Abstract
MutantHuntWGS is a user-friendly pipeline for analyzing Saccharomyces cerevisiae whole-genome sequencing data. It uses available open-source programs to: (1) perform sequence alignments for paired and single-end reads, (2) call variants, and (3) predict variant effect and severity. MutantHuntWGS outputs a shortlist of variants while also enabling access to all intermediate files. To demonstrate its utility, we use MutantHuntWGS to assess multiple published datasets; in all cases, it detects the same causal variants reported in the literature. To encourage broad adoption and promote reproducibility, we distribute a containerized version of the MutantHuntWGS pipeline that allows users to install and analyze data with only two commands. The MutantHuntWGS software and documentation can be downloaded free of charge from https://github.com/mae92/MutantHuntWGS.
Keywords: Mutant hunt, variant calling, Saccharomyces cerevisiae, bulk segregant analysis, lab evolution
Saccharomyces cerevisiae is a powerful model system for understanding the complex processes that direct cellular function and underpin many human diseases (Birkeland et al. 2010; Botstein and Fink 2011; Kachroo et al. 2015; Hamza et al. 2015, 2020; Wangler et al. 2017; Strynatka et al. 2018). Mutant hunts (i.e., genetic screens and selections) in yeast have played a vital role in the discovery of many gene functions and interactions (Winston and Koshland 2016). A classical mutant hunt produces a phenotypically distinct colony derived from an individual yeast cell with at most a small number of causative mutations. However, identifying these mutations using traditional genetic methods (Lundblad 2001) can be difficult and time-consuming (Gopalakrishnan and Winston 2019).
Whole-genome sequencing (WGS) is a powerful tool for rapidly identifying mutations that underlie mutant phenotypes (Smith and Quinlan 2008; Irvine et al. 2009; Birkeland et al. 2010). As sequencing technologies improve, the method is becoming more popular and cost-effective (Shendure and Ji 2008; Mardis 2013). WGS is particularly powerful when used in conjunction with lab-evolution (Goldgof et al. 2016; Ottilie et al. 2017) or mutant-hunt experiments, both with (Birkeland et al. 2010; Reavey et al. 2015) and without (Gopalakrishnan et al. 2019) bulk segregant analysis.
Analysis methods that identify sequence variants from WGS data can be complicated and often require bioinformatics expertise, limiting the number of investigators who can pursue these experiments. There is a need for an easy-to-use, data-transparent tool that allows users with limited bioinformatics training to identify sequence variants relative to a reference genome. To address this need, we created MutantHuntWGS, a bioinformatics pipeline that processes data from WGS experiments conducted in S. cerevisiae. MutantHuntWGS first identifies sequence variants in both control and experimental (i.e., mutant) samples, relative to a reference genome. Next, it filters out variants that are found in both the control and experimental samples while applying a variant quality score-cutoff. Finally, the remaining variants are annotated with information such as the affected gene and the predicted impact on gene expression and function. The program also allows the user to inspect all relevant intermediate and output files.
To enable quick and easy installation and to ensure reproducibility, we incorporated MutantHuntWGS into a Docker container (https://hub.docker.com/repository/docker/mellison/mutant_hunt_wgs). With a single command, users can download and install the software. A second command runs the analysis, performing all steps described above. MutantHuntWGS allows researchers to leverage WGS for the efficient identification of causal mutations, regardless of bioinformatics experience.
Methods
Pipeline overview
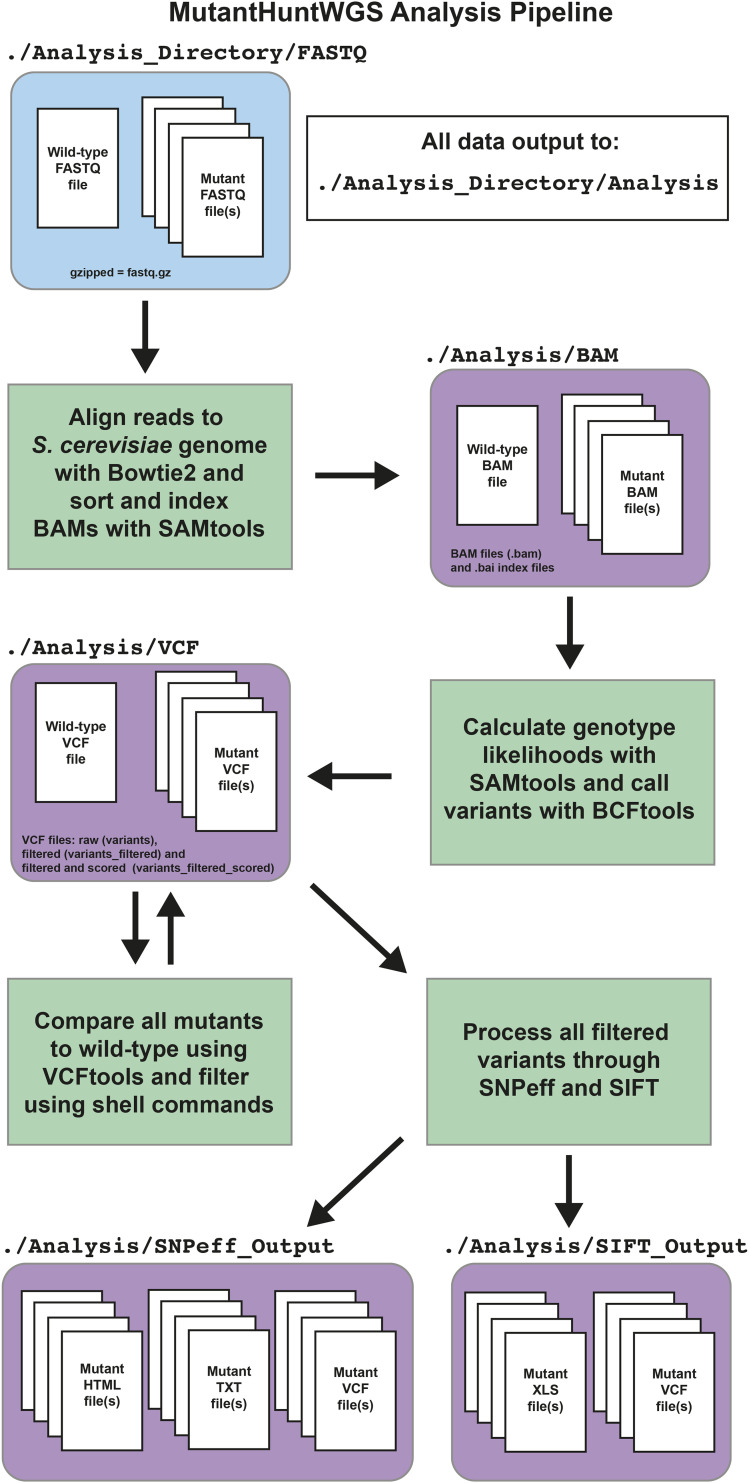
The MutantHuntWGS pipeline integrates a series of open-source bioinformatics tools and Unix commands that accept raw sequencing reads (compressed FASTQ format or .fastq.gz) and a text file containing ploidy information as input, and produces a list of sequence variants as output. The user must provide input data from at least two strains: a control strain and one or more experimental strains. The pipeline uses (1) Bowtie2 to align the reads in each input sample to the reference genome (Langmead and Salzberg 2012), (2) SAMtools to process the data and calculate genotype likelihoods (Li et al. 2009), (3) BCFtools to call variants (Li et al. 2009), (4) VCFtools (Danecek et al. 2011) and custom shell commands to compare variants found in experimental and control strains, and (5) SnpEff (Cingolani et al. 2012) and SIFT (Vaser et al. 2016) to assess where variants are found in relation to annotated genes and the potential impact on the expression and function of the affected gene products (Figure 1). A detailed description of the commands used in the pipeline and all code is available on the MutantHuntWGS Git repository (https://github.com/mae92/MutantHuntWGS; see README.md, Supplemental_Methods.docx files).
Figure 1.
Flow chart of the MutantHuntWGS pipeline. Input data are colored in blue, the various bioinformatics tools in the pipeline are colored in green, and output data are colored in purple. Arrows identify the path of the workflow at each step of the pipeline.
Analysis of previously published data
To demonstrate utility, we used MutantHuntWGS to analyze published datasets from paired-end sequencing experiments with DNA prepared from bulk segregants or lab-evolved strains (Birkeland et al. 2010; Goldgof et al. 2016; Ottilie et al. 2017). These data were downloaded from the sequence read archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra; project accessions: SRP003355, SRP074482, SRP074623) and decompressed using the SRA toolkit (https://github.com/ncbi/sra-tools/wiki). MutantHuntWGS was run from within the Docker container, and each published mutant (experimental) file was compared to its respective published control.
Data availability
All code and supplementary information on the methods used herein are available on the MutantHuntWGS Git repository (https://github.com/mae92/MutantHuntWGS).
Results And Discussion
Installing and running the MutantHuntWGS pipeline
To facilitate distribution and maximize reproducibility, we implemented MutantHuntWGS in a Docker container (Boettiger 2015; Di Tommaso et al. 2015). The container houses the pipeline and all of its dependencies in a Unix/Linux environment. We have successfully installed and run MutantHuntWGS on Mac, Linux, and Windows operating systems. To download and install the MutantHuntWGS Docker container, users need only install Docker Desktop (https://docs.docker.com/get-docker/), open a command-line terminal, and execute the following command:
$ docker run -it -v /PATH_TO_DESKTOP/Analysis_Directory:/Main/Analysis_Directory mellison/mutant_hunt_wgs:version1
After download and installation, the command opens a Unix terminal running in the Docker container so users can begin their analysis. From the Unix terminal running in the Docker container, users need only execute the following command to run the MutantHuntWGS pipeline:
$ MutantHuntWGS.sh \
-n FILENAME \
-g /Main/MutantHuntWGS/S_cerevisiae_Bowtie2_Index_and_FASTA/genome \
-f /Main/MutantHuntWGS/S_cerevisiae_Bowtie2_Index_and_FASTA/genome.fa \
-r single \
-s 100 \
-p /Main/MutantHuntWGS/S_cerevisiae_Bowtie2_Index_and_FASTA/ploidy_n1.txt \
-d /Main/Analysis_Directory \
-o /Main/Analysis_Directory/NAME_YOUR_OUTPUT_FOLDER
-a YES
A detailed description of the pipeline, its installation, and its use is available in the MutantHuntWGS Git repository readme (https://github.com/mae92/MutantHuntWGS/blob/master/README.md).
Utility of the MutantHuntWGS pipeline
MutantHuntWGS processes WGS data through a standard alignment/variant-calling pipeline and compares each experimental strain to a control strain (Figure 1, see Methods). The pipeline’s constituent tools are often used for WGS analysis (Reavey et al. 2015; Gopalakrishnan and Winston 2019; Gopalakrishnan et al. 2019). However, MutantHuntWGS ensures ease of use by assembling these tools in a Docker container and requiring only one command to run them all in sequence. This approach combines the best aspects of previously published pipelines (discussed below) while allowing inexperienced users to install the software and reproducibly apply popular methods.
MutantHuntWGS also ensures that the output data files are well organized and easy to locate. Output files include aligned reads (BAM format), alignment statistics (TXT format), pre- and post-filtering variants (VCF format), SnpEff output (HTML, VCF, and TXT formats), and SIFT output (VCF, XLS formats). The user thus has all the information needed to identify and visually inspect sequence variants, and to generate figures and tables for publication.
MutantHuntWGS combines versatility and simplicity
Our goal in creating MutantHuntWGS was to simplify the installation and usage of robust bioinformatics tools while maintaining flexibility by allowing users to specify certain critical options. Examples of this, discussed below, include (1) enabling use with additional organisms, (2) allowing users to specify ploidy, (3) filtering by a user-specified variant-quality score, and (4) exposing all intermediate and final output files to facilitate additional filtering and quality control.
MutantHuntWGS is designed for use with S. cerevisiae by default but can be adapted to analyze WGS data from any organism. At present, only the necessary reference files for S. cerevisiae are included in the MutantHuntWGS download. Investigators who wish to analyze data from an organism other than S. cerevisiae need to provide, at minimum, new Bowtie2 indices, a genome FASTA file, and a ploidy file. Bowtie2 indices and genome FASTA files for many model organisms are available at https://support.illumina.com/sequencing/sequencing_software/igenome.html. A FASTA index file (genome.fasta.fai) that can be easily converted into a ploidy file is also available at this link. Unfortunately, performing the SnpEff and SIFT analysis would require slight alterations to the SnpEff and SIFT commands in the pipeline script and a copy of the SIFT library for the organism of interest. We chose not to include reference files and SIFT libraries for other organisms within the Docker container due to the large size of these files. If users encounter difficulties when analyzing non-S. cerevisiae WGS data, we encourage them to seek assistance by opening an issue on the MutantHuntWGS Git repository.
Experiments in yeast are often performed in a haploid background, but can also be performed in diploid or occasionally aneuploid backgrounds. The MutantHuntWGS download includes two ploidy files, one for diploids and one for haploids. The user can specify either ploidy file when running the pipeline. MutantHuntWGS will automatically provide this file to BCFtools during the variant-calling step. This may be particularly advantageous for analysis of yeast strains with aneuploid chromosomes. Instructions are provided on the GitHub Readme page explaining how to modify the ploidy file to account for aneuploidy in the analysis.
Users may also set variant-quality-score cutoffs (described in detail on GitHub: https://github.com/mae92/MutantHuntWGS/blob/master/README.md) to tune the stringency of the analysis. They can also toggle the alignment step to save time when resetting the stringency. This option re-subsets variant calls with a higher or lower stringency cutoff, skipping the more time-consuming upstream steps of the pipeline. Although MutantHuntWGS does not allow users to specify additional cutoffs that filter the output per SnpEff/SIFT effect predictions and scores, users can separately apply such filters to the MutantHuntWGS output files after the fact—thus allowing for increased stringency.
Assessing MutantHuntWGS performance using a bulk segregant analysis dataset
To assess MutantHuntWGS performance, we applied it to bulk segregant analysis data (Birkeland et al. 2010) with ploidy set to haploid. MutantHuntWGS identified 188 variants not present in the control strain that passed the variant-quality-score cutoff of 100. Thus only 1.95% of all variants detected in the experimental strain passed the filtering steps (Table 1). Among these was the same PHO81 (VAC6) mutation found in the Birkeland et al. (2010) study, which results in an R701S amino acid substitution in the Pho81 protein (Birkeland et al. 2010). Our pipeline thus identified the same published causal variant described in the original study.
Table 1.
Analysis of previously published bulk-segregant and lab-evolution WGS datasets using MutantHuntWGS
| Birkeland et al. (2010) | ||||||
|---|---|---|---|---|---|---|
| SRA ID | SRR064545 | SRR064546 | ||||
| Total Reads (% Mapped) | 19,782,779 (92.80%) | 20,015,390 (89.57%) | Additional Output Filtering | |||
| Variants Called in Control | 10,022 | Filtering by | Cutoff | Variant count (%) | ||
| Variants Called in Experimental | 9,646 | Variant quality score | >130 | 21 (0.21%) | ||
| Variants Unique to the Experimental strain with Variant Quality Scores > 100 (% of total) | 188 (1.95%) | SnpEff Impact | >Moderate | 55 (0.57%) | ||
| CDS Variants in SIFT output (% of total) | 152 (1.58%) | Variant Quality Score + Impact | >130 + >Moderate | 6 (0.06%) | ||
| Published mutation (s) Identified | Yes | SIFT | Deleterious | 6 (0.06%) | ||
| Ottilie et al. (2017) | |||||
|---|---|---|---|---|---|
| SRA ID | SRR3480136 | SRR3490425 | SRR3490399 | SRR3490397 | SRR3490304 |
| Total Reads (Percent Mapped) | 14,684,843 (93.61%) | 7,935,729 (98.11%) | 3,629,049 (97.53%) | 5,611,439 (97.80%) | 6,904,333 (97.91%) |
| Variants Called in Control | 526 | ||||
| Variants Called in Experimental | 367 | 298 | 336 | 377 | |
| Variants Unique to the Experimental strain with Variant Quality Scores > 100 (% of total) | 11 (3.00%) | 4 (1.34%) | 7 (2.08%) | 8 (2.12%) | |
| CDS Variants in SIFT output (% of total) | 4 (1.09%) | 2 (0.67%) | 4 (1.19%) | 5 (1.33%) | |
| Published Mutation(s) Identified | Yes | Yes | Yes | Yes | |
| Goldgof et al. (2016) | |||||
|---|---|---|---|---|---|
| SRA ID | SRR3480136 | SRR3480251 | SRR3480237 | SRR3480212 | SRR3480267 |
| Total Reads (Percent Mapped) | 14,684,843 (93.61%) | 6,347,816 (94.49%) | 7,480,005 (64.04%) | 3,058,951 (98.11%) | 2,545,805 (97.51%) |
| Variants Called in Control | 526 | ||||
| Variants Called in Experimental | 487 | 316 | 292 | 301 | |
| Variants Unique to the Experimental strain with Variant Quality Scores > 100 (% of total) | 10 (2.05%) | 8 (2.53%) | 6 (2.05%) | 4 (1.33%) | |
| CDS Variants in SIFT output (% of total) | 4 (0.82%) | 3 (0.95%) | 4 (1.37%) | 2 (0.66%) | |
| Published Mutation(s) Identified | Yes | Yes | Yes | Yes |
We were surprised by how many sequence variants (relative to the reference genome) remained after filtering. Given our variant-quality-score cutoff of 100, it is unlikely that these variants were called in error; instead, they likely reflect high sequence heterogeneity in the genetic backgrounds of the experimental and control strains. To further reduce the length of the variant list, we experimented with additional cutoffs, including (1) more stringent variant-quality-score, (2) SIFT score, and (3) SnpEff impact score cutoffs. A SIFT-score cutoff of <0.05 (deleterious) reduced the number of variants in the SIFT output from 152 to 6 (Table 1). An increased variant-quality-score stringency (>130) reduced the number of variants to 21. A SnpEff impact-score cutoff of > Moderate reduced the number of variants to 55. Finally, a variant quality-score cutoff of >130 and a SnpEff score of > Moderate, used together, reduced the number of variants to only 6. All post-hoc tests retained the causal variant. These tests demonstrate how users might similarly narrow their lists of potential candidates. However, we caution readers that filtering by these metrics has the potential to increase the false negative rate in their analysis.
Assessing MutantHuntWGS performance using lab evolution datasets
To test MutantHuntWGS performance on strains that did not undergo bulk-segregant analysis, we analyzed nine datasets from lab evolution experiments (Goldgof et al. 2016; Ottilie et al. 2017), again setting ploidy to haploid and using a variant-quality-score cutoff of 100. In each of these studies, yeast cells were allowed to evolve resistance to a drug and WGS was used to identify mutations (Goldgof et al. 2016; Ottilie et al. 2017).
Shortlists of only 4 to 11 (1.33–3.00%) of the variants originally detected in the experimental strain(s) were obtained for each dataset (Table 1). Out of these variants, only 2 to 5 (0.66–1.37% of called variants in the experimental strain) were present in the SIFT output for each dataset, which contains only protein coding variants. In each case, the list of variants generated by MutantHuntWGS included the mutation identified in the published study (SRR3480237: Pma1 N291K, Yrr1 T623K; SRR3480212: Pma1 P339T, Yrr1 L611F; SRR3480251: Pma1 L290S, Yrr1 T623K; SRR3480267: Pma1 G294S; SRR3490304: Erg11 V154G; SRR3490397: Erg11 T318N; SRR3490399: Erg25 D234E; SRR3490425: Erg25 H156N). These test cases confirm that MutantHuntWGS can identify yeast-sequence variants from WGS sequencing samples and accurately filter out background mutations.
Existing WGS analysis pipelines
Other platforms exist that perform similar analyses. Each possesses a subset of the features enabled by MutantHuntWGS and has notable strengths. MutantHuntWGS is unique in its ability to combine the best attributes of these published tools while including additional functionality and providing output data in standard formats, such as BAM and VCF.
One user-friendly program, Mudi (Iida et al. 2014), uses BWA (Jo and Koh 2015), SAMtools (Li et al. 2009), and ANNOVAR (Wang et al. 2010) for sequence alignment, identification, and annotation of sequence variants, respectively. Like MutantHuntWGS, Mudi performs numerous filtering steps before returning a list of putative causal variants. MutantHuntWGS predicts variant effects and maps variants to annotated S. cerevisiae genes using SnpEff and SIFT instead of ANNOVAR, and also offers access to all intermediate data files.
Another program, VAMP, consists of a series of Perl scripts that build and query an SQL database made from user-provided short-read sequencing data. VAMP identifies sequence variants, including large insertions and deletions. It also has built-in functionality that allows for manual inspection of the data (Birkeland et al. 2010). One advantage of MutantHuntWGS over VAMP is that it adheres to common data formats.
A recent article describing WGS in yeast samples includes a bioinformatics pipeline, referred to as wgs-pipeline (Gopalakrishnan and Winston 2019). It is built in a Snakemake framework (Köster and Rahmann 2012) that runs in a Conda environment (https://docs.conda.io/en/latest/), similar to the container-based analysis environment we used for MutantHuntWGS. This pipeline uses Bowtie2 (Langmead and Salzberg 2012), SAMtools (Li et al. 2009), Picard (Toolkit 2016), and GATK (McKenna et al. 2010) to align, process, and compare datasets. Compared to wgs-pipeline, MutantHuntWGS, which runs both SnpEff and SIFT on the candidate variants, provides a more comprehensive analysis of the predicted effects of the variants.
The Galaxy platform (Giardine et al. 2005; Blankenberg et al. 2010) provides a user-friendly, online interface for building bioinformatics pipelines. Galaxy also offers access to intermediate files. However, analysis with this platform requires the user to select the tools and parameters to incorporate, so some knowledge of the tools themselves is essential. Implementation is straightforward after those decisions are made, and the user need not have any understanding of Unix/Linux. The advantage of MutantHuntWGS over the Galaxy platform and pipelines such as CloudMap (Minevich et al. 2012) is that the user does not need to make decisions about the data analysis workflow.
In summary, the MutantHuntWGS pipeline is among the most user-friendly of these programs. It combines the most useful features of the existing WGS analysis programs while also enabling the user to account for ploidy. Containerization streamlines the installation of MutantHuntWGS and enhances its reproducibility. Thus, MutantHuntWGS offers ease of use, functionality, and data-transparency, setting it apart from other WGS pipelines.
Conclusions
Processing data generated from next-generation sequencing platforms requires significant expertise, and so is inaccessible to many investigators. We have developed a highly effective differential variant-calling pipeline capable of identifying causal variants from WGS data. We demonstrate the utility of MutantHuntWGS by analyzing previously published datasets. In all cases, our pipeline successfully identified the causal variant. We offer this highly reproducible and easy-to-implement bioinformatics pipeline to the Saccharomyces cerevisiae research community (available at https://github.com/mae92/MutantHuntWGS).
Acknowledgments
We would like to thank Margaret Shirra, Sarah Tripplehorn, Alex Francette, and Brendan McShane for careful review of the manuscript. Special thanks to Alex Francette for performing pipeline tests on Linux and Windows operating systems. This research was supported by a National Institutes of Health (NIH) grant R01GM052593 to KMA, a predoctoral fellowship from the NIH (F31GM129917) awarded to MAE, and University of Pittsburgh Central Research Development Funds (CRDF, 2017-2018) to JDD.
Footnotes
Communicating editor: A. Dudley
Literature Cited
- Birkeland S. R., Jin N., Ozdemir A. C., Lyons R. H., Weisman L. S. et al. , 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186: 1127–1137. 10.1534/genetics.110.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R. et al. , 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 89: 10–19. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger C., 2015. An introduction to Docker for reproducible research. Oper. Syst. Rev. 49: 71–79. 10.1145/2723872.2723882 [DOI] [Google Scholar]
- Botstein D., and Fink G. R., 2011. Yeast: an experimental organism for 21st century biology. Genetics 189: 695–704. 10.1534/genetics.111.130765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Coon M., Nguyen T., Wang L. et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L. et al. , 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15: 1451–1455. 10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgof G. M., Durrant J. D., Ottilie S., Vigil E., Allen K. E. et al. , 2016. Comparative chemical genomics reveal that the spiroindolone antimalarial KAE609 (Cipargamin) is a P-type ATPase inhibitor. Sci. Rep. 6: 27806 10.1038/srep27806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R., Marr S. K., Kingston R. E., and Winston F., 2019. A conserved genetic interaction between Spt6 and Set2 regulates H3K36 methylation. Nucleic Acids Res. 47: 3888–3903. 10.1093/nar/gkz119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R., and Winston F., 2019. Whole-genome sequencing of yeast cells. Curr. Protoc. Mol. Biol. 128: e103 10.1002/cpmb.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza A., Tammpere E., Kofoed M., Keong C., Chiang J. et al. , 2015. Complementation of yeast genes with human genes as an experimental platform for functional testing of human genetic variants. Genetics 201: 1263–1274. 10.1534/genetics.115.181099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza A., Driessen M. R. M., Tammpere E., O’Neil N. J., and Hieter P., 2020. Cross-species complementation of nonessential yeast genes establishes platforms for testing inhibitors of human proteins. Genetics 214: 735–747. 10.1534/genetics.119.302971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N., Yamao F., Nakamura Y., and Iida T., 2014. Mudi, a web tool for identifying mutations by bioinformatics analysis of whole-genome sequence. Genes Cells 19: 517–527. 10.1111/gtc.12151 [DOI] [PubMed] [Google Scholar]
- Irvine D. V., Goto D. B., Vaughn M. W., Nakaseko Y., McCombie W. R. et al. , 2009. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 19: 1077–1083. 10.1101/gr.089318.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H., and Koh G., 2015. Faster single-end alignment generation utilizing multi-thread for BWA. Biomed. Mater. Eng. 26: S1791–S1796. 10.3233/BME-151480 [DOI] [PubMed] [Google Scholar]
- Kachroo A. H., Laurent J. M., Yellman C. M., Meyer A. G., Wilke C. O. et al. , 2015. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348: 921–925. 10.1126/science.aaa0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., and Rahmann S., 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., 2001. Cloning yeast genes by complementation. Curr. Protoc. Mol. Biol. 13.8 10.1002/0471142727.mb1308s05 [DOI] [PubMed] [Google Scholar]
- Mardis E. R., 2013. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 6: 287–303. 10.1146/annurev-anchem-062012-092628 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., and Hobert O., 2012. CloudMap: A cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. 10.1534/genetics.112.144204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottilie S., Goldgof G. M., Calvet C. M., Jennings G. K., Lamonte G. et al. , 2017. Rapid Chagas disease drug target discovery using directed evolution in drug-sensitive yeast. ACS Chem. Biol. 12: 422–434. 10.1021/acschembio.6b01037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavey C. T., Hickman M. J., Dobi K. C., Botstein D., and Winston F., 2015. Analysis of polygenic mutants suggests a role for mediator in regulating transcriptional activation distance in Saccharomyces cerevisiae. Genetics 201: 599–612. 10.1534/genetics.115.181164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., and Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26: 1135–1145. 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- Smith D., and Quinlan A., 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 1638–1642 10.1101/gr.077776.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strynatka K. A., Gurrola-Gal M. C., Berman J. N., and McMaster C. R., 2018. How surrogate and chemical genetics in model organisms can suggest therapies for human genetic diseases. Genetics 208: 833–851. 10.1534/genetics.117.300124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Palumbo E., Chatzou M., Prieto P., Heuer M. L. et al. , 2015. The impact of Docker containers on the performance of genomic pipelines. PeerJ 3: e1273 10.7717/peerj.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toolkit P., 2016. Broad Institute
- Vaser R., Adusumalli S., Leng S. N., Sikic M., and Ng P. C., 2016. SIFT missense predictions for genomes. Nat. Protoc. 11: 1–9. 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., and Hakonarson H., 2010. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M. F., Yamamoto S., Chao H.-T., Posey J. E., Westerfield M. et al. , 2017. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207: 9–27. 10.1534/genetics.117.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., and Koshland D., 2016. Back to the future: Mutant hunts are still the way to go. Genetics 203: 1007–1010. 10.1534/genetics.115.180596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All code and supplementary information on the methods used herein are available on the MutantHuntWGS Git repository (https://github.com/mae92/MutantHuntWGS).