ABSTRACT
Many characteristics of cancer such as proliferation, survival, progression, immunogenicity, sensitivity, and resistance to therapy are not just endogenously driven by the tumor cells themselves, but are greatly affected by their interaction with the components of their microenvironment. In our recent report, we comprehensively characterized the bacterial content of solid tumors, which is strongly related to tumor type and subtype, largely presenting as metabolically-active and intra-cellular. Our integration with clinical patient data indicates potential avenues of cross-talk between the tumors and their bacterial counterparts paving the way for a deeper understanding of the physiological/biological context of the tumor and how to harness bacteria in therapy settings.
KEYWORDS: Tumor microbiome, tumor microenvironment, cancer, microbiome, 16S, 5R, bacteria
Tumor microbiome mediated chemoresistance in pancreatic cancer
Tumors represent complex ecosystems that contain, in addition to tumor cells, many other cell types, blood vessels, and extracellular matrix. It is now well appreciated that the microenvironment of the tumor cells can profoundly affect their sensitivity to anti-cancer drugs, including cytotoxic-, targeted- and immuno-therapies. A few years ago, we demonstrated that skin fibroblasts were able to protect colorectal and pancreatic cancer cells from gemcitabine.1 Trying to dissect the mechanism, we surprisingly found that the skin fibroblasts were contaminated with Mycoplasma hyorhinis, and that it was the mycoplasma rather than the skin fibroblasts that protected the cancer cells from chemotherapy.2 We then demonstrated that while human pancreatic tumors do not harbor mycoplasma, other bacteria, mainly proteobacteria, are frequently present in these tumors and that some of these bacteria can degrade and thus inactivate gemcitabine using the long isoform of the bacterial gene cytidine deaminase.2
Characterizing low biomass microbiomes
Intrigued by the finding of bacteria in human pancreatic tumors and being aware of a growing number of papers that demonstrated bacteria in other tumor types,3 we decided to characterize the microbiome of additional solid human tumors.4 We have taken multiple measures to tell apart true signals from contamination of samples with live bacteria or bacterial DNA. To detect medical center-specific contamination, samples were collected from nine medical centers. We included more than 800 negative controls, controlling for every step of sample processing and sequencing as well as for the paraffin in paraffin blocks of embedded tissues. We also applied a stringent 16S rRNA sequencing analysis pipeline involving multiple filters to exclude bacterial taxa that may have been detected due to contaminating DNA. As the generation of metagenomics data from low biomass microbiomes like tumors is still a challenge and as bacterial V4 or V3-V4 16S rRNA sequencing has a limited resolution at the species level, we applied a novel PCR method amplifying five regions (5 R) across the bacterial 16S rRNA gene. This method not only improves the resolution at the species level but is also more suitable for working with degraded DNA as each of the five amplicons is relatively short. Lastly, to strengthen the evidence for the presence of bacteria in different human tumor types we used multiple methods of bacterial visualization. Our study also has a few limitations. First, as multiple hypotheses were tested, in spite of the use of filtering steps, strict P-values, and FDR corrections, our data, by definition, must include some false-positive results. Focused validation screens should be done to validate the presence of specific bacteria in a specific tumor type. Secondly, we used the GreenGenes database to name the hundreds of millions of resulting 16S rRNA sequences that we generated. The incomplete nature of this database as well as the presence of taxonomic naming inaccuracies may be propagated to our results. As the raw reads of our screens were made publicly available, anyone interested in a specific bacterial 16S sequence that is present in a specific cancer type can extract that directly, unconstrained by taxonomic naming.
The tumor microbiome is tumor-type specific and mostly intra-cellular
Profiling the microbiome of human tumors and their adjacent normal tissues in more than 1500 samples, produced a catalog of the bacteria present in seven different tumor types.4 The tumor microbiome displayed a distinct microbial profile in the different tumor types or even subtypes (Figure 1), in line with recent reports.5,6 We found that breast tumors demonstrated a richer and more diverse microbiome than the other tissue types as well as when compared to their adjacent normal tissue or to completely normal breast tissue. We also found differential prevalence of specific bacterial taxa between melanoma tumors from patients that responded or did not respond to immunotherapy.
Figure 1.
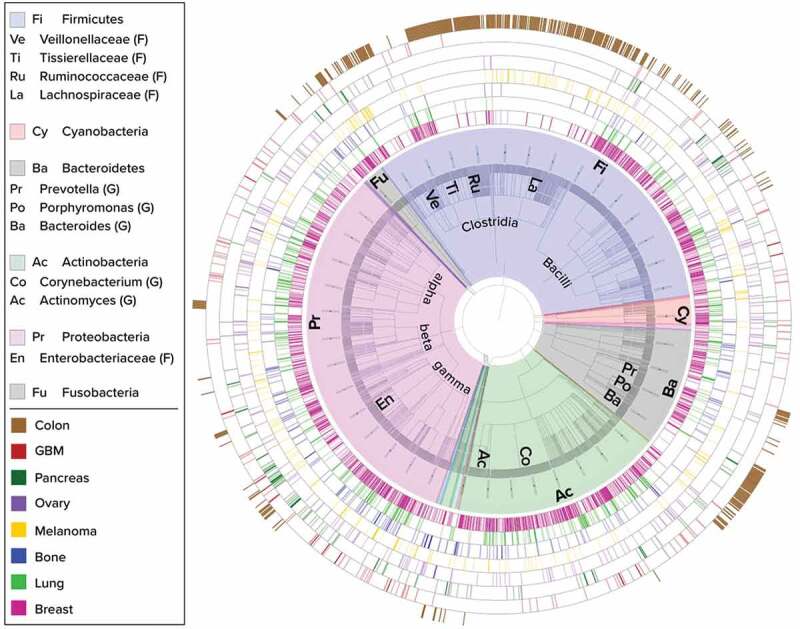
Phylogenetic display of bacteria present in eight solid tumor types. Taxonomic classifications of the bacteria found in tumors are represented as a circular phylogenetic tree with equal branch lengths, overlaid with a color-coded pie-chart according to Phylum classification. Major phyla and sub-clades are annotated with short abbreviations which are expanded in the legend (F – family level, G – genus level). Bacteria at the species level are the outer leaves of the tree and appear as a dense gray outer circle within the pie-chart. Each surrounding ring depicts a different tumor type as indicated by the second legend. Each bar in the rings represents the status of the bacterial species corresponding to its location along the perimeter of the tree, within the specific tumor type: dark color shade indicates that the species passed all filtering stages of our stringent 16S rRNA sequencing analysis pipeline, light color shade indicates species that passed all filters except those related to specific medical center considerations. All tumor types display diverse bacterial profiles spanning several major phyla. Breast tumors (pink inner ring) display high diversity and richness of bacteria. Colon (brown outer ring) was sampled and added as an eighth tumor type of higher bacterial biomass and has a more distinct profile largely comprised of Bacteroidetes and Firmicutes.
We used multiple visualization methods to demonstrate intra-tumor bacteria and found that they can be mostly inside both tumor and immune cells rather than in the extracellular compartment. To show that live bacteria are present in human tumors and not just bacterial components we demonstrated that bacteria can be cultured from the tumors. We also demonstrated that D-alanine, known to be part of the bacterial cell wall but not part of mammalian cells, is incorporated into fresh human breast cancer tissues cultured ex vivo, supporting the presence of metabolically active bacteria in these tissues. Lastly, we used PICRUSt27 to predict metagenome functions of intra-tumoral bacteria and demonstrated, for example, that specific bacterial functions related to the metabolism of cigarette smoke are enriched in lung tumors from smokers as compared to lung tumors from patients who never smoked.
Future directions and unanswered questions
While our study added to the growing catalog of intra-tumor bacteria, it is far from answering many of the open questions related to the tumor microbiome. A few of these open questions include the following: What is the origin of intra-tumor bacteria? Are they getting to the tumor from the blood? Or from the tumors’ immediate surroundings? How early in the tumor transformation process do bacteria colonize tumors and does the composition of the microbiome change with tumor progression? Do some of the intra-tumor bacteria have an active role in the transformation process or are they mostly hitchhikers that reach established tumors? Do bacteria ‘travel’ with cancer cells to metastatic sites, or do metastases have a microbiome that is more related to their new location?
It is also not clear if tumor bacteria adapt their genomes to fit the tumor microenvironment conditions and how exactly do they manage to survive inside the cells. Most importantly, there is still a lot that we do not know about the different effects that intra-tumor bacteria may have on different aspects of tumor biology like response to drugs, effects on tumor immunity, angiogenesis, metastasis, and tumor metabolism. Studies of the microbiome in tumors with well-annotated clinical data would be crucial to better understand these effects, as was recently shown for pancreatic cancer.8
To find answers to these burning questions, there is an urgent need for well-annotated clinical data integrated with better methods of profiling the tumor microbiome. These new methods should allow metagenomics and metatranscriptomics analysis of low biomass microbiomes, single-cell RNA-Seq data that also cover bacterial transcripts and visualization of intra-tumor bacteria with higher resolution than is now available. We hope that the growing interest in the study of tumor bacteria will help to better dissect, in the future, the impact of the tumor microbiome and how manipulation of this microbial community can help provide better treatment options for cancer patients.
Funding Statement
This work was supported by the European Research Council [818086]; Israel Science Foundation [2044/17]; International Collaboration Grant from the Jacki and Bruce Barron Cancer Research Scholars’ Program, and a partnership of the Israel Cancer Research Fund and City of Hope (COH), as supported by The Harvey L. Miller Family Foundation [0]; Fabrikant-Morse Families Research Fund for Humanity [0]; United States-Israel Binational Science Foundation [2013332].
Disclosure of potential conflicts of interest
RS received a grant from Merck EMD Serono and is a paid advisor to Biomica & BiomX
References
- 1.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–3. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodman B, Gardner H.. The microbiome and cancer. J Pathol. 2018;244:667–676. [DOI] [PubMed] [Google Scholar]
- 4.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Rodriguez RM, Hernandez BY, Menor M, Deng Y, Khadka VS. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput Struct Biotechnol J. 2020;18:631–641. doi: 10.1016/j.csbj.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riquelme E, Zhang Y, L Z, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


