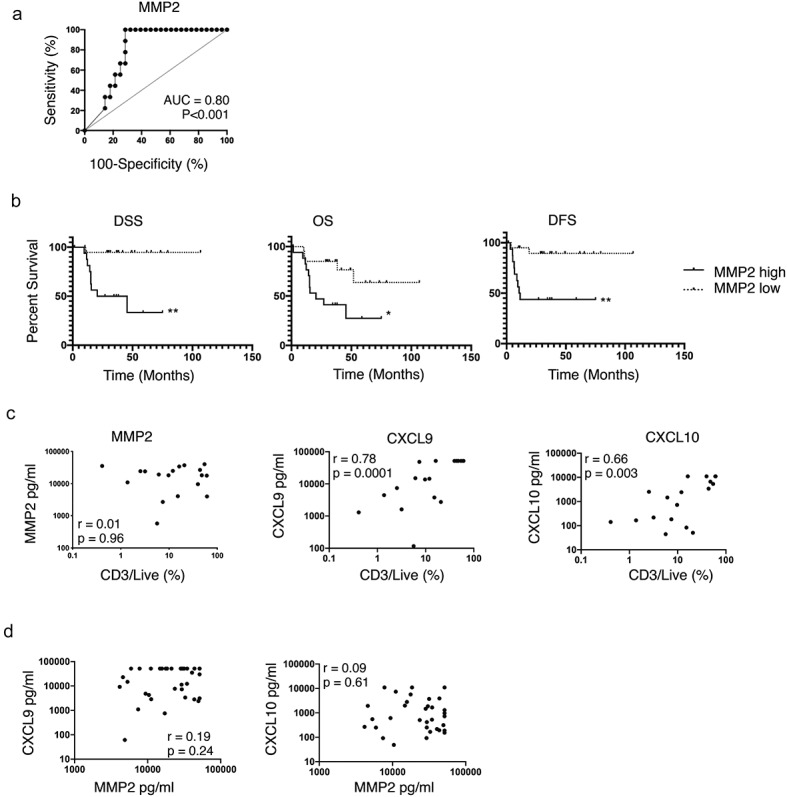
Soluble MMP2 is a prognostic biomarker of OCSCC, independent of T cell infiltration.
Soluble MMP2 levels were independent of T cell infiltration
MMP degrades the extra-cellular matrix and promotes tumor cell invasion.21 Tissue damage may lead to a local increase in danger signals and initiate an innate and then adaptive immune response. Thus, we hypothesized that MMP2 levels might influence T cell infiltration. Paired CD3 and CD8 T cell quantification by flow cytometry, and soluble MMP2 quantification, was available for 18 HNSCC patients. MMP2 was not significantly associated with CD3 nor CD8 infiltration with very weak Spearman correlations (r = 0.01, p = .96, Figure 2c and r = −0.13, p = .61, respectively). Conversely, CD3 and CD8 infiltration were significantly associated, with strong Spearman correlations, to CXCL9 (r = 0.78 and r = 0.79, both p < .0001) and to CXCL10 (both r = 0.66, both p = .003) (Figure 2c, data not shown for CD8). In the secretome analysis of the 37 OCSCC samples, MMP2 was not significantly correlated to CXCL9 and CXCL10 (r = 0.19, weak, p = .24 and r = 0.09, very weak, p = .61), further supporting that MMP2 levels were not associated to T cell infiltration (Figure 2d).
RNA levels of MMP2, CD276, CXCL10, and STAT1 predicted prognosis
To independently validate the prognostic value of MMP2, we measured a 30 genes panel (Table S5) by RTqPCR in a large retrospective cohort of 145 OCSCC patients treated by primary surgery. Gene panel included MMP-2 for validation, MMP-1 and MMP-9 as comparators, a published 18-gene signature predictive of the response to anti-PD-1 immunotherapy,17 and nine other immune-related genes in order to compare the prognostic value of MMP2 to the one of genes related to the immune response and tumor-infiltrating immune cells. These included CD3E and CXCL10 (all T cells), CD8A (CD8 T cells), FUT4 (neutrophils), LAMP3 (mature dendritic cells), CD1 C (type 2 dendritic cells), IL3RA (plasmacytoid dendritic cells), ICOSLG (B cells, immature dendritic cells and positive immune checkpoint), PDCD1 (negative immune checkpoint). Patients’ characteristics are available in Table 1 and the results of the univariate analyses for DSS, OS, and DFS are in Table 2. Among the clinical variables, tumor differentiation index, stage, ENE, LVI, and PNI were significant for both DSS and OS, while only the latter three were significant for DFS. Among the genes, high levels of MMP2 were associated with reduced DSS, OS, and DFS. High levels of CD276 (B7-H3) and low levels of CXCL10 and STAT1 were also among the 5 and 11 genes associated with reduced DSS and OS, respectively (Table 2). This validated the prognostic impact of MMP2, measured by two different methods (protein and mRNA), in a large OCSCC cohort.
Table 1.
Patients characteristics of the RT-qPCR retrospective validation cohort (n = 145).
| Parameter | Percentage (n) | |
|---|---|---|
| Gender | female | 39% (57) |
| male | 61% (88) | |
| Age | 63.8 ± 13.99 (mean ± SD) | |
| Alcohol abuse (n = 121) | absent | 60% (73) |
| present | 40% (48) | |
| Tobacco (n = 137) | non smoker | 43% (59) |
| smoker | 57% (78) | |
| T stage | T1 | 12% (18) |
| T2 | 23% (34) | |
| T3 | 40% (58) | |
| T4 | 24% (35) | |
| N stage | N0 | 51% (74) |
| N1 | 11% (16) | |
| N2 | 16% (23) | |
| N3 | 22% (32) | |
| Stage | I | 11% (16) |
| II | 17% (24) | |
| III | 20% (29) | |
| IVA | 30% (43) | |
| IVB | 23% (33) | |
| Differentiation | verrucous | 3% (5) |
| well | 70% (102) | |
| moderate | 20% (29) | |
| poorly | 6% (8) | |
| basaloid | 1% (1) | |
| Mitotic Index (n = 119) | high | 40% (48) |
| low | 33% (39) | |
| mid | 27% (32) | |
| Perineural invasion (n = 125) | absent | 48% (60) |
| present | 52% (65) | |
| Lymphovascular invasion (n = 126) | absent | 61% (77) |
| present | 39% (49) | |
| ENE | absent | 73% (106) |
| present | 27% (39) | |
| Margins | negative or close | 83% (120) |
| positive | 17% (25) | |
| HPV | negative | 96% (139) |
| positive | 4% (6) | |
| Adjuvant treatment | none | 41% (59) |
| RT | 40% (58) | |
| RT + CT or Cetuximab | 19% (27) | |
| curietherapy | 1% (1) | |
| Recurrence | absent | 61% (88) |
| local | 23% (33) | |
| regional | 19% (27) | |
| metastatic | 13% (19) | |
| Severity | non-severe | 74% (107) |
| severe | 26% (38) | |
Numbers in brackets beside clinical parameters indicate the number of patients for which the information was available.
Table 2.
Prognosis value of the clinical parameters and genes measured by RTqPCR in the validation cohort (univariate analysis, Log–Rank test).
| Parameter |
Mean ± SD |
Poor prognosis if |
p-values per survival (Log-rank) |
||
|---|---|---|---|---|---|
| DSS | OS | DFS | |||
| Gender | ns | 0.8420 | 0.4387 | 0.801 | |
| Age (</> 70) | ns | 0.9460 | 0.9785 | 0.434 | |
| Alcohol | ns | 0.8710 | 0.1860 | 0.848 | |
| Tobacco | ns | 0.7839 | 0.1191 | 0.670 | |
| Stage | III or more | 0.0120 | 0.0036 | 0.053 | |
| Differentiation | moderate or poor | 0.0350 | 0.0434 | 0.117 | |
| Mitotic index | ns | 0.1957 | 0.7066 | 0.928 | |
| Perineural invasion | present | < 0.0001 | < 0.0001 | 0.0046 | |
| Vascular embols | present | 0.0004 | 0.0002 | 0.0130 | |
| ENE | present | < 0.0001 | 0.0004 | 0.003 | |
| Margins | ns | 0.1020 | 0.1484 | 0.193 | |
| HPV | ns | 0.4950 | 0.4536 | 0.823 | |
| MMP2 | 1.84 ± 1.75 | high | 0.0009 | 0.0140 | 0.0440 |
| CD276 | 2.4 ± 1.18 | high | 0.0056 | 0.0340 | 0.0870 |
| CXCL10 | 18.67 ± 27.62 | low | 0.0083 | 0.0008 | 0.0820 |
| STAT1 | 3.72 ± 2.35 | low | 0.0160 | 0.0007 | 0.1300 |
| MMP9 | 8.55 ± 12.93 | high | 0.0190 | 0.0880 | 0.0610 |
| LAMP3 | 7.43 ± 5.59 | low | 0.1500 | 0.0008 | 0.4300 |
| CXCR6 | 1.22 ± 0.92 | low | 0.6200 | 0.0037 | 0.6600 |
| HLA-E | 1.12 ± 0.51 | low | 0.1100 | 0.0056 | 0.0810 |
| CD274 | 3.3 ± 3.25 | low | 0.2100 | 0.0070 | 0.4100 |
| IDO1 | 13.98 ± 20.3 | low | 0.0650 | 0.0095 | 0.1800 |
| PSMB10 | 1.68 ± 0.99 | low | 0.2000 | 0.0270 | 0.2800 |
| CCR7 | 8.41 ± 10.73 | low | 0.4700 | 0.0300 | 0.5900 |
| TIGIT | 3.28 ± 2.8 | ns | 0.8800 | 0.0560 | 0.7700 |
| CCL5 | 2.3 ± 2.41 | ns | 0.7700 | 0.0600 | 0.8800 |
| LAG3 | 3.04 ± 3.28 | ns | 0.4700 | 0.0640 | 0.7900 |
| PDCD1 | 2.19 ± 2.17 | ns | 0.8500 | 0.0670 | 0.5400 |
| CXCL9 | 19.04 ± 30.47 | ns | 0.7000 | 0.0680 | 0.9800 |
| HLA-DQA1 | 1.5 ± 1.2 | ns | 0.5600 | 0.0850 | 0.7200 |
| IL3RA | 0.9 ± 0.69 | ns | 0.6300 | 0.0990 | 0.3700 |
| CD27 | 1.88 ± 2.06 | ns | 0.7700 | 0.0990 | 0.7000 |
| NKG7 | 1.83 ± 2.12 | ns | 0.7900 | 0.1300 | 0.4700 |
| CD3E | 2 ± 1.9 | ns | 0.8100 | 0.1400 | 0.7700 |
| pan_HLA-DRB | 1.35 ± 1.04 | ns | 0.7000 | 0.1500 | 0.6300 |
| PDCD1LG2 | 2.64 ± 2.24 | ns | 0.3100 | 0.2000 | 0.2200 |
| CD8A | 1.74 ± 2.1 | ns | 0.6200 | 0.2800 | 0.4000 |
| ICOSLG | 0.68 ± 0.35 | ns | 0.9400 | 0.4200 | 0.4600 |
| CMKLR1 | 1.13 ± 0.8 | ns | 0.4200 | 0.4300 | 0.4800 |
| MMP1 | 774.76 ± 1051.42 | ns | 0.3000 | 0.6300 | 0.3500 |
| FUT4 | 1.06 ± 0.53 | ns | 0.1600 | 0.8600 | 0.4000 |
| CD1 C | 0.36 ± 0.42 | ns | 0.2300 | 0.9400 | 0.4500 |
Cells highlighted in gray contain significant values at p < 0.05. Mean ± SD values superior to 1 corresponded to an increased expression in tumors as compared to juxtatumor tissues and vice-versa.
MMP2 RNA, ENE, PNI, and stage were independent prognostic factors
To identify clinical and biological parameters significant in multivariate analysis, we performed two Cox proportional hazards models. Model 1 included all the 145 patients and all clinical and biological variables significant in univariate analysis, except PNI and LVI, because of missing values in 21 patients (14%), whereas Model 2 included all significant variables, but was restricted to the 124 patients with complete data (Figure 3a, Table S6). In both models MMP2high was an independent prognostic factor for DSS and DFS (Model 1 DSS: p = .001, DFS: p = .006, Model 2 DSS: p = .034, DFS: p = .016). For DSS, ENE status (p = .006) and PNI (p = .020) were also significant in Model 1 and 2, respectively. For DFS, ENE status was also significant in Model 1 (p = .006), but MMP2 was the only significant parameter in Model 2. For OS, MMP2 (p = .015) and stage (p = .042) were significant in Model 1, and PNI (p = .01) and stage (p = .019) were significant in Model 2 (Figure 3a, Table S6). We defined prognostic groups using the parameters identified in the multivariate analysis by the Model 1 to analyze the largest cohort of 145 patients. MMP2highENE+ patients had the worse DSS and DFS, as compared to MMP2lowENE- patients (p < .001), whereas MMP2highENE- and MMP2lowENE+ had an intermediate DSS and DFS (Figure 3b) (2 by 2 comparisons available in Table S7). MMP2 status induced clinically relevant variations in survival. MMP2high vs MMP2low tumor-bearing patients had a 5-year DSS of 61% versus 88% when ENE was absent, and of 36% versus 52% when ENE was present (Table 3). MMP2high tumors were associated with the presence of metastatic lymph node (p = .031), low or intermediate mitotic index (p = .001) and the presence of PNI (p = .02) (Table S8). Among the 57 patients presenting with recurrences, 20 (35%) presented with a resectable recurrence and underwent salvage surgery (SS) with or without other various associated treatments (Table S9). ENE+ status was associated to reduced rates of SS as compared to ENE- (14% vs 47%, respectively, p = .01, chi2test), whereas MMP2 status was not associated to significantly different rates of SS among all 57 recurrences (MMP2high 26% vs MMP2low 45%, p = .19, chi2test), neither among the 36 recurrences that occurred in patients with ENE-status (MMP2highENE- 38% vs MMP2lowENE- 60%, p = .19, chi2test).
Figure 3.

MMP2, ENE, and stage define prognostic groups with an equivalent expression of an 18-gene signature predictive of response to PD-1 blockade.
Table 3.
Survival durations by prognostic groups defined by the Cox Model 1.
| Survival | Prognostic groups | n (%) | MST (months) | 2-y S | 3-y S | 5-y S |
|---|---|---|---|---|---|---|
| DSS | MMP2 high/ENE- | 50 (34%) | 116.07 | 69.19% | 66.72% | 60.63% |
| MMP2 high/ENE+ | 22 (15%) | 20.04 | 49.23% | 43.76% | 36.47% | |
| MMP2 low/ENE- | 56 (39%) | not reached | 88.44% | 88.44% | 88.44% | |
| MMP2 low/ENE+ | 17 (12%) | not reached | 67.31% | 60.58% | 51.92% | |
| DFS | MMP2 high/ENE- | 50 (34%) | 103.89 | 64.45% | 61.87% | 54.86% |
| MMP2 high/ENE+ | 22 (15%) | 22.57 | 45.85% | 45.85% | 38.21% | |
| MMP2 low/ENE- | 56 (39%) | 172.39 | 79.25% | 77.27% | 73.20% | |
| MMP2 low/ENE+ | 17 (12%) | not reached | 56.31% | 56.31% | 56.31% | |
| OS | MMP2 high/I or II | 17 (12%) | 116.07 | 75.00% | 68.75% | 56.25% |
| MMP2 high/III or more | 55 (38%) | 23.98 | 49.06% | 47.09% | 32.96% | |
| MMP2 low/I or II | 23 (16%) | 135.43 | 86.96% | 82.61% | 82.61% | |
| MMP2 low/III or more | 50 (34%) | 91.83 | 71.49% | 65.16% | 54.47% |
MMP2 may be used as a biomarker to select patients for treatment intensification
MMP2 RNA status was an efficient prognostic biomarker as measured by ROC curves according to severity criteria, in the whole 145 patient cohort (AUC = 0.66, p = .003), and among the ENE negative patients (n = 106, AUC = 0.71, p = .003) (Fig S1). The optimal thresholds were 1.81 and 1.82, which led to high negative predictive values (NPV) of 82% and 88%, respectively, but lower positive predictive values (PPV) of 41% and 36%. For 29 patients, both soluble MMP2 and MMP2 RNA data were available, which allowed us to observe that both biomarkers were significantly associated with moderate Spearman correlation (r = 0.45, p = .016) (Fig S2), suggesting that MMP2 protein or RNA levels can be used as a biomarker.
The expression of an 18-gene signature predictive of response to PD-1 blockade was similar between the different prognostic groups
The proportions of patients expected to respond to immunotherapy may vary between the prognostic groups defined above and have consequences on the type of treatment that could be proposed in a risk-adjusted strategy. Therefore, we measured the expression of an 18-gene signature (18 G)17 that is a predictive biomarker of response to PD-1 blockade. The 18 G signature is composed of a core of 17 highly correlated genes (all p < .0001, 55 strong and 81 moderate Spearman correlations), and CD276 (Fig S3, Fig S4). 18 G score was moderately increased in MMP2high tumors (p = .019) (Fig S5) but was similar whatever the ENE status (p = 0,671) and disease stage (p = .513) (Fig S5). The 18 G score was similar between the prognostic groups defined by MMP2 RNA and ENE status (p = .119), MMP2 RNA status and Stage (p = .051), MMP2 RNA and PNI statuses (p = .089), and stage and PNI status (p = .661) (Figure 3c). This suggests that various prognostic groups may show response to anti-PD-1 therapy, with implications for the design of biomarker-driven trials in untreated resectable OCSCC patient with the goal of limiting early and severe recurrences (Fig S6).
Discussion
In this study, we identified MMP2 as an independent prognostic biomarker for severe outcomes in OCSCC patients treated by primary surgery.
First, we prospectively produced and analyzed tumor and juxtatumor secretomes. Primary tumor-derived supernatant is not a widely applied method for biomarker identification and data on OCSCC secretome are scarce22 if we exclude cancer cell-line derived supernatants. A database for healthy body fluids proteome was created in 2008, highlighting the general interest for such an approach.23 Here, we cannot exclude that tissue handling, although limited to the minimum in our protocol, may have induced or enhanced the production of some proteins, but this limitation was partially overcome by the comparison with paired juxtatumor supernatant. By the mean of an ultrafiltration catheter, interstitial fluid from a single HNSCC patient was analyzed and revealed 525 proteins by mass spectrometry, but the method was not applicable to juxtatumor tissue, which limited the potential to identify candidate biomarkers.24 Another difficulty is that tumor secretome needs to be produced prospectively using fresh tumor samples, which limits the access to large cohorts with sufficient follow-up in order to identify prognostic biomarkers. However, we could overcome these difficulties, and our study illustrates the added value of this approach in providing data with strong biological relevance.
Here, we revealed 29 deregulated soluble molecules, with most of them increased in the tumor tissue, whereas the monocyte attracting chemokines MCP-1, −2, −3 were decreased. Those molecules belonged to various biological classes such as MMPs, chemokines, interleukins, adipokines, and growth factors. One may consider that all these deregulated proteins reflect mechanisms of tumor progression and could be candidate biomarkers. However, only soluble MMP2 was associated with poor prognosis in our study. We confirmed the previously reported association of CXCL9 and CXCL10 with CD3 and CD8 T cell infiltration, 18,25 but we did not find an association with prognosis, as it was expected from former studies.15,16 Additionally, none of the other immune-related interleukins or chemokines measured here had a prognostic impact. This suggests a limited value of the spontaneous immune response to predict prognosis in this specific clinical setting of resectable oral cavity cancers, as compared to the value of soluble MMP2.
For further validation, we designed a homogenous retrospective cohort of patients with the same clinical setting of resectable OCSCC treated by primary surgery, and extracted tumor RNA from biobanked frozen samples to ensure the best quality of RNA.26 Univariate analysis confirmed the prognostic value of MMP2 to predict DSS, OS, and DFS. High levels of CD276 and low levels of CXCL10 and STAT1 were also associated with reduced DSS and OS, but only MMP2 remained significant in multivariate analysis. Several studies have proposed MMP2 as a prognostic biomarker for OCSCC, but all had important limitations, such as the absence of multivariate analysis,27–29 the inclusion of heterogeneous head and neck cancer patients with different tumor locations and treatments,30,31 or retrospective cohorts with less than 60 patients.28,29,32,33 Most of these studies quantified MMP2 by immunohistochemistry (IHC) through semi-quantitative methods. Our study provided unbiased and definite evidence for the independent prognostic role of MMP2, in a large homogeneous OCSCC cohort, within a multivariate prognostic model.
The biological basis explaining why MMP2 is associated with poor prognosis is well known. MMP2 degrades type IV collagen and promotes epithelial–mesenchymal transition and metastasis 21,34 MMP2 is secreted in an inactive form (pro-MMP2) and is activated by MMP135 and MMP14.36 Many cell types may produce MMP2, but fibroblasts seem to be the main source of this molecule in the tumor microenvironment.37,38 From MMP biology, we understand that a high level of MMP is a risk factor for cancer-related events, such as recurrence and disease-related death. This explains why in our study the accuracy of MMP2 as prognostic biomarker was better for DSS than for OS, both in univariate and multivariate analysis. It is well known that HNSCC patients have a reduced cancer-independent life expectancy, which explains the differences observed between OS and DSS.39 In this line, in the TCGA data, MMP2 was co-expressed with MMP1, MMP9, and MMP14 in HNSCC, but the authors did not report the impact of any MMP on OS in HNSCC.40 The absence of DSS evaluation may explain this discrepancy. Beyond prognosis, MMP were also candidate therapeutic targets in cancer, but, so far, most molecules failed in their development because of their toxicities.41 Selective inhibitors are still in development,42 (NCT03486730), as well as other drugs that have an indirect effect on MMP.43
Clinical and histopathological parameters fail to identify around 25% of high-risk patients. Here, we propose that combining MMP2 status to those parameters would improve patients’ risk stratification. MMP2-high tumor-bearing patients could be proposed for an intensified therapeutic plan, as compared to standard of care. MMP2 status may be defined pre-operatively on the initial biopsy, or post-operatively if analyzed on the resection specimen (Fig S6). Pre-operative stratification would guide neoadjuvant treatment such as immunotherapy or chemotherapy, when post-operative stratification would guide adjuvant treatment. The latter setting is particularly important for ENE negative patients who may, in some cases, not be offered any adjuvant treatment. There was no difference in expression of the 18 G score among the different prognostic groups defined by our multivariate analysis for DSS, DFS, and OS. In this line, using soluble CXCL9 and CXCL10 as surrogates for tumor T cell infiltration, or direct measures of frequencies of tumor-infiltrating T cells by flow cytometry, we observed that soluble MMP2 levels were not associated to T cell infiltration. Similar results were previously described for MMP2 measured by IHC in endometrial cancer.44 From these results, we may conclude that there is no direct and constant association between the importance of MMP2 activity and the interferon-gamma response in the TME. However, previous studies have described other interactions between MMP and immune cells at different levels. First, macrophages may promote MMP2 production,45,46 a mechanism enhanced by IL-10.47 Second, MMP2 may directly influence immune cells, such as dendritic cells that promoted the differentiation of naïve CD4 T cells into Th2 cells, via the MMP2-dependent cleavage of IFNAR1 and the subsequent decrease in IL-12 production,48 or natural killer cells that had a reduced cytotoxicity when exposed to MMP2 and MMP9.49 Third, MMPs may indirectly act on immune cells by cleaving chemokines into inactive molecules50 or by converting TNFα.51 Lastly, MMP2 may act as a tumor-associated antigen as shown in melanoma patients.52 Nevertheless, our results also suggest from the clinical perspective that we may estimate a similar proportion of patients expected to respond to PD-1 blockade in the different prognostic groups, leaving immunotherapy as a valid treatment option. Patient stratification in future OCSCC trials and clinical practice would benefit from robust biomarkers used in combination with clinical variables, such as our MMP2/ENE prognostic scoring, and with predictive biomarkers for final treatment decision-making.
Materials and methods
Patients and cohorts
Tumor and juxtatumor samples were obtained from operative specimens from previously untreated head and neck cancer patients. Patients with previous head and neck radiotherapy or chemotherapy were excluded. Juxta-tumor samples were taken on the specimens’ margins, at least 1 cm away from the tumor. Three cohorts of patients treated in our anti-cancer center were included in this study (Table S10). All analyses on secretome presented in Figure 1 were done on a 37 patient cohort including OCSCC patients only, with the exception of the 3 graphs of Figure 1d that show the correlation of CD3 infiltration with soluble MMP2, CXCL9, and CXL10, that was done in an independent 18 patients HNSCC cohort. This 18 patient cohort had paired secretome and flow cytometry data available and included the following tumor locations: 8 oral cavity, 6 oropharynx, 3 larynx, 1 hypopharynx. The third cohort included 145 OCSCC patients and was used to analyze the expression of a panel of 30 genes by RTqPCR. Twenty-nine patients were in common between the n = 37 and n = 145 cohorts and served for the RNA versus soluble protein correlation. Patients were treated between March 2010 and October 2016, for the 37-patient cohort, between January and July 2017 for the 18-patient cohort, and between February 1991 and November 2016 for the 145-patient cohort. The clinical parameters analyzed were all binarized as follows: gender (male/female), HPV status (positive by PCR/negative), Differentiation (well-differentiated or verrucous or basaloid/moderate or poor), Mitotic index (high if ≥10mitoses/field at X400, otherwise low), Perineural invasion (absent/present), Lymphovascular invasion (absent/present), Alcohol (positive if ≥30 g/day), Tobacco (smoker active or former ≥2PY/nonsmoker or former smoker < 2PY), Stage (I or II/III or more) using the pTNM 8th edition AJCC,53 Extranodal extension (absent/present), Margins (negative or close/positive), Age (more or less than 70). For outcomes analyses, we used three survivals: disease-free survival, in which the censoring event was the first occurrence of recurrence, disease-specific survival, in which the censoring event was the occurrence of death caused by the evolution of the cancer (to the exclusion of treatment-related toxicities and post-operative complications), and overall survival. We also used a binary criteria of severity defined as present in cases of DSS < 36 months and/or a DFS < 12 months without subsequent remission (unsuccessful salvage procedures and/or permanent palliative treatment); we considered that these criteria define the population with the most urgent need for prognosis biomarkers.54 This study was done in compliance with the principles of Good Clinical Practice and the Declaration of Helsinki. All patients signed a consent form mentioning that their operative specimens might be used for scientific purposes, and 12 of the 18-patient cohort were also included in the clinical trial NCT03017573.
Tumor and juxta-tumor secretome analyses
Fresh tumor and juxta-tumor were cut into fragments of 17.5 ± 2.5 mg. Each fragment was placed in a 48-well flat bottom plate in 250 µl of RPMI 1640 Medium Glutamax (Life Technologies) enriched with 10% Fetal Calf Serum (Hyclone), 100 U/ml Penicillin/Streptomycin (Gibco), 1% MEM Non-Essential Amino Acids (Gibco), and 1% pyruvate (Gibco), and incubated at 37°C with 5%CO2. After 24 h, supernatants were filtered through a 0,22 µm Millex-GP filter (SLGP033RS, Merck), diluted ½ in the same enriched RPMI Medium and stored at −80°C until the secretome analyses. The 49 analytes measured are listed in Table S3. Analytes concentrations were obtained using Milliplex Map kits used as recommended: Human MMP magnetic Bead panel 2, Human cytokine/chemokine Magnetic Bead panels I, II, III, and Human Adipocyte Magnetic Bead Panel (Millipore), a Bio-Plex 200 plate reader and the Bio-Plex Manager 6.1 software (Bio-Rad Laboratories). Negative control wells (“blank”) were filled with the same culture medium. The final levels of proteins in experimental conditions were obtained by substracting the levels of protein obtained in the negative control condition to their raw measures. All analytes were measured as stored, but MMP1 and MMP9 were also measured after 1/25th dilution for the 18 HNSCC patients with paired flow cytometry data.
Analysis of CD3 and CD8 infiltration by flow cytometry
Details are available at.55 Briefly, single-cell suspensions were obtained from enzymatically digested tumor samples, then filtered, washed, counted, and stained for 15 min with DAPI (Miltenyi Biotec) to exclude dead cells, CD3 (Alexa700, clone UCHT1, from BD, #557943) and CD8b (PC5, clone 2ST8.5H7, from Beckman Coulter, #6607109) antibodies, among other antibodies (data not used in the present paper), before phenotyping by flow cytometry (BD LSRFortessa Analyzer).
Gene expression analysis by real-time RT-PCR
Samples and RNA extraction
Tumor and juxtatumor samples were snap-frozen in liquid nitrogen upon surgical removal after the pathologist’s review and were stored in the corresponding our biological resources center. Samples were sectioned using Tissue-Tek optimal cutting temperature (O.C.T) compound to estimate the percentage of tumor cells and to remove nonmalignant tissue by macrodissection if necessary. Median percentage of tumor cells was 80% (range 40–95). RNA extraction was performed on the same sample, using the miRNeasy miniKit (Qiagen) according to the manufacturer’s protocol. RNA was quantified using Nanodrop spectrophotometer ND-1000 and the integrity and purity were assessed by the Agilent 2100 Bioanalyzer and RNA 6000 Nano Labchip Kit (Agilent Biotechnologies, Palo Alto, CA, USA).
Total RNA was extracted from 145 OCSSC and 31 juxtatumor frozen samples from OCSCC bearing patients by using the acid-phenol guanidium method. RNA samples quality was assessed by electrophoresis through agarose gels and staining with ethidium bromide, and the 18 S and 28 S RNA bands were visualized under UV light.
cDNA synthesis
RNA was reverse transcribed in a final volume of 20 μl containing 1X RT buffer, 0.01 M DTT, 0.5 mM each dNTP, 0.15 µg/µL random primers, 100 U SuperScript™ II Reverse Transcriptase (Life Technologies, Carlsbad, Californie), 20 U RNasin® Ribonuclease Inhibitor (Promega, Madison, Wisconsin) and 1 μg of total RNA. The samples were incubated during 10 min at 25°C 30 min at 42°C, and reverse transcriptase was inactivated by heating 5 min at 99°C and cooling 5 min at 5°C.
PCR amplification and quantification
All of the PCR reactions were performed using an ABI Prism 7900HT Sequence Detection system (Thermo Fisher Scientific, Waltham, Massachusetts). PCR was performed using the Power SYBR™ Green PCR Master Mix (Life Technologies, Carlsbad, Californie). The thermal cycling conditions comprised an initial denaturation step of 10 min at 95°C followed by 50 cycles at 95°C for 15 s and 65°C for 1 min. Cycle Threshold (Ct value) was defined by the cycle number at which the increase in the fluorescence signal associated with the exponential growth of PCR products started to be detected, using Applied Biosystems analysis software according to the manufacturer’s manuals. For quality controls, we quantified the housekeeping gene TBP (Genbank accession NM_003194). Primers for TBP and the 30 target genes were designed with the assistance of Oligo 6.0 computer program (National Biosciences, Plymouth, MN). dbEST and nr databases were used to confirm the total gene specificity of the nucleotide sequences chosen as primers and the absence of single nucleotide polymorphisms. The primer pairs selected were unique relative to the sequences of closely related family member genes and the corresponding retropseudogenes. One of the two primers were placed at the junction between two exons or on two different exons to avoid genomic DNA contaminating. Specificity of PCR amplicons was verified by agarose gel electrophoresis. The oligonucleotide primers sequences used are shown in Table S11.
Data processing
TBP was used for each sample normalization. ΔCt value was equal to the mean Ct value of the target gene minus mean Ct value of TBP. The N-fold differences per sample in target gene expression relative to TBP was equal to 2ΔCt. For each gene, 2ΔCt values of the 31 juxtatumor samples were multiplied by a factor named “k” so that their median was equal to 1. The final values for tumor samples were equal to k2ΔCt and were therefore increased as compared to juxtatumor if they were superior to 1 and decreased if they were inferior to 1. The 30 genes of this study are listed in Table S6. To obtain a score for the 18 genes signature, we standardized each gene separately, and used those values in the formula:
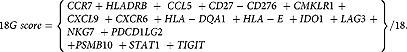 |
Statistical analyses
Descriptive and statistical analyses were performed using GraphPad Prism V8, Xlstat (Addinsoft), and Qlucore softwares. Paired tumor and juxtatumor secretome comparison were done by Wilcoxon test. Univariate unpaired non-parametric comparisons used Mann–Whitney tests and Kruskal–Wallis test for multigroup comparisons. All correlations were evaluated using Spearman method and were qualified as strong (>0.6), moderate (0.3–0.6), weak (0.15–0.3) or very weak (<0.15). Optimal threshold for ROC curves was defined as the value maximizing the sum of sensitivity and specificity. Univariate survival analysis was performed on clinical parameters and biological parameters (soluble molecules or 30 genes measured by RT-PCR) categorized as high or low by cutoff at median, or at optimal threshold when specified. Log-rank tests were used for univariate analysis. For the 145-patient validation cohort, significant variables at the threshold of p < .05 were selected for the Cox proportional hazard models for multivariate analyses. Model 1 included 145 patients and all clinical and biological parameters significant in univariate analysis, but PNI and LVI, because of missing values, whereas Model 2 included all significant parameters, but was restricted to the 124 patients with complete data. The heatmap representing the 18-gene signature in Figure 3a was performed with Qlucore software.
Supplementary Material
Acknowledgments
The authors wish to thank the INSERM U932, and the Institut Curie Flow-Cytometry facility, in particular Olivier Lantz, Zofia Maciorowsky, Annick Viguier, and Sophie Grondin for their technical help and expertise.
Funding Statement
“This work was supported by the Institut National de la Santé et de la Recherche Médicale under Grants [BIO2012-02, BIO2014-08, and HTE201606]; Agence Nationale de la Recherche under Grants [ANR-10- IDEX-0001-02 PSL*, ANR-11-LABX-0043 CIC IGR-Curie 1428]; Institut National du Cancer under Grant Cancéropole INCA PhD grant to CH, and INCA PLBio Grant [INCA 2016-1-PL BIO-02-ICR-1]; Ligue nationale contre le cancer (labellisation EL2016.LNCC/VaS); and Institut Curie, in particular the PIC TME.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Disclosure Statement
The authors declare no potential conflicts of interest.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- 1.NCCN . https://www.nccn.org/professionals/physician_gls/default.aspx.
- 2.Gañán L, López M, García J, Esteller E, Quer M, León X.. Management of recurrent head and neck cancer: variables related to salvage surgery. Eur Arch Oto-Rhino-Laryngol Off J Eur Fed Oto-Rhino-Laryngol Soc EUFOS Affil Ger Soc Oto-Rhino-Laryngol - Head Neck Surg. 2016. December;273(12):4417–12. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Karamouzis MV, Raben D, Ferris RL.. Head and neck cancer. Lancet Lond Engl. 2008. May 17;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tam S, Araslanova R, Low T-H-H, Warner A, Yoo J, Fung K, MacNeil SD, Palma DA, Nichols AC. Estimating survival after salvage surgery for recurrent oral cavity cancer. JAMA Otolaryngol– Head Neck Surg. 2017. 1;143(7):685–690. doi: 10.1001/jamaoto.2017.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ord RA, Kolokythas A, Reynolds MA. Surgical salvage for local and regional recurrence in oral cancer. J Oral Maxillofac Surg Off J Am Assoc Oral Maxillofac Surg. 2006. September;64(9):1409–1414. doi: 10.1016/j.joms.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Sacco AG, Cohen EE. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol Off J Am Soc Clin Oncol. 2015. October 10;33(29):3305–3313. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 7.Liao C-T, Chang JT-C, Wang H-M, Ng S-H, Hsueh C, Lee L-Y, Lin C-H, Chen I-H, Huang S-F, Cheng A-J. Salvage therapy in relapsed squamous cell carcinoma of the oral cavity: how and when? Cancer. 2008. January 1;112(1):94–103. doi: 10.1002/cncr.23142. [DOI] [PubMed] [Google Scholar]
- 8.Janot F, de Raucourt D, Benhamou E, Ferron C, Dolivet G, Bensadoun R-J, Hamoir M, Géry B, Julieron M, Castaing M, et al. Randomized trial of postoperative reirradiation combined with chemotherapy after salvage surgery compared with salvage surgery alone in head and neck carcinoma. J Clin Oncol Off J Am Soc Clin Oncol. 2008. December 1;26(34):5518–5523. doi: 10.1200/JCO.2007.15.0102. [DOI] [PubMed] [Google Scholar]
- 9.Zorat PL, Paccagnella A, Cavaniglia G, Loreggian L, Gava A, Mione CA, Boldrin F, Marchiori C, Lunghi F, Fede A, et al. Randomized phase III trial of neoadjuvant chemotherapy in head and neck cancer: 10-year follow-up. J Natl Cancer Inst. 2004. November 17;96(22):1714–1717. doi: 10.1093/jnci/djh306. [DOI] [PubMed] [Google Scholar]
- 10.Bossi P, Lo Vullo S, Guzzo M, Mariani L, Granata R, Orlandi E, Locati L, Scaramellini G, Fallai C, Licitra L, et al. Preoperative chemotherapy in advanced resectable OCSCC: long-term results of a randomized phase III trial. Ann Oncol Off J Eur Soc Med Oncol. 2014. February;25(2):462–466. doi: 10.1093/annonc/mdt555. [DOI] [PubMed] [Google Scholar]
- 11.Uppaluri R, Zolkind P, Lin T, Nussenbaum B, Jackson RS, Rich J. Neoadjuvant pembrolizumab in surgically resectable, locally advanced HPV negative head and neck squamous cell carcinoma (HNSCC). J Clin Oncol. 2017;35(suppl; abstr 6012). doi: 10.1200/JCO.2017.35.15_suppl.6012. [DOI] [Google Scholar]
- 12.Ferris RL, Gonçalves A, Baxi S, Martens A, Gauthier H, Langenberg M, Spanos WC, Leidner RS, Kang H, Russell J. An open-label, multicohort, phase 1/2 study in patients with virus-associated cancers (checkmate 358): safety and efficacy of neoadjuvant nivolumab in squamous cell carcinoma of the head and neck. Annals of oncology. 2017;28(suppl_5): v605-v649. doi: 10.1093/annonc/mdx440 [Google Scholar]
- 13.Wise-Draper TM, Old MO, Worden FP, O’Brien PE, Cohen EEW, Dunlap N, Mierzwa ML, Casper K, Palackdharry S, Hinrichs B, et al. Phase II multi-site investigation of neoadjuvant pembrolizumab and adjuvant concurrent radiation and pembrolizumab with or without cisplatin in resected head and neck squamous cell carcinoma. J Clin Oncol. 2018;36(suppl; abstr 6017). [Google Scholar]
- 14.Rivera C, Oliveira AK, Costa RAP, De Rossi T, Paes Leme AF. Prognostic biomarkers in oral squamous cell carcinoma: A systematic review. Oral Oncol. 2017;72:38–47. doi: 10.1016/j.oraloncology.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 15.de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology. 2017;6(11):e1356148. doi: 10.1080/2162402X.2017.1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017. December;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 17.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018. 12;362:6411. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbachev AV, Kobayashi H, Kudo D, Tannenbaum CS, Finke JH, Shu S, Farber JM, Fairchild RL. CXC chemokine ligand 9/monokine induced by IFN-gamma production by tumor cells is critical for T cell-mediated suppression of cutaneous tumors. J Immunol Baltim Md 1950. 2007. February 15;178(4):2278–2286. [DOI] [PubMed] [Google Scholar]
- 19.Zhang YJ, Rutledge BJ, Rollins BJ. Structure/activity analysis of human monocyte chemoattractant protein-1 (MCP-1) by mutagenesis. Identification of a mutated protein that inhibits MCP-1-mediated monocyte chemotaxis. J Biol Chem. 1994. June 3;269(22):15918–15924. [PubMed] [Google Scholar]
- 20.Van Damme J, Proost P, Lenaerts JP, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992. July 1;176(1):59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010. April 2;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gromov P, Gromova I, Olsen CJ, Timmermans-Wielenga V, Talman M-L, Serizawa RR, Moreira JMA. Tumor interstitial fluid - a treasure trove of cancer biomarkers. Biochim Biophys Acta. 2013. November;1834(11):2259–2270. doi: 10.1016/j.bbapap.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Li S-J, Peng M, Li H, Liu B-S, Wang C, Wu J-R, Li Y-X, Zeng R. Sys-BodyFluid: a systematical database for human body fluid proteome research. Nucleic Acids Res. 2009. January;37(Database issue):D907–912. doi: 10.1093/nar/gkn849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone MD, Odland RM, McGowan T, Onsongo G, Tang C, Rhodus NL, Jagtap P, Bandhakavi S, Griffin TJ. Novel in situ collection of tumor interstitial fluid from a head and neck squamous carcinoma reveals a unique proteome with diagnostic potential. Clin Proteomics. 2010. September;6(3):75–82. doi: 10.1007/s12014-010-9050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Chu Y, Wang Y, Zhang R, Xiong S. Targeted in vivo expression of IFN-gamma-inducible protein 10 induces specific antitumor activity. J Leukoc Biol. 2006. December;80(6):1434–1444. doi: 10.1189/jlb.0306212. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Fu C, Speed TP, Wang W, Symmans WF. Accurate RNA sequencing from formalin-fixed cancer tissue to represent high-quality transcriptome from frozen tissue. JCO Precis Oncol. 2018:2, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C-H, Chien C-Y, Huang -C-C, Hwang C-F, Chuang H-C, Fang F-M, Huang H-Y, Chen C-M, Liu H-L, Huang C-Y. Expression of FLJ10540 is correlated with aggressiveness of oral cavity squamous cell carcinoma by stimulating cell migration and invasion through increased FOXM1 and MMP-2 activity. Oncogene. 2009. July 30;28(30):2723–2737. doi: 10.1038/onc.2009.128. [DOI] [PubMed] [Google Scholar]
- 28.Fan H-X, Li H-X, Chen D, Gao Z-X, Zheng J-H. Changes in the expression of MMP2, MMP9, and ColIV in stromal cells in oral squamous tongue cell carcinoma: relationships and prognostic implications. J Exp Clin Cancer Res CR. 2012. October 29;31:90. doi: 10.1186/1756-9966-31-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aparna M, Rao L, Kunhikatta V, Radhakrishnan R. The role of MMP-2 and MMP-9 as prognostic markers in the early stages of tongue squamous cell carcinoma. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2015. May;44(5):345–352. [DOI] [PubMed] [Google Scholar]
- 30.Ruokolainen H, Pääkkö P, Turpeenniemi-Hujanen T. Tissue and circulating immunoreactive protein for MMP-2 and TIMP-2 in head and neck squamous cell carcinoma–tissue immunoreactivity predicts aggressive clinical course. Mod Pathol Off J U S Can Acad Pathol Inc. 2006. February;19(2):208–217. [DOI] [PubMed] [Google Scholar]
- 31.Gunawardena I, Arendse M, Jameson MB, Plank LD, Gregor RT. Prognostic molecular markers in head and neck squamous cell carcinoma in a New Zealand population: matrix metalloproteinase-2 and sialyl Lewis x antigen. ANZ J Surg. 2015. November;85(11):843–848. doi: 10.1111/ans.12424. [DOI] [PubMed] [Google Scholar]
- 32.Gontarz M, Wyszyńska-Pawelec G, Zapała J, Czopek J, Lazar A, Tomaszewska R. Immunohistochemical predictors in squamous cell carcinoma of the tongue and floor of the mouth. Head Neck. 2016;38(Suppl 1):E747–753. doi: 10.1002/hed.24087. [DOI] [PubMed] [Google Scholar]
- 33.Katayama A, Bandoh N, Kishibe K, Takahara M, Ogino T, Nonaka S, Harabuchi Y. Expressions of matrix metalloproteinases in early-stage oral squamous cell carcinoma as predictive indicators for tumor metastases and prognosis. Clin Cancer Res Off J Am Assoc Cancer Res. 2004. January 15;10(2):634–640. doi: 10.1158/1078-0432.CCR-0864-02. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama K, Kamata N, Fujimoto R, Tsutsumi S, Tomonari M, Taki M, Hosokawa H, Nagayama M. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int J Oncol. 2003. April;22(4):891–898. [PubMed] [Google Scholar]
- 35.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994. July 7;370(6484):61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 36.Jobin PG, Butler GS, Overall CM. New intracellular activities of matrix metalloproteinases shine in the moonlight. Biochim Biophys Acta Mol Cell Res. 2017. November;1864(11Pt A):2043–2055. doi: 10.1016/j.bbamcr.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017. December 14;171(7):1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Liu R, Jin R, Fan Y, Li T, Shuai Y, Li X, Wang X, Luo J. Integrating clinical and genetic analysis of perineural invasion in head and neck squamous cell carcinoma. Front Oncol. 2019;9:434. doi: 10.3389/fonc.2019.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massa ST, Cass LM, Osazuwa-Peters N, Christopher KM, Walker RJ, Varvares MA. Decreased cancer-independent life expectancy in the head and neck cancer population. Head Neck. 2017;39(9):1845–1853. doi: 10.1002/hed.24850. [DOI] [PubMed] [Google Scholar]
- 40.Gobin E, Bagwell K, Wagner J, Mysona D, Sandirasegarane S, Smith N, Bai S, Sharma A, Schleifer R, She J-X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer. 2019. June 14;19(1):581. doi: 10.1186/s12885-019-5768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong Y, Lu Y-T, Sun Y, Shi Z-H, Li N-G, Tang Y-P, Duan J-A. Recent opportunities in matrix metalloproteinase inhibitor drug design for cancer. Expert Opin Drug Discov. 2018;13(1):75–87. doi: 10.1080/17460441.2018.1398732. [DOI] [PubMed] [Google Scholar]
- 42.Scannevin RH, Alexander R, Haarlander TM, Burke SL, Singer M, Huo C, Zhang Y-M, Maguire D, Spurlino J, Deckman I. Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation. J Biol Chem. 2017. 27;292(43):17963–17974. doi: 10.1074/jbc.M117.806075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Tubergen EA, Banerjee R, Liu M, Vander Broek R, Light E, Kuo S, Feinberg SE, Willis AL, Wolf G, Carey T, et al. Inactivation or loss of TTP promotes invasion in head and neck cancer via transcript stabilization and secretion of MMP9, MMP2, and IL-6. Clin Cancer Res Off J Am Assoc Cancer Res. 2013. March 1;19(5):1169–1179. doi: 10.1158/1078-0432.CCR-12-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jedryka M, Chrobak A, Chelmonska-Soyta A, Gawron D, Halbersztadt A, Wojnar A, Kornafel J. Matrix metalloproteinase (MMP)-2 and MMP-9 expression in tumor infiltrating CD3 lymphocytes from women with endometrial cancer. Int J Gynecol Cancer Off J Int Gynecol Cancer Soc. 2012. October;22(8):1303–1309. doi: 10.1097/IGC.0b013e318269e27b. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Huang N, Zhu W, Wu J, Yang X, Teng W, Tian J, Fang Z, Luo Y, Chen M. Modulation the crosstalk between tumor-associated macrophages and non-small cell lung cancer to inhibit tumor migration and invasion by ginsenoside Rh2. BMC Cancer. 2018. May 22;18(1):579. doi: 10.1186/s12885-018-4299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinoza-Sánchez NA, Chimal-Ramírez GK, Mantilla A, Fuentes-Pananá EM. IL-1β, IL-8, and matrix metalloproteinases-1, −2, and −10 are enriched upon monocyte-breast cancer cell cocultivation in a matrigel-based three-dimensional system. Front Immunol. 2017;8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardoso AP, Pinto ML, Pinto AT, Pinto MT, Monteiro C, Oliveira MI, Santos SG, Relvas JB, Seruca R, Mantovani A, et al. Matrix metalloproteases as maestros for the dual role of LPS- and IL-10-stimulated macrophages in cancer cell behaviour. BMC Cancer. 2015. June 5;15(1):456. doi: 10.1186/s12885-015-1466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godefroy E, Manches O, Dréno B, Hochman T, Rolnitzky L, Labarrière N, Guilloux Y, Goldberg J, Jotereau F, Bhardwaj N, et al. Matrix metalloproteinase-2 conditions human dendritic cells to prime inflammatory T(H)2 cells via an IL-12- and OX40L-dependent pathway. Cancer Cell. 2011. March 8;19(3):333–346. doi: 10.1016/j.ccr.2011.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B-K, Kim M-J, Jang H-S, Lee H-R, Ahn K-M, Lee J-H, Choung PH, Kim MJ. A high concentration of MMP-2/gelatinase A and MMP-9/gelatinase B reduce NK cell-mediated cytotoxicity against an oral squamous cell carcinoma cell line. Vivo Athens Greece. 2008. October;22(5):593–597. [PubMed] [Google Scholar]
- 50.McQuibban GA, Gong J-H, Wong JP, Wallace JL, Clark-Lewis I, Overall CM. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood. 2002. August 15;100(4):1160–1167. doi: 10.1182/blood.V100.4.1160.h81602001160_1160_1167. [DOI] [PubMed] [Google Scholar]
- 51.Goetzl EJ, Banda MJ, Leppert D. Matrix metalloproteinases in immunity. J Immunol Baltim Md 1950. 1996. January 1;156(1):1–4. [PubMed] [Google Scholar]
- 52.Godefroy E, Moreau-Aubry A, Diez E, Dreno B, Jotereau F, Guilloux Y. alpha v beta3-dependent cross-presentation of matrix metalloproteinase-2 by melanoma cells gives rise to a new tumor antigen. J Exp Med. 2005. July 4;202(1):61–72. doi: 10.1084/jem.20042138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, et al. AJCC cancer staging manual. 8th. New York, NY, USA; Springer; 2017. [Google Scholar]
- 54.Agra IMG, Carvalho AL, Ulbrich FS, de Campos OD, Martins EP, Magrin J, Kowalski LP. Prognostic factors in salvage surgery for recurrent oral and oropharyngeal cancer. Head Neck. 2006. February;28(2):107–113. doi: 10.1002/hed.20309. [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann C, Noel F, Grandclaudon M, Michea P, Surun A, Faucheux L, Philemon Sirven P, Lantz O, Rochefort J, Klijanienko J. PDL1 and ICOSL discriminate human secretory and helper dendritic cells. bioRxiv [Internet]. 2019. August 1 [cited 2019 August2]. doi: 10.1101/721563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


