Abstract
The diatom, Cyclotella cryptica, is a well-established model species for physiological studies and biotechnology applications of diatoms. To further facilitate its use as a model diatom, we report an improved reference genome assembly and annotation for C. cryptica strain CCMP332. We used a combination of long- and short-read sequencing to assemble a high-quality and contaminant-free genome. The genome is 171 Mb in size and consists of 662 scaffolds with a scaffold N50 of 494 kb. This represents a 176-fold decrease in scaffold number and 41-fold increase in scaffold N50 compared to the previous assembly. The genome contains 21,250 predicted genes, 75% of which were assigned putative functions. Repetitive DNA comprises 59% of the genome, and an improved classification of repetitive elements indicated that a historically steady accumulation of transposable elements has contributed to the relatively large size of the C. cryptica genome. The high-quality C. cryptica genome will serve as a valuable reference for ecological, genetic, and biotechnology studies of diatoms.
Keywords: algal biofuels, horizontal gene transfer, lipids, nanopore, transposable elements
The diatom Cyclotella cryptica Reimann, J.C.Lewin & Guillard has a range of properties that have made it a valuable experimental model in studies dating back to the 1960s (Lewin and Lewin 1960). Cyclotella cryptica can grow across a broad range of salinities, and its responses to altered salinity offer opportunities to study several important aspects of diatom biology. For example, salinity shifts can induce gamete production (Schultz and Trainor 1970) and cause cells to alternate between cell wall morphologies resembling C. cryptica and the closely related freshwater species, Cyclotella meneghiniana Kützing (Schultz 1971). Later studies demonstrated the utility of C. cryptica for understanding cell wall morphogenesis in diatoms (Tesson and Hildebrand 2010). Cyclotella cryptica has other properties that make it an attractive candidate for biotechnology applications, including the ability to grow heterotrophically (Hellebust 1971; White 1974; Pahl et al. 2010) and produce high levels of lipids for use as biofuels or nutraceuticals (Roessler 1988; Traller and Hildebrand 2013; Slocombe et al. 2015).
A draft genome assembly for C. cryptica revealed a large, gene- and repeat-rich genome (Traller et al. 2016). The genome was sequenced without the benefit of long-read sequencing platforms, which enable short contigs—particularly those containing repetitive DNA—to be joined into large contiguous scaffolds. Consequently, the version 1.0 genome assembly of C. cryptica was highly fragmented, with most fragments measuring <1 kb in length. Although the gene space appeared to be well characterized and the size accurately estimated, highly fragmented assemblies can suffer from overestimation of gene number (Denton et al. 2014) and hinder insights into genome structure. It is also challenging to fully characterize intergenic regions, which hold noncoding RNAs, promoter regions, and allow comparisons of genomic synteny across species. This is especially challenging for historically understudied groups, such as diatoms, in which the pace of genomic sequencing has lagged behind other groups such as animals and flowering plants. The relatively small number of sequenced genomes from distantly related diatom species gives the impression that each newly sequenced diatom genome contains a large fraction of unique, species-specific sequence. As small fragments, the origin and identity of these sequence fragments are especially challenging to characterize. Diatoms maintain intimate relationships with bacteria both in nature (Amin et al. 2012) and in cell culture (Johansson et al. 2019). In addition, some diatom genomes appear to contain bacterial-derived genes (Bowler et al. 2008). With long contiguous scaffolds, the proximal source of bacterial-like genes should be much easier to determine in genome assemblies that contain a mix of DNA from both the diatom and its associated bacteria.
We combined short and long sequencing reads to produce a more contiguous version 2.0 genome assembly for C. cryptica CCMP332. The addition of long direct sequencing reads allowed us to improve the gene models, remove contaminant sequences, and better characterize the structure of this relatively large genome. As a result, the improved assembly provides a better resource for functional studies of C. cryptica such as genome-enabled reverse genetics and read-mapping for resequencing and experimental transcriptomics.
Materials And Methods
Strain information and sequencing
We acquired Cyclotella cryptica strain CCMP332 from the National Center for Marine Algae and Microbiota (NCMA). This strain was originally isolated from Martha’s Vineyard, MA, USA, by R. Guillard in 1956. We grew the culture in L1 marine medium (Guillard 1975) at 22° on a 12:12 light: dark cycle.
We harvested non-axenic cells during late exponential-phase growth, filtered them through 5.0 µm Millipore membrane filters to reduce the bacterial load, rinsed cells from the filter before pelleting them by centrifugation at 2500 × g for 10 min, and stored the cell pellets at –80°. We extracted DNA using the DNeasy Plant Kit (Qiagen) or a modified CTAB protocol (Doyle and Doyle 1987). For the CTAB protocol, we resuspended cell pellets in 3X CTAB buffer (CTAB, 3% w/v; 1.4 M NaCl; 20 mM EDTA, pH 8.0; 100 mM Tris-HCl, pH 8.0; 0.2% β-mercaptoethanol), disrupted them by vortexing briefly with 1.0 mm glass beads, and incubated them at 65° for 1 hr. We then extracted the DNA twice with 1X volume of 24:1 chloroform:isoamyl alcohol and precipitated the DNA with 1X volume of isopropanol and 0.8X volume of 7.5 M ammonium acetate. We assessed the quality and quantity of the DNA with 0.8% agarose gels, a Nanodrop 2000 (Thermo Fisher Scientific), and a Qubit 2.0 Fluorometer (dsDNA BR kit; Thermo Fisher Scientific).
For DNA samples with high molecular weight and sufficient quantity (1–3 µg), we prepared libraries for long-read sequencing using the ligation sequencing kit SQK-LSK108 (Oxford Nanopore Technologies, ONT). We sequenced these libraries on the MinION platform with FLO-MIN106 (R9.4.1) flowcells (Table S1). We used Guppy (version 2.3.5) (ONT) with default settings to convert raw signal intensity data into base calls. We kept all nanopore reads with a length greater than 500 bp and trimmed them for adapter sequences with NanoPack (De Coster et al. 2018). We used Canu (version 1.7) (Koren et al. 2017) to correct low-quality base calls in the nanopore raw reads.
We prepared short-read Illumina sequencing libraries using the Kapa HyperPlus Kit (Roche) with 300–400 bp insert sizes and barcoded the libraries with dual indices. These libraries were sequenced using the Illumina HiSeq4000 at the University of Chicago Genomics Facility. Twelve libraries were sequenced for 50 bp single end (SE) reads and three libraries were sequenced for 100 bp paired-end (PE) reads (Table S1). We quality trimmed the short-reads using Trimmomatic (version 0.36) (Bolger et al. 2014) with options ‘ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50’.
Genome assembly, error correction, and scaffolding
To estimate the haploid genome size of C. cryptica, we first mapped the PE Illumina reads to the C. cryptica version 1.0 assembly using BWA-MEM (version 0.7.17-r1188) (Li and Durbin 2009) and extracted the mapped reads using SAMTOOLS (version 1.9) (Li et al. 2009). We then counted k-mers and generated histograms for k-mer sizes 17, 19, 21, 23, and 25 bp using Jellyfish (version 2.3.0) (Marçais and Kingsford 2011). We estimated the haploid genome size for each k-mer size by dividing the total number of k-mers by the mean k-mer coverage. The average genome size estimated from all k-mer sizes was 164.6 Mb [160.3–170.5], slightly larger than the 161.7 Mb size of the C. cryptica version 1.0 assembly (Traller et al. 2016). We used an estimated genome size of 165 Mb for genome assembly.
We assembled the raw nanopore reads from C. cryptica and associated bacteria using Flye (version 2.4.2) (Kolmogorov et al. 2019a, 2019b) with options ‘–meta –plasmids --iterations 1 –genome-size 165m’. We then mapped the corrected nanopore reads back to the assembled contigs with Minimap2 (version 2.10-r761) (Li 2018), using the settings recommended for nanopore reads. We used these mappings for error correction of the initial draft assembly with Racon (version 1.3.3) (Vaser et al. 2017) using default settings. We then used the r941_flip935 model in Medaka (version 0.8.1) (https://github.com/nanoporetech/medaka) for a second round of error correction using the Racon-corrected contigs and the corrected nanopore reads. After contig correction, we separately aligned the SE and PE Illumina reads to the corrected contigs with BWA-MEM. We merged and sorted the alignment BAM files with SAMTOOLS and used the merged BAM file for sequence polishing of all variant types (‘–changes –fix all’) with Pilon (version 1.23) (Walker et al. 2014). We performed three iterative rounds of Illumina read mapping and Pilon polishing.
We scaffolded the polished contigs using the corrected nanopore sequences with SSPACE-LongRead (version 1-1) (Boetzer and Pirovano 2014), requiring at least three overlapping sequences to connect any two contigs (‘-l 3′). We used the corrected nanopore reads to extend contigs and fill scaffold gaps using LR_Gapcloser (Xu et al. 2019) with default settings and a total of ten iterative rounds. Finally, we aligned the corrected nanopore reads to the scaffolds with Minimap2 and used these alignments to remove redundant scaffolds from the assembly using Purge Haplotigs (version 1.0.0) (Roach et al. 2018). We evaluated each stage of the assembly for quality and completeness with QUAST (version 5.0.0) (Gurevich et al. 2013) and BUSCO (version 4.0.6; genome mode, eukaryote_odb10 dataset) (Simão et al. 2015) (Table S2).
Contaminant identification and removal
We used the Blobtools pipeline (version 1.1.1) (Laetsch and Blaxter 2017) to identify and remove contaminant scaffolds. Blobtools uses a combination of BLAST-based taxonomic assignment, GC content, and read coverage to identify contaminants. We assigned the taxonomy of each scaffold from a Diamond BLASTX search (version 0.9.21) (Buchfink et al. 2015) against the UniProt Reference Proteomes database (release 2019_06) (UniProt Consortium 2018) using options ‘–max-target-seqs 1 –sensitive –evalue 1e-25 –outfmt 6’. We estimated read coverage using all reads (corrected nanopore and Illumina) mapped to the scaffolds with Minimap2, and merged and sorted the alignments using SAMTOOLS. We flagged and removed scaffolds that met the following criteria: (1) taxonomic assignment to bacteria, archaea, or viruses, (2) low GC percentage indicative of organellar scaffolds, and (3) no taxonomic assignment for scaffolds < 1 kb in length. After removing these scaffolds, we performed two additional rounds of Pilon polishing as described above.
Chloroplast and mitochondrial genome assembly
For the chloroplast genome, we mapped the uncorrected nanopore reads against a set of diatom chloroplast genomes (GenBank accessions NC_025314.1, NC_025312.1, NC_014808.1, NC_008589.1, and NC_038005.1) with Minimap2 and extracted the mapped reads using SAMTOOLS. We assembled the chloroplast-mapped reads using Flye and options ‘–genome-size 132k –iterations 1’, which resulted in a single circular-mapping contig. We mapped the PE and SE Illumina reads and polished the circular contig with Pilon as described above. We performed three iterations of this mapping and polishing procedure.
We followed a similar procedure for the mitochondrial genome. We mapped the uncorrected nanopore reads against a small set of diatom mitochondrial genomes (GenBank accessions NC_007405.1 and NC_028615.1) using Minimap2 and extracted the mapped reads using SAMTOOLS. We assembled these mitochondria-mapped reads using Flye and options ‘–genome-size 58k –iterations 1’, resulting in a single circular-mapping contig. We then performed the same polishing procedure as we did for the chloroplast genome.
RNA sequencing and assembly
We used the RNA-seq reads and transcriptome assemblies for C. cryptica CCMP332 from Nakov et al. (2020). For that study, total RNA was extracted from cells grown in five different salinity treatments (0, 2, 12, 24, 36 parts per thousand salinity) using the RNeasy Plant Kit (Qiagen), and 15 Illumina libraries were prepared using the Kapa mRNA HyperPrep kit (Roche) and sequenced on the Illumina HiSeq2000 platform at the Beijing Genomics Institute. The RNA-seq reads were corrected for sequencing errors using Rcorrector (Song and Florea 2015), trimmed for adapters and low quality bases with Trimmomatic, and assembled using Trinity (Grabherr et al. 2011).
Gene annotation
We used the MAKER software package (version 2.31.10) to identify protein-coding genes in the genome (Cantarel et al. 2008; Holt and Yandell 2011). We used the C. cryptica transcriptome as expressed sequence tag (EST) evidence (est2genome = 1) and the protein sequences from Cyclotella nana, Thalassiosira oceanica, Phaeodactylum tricornutum, and Fragilariopsis cylindrus as protein evidence (protein2genome = 1) for the MAKER pipeline. Protein sequences were downloaded from the Joint Genome Institutes (JGI) PhycoCosm resource (https://phycocosm.jgi.doe.gov/phycocosm/home; last accessed 2 Jan 2020). We also allowed MAKER to predict single exon genes (single_exon = 1) and search for alternative splicing (alt_splice = 1). Repetitive elements identified during the repeat analysis (see below) were used to mask the repetitive regions for this analysis. After the first round of MAKER using EST and protein evidence, we used the predicted genes with annotation edit distance (AED) scores less than 0.5 to train gene prediction models in SNAP (version 2006-07-28) (Korf 2004) and Augustus (version 3.3.2) (Stanke et al. 2008). We then performed two subsequent rounds of MAKER annotation using the trained SNAP and Augustus models. We retrained SNAP after the second round of MAKER. We evaluated the completeness and quality of the MAKER proteins after each round using BUSCO (protein mode against the eukaryota_odb9 dataset) and AED scores (Table S3).
To identify protein families, domains, and gene ontology (GO) terms, we searched the predicted protein sequences against the Pfam (version 32.0) (El-Gebali et al. 2019), PRINTS (version 42.0) (Attwood et al. 2012), PANTHER (version 14.1) (Thomas et al. 2003), SMART (version 7.1) (Letunic et al. 2012), SignalP (version 4.1) (Petersen et al. 2011), and TMHMM (version 2.0c) (Krogh et al. 2001) databases using InterProScan (version 5.36-75.0) (Jones et al. 2014). We also searched the proteins against the SwissProt (release 2019_06) and UniProt Reference Proteomes (release 2019_06) databases using NCBI BLASTP (version 2.4.0+) (Camacho et al. 2009) using options ‘-evalue 1e-6 -outfmt 6 -num_alignments 1 -seg yes -soft_masking true -lcase_masking -max_hsps 1’.
We predicted non-coding RNAs (ncRNAs) in the genome using Infernal (version 1.1.2) (Nawrocki and Eddy 2013) against the Rfam database (version 14.1) (Kalvari et al. 2018). We used tRNAscan-SE (version 2.0.5) (Chan and Lowe 2019) for tRNA annotation and RNAmmer (version 1.2) (Lagesen et al. 2007) for rRNA annotation.
Chloroplast and mitochondrial genomes were annotated with GeSeq (Tillich et al. 2017). Gene and inverted repeat boundaries from GeSeq were manually curated as necessary by comparison to annotations from other diatom organellar genomes.
Repetitive element annotation
We built custom repeat libraries to identify repetitive elements across the genome. We searched for long terminal repeat (LTRs) retrotransposons using the program LTRharvest (version 1.5.8) (Ellinghaus et al. 2008) with options ‘-minlenltr 100 -maxlenltr 6000 -mindistltr 1500 -maxdistltr 25000 -motif tgca -similar 85 -mintsd 5 -maxtsd 5 -vic 10’. We filtered the candidate LTRs from LTRharvest using LTRdigest (version 1.5.8) (Steinbiss et al. 2009) to keep elements with polypurine tracts (PPT) and primer binding sites (PBS) inside the predicted LTR sequence region. We further filtered LTR elements to remove those with nested insertions and select representative (exemplar) elements using Perl scripts (available from https://weatherby.genetics.utah.edu/MAKER/wiki/index.php/Repeat_Library_Construction-Advanced) (Campbell et al. 2014). We identified miniature inverted transposable elements (MITEs) with MITE-Hunter (Han and Wessler 2010). We then masked the genome with the combined LTR and MITE libraries using RepeatMasker (version 4.0.5) (http://www.repeatmasker.org/). After masking, we identified other repetitive elements using RECON (version 1.08) (Bao and Eddy 2002) and RepeatScout (version 1.06) (Price et al. 2005) as implemented within the RepeatModeler package (version 2.0) (Flynn et al. 2020). We combined all candidate exemplar elements and searched them against the UniProt Reference Proteomes database with NCBI BLASTX using settings ‘-evalue 1e-10 -num_descriptions 10’. We removed elements from the final repeat library that contained overlaps with any predicted proteins using ProtExcluder (version 1.2) (Campbell et al. 2014).
We used RepeatMasker and the final repeat library to annotate repetitive elements in the genome. We ran RepeatMasker with the NCBI RMBLAST (version 2.6.0+) search engine (‘-e ncbi’), the sensitive option (‘-s’), and the ‘-a’ option to obtain the alignment file. We then used the provided parseRM.pl script (version 5.8.2) (downloaded from https://github.com/4ureliek/Parsing-RepeatMasker-Outputs) on the alignment files from RepeatMasker to generate the repeat landscape with the ‘-l’ option (Kapusta et al. 2017). This script collects the percent divergence from the repeat library for each TE element, correcting for higher mutation rates at CpG sites and using Kimura 2-Parameter distance output by RepeatMasker. The percent divergence to the repeat library is a proxy for age (older TE elements will have accumulated more nucleotide substitutions), and the script splits TEs into bins of 1% divergence.
Assembly comparisons
We downloaded the C. cryptica version 1.0 genome assembly and gene models from http://genomes.mcdb.ucla.edu/Cyclotella/download.html. We downloaded the P. tricornutum version 2.0, F. cylindrus version 1.0, and C. nana version 3.0 genome assemblies from GenBank (Table 1). We performed QUAST and BUSCO analyses on these genomes to allow for comparisons with the C. cryptica version 2.0 assembly.
Table 1. Genome characteristics for P. tricornutum, F. cylindrus, C. nana, and C. cryptica.
| PHAEODACTYLUM TRICORNUTUM VERSION 2.0 | FRAGILARIOPSIS CYLINDRUS VERSION 1.0 | CYCLOTELLA NANA VERSION 3.0 | CYCLOTELLA CRYPTICA VERSION 1.0 | CYCLOTELLA CRYPTICA VERSION 2.0 | |
|---|---|---|---|---|---|
| GENOME SIZE, MB | 27.4 | 61.1 | 32.4 | 161.8 | 171.1 |
| NUMBER OF SCAFFOLDS | 33 | 271 | 27 | 116,815 | 662 |
| N50 LENGTH, KB | 945 | 1295.6 | 1,992 | 12 | 494 |
| MEDIAN SCAFFOLD LENGTH, KB | 703.2 | 17.2 | 965.0 | 0.2 | 139.0 |
| GC CONTENT, % | 49 | 39 | 47 | 43 | 43 |
| REPETITIVE ELEMENTS, % | 12 | Not available | 2 | 54 | 59 |
| COMPLETE EUKARYOTIC BUSCO COUNT (%)a | 200 (78.5%) | 199 (78.0%) | 191 (74.9%) | 183 (71.8%) | 191 (74.9%) |
| GENBANK ACCESSION NUMBER | GCA_000150955.2 | GCA_001750085.1 | GCA_000149405.2 | Noneb | GCA_013187285.1 |
| PLASTID GENOME SIZE, BP | 117,369 | 123,275 | 128,814 | 129,320 | 129,328 |
| MITOCHONDRIAL GENOME SIZE, BP | 77,356 | 58,295 | 43,827 | 58,021 | 46,485 |
| REFERENCE | Bowler et al. (2008) | Mock et al. (2017) | Armbrust et al. (2004) | Traller et al. (2016) | This study |
Genome mode against the eukaryota_odb10 dataset.
Available from http://genomes.mcdb.ucla.edu/Cyclotella/download.html
We identified putative contaminant scaffolds in the C. cryptica version 1.0 assembly using the same Blobtools procedure described above and with read-mapping information from our Illumina reads. To compare functional information between the C. cryptica versions 1.0 and 2.0 annotations, we searched the proteins from the version 1.0 assembly against the Pfam, PRINTS, PANTHER, SMART, SignalP, and TMHMM databases using InterProScan. We also searched the proteins against the SwissProt and UniProt Reference Proteomes databases using NCBI BLASTP.
To assess overlap between the C. cryptica versions 1.0 and 2.0 annotations, we aligned predicted protein sequences from the two genomes to one another using NCBI BLASTP with options ‘-evalue 1e-6 -max_target_seqs 1 -max_hsps 1 -outfmt 6’. We parsed these results to count those with the same length (qlen == slen), those with 100% identity (pident == 100), those with high similarity (pident ≥ 90), and those with full alignment lengths (qcovs == 100).
Data availability
The genome assembly and sequence data are available from NCBI BioProject PRJNA628076. RNAseq data are available through the NCBI Short Read Archive under BioProject PRJNA589195. A genome browser and gene annotations are available through the Comparative Genomics (CoGe) web platform (https://genomevolution.org/coge/) under genome ID 57836. File S1 contains Tables S1–S6. File S2 contains Figures S1–S3. File S3 contains the genome annotation GFF3, protein fasta, and transcript fasta files. File S4 contains the non-coding RNA annotation files. File S5 contains the repeat element annotation files. Supplemental material available at figshare: https://doi.org/10.25387/g3.12341072.
Results And Discussion
Genome assembly
We sequenced five libraries on the MinION platform and base called >5.9 million reads that totaled 9.42 Gb of sequence, where the read length N50 was 4.4 kb and the median quality score per read was 9.9 (Table S1 and Figure S1). After trimming and filtering, we used a total of 2,941,466 nanopore reads for genome assembly (Figure S1). We also sequenced 15 short-read libraries on the Illumina platform which provided nearly 450 million reads, totaling 35.3 Gb of sequence data (Table S1). The transcriptome was assembled into 35,726 transcripts that were used for genome annotation (Nakov et al. 2020).
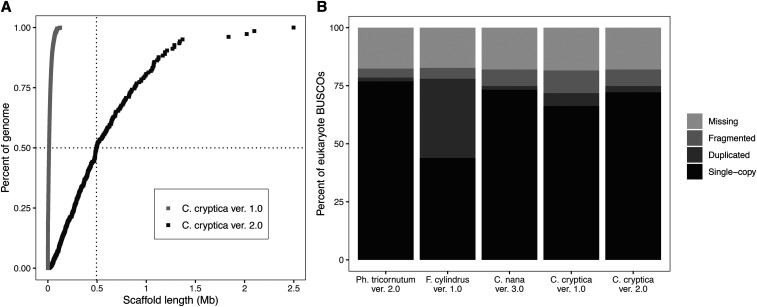
On average, each position in the version 2.0 genome was covered by 152 reads, including both Nanopore (74X) and Illumina (78X) reads. The new assembly represents a substantial improvement over the original version 1.0 genome assembly, which was generated from Illumina short-read sequencing data only. The application of long-read sequencing data resulted in several important changes or improvements, including: (1) an increase in the estimated genome size, from 161.7 Mb to 171.1 Mb; (2) a 176-fold decrease in the number of scaffolds, from 116,815 in version 1.0 to 662 in version 2.0; (3) a 41-fold increase in the scaffold N50, from 12 kb in version 1.0 to 494 kb in version 2.0; (4) a substantial decrease in the number of N’s linking contigs into scaffolds, from 5,360 N’s per 100 kb in version 1.0 to 52 N’s per 100 kb in version 2.0; and (5) increased detection of conserved eukaryotic orthologs, from 183/255 (72%) complete BUSCO genes in version 1.0 to 191/255 (75%) in version 2.0 (Table 1 and Figure 1). The BUSCO count for C. cryptica is now on par with those of C. nana (75%), F. cylindrus (78%), and P. tricornutum (78.5%) (Table 1 and Figure 1).
Figure 1.
Improved genome assembly for Cyclotella cryptica. (A) Cumulative scaffold length and N50 comparison in the version 1.0 and version 2.0 assemblies. Summary statistics for each assembly are given in Table 1. (B) BUSCO analysis of selected diatom genomes using the set of 255 conserved eukaryotic single-copy orthologs. Bars show the proportions of genes found in each assembly as a percentage of the total gene set.
The plastid genome assembly was 129,328 bp in total length (2,707X coverage), which did not differ significantly from the 129,320 bp plastid genome size reported in the version 1.0 assembly (Table 1). The mitochondrial genome was assembled to a total size of 46,485 bp (2,520X coverage), which was nearly 12 kb shorter than the 58,021 bp genome assembled previously (Table 1). This 12 kb difference reflects the size of the complex repeat region present in many diatom mitochondrial genomes (Oudot-Le Secq and Green 2011). We were able to fully span this region with long sequencing reads.
Bacterial co-assembly vs. horizontal gene transfer
Microbial eukaryotic cultures often contain diverse bacterial communities. As a result, genome sequencing projects can generate data from both the target (host) and non-target (bacterial) genomes. Identifying and removing contaminant contigs from these metagenome assemblies can be challenging, particularly for those based only on short-read Illumina data. Illumina-only assemblies can result in many short contigs (Figure 1A) that contain one or few fragmented genes that may or may not belong to the target genome. In contrast, assemblies from long-read sequencing platforms can produce contigs and scaffolds with hundreds or thousands of genes or even entire bacterial genomes, making it much easier to identify and remove non-target sequences from the final assembly.
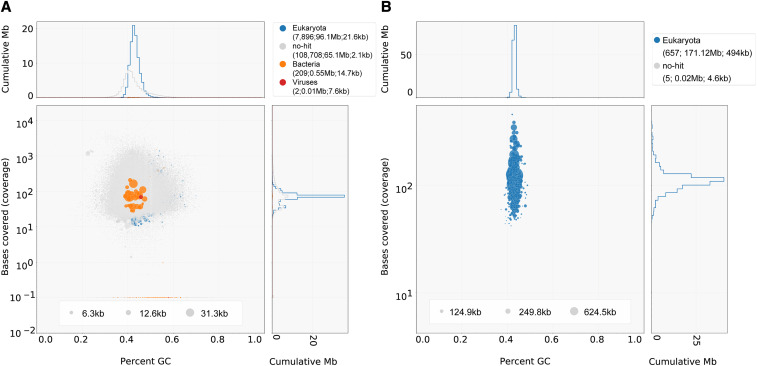
Contaminant scaffolds can be identified using the Blobtools pipeline on the basis of GC content, sequencing coverage, and taxonomic assignment via BLAST searches to reference protein databases. This pipeline has been used to identify and remove contaminants from other microbial eukaryotic genome projects (Koutsovoulos et al. 2016; Nowell et al. 2018; Yubuki et al. 2020). During the construction of the version 2.0 assembly, we used the Blobtools pipeline to identify and remove all scaffolds from the metagenome assembly with lengths less than 1 kb or with a taxonomic assignment to bacteria, archaea, or viruses (Figure S2). These criteria resulted in the removal of 1,974 contaminant scaffolds, leaving a total of 662 scaffolds in the version 2.0 assembly (Figure 2). We also applied the Blobtools pipeline and the same filtering criteria to the version 1.0 assembly and found 99,200 contigs that were less than 1 kb in length with no taxonomic assignment and 211 contigs that were assigned to bacteria or viruses (Figure 2). Of these 211 bacterial or viral scaffolds, a majority (169, or 80%) were less than 1 kb in length, whereas 36 of them had lengths greater than 5 kb, and 21 were larger than 10 kb in length. After removing short and contaminant contigs, the size of the version 1.0 assembly was reduced to 143.4 Mb (161.8 Mb original) and 30,667 scaffolds (116,815 original) (Figure S3).
Figure 2.
The updated assembly of Cyclotella cryptica is highly contiguous and contaminant-free. Blobplots showing the taxon-annotated GC content and coverage of (A) the version 1.0 assembly, and (B) the version 2.0 genome assembly after contaminant filtering. Legend format: “superkingdom (number of scaffolds; length of scaffolds; scaffold N50 length)”.
Confidently removing potential contaminant sequences has important implications for the identification of genes that arose by horizontal gene transfer (HGT) (Koutsovoulos et al. 2016). This is especially complicated for a group like diatoms, which are thought to contain hundreds of genes acquired by HGT from bacteria (Bowler et al. 2008). The version 1.0 assembly included 368 foreign genes (1.7% of the 21,121 genes) from bacteria (n = 340 genes), archaea (n = 12 genes), and viruses (n = 16) (Traller et al. 2016). Application of our filtering routine to the version 1.0 assembly showed that 31 of the 368 HGT genes (8.4%) originally identified as foreign were located on one or more of the 211 contigs that were flagged and removed as contaminants by our filtering criteria. Repeating the Blobtools pipeline to use either 20 or 50 of the top BLASTX hits to each contig for taxonomic assignment, we flagged 540 and 699 contigs in the version 1.0 assembly as contaminants, respectively. These contaminant scaffolds contained a total of 1037 and 1639 genes, respectively, with 67 (18.2%) and 73 (19.8%) of those genes present in the set of 368 HGT genes in the version 1.0 assembly.
These results show that long-read sequencing, combined with better tools to identify and remove contaminant sequences, can greatly improve genome assemblies, particularly for repeat-rich genomes that contain a mix of eukaryotic and bacterial sequences. Applying our pipeline to both assemblies, we found that the version 1.0 assembly of C. cryptica contained hundreds of scaffolds matching bacterial or viral proteins, whereas the version 2.0 assembly appears to be free of contaminants (Figure 2).
Updated gene annotation of the Cyclotella cryptica genome
The version 2.0 assembly includes an updated and more thorough set of gene models. The updated annotation contains 21,250 gene models and 31,409 transcript isoforms (Table 2). The version 2.0 gene models contain more annotated features, including predicted genes, exons, introns, CDS (coding sequences), mRNAs (messenger RNAs), and UTRs (untranslated regions) (File S3). Our annotations of the version 2.0 assembly led to substantial increases in: (1) the mean predicted gene size [from 1.47 kb in version 1.0 to 2.09 kb in version 2.0], (2) mean exon length [608 vs. 722 bp], (3) mean intron length [125 vs. 152 bp], and (4) total length of the coding regions [27.96 vs. 41.84 Mb] (Table 2).
Table 2. Summary of the Cyclotella cryptica genome annotations.
| VERSION 1.0 | VERSION 2.0 | |
|---|---|---|
| TOTAL GENE MODELS | 21,121 | 21,250 |
| TOTAL GENE LENGTH, MB (%) | 31.07 (19.2%) | 44.35 (25.9%) |
| GENE DENSITY (GENES PER MB) | 131 | 124 |
| MEAN GENE SIZE, BP | 1,471 | 2,087 |
| TOTAL CODING LENGTH, MB (%) | 27.96 (17.3%) | 41.84 (24.3%) |
| EXONS PER GENE | 2.18 | 4.30 |
| MEAN EXON LENGTH, BP | 608 | 722 |
| MEAN INTRON LENGTH, BP | 125 | 152 |
| TOTAL TRANSCRIPT ISOFORMS | 23,235 | 31,409 |
| AVERAGE TRANSCRIPT ISOFORMS PER GENE | 1.10 | 1.48 |
| PROTEINS WITH PFAM DOMAIN (%) | 10,384 (44.7%) | 14,518 (46.2%) |
| PROTEINS WITH INTERPROSCAN HIT (%) | 14,565 (62.7%) | 19,690 (62.7%) |
| PROTEINS WITH SWISSPROT HIT (%) | 6,219 (26.8%) | 13,054 (41.6%) |
| PROTEINS WITH UNIPROT HIT (%) | 16,495 (71.0%) | 23,530 (74.9%) |
| GENE MODELS WITH AED < 0.5 (%) | Not determined | 20,506 (96.5%) |
| COMPLETE EUKARYOTIC BUSCO COUNT (%)a | 184 (72.2%) | 192 (75.3%) |
Protein mode against the eukaryota_odb10 dataset.
More importantly, we saw an increase in support for the protein gene models in the version 2.0 assembly, with a higher proportion of proteins containing Pfam protein domains (from 44.7% in version 1.0–46.2% in version 2.0) and matches to SwissProt (26.8% vs. 41.6%) or UniProt proteins (71.0% vs. 74.9%) (Table 2 and Table S4). These increases were possibly due to longer lengths of transcript isoforms in version 2.0 (Table 2). We also identified 188 tRNAs and 36 ncRNAs (File S4). These updated models should better enable physiological, metabolomic, and evolutionary studies of C. cryptica.
Fully 96.5% of the models had AED scores less than 0.5 (Table 2), indicating that the updated gene annotations were highly concordant with the input evidence (transcripts and proteins). Additionally, the 31,409 annotated isoforms included 192/255 (75.3%) of the BUSCO conserved single-copy orthologs in eukaryotes, which represents an increase from 184/255 (72.2%) in the version 1.0 assembly (Table 2). The BUSCO counts for the updated C. cryptica protein models are now comparable to those of the model diatoms, C. nana (70.2%), F. cylindrus (73.7%), and P. tricornutum (76.5%) (Table S5).
We compared the non-redundant protein sets of versions 1.0 and 2.0 using NCBI BLASTP. Protein sets were similar overall, with 19,333 (83.2%) of the version 1.0 proteins aligned to version 2.0 proteins. Of these, 4,949 (25.6%) were perfect matches (same length, 100% identity, and full length) and 6,337 (32.8%) were the same length with high similarity (> 90% identity). The remaining 8,047 (41.6%) alignments were not the same length, but 4,221 (21.8%) of these had 100% identity.
Repeat landscape of the Cyclotella cryptica genome
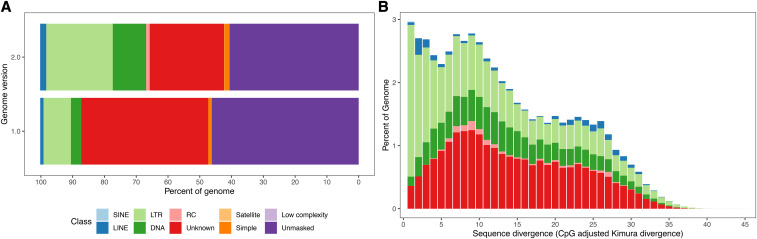
We revisited the characterization of repetitive elements in the C. cryptica genome by applying a more robust set of structural and de novo discovery approaches (File S5). Repeats collectively comprised 59.3% (101.5 Mb) of the version 2.0 assembly, which was slightly greater than the version 1.0 assembly (53.8%, 98.3 Mb) (Table 1 and Figure 3). We also classified a greater fraction of the genome as transposable elements (TEs) in the version 2.0 (32.4%) than version 1.0 (12.9%) assemblies (Figure 3). Additionally, the number of unclassified repeat elements decreased from 40 to 24% between the version 1.0 and version 2.0 assemblies (Figure 3). Repeats represent just 2% and 12% of the genomes of C. nana and P. tricornutum (Armbrust et al. 2004; Maumus et al. 2009; Rastogi et al. 2018) (Table 1).
Figure 3.
Repeat content of the Cyclotella cryptica genome. (A) Repeat content in the version 1.0 and version 2.0 assemblies. Bars show the proportions of the genome assemblies masked and annotated by RepeatMasker. (B) Age distribution of transposable elements in the C. cryptica version 2.0 genome. The total amount of DNA in each TE class was split into bins of 1% Kimura divergence, shown on the X axis (see Methods). Abbreviations: DNA, DNA transposon; LINE, long interspersed nuclear element; LTR, long terminal repeat retrotransposon; RC, rolling circle transposons (Helitron); SINE, small interspersed nuclear element.
Among Class I retrotransposons, we identified short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), and long terminal repeats (LTRs) (Figure 3 and Table S6). SINEs were not identified in the C. cryptica version 1.0 assembly and have only been identified in later annotations of the P. tricornutum genome (Rastogi et al. 2018). SINEs are known for their impacts on mRNA splicing, protein translation, and allelic expression (Kramerov and Vassetzky 2011) and were previously thought to be absent from unicellular eukaryotes (Kramerov and Vassetzky 2011). Their functional roles, if any, in diatoms remain poorly understood. Similar numbers of LINEs were identified in C. cryptica versions 1.0 and 2.0 (2,626 vs. 2,350) (Table S6). We detected fewer numbers of LTRs in C. cryptica version 2.0 than version 1.0 (26,418 vs. 43,176), but these elements appear to represent a larger fraction of the genome (20.9%) than previously thought (8.6%) (Figure 3 and Table S6). Comparative genomics has established that diatom genomes contain diatom-specific Copia-like LTR elements called CoDis (Maumus et al. 2009). Gypsy-type LTR elements were predominant in C. cryptica and covered approximately 22.7 Mb of the genome, whereas Copia-type LTR elements covered 9.5 Mb (Table S6). Both Gypsy- and Copia-type LTRs have been identified in C. nana (Armbrust et al. 2004; Maumus et al. 2009), whereas only Copia-type LTRs have been found in P. tricornutum (Rastogi et al. 2018).
We also identified higher numbers of Class II DNA transposons in the version 2.0 assembly than version 1.0 (52,786 vs. 15,402), constituting a higher proportion of the genome (11.5%) than the previous assembly (3.2%) (Figure 3 and Table S6). These elements in the version 2.0 genome were classified into 12 superfamilies: Crypton, Ginger, EnSpm, hAT, Helitron, Kolobok, MuDr, PiggyBac, PIF-Harbinger, Polintron, Sola, and TcMar (Table S6). The age distribution of TEs, based on sequence divergence from exemplar elements in the repeat library, indicates that there has been a steady accumulation of DNA TEs over time in the C. cryptica genome (Figure 3). By comparison, DNA TEs make up less than 1% of the genome in both C. nana and P. tricornutum (Maumus et al. 2009).
With the improved genome assembly, we can infer that the large genome of C. cryptica is due to recent and historically gradual accumulation of repetitive elements, particularly LTR and DNA TEs (Figure 3), similar to what has been found in flowering plants (Piegu et al. 2006; International Peach Genome Initiative et al. 2013). TEs can impact gene function and regulation and may contribute to the emergence of novel phenotypes (Kazazian 2004; Veluchamy et al. 2013). They have previously been investigated in diatoms for their roles in stress response and environmental adaptation (Maumus et al. 2009; Oliver et al. 2010; Norden-Krichmar et al. 2011). The expanded repeat classification of C. cryptica contributes to our growing knowledge of TE diversity in diatoms and their role in diatom genome evolution.
Conclusions
Cyclotella cryptica is one of a growing list of diatoms with a high-quality sequenced genome. The addition of long-read sequencing data improved the contiguity, completeness, and overall quality of the genome. The version 2.0 assembly allowed for new mechanistic insights into the large size of the genome, namely the historically steady and ongoing accumulation of TEs. The combination of long- and short-read sequencing data provides an effective and relatively inexpensive approach for sequencing modestly sized diatom genomes that will hopefully accelerate the pace of genomic sequencing in diatoms. The improved genome and genome annotation should also help facilitate the continued use of C. cryptica as a model for addressing a wide range of basic and applied research questions in diatoms.
Acknowledgments
This work was supported by the National Science Foundation (grant no. DEB-1651087, AJA) and by a grant from the Simons Foundation (403249, AJA).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12341072.
Communicating editor: A. Rokas
Literature Cited
- Amin S. A., Parker M. S., and Armbrust E. V., 2012. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76: 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D. et al. , 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Attwood T. K., Coletta A., Muirhead G., Pavlopoulou A., Philippou P. B. et al. , 2012. The PRINTS database: A fine-grained protein sequence annotation and analysis resource—its status in 2012. Database (Oxford) 2012 10.1093/database/bas019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z., and Eddy S. R., 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12: 1269–1276. 10.1101/gr.88502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., and Pirovano W., 2014. SSPACE-LongRead: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15: 211 10.1186/1471-2105-15-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K. et al. , 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., and Huson D. H., 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: Architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. S., Law M., Holt C., Stein J. C., Moghe G. D. et al. , 2014. MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. 10.1104/pp.113.230144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Korf I., Robb S. M. C., Parra G., Ross E. et al. , 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. P., and Lowe T. M., 2019. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962: 1–14. 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W., D’Hert S., Schultz D. T., Cruts M., and Van Broeckhoven C., 2018. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 34: 2666–2669. 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton J. F., Lugo-Martinez J., Tucker A. E., Schrider D. R., Warren W. C. et al. , 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLOS Comput. Biol. 10: e1003998 10.1371/journal.pcbi.1003998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., and Doyle J. L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A. et al. , 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47: D427–D432. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., and Willhoeft U., 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. M., Hubley R., Goubert C., Rosen J., Clark A. G. et al. , 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 117: 9451–9457. 10.1073/pnas.1921046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A. et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard R. R. L., 1975. Culture of Phytoplankton for Feeding Marine Invertebrates, pp. 29–60 in Culture of Marine Invertebrate Animals: Proceedings — 1st Conference on Culture of Marine Invertebrate Animals Greenport, edited by Smith W. L. and Chanley M. H.. Springer US, Boston, MA. [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., and Tesler G., 2013. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., and Wessler S. R., 2010. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38: e199 10.1093/nar/gkq862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebust J. A., 1971. Kinetics of glucose transport and growth of Cyclotella cryptica Reimann, Lewin and Guillard. J. Phycol. 7: 1–4. [Google Scholar]
- Holt C., and Yandell M., 2011. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson O. N., Pinder M. I. M., Ohlsson F., Egardt J., Töpel M. et al. , 2019. Friends with benefits: Exploring the phycosphere of the marine diatom Skeletonema marinoi. Front. Microbiol. 10: 1828 10.3389/fmicb.2019.01828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W. et al. , 2014. InterProScan 5: Genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E. P., Rivas E. et al. , 2018. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46: D335–D342. 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A., Suh A., and Feschotte C., 2017. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA 114: E1460–E1469. 10.1073/pnas.1616702114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kolmogorov, M., M. Rayko, J. Yuan, E. Polevikov, and P. Pevzner, 2019a metaFlye: Scalable long-read metagenome assembly using repeat graphs. bioRxiv. doi:10.1101/637637 (Preprint posted May 15, 2019). [DOI] [PMC free article] [PubMed]
- Kolmogorov M., Yuan J., Lin Y., and Pevzner P. A., 2019b Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. et al. , 2017. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsovoulos G., Kumar S., Laetsch D. R., Stevens L., Daub J. et al. , 2016. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc. Natl. Acad. Sci. USA 113: 5053–5058. 10.1073/pnas.1600338113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., and Vassetzky N. S., 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity 107: 487–495. 10.1038/hdy.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L., 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Laetsch D. R., and Blaxter M. L., 2017. BlobTools: Interrogation of genome assemblies. F1000 Res. 6: 1287 10.12688/f1000research.12232.1 [DOI] [Google Scholar]
- Lagesen K., Hallin P., Rødland E. A., Staerfeldt H.-H., Rognes T. et al. , 2007. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., and Bork P., 2012. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin J. C., and Lewin R. A., 1960. Auxotrophy and heterotrophy in marine littoral diatoms. Can. J. Microbiol. 6: 127–134. 10.1139/m60-015 [DOI] [PubMed] [Google Scholar]
- Li H., 2018. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., and Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F., Allen A. E., Mhiri C., Hu H., Jabbari K. et al. , 2009. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics 10: 624 10.1186/1471-2164-10-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T., Otillar R. P., Strauss J., McMullan M., Paajanen P. et al. , 2017. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541: 536–540. 10.1038/nature20803 [DOI] [PubMed] [Google Scholar]
- Nakov T., Judy K. J., Downey K. M., Ruck E. C., and Alverson A. J., 2020. Transcriptional response of osmolyte synthetic pathways and membrane transporters in a euryhaline diatom during long-term acclimation to a salinity gradient. J. Phycol. (in press). [DOI] [PubMed] [Google Scholar]
- Nawrocki E. P., and Eddy S. R., 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden-Krichmar T. M., Allen A. E., Gaasterland T., and Hildebrand M., 2011. Characterization of the small RNA transcriptome of the diatom, Thalassiosira pseudonana. PLoS One 6: e22870 10.1371/journal.pone.0022870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell R. W., Almeida P., Wilson C. G., Smith T. P., Fontaneto D. et al. , 2018. Comparative genomics of bdelloid rotifers: Insights from desiccating and nondesiccating species. PLoS Biol. 16: e2004830 10.1371/journal.pbio.2004830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver M. J., Schofield O., and Bidle K., 2010. Density dependent expression of a diatom retrotransposon. Mar. Genomics 3: 145–150. 10.1016/j.margen.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M.-P., and Green B. R., 2011. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476: 20–26. 10.1016/j.gene.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Pahl S. L., Lewis D. M., Chen F., and King K. D., 2010. Heterotrophic growth and nutritional aspects of the diatom Cyclotella cryptica (Bacillariophyceae): Effect of some environmental factors. J. Biosci. Bioeng. 109: 235–239. 10.1016/j.jbiosc.2009.08.480 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., and Nielsen H., 2011. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Piegu B., Guyot R., Picault N., Roulin A., Sanyal A. et al. , 2006. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16: 1262–1269. 10.1101/gr.5290206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Jones N. C., and Pevzner P. A., 2005. De novo identification of repeat families in large genomes. Bioinformatics 21: i351–i358. 10.1093/bioinformatics/bti1018 [DOI] [PubMed] [Google Scholar]
- Rastogi A., Maheswari U., Dorrell R. G., Vieira F. R. J., Maumus F. et al. , 2018. Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 8: 4834 10.1038/s41598-018-23106-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach M. J., Schmidt S. A., and Borneman A. R., 2018. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19: 460 10.1186/s12859-018-2485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler P. G., 1988. Effects of silicon deficiency on lipid composition and metabolism in the diatom Cyclotella cryptica. J. Phycol. 24: 394–400. 10.1111/j.1529-8817.1988.tb00189.x [DOI] [Google Scholar]
- Schultz M. E., 1971. Salinity-related polymorphism in the brackish-water diatom Cyclotella cryptica. Can. J. Bot. 49: 1285–1289. 10.1139/b71-182 [DOI] [Google Scholar]
- Schultz M. E., and Trainor F. R., 1970. Production of male gametes and auxospores in a polymorphic clone of the centric diatom Cyclotella. Can. J. Bot. 48: 947–951. 10.1139/b70-133 [DOI] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Slocombe S. P., Zhang Q., Ross M., Anderson A., Thomas N. J. et al. , 2015. Unlocking nature’s treasure-chest: Screening for oleaginous algae. Sci. Rep. 5: 9844 10.1038/srep09844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., and Florea L., 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. Gigascience 4: 48 10.1186/s13742-015-0089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., and Haussler D., 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. 10.1093/bioinformatics/btn013 [DOI] [PubMed] [Google Scholar]
- Steinbiss S., Willhoeft U., Gremme G., and Kurtz S., 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37: 7002–7013. 10.1093/nar/gkp759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson B., and Hildebrand M., 2010. Dynamics of silica cell wall morphogenesis in the diatom Cyclotella cryptica: Substructure formation and the role of microfilaments. J. Struct. Biol. 169: 62–74. 10.1016/j.jsb.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B. et al. , 2003. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 13: 2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E. S., Fischer A. et al. , 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45: W6–W11. 10.1093/nar/gkx391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traller J. C., Cokus S. J., Lopez D. A., Gaidarenko O., Smith S. R. et al. , 2016. Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 9: 258 10.1186/s13068-016-0670-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traller J. C., and Hildebrand M., 2013. High throughput imaging to the diatom Cyclotella cryptica demonstrates substantial cell-to-cell variability in the rate and extent of triacylglycerol accumulation. Algal Res. 2: 244–252. 10.1016/j.algal.2013.03.003 [DOI] [Google Scholar]
- UniProt Consortium , 2018. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 46: 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Sović I., Nagarajan N., and Šikić M., 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27: 737–746. 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy A., Lin X., Maumus F., Rivarola M., Bhavsar J. et al. , 2013. Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 4: 2091 10.1038/ncomms3091 [DOI] [PubMed] [Google Scholar]
- Verde I., A. G., Abbott S., Scalabrin S., Jung S., Shu, et al. , 2013. International Peach Genome Initiative The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. 10.1038/ng.2586 [DOI] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A. et al. , 2014. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. W., 1974. Growth of two facultatively heterotrophic marine centric diatoms. J. Phycol. 10: 292–300. [Google Scholar]
- Xu G.-C., Xu T.-J., Zhu R., Zhang Y., Li S.-Q. et al. , 2019. LR_Gapcloser: A tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience 8: giy157 10.1093/gigascience/giy157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubuki N., Galindo L. J., Reboul G., López-García P., Brown M. W. et al. , 2020. Ancient adaptive lateral gene transfers in the symbiotic Opalina-Blastocystis stramenopile lineage. Mol. Biol. Evol. 37: 651–659. 10.1093/molbev/msz250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome assembly and sequence data are available from NCBI BioProject PRJNA628076. RNAseq data are available through the NCBI Short Read Archive under BioProject PRJNA589195. A genome browser and gene annotations are available through the Comparative Genomics (CoGe) web platform (https://genomevolution.org/coge/) under genome ID 57836. File S1 contains Tables S1–S6. File S2 contains Figures S1–S3. File S3 contains the genome annotation GFF3, protein fasta, and transcript fasta files. File S4 contains the non-coding RNA annotation files. File S5 contains the repeat element annotation files. Supplemental material available at figshare: https://doi.org/10.25387/g3.12341072.