ABSTRACT
Accumulating evidence from preclinical studies and human trials demonstrated the crucial role of the gut microbiota in determining the effectiveness of anticancer therapeutics such as immunogenic chemotherapy or immune checkpoint blockade. In summary, it appears that a diverse intestinal microbiota supports therapeutic anticancer responses, while a dysbiotic microbiota composition that lacks immunostimulatory bacteria or contains overabundant immunosuppressive species causes treatment failure. In this review, we explore preclinical and translational studies highlighting how eubiotic and dysbiotic microbiota composition can affect progression-free survival in cancer patients.
KEYWORDS: Anticancer therapeutics, gut microbiota, Cancer
Introduction
The rise of cancer immunotherapy over the past decade has revolutionized the clinical management of a wide array of malignancies that were previously associated with poor prognosis.1 Immune checkpoint inhibitors (ICIs) targeting the PD-1/PD-L1/PD-L2 and CTLA-4/CD86 axis are at the forefront of current implementations in various indications, alone or in combination, for advanced, metastatic, neoadjuvant, and adjuvant settings. Given the broad bioactivity across multiple histological tumor types, the durability of response, and therapeutic success in second or third line chemo-resistant diseases, ICIs are now positioned as a first-in-class drug and thus constitute major pillar in the oncological armamentarium.2–8 As such, ICIs have been approved by multiple regulatory agencies worldwide and are now considered the standard of care in a wide range of solid and hematologic neoplastic diseases including advanced-stage melanoma, non-small-cell lung cancer (NSCLC), head and neck cancer, bladder cancer, or renal cell carcinoma (RCC).9
Figure 1.
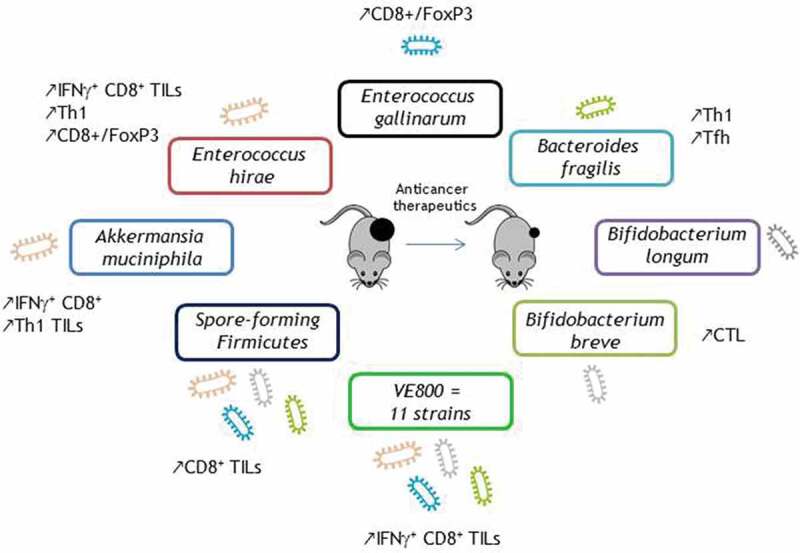
Key bacteria safely boosting the efficacy of anticancer therapeutics in vivo.
Despite the exceptional improvement in objective response rates and overall survival (OS) benefits, ICI responses are currently only observed in a minority (~30%) of patients.5,7 Indeed, most patients manifest primary or secondary resistance to ICIs or even acceleration of the disease called “hyperprogression.”10 Large efforts are being dedicated to identify the “cancer immune set-point,” a notion defined as the point which determines the parameters that govern the strength, timing, and threshold beyond which an effective immune response can occur in a given individual.11,12 Besides tumor intrinsic factors, many non-cell autonomous parameters control primary resistance to ICIs. Recent evidence points to the biological significance of the composition of the gut microbiota in influencing peripheral immune tonus and the effectiveness of immunotherapy in cancer patients.13–16
The human gut microbiota modulates many host processes, including metabolism, inflammation, peristalsis, immune functions, and intestinal epithelial barrier fitness.17–19 In the last decade, major progress has been made in the comprehension of colon cancer development in interaction with the local microbiota.20 Surprisingly, a ‘deviated’ repertoire of the gut microbiota, called ‘intestinal dysbiosis,’ has been epidemiologically – and sometimes causally – associated with a variety of chronic inflammatory disorders including neoplasia, located at sites distant from the gut. In parallel, discoveries made in preclinical tumor models and in cancer patients have demonstrated that the composition of the intestinal microbiota influences the effectiveness of anticancer agents (such as immunogenic chemotherapies and ICIs) and regulates tumor immunosurveillance.21–28 Several lines of evidence have unraveled the link between the gut microbiota composition and ICI-mediated anti-tumor immune responses. This review will summarize arguments supporting the links between the intestinal ecosystem and tumor immunosurveillance, overviewing the deleterious effects of antibiotics on the clinical benefits to be expected from ICIs, the metagenomics-based fingerprints dictating survival, and the key regulatory bacteria associated with tumor control during treatment with immunotherapy.
Antibiotics hinder the efficacy of ICIs
Preclinical studies performed in axenic (gnotobiotic) or broad-spectrum antibiotic (ATB)-treated mice have supported a cause–effect relationship between dysbiosis and the failure of anticancer therapeutics.21–23,26,28 Several independent retrospective studies in advanced cancer patients across a diverse range of malignancies (NSCLC, RCC, bladder cancer, melanoma, and geographic locations) revealed that antibiotic treatment taken 1 month before anticancer therapeutics dampens the clinical efficacy of ICIs and immunogenic chemotherapy. These observations specifically highlighted that the disruption of a homeostatic microbiota (i.e., a switch from eubiosis to dysbiosis) and the loss of specific bacterial species may be detrimental for the success of anticancer therapies.28–36 Recently, corroborating this notion, Derosa et al. confirmed in a prospective trial investigating the composition of the gut microbiota through shotgun metagenomics that antibiotics prior to second-line PD-1 blockade in advanced RCC patients had a deleterious clinical impact, reducing the microbiota diversity, and increasing Clostridium hathewayi, a species associated with immune tolerance.37 In parallel, microbiota profiling from 70 Japanese NSCLC patients also showed that ATB prior to ICIs decreases bacterial diversity and increases Clostridium hathewayi.38 Intrinsically, identification of key bacteria driving the sensitivity/primary resistance to anticancer treatments is crucial to unravel the role of the gut microbiota in this scenario. Many of the recently published studies in this area highlight the deleterious effect of antibiotics in patients with more advanced disease and with multifariousness incidents that may influence ICI responses.28,37 However, a recent analysis showed that patients with non-metastatic melanoma, a “best-prognosis” subgroup receiving ICIs in the adjuvant setting, also had a survival detriment if exposed to antibiotics. Clearly, harm from antibiotics is not limited to cancer patients with advanced metastatic disease.39 Nevertheless, antibiotic classes should be carefully considered. Some antibiotics can provide a positive ‘eubiotic’ effect on the gut microbiota by reducing the abundance of unfavorable gut bacteria. Vancomycin, mostly targeting gram-positive bacteria, including butyrate-producing bacteria and decreasing short-chain fatty acids (SCFA) concentrations, in combination with radiotherapy was able to potentiate the abscopal antitumor immune effect and tumor growth inhibition in mice. Notably, butyrate, a metabolite produced by the vancomycin-depleted gut bacteria, abrogated the vancomycin effect.40 In fact, high levels of butyrate and propionate in the blood are associated with resistance to CTLA-4 blockade and an increase in the abundance of Treg cells.41 However, these results are in contrast with a small Japanese study (52 patients suffering from a broad range of cancer types) showing that high concentrations of fecal and plasma SCFAs were associated with a response to PD-1 treatment and longer progression-free survival (PFS).42 Additional research is needed to clarify the association between fecal and plasma SCFAs and the efficacy of ICIs. Conversely, there is strong evidence indicating that antibiotics-induced dysbiosis is associated with poor therapeutic efficacy of ICI-based immunotherapy, suggesting a causal link between dysbiosis and poor therapeutic outcome.28–33,36,37
Gut oncomicrobiota signatures associated with response to ICIs
Recent advances in next-generation sequencing (NGS) approaches, allowing for the in-depth study of the intestinal microbiota composition, facilitated the discovery of correlations between specific fingerprints of the gut microbiota with the onset and course of certain pathologies.43 Accordingly, the exploration of the composition of the gut microbiota in cancer patients through 16S rRNA gene sequencing or shotgun metagenomics has demonstrated a major impact of the gut microbiota on the clinical activity of ICIs. These analyses led to the hypothesis that the intestinal microbiota can be used to categorize patients receiving ICIs in responders (R) and non-responders (NR) as defined by standardized response evaluation criteria in solid tumors (RECIST 1.1 criteria) regardless of methodologies for DNA extraction and sequencing, geo-distributions of patient populations, and therapies (Table 1).
Table 1.
Studies highlighting the role of the gut microbiota in the clinical efficacy of anticancer therapeutics.
| Cancer | Study | ICI | N = | Tech. | Diversity | Results (good) | Results (bad) | Country | Sample |
|---|---|---|---|---|---|---|---|---|---|
| HCC | Zheng et al., Journal for ImmunoTherapy of Cancer, 2019 | aPD-1 | 8 | MGN | Increased in R | Akkermansia muciniphila | Bacteroides nordii | China | Stool |
| Ruminococcaceae spp. | Fusobacterium varium | ||||||||
| Bifidobacterium dentium | |||||||||
| Dialister invisus | |||||||||
| Coprococcus comes | |||||||||
| MM | Frankel et al., Neoplasia, 2017 | aCTLA-4± aPD-1 | 39 | MGN | No difference | Bacteroides caccae | Coriobacteriaceae | Texas, USA | Stool |
| Streptococcus parasanguinis | Atopobium parvulum | ||||||||
| Faecalibacterium prausnitzi | Acidaminococcaceae | ||||||||
| Bacteroides thetaiotamicron | |||||||||
| Holdemania filiformis | |||||||||
| Dorea formicogenerans | |||||||||
| MM | Chaput et al., Ann. Oncol., 2017 | aCTLA-4 | 26 | 16S rRNA | Not adressed | Faecalibacterium prausnitzii | Bacteroides | France | Stool |
| Unclassified Ruminococcaceae | |||||||||
| Clostridium XIVa | |||||||||
| Blautia | |||||||||
| MM | Gopalakrishnan et al., Science, 2018 | aPD-1 | 43 | 16S rRNA | Increased in R | Clostridiales | Bacteroidales | Texas, USA | Stool |
| Ruminococcaceae | |||||||||
| Faecalibacterium | |||||||||
| MM | Matson et al., Science, 2018 | aPD-1 | 42 | 16S rRNA | Not adressed | Bifidobacterium longum | Ruminococcus obeum | Chicago, USA | Stool |
| Collinsella aerofaciens | Roseburia intestinalis | ||||||||
| Enterococcus faecium | |||||||||
| MM | Coutzac et al., Nature Communications, 2020 | aCTLA-4 | 38 | MGN | Not adressed | Faecalibacterium | France | Stool | |
| Gemminger | |||||||||
| MM | Wind et al., Melanoma Research, 2020 | aCTLA-4± aPD-1 | 25 | MGN | No difference | Streptococcus parasanguinis | Peptostreptococcaceae | Netherlands | Stool |
| Bacteroides massiliensis | |||||||||
| Akkermansia muciniphila | |||||||||
| NSCLC/ gastric |
Fukuoka et al., ASCO, 2018 | aPD-1 | 38 | 16S rRNA | Increased in R | Clostridiales | Japan | Stool | |
| NSCLC/ RCC |
Routy et al., Science, 2018 | aPD-1 | 100 | MGN | NA | Firmicutes | Parabacteroides distasonis | France | Stool |
| Akkermansia muciniphila | Bacteroides nordi | ||||||||
| Alistipes indistinctus | |||||||||
| NSCLC | Jin et al., Journal of Thoracic Oncology, 2019 | aPD-1 | 37 | 16S rRNA | Increased in R | Alistipes putredinis | Ruminococcus unclassified | China | Stool |
| Bifidobacterium longum | |||||||||
| Prevotella copri | |||||||||
| NSCLC | Katayama et al., Transl Lung Cancer Research 2019 | aPD-1 | 17 | 16S rRNA | NA | Lactobacillus | Bilophila | Japan | Stool |
| Clostridium | Sutterella | ||||||||
| Syntrophococcus | |||||||||
| NSCLC | Hakozaki et al., ASCO, 2020 | aPD-(L)1 | 70 | 16S rRNA | Increased in R | Clostridiales | Japan | Stool | |
| Ruminococcaceae UCG 13 | |||||||||
| NSCLC | Song et al., Thorac Cancer., 2020 | aPD-1 | 63 | MGN | Increased in R | Parabacteroides | Veillonella | China | Stool |
| Methanobacteriaceae | Selenomonadales | ||||||||
| Negativicutes | |||||||||
| NSCLC | Botticelli J Transl Med. 2020 | a-PD-1 | 11 | NA | NA | Propionate | 2-Pentanone | Italy | Stool |
| Butyrate | Tridecane | ||||||||
| Lysine | |||||||||
| Nicotinic acid | |||||||||
| PC | Riquelme et al., Cell, 2019 | Surgery | 43 | 16S rRNA | Increased in R | Pseudoxanthomonas | Tumor | ||
| Saccharopolyspora | |||||||||
| Streptomyces | |||||||||
| Rectal cancer | Jang et al., International Journal of Radiation Oncology • Biology • Physics (2020) | Preoperative Chemoradiation | 45 | 16S rRNA | Increased in R | Duodenibacillus massiliensis | Bacteroidales | Korea | Stool |
| RCC | Derosa et al., European Urology, 2020 | aPD-1 | 58 | MGN | Increased in R | Akkermansia muciniphila | Erysipelotrichaceae bacterium_2_2_44A | France | Stool |
| Bacteroides salyersiae | Clostridium hathewayi | ||||||||
| Eubacterium siraeum | Clostridium clostridioforme | ||||||||
| RCC | Agarwal et al., JCO, 2020 | aPD-1 | 22 | 16S rRNA | Increased in R | Akkermansia muciniphila | USA | Stool | |
| Solid cancers | Nomura et al., JAMA Netw Open. 2020 | aPD-1 | 52 | NA | NA | Acetic acid | Japan | Stool | |
| Propionic acid | |||||||||
| Butyric acid | |||||||||
| Valeric acid | |||||||||
| Solid cancers |
Heshiki et al., Microbiome, 2020 | Chemotherapy/ | 26 | MGN | Increased in R | Bacteroides xylanisolvens | Clostridium symbiosum | NA | Stool |
| immunotherapy | Bacteroides ovatus | Ruminococcus gnavus | |||||||
| Prevotella copri | |||||||||
| Alistipes spp. |
MM: metastatic melanoma; NSCLC: non-small cell lung cancer; PC: pancreatic cancer; RCC: renal cell carcinoma; R: responder; NR: non-responder; MGN: metagenomic; NA: not applicable.
The first evidence came from a French cohort of metastatic melanoma (MM) patients treated with the anti-CTLA-4 antibody ipilimumab. Twenty-six MM patients were prospectively enrolled to analyze the impact of gut microbiota composition at baseline on clinical response to ipilimumab.44 Interestingly, the authors could segregate cancer patients into clusters driven by specific bacterial fingerprints found using 16S rRNA gene sequencing. Patients belonging to cluster A harbored Faecalibacterium spp. and were associated with longer PFS than Bacteroides spp.-driven cluster B patients. Moreover, patients from cluster A exhibited lower circulating CD4+ Tregs.41,44 An additional study including 39 patients focusing on various ICI regimens (anti-CTLA-4, anti-PD-1, or the combination of both) corroborated the finding that the metagenomics-based analysis of the gut microbiota composition can predict clinical outcome of immune checkpoint blockade in MM patients. It also showed with the bias of a limited number of patients in each immunotherapy arm that the best species predictive for response are different in each regimen.45 Here again, a relative enrichment in Faecalibacterium prausnitzii was strongly associated with responses to a combination of both nivolumab and ipilimumab while Dorea formicigenerans correlated with a favorable clinical outcome during the course of pembrolizumab.45 A study published by Gopalakrishnan et al. revealed that MM patients, from Texas (USA), who responded to anti-PD-1 therapy, had a significantly higher diversity of bacteria in their stool at diagnosis compared to NR. Moreover, a higher relative abundance of Clostridiales, Ruminococcaceae, and Faecalibacterium was observed in individuals with a good prognosis while NR cancer patients had a higher abundance of Bacteroidales.27 The relationship between the dominance of distinct intestinal bacteria and tumor immunosurveillance was discussed when correlating tumor-infiltrating lymphocyte (TIL) phenotyping and 16S rRNA-based bacterial enrichment. The authors showed, in 25 patients, that CD8+, CD3+, FOXP3+, PD1+, and Granzyme B+ TILs were associated with the Faecalibacterium genus, the Ruminococcaceae family, and the Clostridiales order, suggesting the impact of distinct commensals on cytolytic T cells entailing tumor progression.27 Another US report, from Chicago, also demonstrated significant microbiota-related differences in the response to treatment with PD-1 blocking antibodies. 16S rRNA sequencing of gene amplicons in fecal materials of 42 MM patients at baseline demonstrated that R had enrichment in Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium.25 Moreover, a study performed in 25 Dutch MM patients showed that differences in taxa abundance contrasted R and NR, with similar results with previous studies. Indeed, carriers of Streptococcus parasanguinis or Bacteroides massiliensis exhibited prolonged PFS while individuals harboring Peptostreptococcaceae (unclassified species) exhibited a shorter OS and PFS compared to non-carriers.46 Taken together, these epidemiological studies described the association between the composition of the intestinal ecosystem at diagnosis and the clinical outcome of MM patients treated with ICIs.
These particular findings are not restricted to MM. Indeed, the fecal bacteria repertoire has been found to also critically influence the prognosis of advanced NSCLC and RCC cancer patients during the course of ICI-based therapies in France. Quantitative metagenomics analysis performed prior to anti-PD-1 blockade identified a distinct gut metagenomic fingerprint (centered around Akkermansia muciniphila and Alistipes spp.) in stools of 100 patients who benefited from PD-1 inhibition; considering response rates or PFS at 3 months.28 Interestingly, the role of the gut microbiota has also been addressed in an East-Asian NSCLC population.47 In this cohort, 16S rRNA gene sequencing of 37 stools demonstrated that higher diversity of the gut microbiota paved the way to prolonged PFS. Differential gut microbiota signatures contrasted R versus NR cancer patients. Here again, Alistipes putredinis, Prevotella copri, or Bifidobacterium longum were enriched in R patients. The Shannon diversity index of the taxonomic composition was positively correlated with circulating immune antigen-primed–cytotoxic T cells (such as GZMB+CD45RO+CD27+ CD8+ T cells or GZMB+CD45RO+CD27− CD8+ T cells).47 Two additional Japanese studies performed 16S rRNA gene sequencing of fecal materials from NSCLC (n = 70) and NSCLC (n = 14) as well as gastric cancer (n = 24) patients confirmed that higher diversity of the bacterial community and enrichment of the Ruminococcaceae and Clostridiales order predicted benefit to PD-1 blockade.38,48 Furthermore, the relative abundance of members of the Ruminococcaceae family48 correlated with the density of PD-1+CD8+ T cells among (TILs). Again, another report analyzing the gut microbiota composition from 17 NSCLC patients revealed that Lactobacillus, Clostridium, and Syntrophococcus were overrepresented in R, while Bilophila or Sutterella49 was dominant in NR. Of note, the presence of Bilophila drastically shortened the time to treatment failure.49 A Chinese prospective study including 63 NSCLC cancer patients revealed Parabacteroides and Methanobacteriaceae as species and family members associated with PFS >6 months while stool enriched in Veillonella, Selenomonadales, and Negativicutes50 predicted shorter PFS during PD-1 blockade. In sharp contrast with these findings, ileal enrichment with Veillonella, Selenomonadales, and Negativicutes was found to be associated with increased TIL and favorable prognosis during oxaliplatin-based chemotherapy in proximal colon cancer patients.51
Several teams have confirmed the potential clinical significance of Akkermansia muciniphila in driving a therapeutic benefit to ICIs, more specifically in NSCLC,28 melanoma,46 HCC patients,52 and recently in RCC.37,53 In brief, Derosa et al. reported in 58 RCC cancer patients treated in 2 L with nivolumab that a significant bacterial composition contrasted R versus NR with an overrepresentation of distinct species including Akkermansia muciniphila, Bacteroides salyersiae, or Eubacterium siraeum in patients disposed to becoming R.37
In a parallel study, all RCC cancer patients exhibiting a complete response to ICIs (n = 3) harbored Akkermansia muciniphila although the number of patients was not sufficient to draw definitive conclusions.53 Fecal metagenomics analysis performed in 8 HCC cancer patients identified Akkermansia muciniphila and Ruminococcaceae spp. in the 20 enriched spp. characterizing R patients and B. nordii in a 15 spp-fingerprint associated with NR as already reported.28,52 In Dutch MM patients, A. muciniphila was also listed in the favorable commensals associated with objective responses to ICI therapy.46
Interestingly, focusing on describing also negative species by applying various bioinformatic and clinical subgroup analyses (LEfSe, PLS-DA VIP, networks), Derosa et al. identified a set of species (phylum Firmicutes, family Clostridiaceae, Clostridium clostridioforme, Clostridium hathewayi) as associated with primary resistance to ICIs, enriched by ATB use and metastatic cancer status.37
Although ICIs have revolutionized therapeutic approaches across various malignancies, conventional anticancer regimens such as chemotherapy or radiotherapy still represent the cornerstone of oncological arsenal. Numerous studies have addressed the putative influence of the gut microbiota repertoire in the prediction of clinical responses to these cytotoxic agents. Twenty-six cancer patients diagnosed with miscellaneous malignancies, treated either with cytotoxic compounds or targeted medicine or a combination of the latter drugs with immunotherapy, were enrolled in a prospective study aimed at segregating patients according to their intestinal commensalism. Bacteroides xylanisolvens, Bacteroides ovatus, and Prevotella copri were significantly overrepresented in R compared to NR defined using the RECIST1.1 criterion. In contrast, Clostridium symbiosum and Ruminococcus gnavus were enriched in NR.54 A second study analyzing fecal composition prior to preoperative concurrent chemoradiations in 45 rectal cancer patients concluded that Duodenibacillus massiliensis was linked to complete responses.55
Altogether, the emerging field of oncoimmunomicrobiology is progressively integrating the gut microbiota into the parameters that determine the cancer immune set-point governing the clinical efficacy of immuno-chemo-radio-therapy. First, low alpha diversity of the intestinal ecosystem is associated with dismal prognosis in advanced cancer patients, as also shown in several chronic inflammatory disorders (such as obesity).56 Secondly, some bacteria species arise to be repeatedly associated with favorable clinical outcomes (namely Akkermansia muciniphila, Ruminococcaceae including Faecalibacterium prausnitzi, Bifidobacterium spp.) although variabilities in the main commensal fingerprints associated with a specific pattern of responses appear obvious within analogous patients’ populations and therapies. These variabilities could be explained by many factors such as DNA extraction and sequencing methodologies,55 cohort size, age and gender, geography,57,58 and confounding factors (including diet, lifestyle, exposure to xenobiotics, antibiotic class and window, comedications and comorbidities).59,60 Other important players that affect intestinal barrier integrity are the tumor itself, disease stage, ECOG performance status, medication, peripheral inflammatory tone, and pro-cachexia signs.61,62 Finally, aside from the basal composition of the gut commensalism, dictated by the original network of bacterial co-occurrence, the treatment itself may impact on the relative abundance of microbes, as shown with ipilimumab23 and tyrosine kinase inhibitors (TKIs).37 Overall, TKIs induced a significant and characteristic microbiota shift promoting a higher abundance of immunostimulatory commensals that could be used to improve the efficacy of ICIs in RCC patients such as A. senegalensis and A. muciniphila.
In addition, a different study paved the way in understanding the reciprocal relationship between the intratumoral microbiota and the clinical outcome of resected pancreatic ductal adenocarcinoma (PDAC) cancer patients. Although most of the patients died at an advanced stage with an OS of 9% at 5 y, a minor subset of patients survives longer.63 Interestingly, alpha-diversity of the tumor microbiota was significantly higher in the long-term survivor of PDAC.64 In fact, an enrichment on Proteobacteria (Pseudoxanthomonas) and Actinobacteria (Saccharopolyspora and Streptomyces) was observed in this subset of patients.64 However, intra-tumoral microbes in pancreatic cancer may also be harmful. Pushalkar et al. demonstrated the negative impact of microbes on antitumor immunity with evidence for possible migration of bacteria from the gut to the pancreas.65 Nevertheless, these emerging findings indicate there is a cross-talk between gut microbiota and local microbiota (as exemplified by pancreatic cancer) and that also local microbiota may contribute positively or negatively to carcinogenesis and therapeutic responses.
Identification of key bacteria boosting the antitumoral efficacy of anticancer treatments
Modulating the composition of the gut microbiota and harnessing the immunogenicity of the intestinal microbiota may be a promising strategy with which to circumvent primary resistance to anticancer therapeutics. Several bacterial candidates have been identified, isolated, characterized, and are currently or on the verge to be tested in clinical trials in combination with anticancer treatments or as a standalone therapy.66
Cause-effects relationships between the presence of distinct microbial commensals and antitumor activity have been examined primarily in preclinical models. So far, investigators have performed oral gavages in germ-free or broad ATB-treated mice using a complete human or mouse ecosystem or a complex mixture of several bacteria or “monoclonal” strains, into immunocompetent syngeneic hosts inoculated with ortho-or hetero-topic cancers. The concept of “avatar” mice which consists in colonizing gut-sterilized mice with patients’ stools has proven useful to recapitulate human dysbiosis across various diseases.67,68 In the setting of cancer, avatar mice transferred with feces from patients bearing melanoma, NSCLC, RCC, or colon cancer and transplanted with orthotopic tumors could convey the phenotype of R versus NR following immunotherapy with anti-PD1 and/or anti-CTLA-4 Ab, in 100% cases after oral gavage with R fecal material and in 75% cases when supplementing with NR derived feces.23,25,27,28
Akkermansia muciniphila is a strictly anaerobic Gram-negative bacterium from the phylum Verrucomicrobia displaying a multifaceted mode of action.69,70 Indeed, a Phase I trial conducted in 32 overweight/obese insulin-resistant volunteers demonstrated that supplementation with Akkermansia muciniphila is safe and capable of improving the metabolic fitness.71 Further studies have emphasized its capacity to prolong lifespan in progeroid mice72 while ameliorating the symptoms of amyotrophic lateral sclerosis through nicotinamide accumulation in the central nervous system.73 A recent report showed that A. muciniphila prevents colitis-induced colon cancer by mobilizing TNF producing CTL primed in the mesenteric lymph nodes and expressing low levels of PD1 despite their lytic potential.74 We highlighted the capacity of A. muciniphila to boost immune responses during the course of PD-1 blockade, both in tumor-bearing rodents and humans.28,37 Supplementation of NR-FMT treated avatar mice with A. muciniphila rescued the antitumoral efficacy of PD1-blockade in an IL-12-dependent manner demonstrating that A. muciniphila dictates the clinical outcome of ICIs. In addition, the bacterium dampened the recruitment of immunosuppressive Tregs cells into the tumor microenvironment while eliciting the accumulation of CC-chemokine receptor 9 (CCR9)-expressing Th1 cells in the tumor bed. Accordingly, memory Th1 and Tc1 cell reactivity against A. muciniphila correlated with a clinical benefit of PD-1 blockade in NSCLC and RCC cancer patients.28
Enterococcus hirae has been one of the first bacterial isolates that show antitumoral potential in combination with chemotherapy. This Gram+ bacterium is essential to mediate the antitumoral efficacy of cyclophosphamide (CTX), a prominent alkylating anticancer agent.21,75 CTX promotes the translocation of E. hirae in secondary lymphoid organs (mLN and spleen), inducing FNγ (and IL-17) producing CD4 + T cells and Tc1 cells. Moreover, the combination of E. hirae and CTX reduced Treg numbers in sarcomas, culminating in a significant rise of the CD8/Foxp3 ratio, which in turn anticorrelated with tumor size.21 Hence, oral gavage with E. hirae restored the antitumoral efficacy of CTX lost in ATB-treated mice. In advanced cancer patients, memory CD4+ Th1 cell responses against E. hirae were associated with survival in CTX- or anti-PD-127 antibody-treated individuals. While the prevalence of E. hirae is minimally detected using shotgun metagenomics-based analyses of patient stool, culturomics allowed for the isolation of E. hirae colonies in 20% cancer patients. Diagnosis of E. hirae in stool culturomics of NSCLC patients at diagnosis before starting second-line PD-1 blockade predicted prolonged survival.28 An independent study revealed that the frequency of circulating T cells recognizing E. hirae correlated with robust CD8+ T cell responses and better prognosis in HBV-related hepatocellular carcinoma,76 suggesting the clinical significance of this particular bacterium across different malignancies.
In addition to E. hirae, other Enterococci spp. have been isolated and characterize for their immunomodulatory potential against cancer cells. A strain of Enterococcus gallinarum, isolated from a healthy human gut, has demonstrated its antiproliferative effects against EMT6 breast, RENCA renal, and LLC1 lung carcinoma.77 This microbial product caused changes in the tumor immune microenvironment and increased the CD8+/FoxP3 ratio. In addition, a TLR5 dependent immuno-stimulatory phenotype of this strain was monitored using reporter cell lines. The authors identified flagellin as the active component of Enterococcus gallinarum.78 Therefore, the antitumoral potential of this strain is currently under investigation in cancer patients amenable to ICI-based therapy in advanced diseases, as well as in neoadjuvant settings to determine its property to modulate the tumor microenvironment before tumor resection (NCT03934827/NCT04193904).
Bifidobacterium is a gram-positive, non-spore-forming, non-motile, non-filamentous polymorphic rod bacterium. Pioneering studies demonstrated that the growth kinetics of B16.SIY melanoma as well as the intratumoral CD8+ T cell accumulation were completely different in mice purchased from different vendors (Jackson Laboratories (JAX) versus Taconic Farms (TAC)) harboring distinct commensal microbiota.24 These differences were ablated when the two mouse colonies were cohoused, demonstrating that the normalization of the gut microbiota could boost anti-cancer immune responses. Further investigations characterizing the composition of the gut microbiota between JAX and TAC highlighted that certain Bifidobacterium species could induce tumor-infiltrating CD8+ T cells. Transfer of Bifidobacterium breve or Bifidobacterium longum or fecal material from JAX mice into TAC mice could all reduce melanoma growth and restore anti-melanoma cytotoxic T lymphocyte (CTL) responses.24 A recent study unveiled that the SIY antigen (TAA) of B1610 displayed antigen mimicry with an epitope belonging to Bifidobacterium breve, accounting for the T cell–mediated antitumor responses achieved by oral supplementation with this probiotic.79 Accordingly, T cells targeting the microbial antigen recognized melanoma tumor cells expressing the SIY tumor-associated antigen. Conversely, tumors expressing the TAA also grew faster in mice lacking Bifidobacterium breve bacterium.79 Of note, memory immune reactivity against B. longum also correlated with robust CD8+ T cell responses and better prognosis in HBV-related hepatocellular carcinoma patients.76
The first bacterial species known to harbor “zwitterionic” peptides capable of engaging CD4+ T cell receptors was Bacteroides fragilis.80–82 B. fragilis was very effective in boosting immune responses primed in the setting of sarcoma tumors treated with anti-CTLA4 Ab23 as well as colon carcinoma treated with oxaliplatin-based immunogenic chemotherapy.51 Antibiotics blunted the anticancer efficacy of CTLA-4 blockade against various transplantable tumors unless oral supplementation with B. fragilis was performed, which reinstated IL-12-dependent Th1 immune responses. Interestingly, anti-CTLA-4 Abs administered to tumor-bearing avatar mice reconstituted with FMT from melanoma patients foster the overrepresentation of distinct Bacteroides spp. (Bacteroides fragilis or Bacteroides thetaiotaomicron) and recapitulated the phenotype of response observed in patient.23 Interestingly, oral supplementation with B. fragilis (as opposed to Fusobacterium nucleatum or Paraprevotella clara) turned chemotherapy-induced tolerogenic ileal apoptosis into immunogenic cell demise capable of eliciting PD1high follicular helper T cells and B cell responses and of promoting the efficacy of anti-PD1 Abs against established colon cancers.51 Hence, the ileal microbiota enriched in commensals playing the role of adjuvant for ileal apoptosis triggered TFH and the efficacy of PD-1 blockade, even in tumors devoid of neoantigens.
In contrast to the aforementioned approaches, using very common commensals to compensate gut dysbiosis, another study demonstrated that a mixture of several rare strains isolated from fecal materials from healthy Japanese individuals was effective in shaping immunity in the colonic mucosae and tumor microenvironment.83 The authors identified a cocktail of 11 human bacterial strains capable of promoting tent IFNγ producing CD8+ T cells that are not only crucial for combatting infectious pathogens but also for dampening cancer progression.83 Supplementation of germ-free mice with these 11 strains (composed of 7 Bacteroidales spp. and 4 non-Bacteroidales spp.) resulted in the robust induction of IFNγ-CD8+ T cells in the colon through a mechanism requiring Batf3 dependent CD103+ CD11b− dendritic cells. Next, they showed the capacity of the cocktail to ameliorate the efficacy of PD-1 blockade in axenic MC38 adenocarcinoma bearing mice. It significantly improved the efficacy of ICIs while increasing the frequency IFNγ+ CD8 TILs phenotypically distinct from the colonic IFNγ+ CD8 T cell subsets. Of note, the bacterial cocktail could reduce tumor growth as a standalone therapy (in the absence of PD-1 blockade).83 This microbial product is currently tested in combination with PD-1 blockade in advanced cancer patients after vancomycin sensitization (NCT04208958).
It is well known that bacteria can sporulate under life-threatening circumstances84,85 offering an advantage over non-sporulating commensals for the long-lasting colonization of their hosts. This property has been exploited by other investigators in the setting of PD-1 blockade. Firmicutes spores fraction isolated from a healthy donor stool was capable of rescuing the antitumoral efficacy of PD-1 blockade in both conventional mice treated with antibiotics and axenic mice by increasing CD8+ TILs.86
Needless to say that most of the antitumoral efficacy described in all these preclinical studies appear to be strain-specific,22 urging for delineating precise modes of action for each single isolate.
Concluding remarks
Several clinical trials are evaluating the capacity of harnessing the gut microbiota to improve cancer treatments from different angles such as examining the ability to prevent primary resistance to various anticancer treatment modalities, transforming “cold into hot” tumor microenvironment, and mitigating toxicities associated with a single line or combination ICIs.66 Several important issues need to be addressed in the clinical development of live biotherapeutic products or their derivatives (metabolites, antigens, or adjuvants). First, robust preclinical datasets are mandatory to characterize the mechanisms of action of each strain or microbial products to design the most suitable clinical strategy and indications. This will allow for the design of appropriate pharmacodynamic parameters to follow compliance and transient colonization of the patient. Secondly, patient stratification will be necessary to avoid treating patients without overt intestinal dysbiosis, and for whom primary resistance to ICIs may be related to tumor intrinsic factors. Third, compatibility networks between the indigenous microflora and the live biotherapeutic product may be crucial for a long-lasting benefit of repetitive courses of anticancer probiotics. Pre-sensitization with antibiotics or other innovative approaches aimed at eliminating pathobionts associated with ICI failure or precluding colonization or bioactivity of immunogenic commensals may be important to optimize clinical regimen. Regardless of these considerations, this emerging field will benefit from pioneering trials showing the efficacy of FMT from complete responders into patients experiencing primary resistance to PD-1 blockade.
Acknowledgments
LZ and GK are supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; AMMICa US23/CNRS UMS3655; Association pour la recherche sur le cancer (ARC); Association “Le Cancer du Sein, Parlons-en!”; Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); Gustave Roussy Odyssea, the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; High-end Foreign Expert Program in China (GDW20171100085), Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology (ANR-18-IDEX-0001); the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and the SIRIC Cancer Research and Personalized Medicine (CARPEM). A.P.C. is supported by the CPRIT Research Training Program (RP170067). LD is supported by Fondation Philanthropia, Gustave Roussy.
Conflicts of interest
RD is a full-time employee of everImmune, a biotech company dedicated to immunostimulatory bacteria. RD, GK, and LZ are the scientific cofounders of everImmune.
Supplementary
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Pardoll D. Cancer and the immune system: basic concepts and targets for intervention. Semin Oncol. 2015;42(4):523–11. doi: 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain J-F, Testori A, Grob -J-J, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364(26):2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 3.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu W-J, Weber JS, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315(15):1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 7.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu JX, Hubbard-Lucey VM, Tang J. Immuno-oncology drug development goes global. Nat Rev Drug Discov. 2019;18(12):899–900. doi: 10.1038/d41573-019-00167-9. [DOI] [PubMed] [Google Scholar]
- 10.Champiat S, Ferrara R, Massard C, Besse B, Marabelle A, Soria J-C, Ferté C. Hyperprogressive disease: recognizing a novel pattern to improve patient management. Nat Rev Clin Oncol. 2018;15(12):748–762. doi: 10.1038/s41571-018-0111-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 12.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359(6382):1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 14.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 15.Kroemer G, Zitvogel L. Cancer immunotherapy in 2017: the breakthrough of the microbiota. Nat Rev Immunol. 2018;18(2):87–88. doi: 10.1038/nri.2018.4. [DOI] [PubMed] [Google Scholar]
- 16.Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15(8):465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 17.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 19.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Vos WM, de Vos EAJ. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–56. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 21.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45(4):931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 29.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, Long N, Plodkowski AJ, Arbour KC, Chaft JE, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29(6):1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinato DJ, Howlett S, Ottaviani D, Urus H, Patel A, Mineo T, Brock C, Power D, Hatcher O, Falconer A, et al. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol. 2019. September 12;5(12):1774. doi: 10.1001/jamaoncol.2019.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkrief A, El Raichani L, Richard C, Messaoudene M, Belkaid W, Malo J, Belanger K, Miller W, Jamal R, Letarte N, et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology. 2019;8(4):e1568812. doi: 10.1080/2162402X.2019.1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Gao G, Li W, Li X, Zhao C, Jiang T, Jia Y, He Y, Li A, Su C, et al. Antibiotics are associated with attenuated efficacy of anti-PD-1/PD-L1 therapies in Chinese patients with advanced non-small cell lung cancer. Lung Cancer. 2019;130:10–17. doi: 10.1016/j.lungcan.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Lalani A-KA, Xie W, Braun DA, Kaymakcalan M, Bossé D, Steinharter JA, Martini DJ, Simantov R, Lin X, Wei XX, et al. Effect of antibiotic use on outcomes with systemic therapies in metastatic renal cell carcinoma. Eur Urol Oncol. 2019. September 24; doi: 10.1016/j.euo.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pflug N, Kluth S, Vehreschild JJ, Bahlo J, Tacke D, Biehl L, Eichhorst B, Fischer K, Cramer P, Fink A-M, et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5(6):e1150399. doi: 10.1080/2162402X.2016.1150399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nenclares P, Bhide SA, Sandoval-Insausti H, Pialat P, Gunn L, Melcher A, Newbold K, Nutting CM, Harrington KJ. Impact of antibiotic use during curative treatment of locally advanced head and neck cancers with chemotherapy and radiotherapy. Eur J Cancer. 2020;131:9–15. doi: 10.1016/j.ejca.2020.02.047. [DOI] [PubMed] [Google Scholar]
- 36.Wilson BE, Routy B, Nagrial A, Chin VT. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: a systematic review and meta-analysis of observational studies. Cancer Immunol Immunother. 2020;69(3):343–354. doi: 10.1007/s00262-019-02453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, Segata N, Desnoyer A, Pietrantonio F, Ferrere G, et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol. 2020. May 3; doi: 10.1016/j.eururo.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 38.Meeting Library Gut microbiome to predict efficacy and immune-related toxicities in patients with advanced non-small cell lung cancer treated with anti-PD-1/PD-L1 antibody-based immunotherapy. [accessed May25, 2020]. https://meetinglibrary.asco.org/record/188511/abstract
- 39.Mohiuddin JJ, Chu B, Facciabene A, Poirier K, Wang X, Doucette A, Zheng C, Xu W, Anstadt EJ, Amaravadi RK, et al. Association of antibiotic exposure with survival and toxicity in patients with melanoma receiving immunotherapy. J Natl Cancer Inst. 2020. April 15; doi: 10.1093/jnci/djaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gómez L, Verginadis I, Bittinger K, Pustylnikov S, Pierini S, Perales-Linares R, Blair IA, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest. 2020;130(1):466–479. doi: 10.1172/JCI124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutzac C, Jouniaux J-M, Paci A, Schmidt J, Mallardo D, Seck A, Asvatourian V, Cassard L, Saulnier P, Lacroix L, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. 2020;11(1):1–13. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura M, Nagatomo R, Doi K, Shimizu J, Baba K, Saito T, Matsumoto S, Inoue K, Muto M. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw Open. 2020;3(4):e202895. doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claesson MJ, Clooney AG, O’Toole PW. A clinician’s guide to microbiome analysis. Nat Rev Gastroenterol Hepatol. 2017;14(10):585–595. doi: 10.1038/nrgastro.2017.97. [DOI] [PubMed] [Google Scholar]
- 44.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 45.Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19(10):848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wind TT, Gacesa R, Vich Vila A, de Haan JJ, Jalving M, Weersma RK, Hospers GAP. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res. 2020;30(3):235–246. Publish Ahead of Print. doi: 10.1097/CMR.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 47.Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in chinese patients with NSCLC. J Thorac Oncol. 2019;14(8):1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Fukuoka S, Daisuke M, Togashi Y, Sugiyama E, Udagawa H, Kirita K, Kamada T, Kawazoe A, Goto K, Doi T, et al. Association of gut microbiome with immune status and clinical response in solid tumor patients who received on anti-PD-1 therapies. JCO. 2018;36(15_suppl):3011. doi: 10.1200/JCO.2018.36.15_suppl.3011. [DOI] [Google Scholar]
- 49.Katayama Y, Yamada T, Shimamoto T, Iwasaku M, Kaneko Y, Uchino J, Takayama K. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl Lung Cancer Res. 2019;8(6):847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song P, Yang D, Wang H, Cui X, Si X, Zhang X, Zhang L. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese NSCLC patients. Thoracic Cancer. 2020. doi: 10.1111/1759-7714.13442 [DOI] [PMC free article] [PubMed]
- 51.Roberti MP, Yonekura S, Duong CPM, Picard M, Ferrere G, Tidjani Alou M, Rauber C, Iebba V, Lehmann CHK, Amon L, et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat Med. 2020. May;25:1–13. doi: 10.1038/s41591-020-0882-8. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J ImmunoTher Cancer. 2019;7(1):193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal A, Modliszewski J, Davey L, Reyes-Martinez M, Runyambo D, Corcoran D, Dressman H, George DJ, Valdivia R, Armstrong AJ, et al. Investigating the role of the gastrointestinal microbiome in response to immune checkpoint inhibitors (ICIs) among patients (pts) with metastatic renal cell carcinoma (mRCC). JCO. 2020;38(6_suppl):730. doi: 10.1200/JCO.2020.38.6_suppl.730. [DOI] [Google Scholar]
- 54.Heshiki Y, Vazquez-Uribe R, Li J, Ni Y, Quainoo S, Imamovic L, Li J, Sørensen M, Chow BKC, Weiss GJ, et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome. 2020;8. doi: 10.1186/s40168-020-00811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang B-S, Chang JH, Chie EK, Kim K, Park JW, Kim MJ, Song E-J, Nam Y-D, Kang SW, Jeong S-Y, et al. Gut microbiome composition is associated with a pathologic response after preoperative chemoradiation in rectal cancer patients. Int J Radiat Oncol Biol Phys. 2020. April 18;107(4):736–746. doi: 10.1016/j.ijrobp.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 56.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 57.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senghor B, Sokhna C, Ruimy R, Lagier J-C. Gut microbiota diversity according to dietary habits and geographical provenance. Hum Microbiome J. 2018;7–8:1–9. doi: 10.1016/j.humic.2018.01.001. [DOI] [Google Scholar]
- 59.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. Isme J. 2011;5(2):220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012;33(9):459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bindels LB, Neyrinck AM, Salazar N, Taminiau B, Druart C, Muccioli GG, François E, Blecker C, Richel A, Daube G, et al. Non digestible oligosaccharides modulate the gut microbiota to control the development of leukemia and associated cachexia in mice. PLoS ONE. 2015;10(6):e0131009. doi: 10.1371/journal.pone.0131009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bindels LB, Neyrinck AM, Claus SP, Le Roy CI, Grangette C, Pot B, Martinez I, Walter J, Cani PD, Delzenne NM. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. Isme J. 2016;10(6):1456–1470. doi: 10.1038/ismej.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 64.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daillère R, Derosa L, Bonvalet M, Segata N, Routy B, Gariboldi M, Budinská E, Vries IJMD, Naccarati AG, Zitvogel V, et al. Trial watch: the gut microbiota as a tool to boost the clinical efficacy of anticancer immunotherapy. OncoImmunology. 2020;9(1):1774298. doi: 10.1080/2162402X.2020.1774298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharon G, Cruz NJ, Kang D-W, Gandal MJ, Wang B, Kim Y-M, Zink EM, Casey CP, Taylor BC, Lane CJ, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. 2019;177(6):1600–1618.e17. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 69.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Cani PD, de Vos WM. Next-generation beneficial microbes: the case of akkermansia muciniphila. Front Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25(7):1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bárcena C, Valdés-Mas R, Mayoral P, Garabaya C, Durand S, Rodríguez F, Fernández-García MT, Salazar N, Nogacka AM, Garatachea N, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med. 2019;25(8):1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- 73.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572(7770):474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 74.Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, Yuan G, Zhu J, Cao S, Wu Q, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8 + T cells in mice. Gut. 2020. March 13:gutjnl-2019-320105. doi: 10.1136/gutjnl-2019-320105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sistigu A, Viaud S, Chaput N, Bracci L, Proietti E, Zitvogel L. Immunomodulatory effects of cyclophosphamide and implementations for vaccine design. Semin Immunopathol. 2011;33(4):369–383. doi: 10.1007/s00281-011-0245-0. [DOI] [PubMed] [Google Scholar]
- 76.Rong Y, Dong Z, Hong Z, Jin Y, Zhang W, Zhang B, Mao W, Kong H, Wang C, Yang B, et al. Reactivity toward Bifidobacterium longum and Enterococcus hirae demonstrate robust CD8+ T cell response and better prognosis in HBV-related hepatocellular carcinoma. Exp Cell Res. 2017;358(2):352–359. doi: 10.1016/j.yexcr.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 77.Stevenson A, Panzica A, Holt A, Laute Caly D, Ettore A, Delday M, Hennessy E, Cowie P, Pradhan M, Jeffery I, et al. Host-microbe interactions mediating antitumorigenic effects of MRX0518, a gut microbiota-derived bacterial strain, in breast, renal and lung carcinoma. JCO. 2018;36(15_suppl):e15006. doi: 10.1200/JCO.2018.36.15_suppl.e15006. [DOI] [Google Scholar]
- 78.Lauté-Caly DL, Raftis EJ, Cowie P, Hennessy E, Holt A, Panzica DA, Sparre C, Minter B, Stroobach E, Mulder IE. The flagellin of candidate live biotherapeutic Enterococcus gallinarum MRx0518 is a potent immunostimulant. Sci Rep. 2019;9(1):801. doi: 10.1038/s41598-018-36926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bessell CA, Isser A, Havel JJ, Lee S, Bell DR, Hickey JW, Chaisawangwong W, Bieler JG, Srivastava R, Kuo F, et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight. 2020;5(8):8. doi: 10.1172/jci.insight.135597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker H-C, Kasper DL. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15(4):413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stingele F, Corthésy B, Kusy N, Porcelli SA, Kasper DL, Tzianabos AO. Zwitterionic polysaccharides stimulate T cells with no preferential V beta usage and promote anergy, resulting in protection against experimental abscess formation. J Immunol. 2004;172(3):1483–1490. doi: 10.4049/jimmunol.172.3.1483. [DOI] [PubMed] [Google Scholar]
- 83.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 84.Cutting SM, Ricca E. Bacterial spore-formers: friends and foes. FEMS Microbiol Lett. 2014;358(2):107–109. doi: 10.1111/1574-6968.12572. [DOI] [PubMed] [Google Scholar]
- 85.Hutchison EA, Miller DA, Angert ER. Sporulation in bacteria: beyond the standard model. Microbiol Spectr. 2014;2(5): doi: 10.1128/microbiolspec.TBS-0013-2012. [DOI] [PubMed] [Google Scholar]
- 86.Jayaraman L, Sceneay J, Srinivasan S, Halley K, Bist M, Cieciuch K, Marnellos G, Desjardins C, Wortman J, Henn M, et al. Abstract B063: leveraging gut microbiota networks to impact tumor immunotherapy. Cancer Immunol Res. 2019;7(2 Supplement):B063. doi: 10.1158/2326-6074.CRICIMTEATIAACR18-B063. [DOI] [Google Scholar]


