Abstract
The USDA-ARS National Clonal Germplasm Repository (NCGR) in Corvallis, Oregon, maintains one of the world’s largest and most diverse living Pyrus collection. A thorough genetic characterization of this germplasm will provide relevant information to optimize the conservation strategy of pear biodiversity, support the use of this germplasm in breeding, and increase our knowledge of Pyrus taxonomy, evolution, and domestication. In the last two decades simple sequence repeat (SSR) markers have been used at the NCGR for cultivar identification and small population structure analysis. However, the recent development of the Applied Biosystems Axiom Pear 70K Genotyping Array has allowed high-density single nucleotide polymorphism (SNP)-based genotyping of almost the entire collection. In this study, we have analyzed this rich dataset to discover new synonyms and mutants, identify putative labeling errors in the collection, reconstruct the largest pear cultivar pedigree and further elucidate the genetic diversity of Pyrus.
Keywords: population structure, germplasm characterization, single nucleotide polymorphism markers, biodiversity conservation, pear breeding
The USDA-ARS National Clonal Germplasm Repository (NCGR) in Corvallis, Oregon, maintains one of the world’s largest and most diverse collection of Pyrus (Postman 2008a). It includes 2,300 clonal pear accessions and 364 seed lots, encompassing 36 different species or interspecific hybrids from 55 countries (Postman 2008b). This collection represents, therefore, a useful tool for population and evolutionary genetic studies in Pyrus, as well as a valuable source of material for breeding purposes. An understanding of the genetic diversity of this collection will better support the use of this germplasm for the improvement of scion and rootstock pear cultivars (Volk et al. 2019). Additionally, since in the past germplasm to be preserved at the NCGR was selected based on morphological, geographical and passport data, there is a need to implement molecular marker screening to verify the trueness-to-type of the accessions maintained, as well as to eliminate redundancies. Finally, the use of molecular markers will aid in the elucidation of parentage for hundreds of cultivars and breeding material in the collection. Accurate pedigree information is not only essential for proper parental selection in breeding programs, but it also allows geneticists to infer trait heritability, understand genetic correlations among phenotypes of interest, and estimate breeding values (Kouassi et al. 2009; Cellon et al. 2018; Piaskowski et al. 2018). Such validated pedigree information also enables more powerful marker association studies (Bink et al. 2007; Kumar et al. 2013).
In the last two decades numerous studies implemented the use of simple sequence repeat (SSR) markers at the NCGR for cultivar identification and germplasm characterization (Bassil et al. 2005; Volk et al. 2006; Bassil and Postman 2010; Evans et al. 2015). However, single nucleotide polymorphism (SNP) markers are more abundant in the genomes (Rafalski 2002) and, when applied in large numbers, they have been shown to outperform SSRs for population structure and genetic relatedness studies (Lemopoulos et al. 2019). Thanks to many recent technological advances, it is today possible to carry out high-density SNP-based genotyping of a large number of samples at a low cost per data point. Therefore, while SSRs are still the markers of choice for routine fingerprinting analyses, the use of SNPs for germplasm characterization has recently increased (Hinze et al. 2017; Arab et al. 2019; Rufo et al. 2019; Xia et al. 2019). Additionally, public SNP arrays could be used as common marker sets in separate studies of different germplasm collections, thereby providing an opportunity to have comparable analysis at a global level. Such knowledge would be instrumental for the identification of gaps and for the optimization of the conservation strategy at genebank collections worldwide (Urrestarazu et al. 2015).
In the past decade there has been a flurry of studies on the genetic diversity, population structure and phylogeny of subsets of local Pyrus germplasm collections (Chevreau et al. 2020). However only recently did the application of next-generation sequencing technologies and high density SNP-based genotyping lead to important discoveries about the degree of diversity among Pyrus species and increase our understanding of the evolution and domestication of this genus (Kumar et al. 2017; Wu et al. 2018; Kim et al. 2019).
In this study, we used high-density genotypic data generated with the recently developed Applied Biosystems Axiom Pear 70 K Genotyping Array (Montanari et al. 2019) to begin the characterization of the Pyrus collection held at the NCGR. By genotyping almost 2,000 samples, we discovered new synonyms and mutants, identified putative labeling errors at the collection, reconstructed the largest pedigree of pear cultivars, and further elucidated the genetic diversity of Pyrus.
Material and Methods
Plant material and genotyping
A total of 1,890 diploid Pyrus spp. samples, two haploids, and five intergeneric hybrids (Pyronia Pyrus Cydonia; Sorbopyrus Sorbus Pyrus), were used for the analysis performed in this work (Table S1). Specifically, this list of 1,897 samples consisted of: i) 288 (including biological and technical replicates) that were screened with the draft Axiom Pear 700 K Genotyping Array by Montanari et al. (2019) (hereafter called Screening Panel); ii) 1,415 (including biological and technical replicates and the two haploids, two Pyronia and three Sorbopyrus accessions) genotyped with the Axiom Pear 70 K Genotyping Array by Montanari et al. (2019) (hereafter called Genotyping 1 Panel); iii) 194 additional accessions screened in this work with the Axiom Pear 70 K Genotyping Array (hereafter called Genotyping 2 Panel). From the Screening Panel, only the 275 samples that had passed genotyping standards were kept (Montanari et al. 2019), and only the genotypic information for the 71,363 SNPs that were also included in the 70 K array were used for the following analysis. For all other samples, the raw data were merged and re-analyzed, using a QC CR threshold of 96.385 (for details see Affymetrix Axiom Genotyping Solution – Data Analysis Guide, https://assets.thermofisher.com/TFS-Assets/LSG/manuals/axiom_genotyping_solution_analysis_guide.pdf).
The 71,363 SNPs of the 70 K array were aligned to the new Double Haploid (DH) Bartlett Genome (Linsmith et al. 2019) using BLAST (Altschul et al. 1990) as explained in Montanari et al. (2019), except with an identity threshold of 90%.
Identification of duplicated samples
Among the SNPs that had high quality and unique alignments to the new pear genome, only the PolyHighResolution (PHR, according to the Affymetrix default parameters for diploid samples) were used for pairwise comparison of all 1,897 samples. Identity by state (IBS) values were computed using plink v1.90 (options--allow-extra-chr--distance square0 ibs). The available biological and technical replicates were used to set the IBS threshold for the identification of the duplicated samples. For each group of duplicates, the genotype with the lowest number of missing data were selected for subsequent analysis.
Pedigree reconstruction
Five F1 crossing populations from the Washington State University (Guzman 2018) and the USDA-ARS Appalachian Fruit Research Laboratory (Zurn et al. 2020) pear breeding programs were used to identify erroneous SNPs based on Mendelian inheritance and then aid in the pedigree reconstruction of the Pyrus accessions. These populations consisted of: 63 offspring derived from ‘Bartlett’ × ‘Anjou’, 82 from ‘Bartlett’ × ‘Doyenne du Comice’, 97 from ‘Old Home’ × ‘Bartlett’, 83 from ‘Potomac’ × ‘El Dorado’, and 85 from NJA2R59T69 × ‘Bartlett’, for a total of 410 trios. They were added to 260 known trios/duos from the sample set of this study. SNPs were filtered for missing data (removed if >2%), and then a Mendelian test was run on the 670 known trios using trio.check as described in Montanari et al. (2019), and SNPs with an error rate > 5% were removed. This marker dataset was used to compute the relationship inference between each pair of samples with the KING-robust method (Manichaikul et al. 2010), implemented in the R package SNPRelate v1.14.0 (Zheng et al. 2012). As demonstrated by Manichaikul et al. (2010), Linkage Disequilibrium (LD) pruning is not necessary for application of this method. The computed kinship coefficients (k), which represents the probability that two random alleles from the two samples are identical by descent, can be used to identify first-degree relationships. Within those, the value of IBD0, i.e., the probability that the two samples share zero alleles identical by descent, can be used to distinguish parent-offspring (PO) from full-sib (FS) relationships. The theoretical value of k in first-degree relationships is 0.25, and the IBD0 is 0 in PO and 0.25 in FS. However, in practice such values deviate from the theoretical ones and depend on the characteristics of the specific population under study. The values of k and IBD0 for 90% of the pairwise combinations in the known trios, confirmed by Mendelian test (< 10% error rate), were used to set the thresholds to apply in this study. All new trios and duos that were identified upon applying the set thresholds for k and IBD0 were again tested for Mendelian errors, and those with error rate < 1.5% were considered true. A second search of PO and FS was carried out by refining the inference criteria (in this case using k and IBD0 values for 95% of the newly confirmed relationships), and were again confirmed by Mendelian test (<1.5% error rate). New PO and FS relationships were compared with the literature (Hedrick et al. 1921; Jacob 1998; Mielke and Smith 2002; Simard and Michelesi 2002; Pasqualini et al. 2006; Sawamura et al. 2008; Bassil and Postman 2010; Postman et al. 2013; Bell et al. 2014; Morgan 2015) and the information stored at the NCGR website (https://www.ars.usda.gov/ARSUserFiles/20721500/catalogs/pyrcult.html) regarding year and country of origin and believed parentage, when available. Pedigree networks were designed with the R package network v1.13 (Butts 2008) and with the software Helium (Shaw et al. 2014).
Population structure analysis
The SNP dataset was pruned for LD using an r2 threshold of 0.80 in plink v1.90 (options--allow-extra-chr--indep-pairwise 50 5 0.80), but not filtered for MAF, as the Axiom 70K SNPs were carefully chosen to include rare alleles that would correctly depict population structure (Montanari et al. 2019). A Principal Component Analysis (PCA) was run using the R package SNPRelate, and the graph for the first two PCs plotted with ggplot2 and using a species-based color-coding for the samples. The software fastSTRCTURE (Raj et al. 2014) was then run to infer the population structure, using a hierarchical approach. First, inferences were performed for K = 2 to 30, with 15 replicates per K, and then both the fastSTRUCTURE algorithm for multiple choices of K and the Evanno’s ad hoc procedure (Evanno et al. 2005) were performed in an attempt to choose the optimal number of subpopulations. Because of the complexity of the structure, another round of structure inference was run separately on the subpopulations and the admixed group identified at K = 2; up to K = 22 was used in this second round. Finally, Clumpp (Jakobsson and Rosenberg 2007) was used to summarize data from the 15 replicates and obtain mean Q-values. Samples with Q 0.75 were assigned to the relative subpopulation, and plots were designed with the program Structure Plot v2.0 (Ramasamy et al. 2014). Additionally, a PCA was run again for each of the subpopulations and the admixed identified at K = 2 in the initial structure analysis. Results from the PCA and the structure analysis were compared, and used to identify samples that had been likely assigned to the wrong species and propose a new classification. PC1 vs. PC2 plots for each subpopulation used a color-coding based on the new proposed classification.
In an attempt to further resolve the complexity of one of the subpopulations identified (the Occidental group), a discriminant analysis of principal components (DAPC) was also carried out (Jombart et al. 2010). DAPC was performed using the R package adegenet v2.1.2 (Jombart 2008; Jombart and Ahmed 2011). The optimal number of clusters was chosen running the find.clusters function for up to 90 clusters, and then examining the values of Bayesian Information Criterion (BIC) for each number of clusters. The function dapc was then run on the groups inferred with find.clusters at the chosen number of clusters and using the first 500 PCs and four discriminant functions. Results were plotted with ggplot2.
Data availability
Supplemental data (Tables S1-S4; Figure S1; Files S1-S5) are provided through figshare. The genotyping data for the 1,749 samples that passed genotyping standards and 64,571 SNPs that had unique, high-quality alignment to the DH Bartlett Genome and that were classified as PHR are provided through the Genome Database for Rosaceae (GDR, https://www.rosaceae.org/, accession number tfGDR1042). Supplemental material available at figshare: https://doi.org/10.25387/g3.12186105.
Results
SNP genotyping and BLAST on the new ‘Bartlett’ genome assembly
A total of 1,474 samples from Genotyping1 and 2 Panels passed genotyping standards, which, together with the passed samples from the Screening Panel, summed up to 1,749 samples.
All 71,363 SNPs aligned to the DH Bartlett Genome, as expected. However, 965 SNPs were discarded after quality filtering of the alignments. Additionally, 4,638 SNPs aligned to multiple locations, and therefore were eliminated. Of the remaining 65,760 SNPs, 64,571 were classified as PHR and used for the subsequent analysis.
Identification of mutants, synonyms and labeling errors, and pedigree reconstruction
In this study, a large number of replicates (77) was used as controls among the different plates and genotyping panels. The IBS threshold above which two samples were considered identical was set to 97.7%. A total of 1,113 genotypes were unique (i.e., did not have any duplicate). Excluding the 77 replicated samples, 218 groups of identical genotypes were found, encompassing a total of 534 samples. Most of the groups included just two samples, but some others had 10 or more. The group with the largest number of identical genotypes included the duplicates of ‘Bartlett’ (a.k.a. ‘Williams’ Bon Chretien’) and consisted of 31 samples. Table S2 reports all the samples with identical genotypes found in this study, with notes about whether they were already known (as reported in Hedrick et al. (1921); Morgan (2015) and from NCGR passport data available through the GRIN-Global website), if they were biological or technical replicates, or if they are suspected to be sampling or labeling errors, based on the following results from pedigree reconstruction and structure analysis.
Removal of duplicates resulted in a number of 1,331 unique genotypes. After filtering for missing data and Mendelian error, 62,673 SNPs were left and 13 trios with > 10% error rate were eliminated, leaving 657 trios. In the first search, pairs of samples were assigned first-degree relationship if they had values of k 0.133, and among those PO were identified when IBD0 0.005. In the second search, the thresholds for k and IBD0 were refined to 0.136 and 0.002, respectively. PO relationships were found for 723 accessions, across 13 species or interspecific hybrids (Figure 1); only 90 founders were identified (Table 1). In total, 139 trios/duos that were known before this study were hereby confirmed (at a more stringent threshold of 1.5% Mendelian error rate), and 498 new ones were identified. These numbers refer only to the Pyrus accessions evaluated in this study and do not include the five F1 crossing populations. Full information about discovered parentages can be found in Table S3, with relevant literature citations.
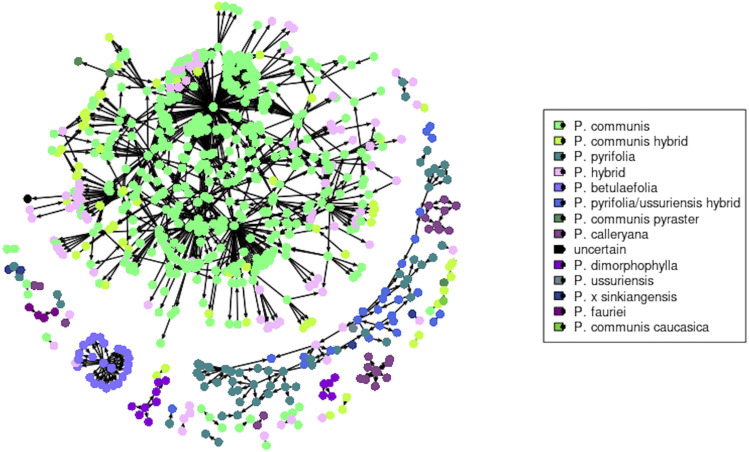
Figure 1.
Pedigree network for all the trios and duos identified in this study. Each dot represents an accession and a color-coding based on the species is used, as shown in the legend on the right-hand side. Relationships are shown with an arrow from the parent to the offspring accession.
Table 1. List of the 90 founders from the inferred pedigree.
| Accession ID | Sample name | Taxon | Plant name |
|---|---|---|---|
| PI 542023 | CPYR_1177.001 | Pyrus ×bretschneideri | Tsu Li |
| PI 665781 | CPYR_2638.002 | Pyrus ×bretschneideri | Tsu Li 1 |
| PI 542022 | CPYR_1617.002 | Pyrus ×bretschneideri | Xiangshui Li [Hsiang Sui-Li] |
| Q 27647 | CPYR_2681.002 | Pyrus ×bretschneideri | Xuehuali (Snowflake) |
| PI 506362 | CPYR_1678.001 | Pyrus ×bretschneideri | Ya Li |
| PI 665771 | CPYR_2989.003 | Pyrus ×sinkiangensis | Chinese Fragrant Pear |
| PI 540943 | CPYR_653.001 | Pyrus betulaefolia | OPR-110 P. betulifolia No. 1 |
| PI 540946 | CPYR_656.001 | Pyrus betulaefolia | OPR-114 P. betulifolia No. 5 |
| PI 540973 | CPYR_1263.001 | Pyrus betulaefolia | P. betulifolia OSU-3 |
| PI 541108 | CPYR_2189.001 | Pyrus calleryana | Aristocrat (P. calleryana) |
| PI 617646 | CPYR_2577.001 | Pyrus calleryana | Bradford (P. calleryana) |
| PI 541083 | CPYR_1601.001 | Pyrus calleryana | P. calleryana OSU-10 |
| PI 541053 | CPYR_1264.003 | Pyrus calleryana | P. calleryana OSU-2 |
| PI 541018 | CPYR_673.001 | Pyrus calleryana | P. calleryana PC-5 |
| PI 617505 | CPYR_674.001 | Pyrus calleryana | P. calleryana PC-6 |
| PI 324124 | CPYR_12.002 | Pyrus communis | Akca |
| PI 264694 | CPYR_23.002 | Pyrus communis | Arganche |
| PI 654945 | CPYR_2757.001 | Pyrus communis | Bellissime d’Hiver |
| PI 541128 | CPYR_52.002 | Pyrus communis | Bergamote d’Ete |
| PI 541127 | CPYR_51.002 | Pyrus communis | Bergamotte d’Automne |
| PI 541523 | CPYR_2131.001 | Pyrus communis | Bergamotte de Baillargues |
| PI 260153 | CPYR_53.001 | Pyrus communis | Bergamotte Esperen |
| PI 541130 | CPYR_56.002 | Pyrus communis | Besi d’Hery |
| PI 654936 | CPYR_2706.001 | Pyrus communis | Bessemianka |
| PI 295083 | CPYR_64.003 | Pyrus communis | Beurré d’Arenberg |
| PI 541145 | CPYR_78.002 | Pyrus communis | Beurré Gris |
| PI 307539 | CPYR_83.002 | Pyrus communis | Beurré Inflancka |
| PI 541148 | CPYR_86.002 | Pyrus communis | Beurré Millet |
| PI 617587 | CPYR_2510.002 | Pyrus communis | Blanquilla (=Spadona) |
| PI 541387 | CPYR_1165.001 | Pyrus communis | Bosc - OP-5 |
| PI 541305 | CPYR_103.001 | Pyrus communis | Brandy |
| PI 541163 | CPYR_139.004 | Pyrus communis | Citron de Carmes (Madeleine) |
| PI 654920 | CPYR_2449.001 | Pyrus communis | Colmar d’Ete |
| PI 541168 | CPYR_156.001 | Pyrus communis | Conference |
| PI 541183 | CPYR_202.003 | Pyrus communis | Early Harvest (=Chambers) |
| PI 392319 | CPYR_205.002 | Pyrus communis | Ecmianka |
| PI 231889 | CPYR_230.001 | Pyrus communis | Fondante de Charneu |
| PI 541191 | CPYR_233.001 | Pyrus communis | Forelle |
| PI 264194 | CPYR_244.004 | Pyrus communis | Gieser Wildeman |
| PI 260161 | CPYR_490.004 | Pyrus communis | King Sobieski |
| CPYR 2992 | CPYR_2992.001 | Pyrus communis | Kings Valley Pear 1 |
| PI 541215 | CPYR_346.001 | Pyrus communis | Lemon |
| PI 130990 | CPYR_1113.001 | Pyrus communis | Madame Verte |
| Q 24302 | CPYR_2978.001 | Pyrus communis | Malti |
| PI 541233 | CPYR_393.003 | Pyrus communis | Messire Jean |
| PI 255616 | CPYR_410.001 | Pyrus communis | Napoleon |
| PI 541456 | CPYR_431.001 | Pyrus communis | Old Home |
| PI 541242 | CPYR_451.002 | Pyrus communis | Petit Blanquet |
| PI 541245 | CPYR_466.002 | Pyrus communis | President Loubet |
| PI 541256 | CPYR_496.001 | Pyrus communis | Rousselet de Reims |
| PI 541444 | CPYR_1516.002 | Pyrus communis | Stuttgarter-Geishirtle (= Zuckerbirne) |
| PI 260162 | CPYR_578.001 | Pyrus communis | Tonkowietka |
| PI 541281 | CPYR_602.004 | Pyrus communis | White Doyenne |
| PI 541282 | CPYR_603.002 | Pyrus communis | White Star |
| PI 638016 | CPYR_2826.001 | Pyrus communis | Yaquina (Payson) |
| PI 665773 | CPYR_2859.001 | Pyrus communis | Zutica |
| PI 337437 | CPYR_687.001 | Pyrus communis subsp. caucasica | P. communis ssp. caucasica - Stavropol |
| PI 483401 | CPYR_1551.002 | Pyrus communis subsp. pyraster | Crna Poloska |
| PI 325930 | CPYR_1390.001 | Pyrus dimorphophylla | P. dimorphophylla - Japan |
| PI 617507 | CPYR_776.001 | Pyrus fauriei | P. fauriei MSU5768 |
| PI 260200 | CPYR_1275.001 | Pyrus hybrid | Cherry Pear |
| PI 541711 | CPYR_239.002 | Pyrus hybrid | Garber |
| PI 483372 | CPYR_1526.002 | Pyrus hybrid | Ilinka |
| PI 312503 | CPYR_2386.001 | Pyrus hybrid | Michurin Beurré Zimnaya (Winter) |
| PI 541239 | CPYR_433.002 | Pyrus hybrid | Orel No. 15 |
| PI 617526 | CPYR_1494.001 | Pyrus hybrid | P. betulifolia 2 x P. call. 2 |
| PI 541768 | CPYR_1239.001 | Pyrus hybrid | P. pashia x P. calleryana |
| PI 541776 | CPYR_1315.001 | Pyrus hybrid | P. ussuriensis x P. calleryana |
| PI 541812 | CPYR_1702.001 | Pyrus hybrid | South Dakota E-31 |
| PI 134606 | CPYR_573.002 | Pyrus hybrid | Tioma |
| PI 541859 | CPYR_725.002 | Pyrus nivalis | P. nivalis P-91 (pure) |
| PI 228012 | CPYR_178.002 | Pyrus pyrifolia | Doitsu |
| PI 541897 | CPYR_270.001 | Pyrus pyrifolia | Hawaii |
| PI 352641 | CPYR_294.001 | Pyrus pyrifolia | Imamura Aki |
| PI 228013 | CPYR_296.002 | Pyrus pyrifolia | Ishiiwase |
| PI 541898 | CPYR_303.003 | Pyrus pyrifolia | Japanese Golden Russet |
| PI 97348 | CPYR_1119.001 | Pyrus pyrifolia | Meigetsu |
| PI 654923 | CPYR_2642.002 | Pyrus pyrifolia | Nepal 5053 |
| PI 224196 | CPYR_413.001 | Pyrus pyrifolia | Nijisseiki |
| PI 392318 | CPYR_428.001 | Pyrus pyrifolia | Okusankichi |
| PI 541927 | CPYR_1018.001 | Pyrus pyrifolia | P. pyrifolia from A. Donovan house |
| PI 278731 | CPYR_533.001 | Pyrus pyrifolia | Sivaganga Estate |
| CPYR 2892 | CPYR_2892.002 | Pyrus sachokiana | P. sachokiana GE-2006-114 |
| PI 541985 | CPYR_27.002 | Pyrus ussuriensis | Ba Li Xiang [Ba Li Hsiang] |
| PI 617537 | CPYR_2338.001 | Pyrus ussuriensis | Chien Li |
| PI 315064 | CPYR_268.001 | Pyrus ussuriensis | Hang Pa Li |
| PI 541990 | CPYR_288.002 | Pyrus ussuriensis | Huangxianshui Li [Huang Hsing Sui Li] |
| PI 541993 | CPYR_291.001 | Pyrus ussuriensis | Hung Li |
| PI 267863 | CPYR_455.002 | Pyrus ussuriensis | Ping Guo Li [Pingo Li] |
| PI 542007 | CPYR_1157.002 | Pyrus ussuriensis | Tzu Ma Li |
A small number of accessions appeared to be the main founders in P. communis, and they include ancient and commercially important cultivars. For example, ‘White Doyenne’, believed to be the ancient cultivar ‘Doyenné Blanc’ originated in 1652, and possibly the same as the earlier ‘Pera Ghiacciuola’ described in 1559 (Hedrick et al. 1921), is the parent of 56 accessions, which are themselves involved in four more pedigree generations. ‘White Doyenne’ offspring of note are ‘Duchesse d’Angouleme’, ‘Bartlett’, ‘Comtesse de Paris’, ‘Anjou’ and ‘Coscia’. ‘Duchesse d’Angouleme’, first reported in 1808 (Hedrick et al. 1921), is itself the parent of 30 cultivars, including ‘Doyenne du Comice’ (second parent inferred is ‘Glou Morceau’), ‘Roi Charles de Wurttemberg’ and ‘Beurré Clairgeau’, this last one also parent of 33 accessions. ‘Roi Charles de Wurttemberg’ (presumed origin 1886) appeared to be a backcross of ‘Beurré Clairgeau’ × ‘Duchesse d’Angouleme’ (Figure S1a). ‘Bartlett’, found in Berkshire (UK) in 1770 (Hedrick et al. 1921), is the parent of the largest number of accessions (156), as expected, including ‘Clapp Favorite’ and ‘Kieffer’. ‘Clapp Favorite’, whose parentage ‘Flemish Beauty’ (syn. ‘Lesnaya Krasavitza’) × ‘Bartlett’ was confirmed, is the founder for 17 accessions, eight of which appeared to have a hybrid ancestry between P. communis and subsp. pyraster or caucasica (Figure S1b). ‘Kieffer’, a US hybrid cultivar which was reported to have first fruited in 1863, is itself the parent of 15 accessions (Figure S1c). One of ‘Kieffer’’s offspring is BP-2, a rootstock selection that originated in 1928 in South Africa. A number of accessions indistinguishable from ‘Kieffer’ were identified at the repository, including ‘Burford Pear’, ‘Campas No. 2’, and ‘Hermit’. Also, five accessions that were collected together in Pakistan (namely Nak I, Khan Tangoo I, India IC 20821, Kharnak I and Kharnak II) had the same genotype as ‘Kieffer’ (Table S2). The old Belgian cultivar ‘Comtesse de Paris’ had the same genotype as ‘Flemish Beauty’ (here its known synonym ‘Lesnaya Krasavitza’ was used). The Romanian cultivar ‘Rosii Untoase’, ‘Parker’ (claimed to be selected in Minnesota) and ‘Southworth’ also turned out to be identical to ‘Comtesse de Paris’. ‘Southworth’ was reported to be a synonym of ‘Vermont Beauty’ (Morgan 2015), which was not confirmed here. However, ‘Southworth’ and ‘Parker’ were donated to the NCGR by the same nursery, thus ‘Southworth’ might be a labeling error. ‘Comtesse de Paris’ is the founder to 23 accessions, including P. communis and hybrids. ‘Anjou’, whose first record was in the UK in the early XIX Century, turned out to have originated from ‘White Doyenne’ × ‘Sucre Verte’, this last one being an old cultivar known since 1670 and an inferred offspring of ‘Bergamotte d’Automne’ (Figure S1a). ‘Anjou’ is itself the parent of 30 accessions. It appeared that the labels for ‘Coscia’ and ‘Coscia Tardive’ had been swapped at the repository. ‘Coscia’ was reported to have originated in the late XVII Century, while ‘Coscia Tardive’ is known only since 1910 (Morgan 2015); they turned out to be connected by a PO relationship. Taking into account the swapped identity, ‘Coscia’ was inferred as offspring of ‘Blanquilla’ (syn. ‘Spadona’) × ‘White Doyenne’, and ‘Coscia Tardive’ originated from ‘Coscia’ × ‘Beurré Giffard’, this last one also a descendent of ‘White Doyenne’. Additionally, a number of cultivars known to be offspring of ‘Coscia’ were confirmed, in particular ‘Coscia Precoce’, ‘Butirra Precoce Morettini’ (‘Coscia’ × ‘Bartlett’), ‘Santa Maria’ (‘Coscia’ × ‘Bartlett’), ‘Etrusca’ (‘Coscia’ × ‘Ilinka’), ‘Butirra Rosata Morettini’ (‘Coscia’ × ‘Beurré Clairgeau’), ‘Tosca’ (‘Coscia’ × ‘Bartlett’) and ‘Leopardo Morettini’ (‘Coscia’ × ‘Beurré Easter’) (Figure S1d). These provided further evidence to support the swapped identity of ‘Coscia’ and ‘Coscia Tardive’ at the repository.
The old cultivars ‘Beurré Gris’ and ‘Glou Morceau’ showed a PO relationship, however it is unclear which one originated first. ‘Beurré Gris’ (syn. ‘Beurré Brown’) might be as early as 1628 or could have originated in 1867 in France (Morgan 2015), while ‘Glou Morceau’ was released in 1759 and introduced to France in 1806 (Hedrick et al. 1921; Morgan 2015). ‘Glou Morceau’ is the parent of 43 accessions, which gave rise to two more generations of cultivars. ‘Rousselet de Reims’ (inferred as a synonym of ‘Petite Rousselet’) is the founder of a five generation-pedigree. This cultivar is centuries old, it may even date back to the Roman age (Hedrick et al. 1921). The old cultivar ‘Verte Longue d’Automne’, first mentioned in 1628, appeared to be an offspring of ‘Rousselet de Reims’ and ‘Bergamotte d’Automne’. This last one was first reported in 1536 and is the parent of ten cultivars and the founder of a four generation-pedigree. ‘Seckel’, found in the USA in the mid XVIII Century, was inferred as an offspring of ‘Rousselet de Reims’ and ‘White Doyenne’, and is itself a parent of 19 accessions. ‘Winter Nelis’, a Belgian cultivar from the early XIX Century, turned out to be an offspring of ‘Besi de La Motte’ (first reported in 1685), which originated from ‘Bergamotte d’Automne’. ‘Winter Nelis’ is the parent of 17 accessions.
‘Old Home’ was confirmed as the parent of the erroneously named rootstock series ‘Old Home × Farmingdale’ (OH×F), as well as of ‘Pyrodwarf’ (‘Old Home’ × ‘Conference’), OH 20 and OH 50 (‘Old Home’ × ‘Bartlett’), BU 2/33 –Pyro II (‘Old Home’ × ‘Glou Morceau’), OH 11 – Pyriam, and QR 708-2, QR 708-12 and QR 708-36 (BP-2 × ‘Old Home’). The pollen parent of the OH×F rootstocks was again confirmed to be ‘Bartlett’, as already reported by Postman et al. (2013), except for OH×F 247 and 512 that resulted from a cross between ‘Old Home’ × ‘Anjou’.
In P. pyrifolia, a high degree of inbreeding from the cultivar ‘Nijisseiki’ was observed, as previously reported (Nishio et al. 2016) (Figure S1e). Furthermore, several accessions here identified as hybrids between P. pyrifolia and P. ussuriensis are related to each other, with the cultivar ‘Hau Kai’ having a central role in their pedigree (Figure S1f). ‘Hau Kai’ is a very old cultivar from Liaoning (Northeast) China that turned out to be an offspring of ‘Tzu Ma Li’ × ‘Ba Li Shian’. ‘Man Yuan Xiang’, also an old cultivar from Northeast China, resulted a synonym of ‘Hau Kai’ (Table S2). Finally, two accessions were inferred to be the founders of all the P. betulaefolia held at the NCGR: P. betulaefolia OSU-3 (CPYR 1263.001) and OPR-114 P. betulaefolia No. 5 (identical to OPR-111 P. betulaefolia No. 2), parents of 15 and 19 accessions, respectively. Both of these accessions are seedling selections from seeds collected in China and brought to Oregon and implemented in the pear rootstock breeding program there. Accession CPYR 1255.001 was given the same name of P. betulaefolia OSU-3, however its genotype was identical to that of P. betulaefolia OPR-260 and it was inferred to be an offspring of CPYR 1263.001 (Table S2, Table S3).
Uncertainties remain for cultivars of commercial or breeding importance. For example, the pedigree of ‘Bosc’, one of the main cultivars in the US Pacific Coast, was not resolved, and doubts persist about the identity of ‘Louise Bonne d’Avranches’ (a.k.a. ‘Louise Bonne Jersey’) at the NCGR. Two accessions of ‘Louise Bonne d’Avranches’ and its panachee mutant were analyzed, and they all turned out to be different from each other. Accessions CPYR 2106.001 and CPYR 2106.002 are likely to be either sampling errors at the time of leaf collection, or mis-labeling at the repository, while the accession of the mutant panachee (CPYR 2491.001) was inferred to be parent of the cultivar ‘Princess’, which was indeed thought to be a seedling of ‘Louise Bonne d’Avranches’. However, it also appeared to be identical to ‘Marie Louise’. The identity of ‘Marie Louise’ is also uncertain, since it was confirmed as parent of ‘Laxton’s Early Market’, but not of ‘Marie Louise d’Uccle’ (Table S2).
Population structure
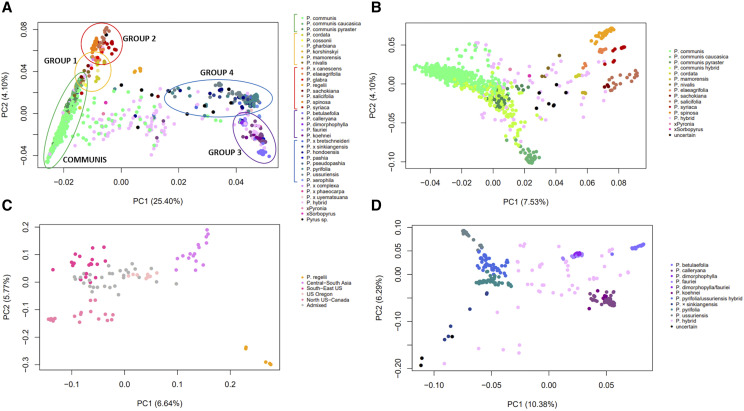
A total of 60,866 SNPs passed LD pruning and were used to examine the population structure and its consistency with the geographic-based grouping of the Pyrus species as reported in Challice and Westwood (1973) and in Montanari et al. (2019) (Table 2). The first four PCs explained, respectively, 25.40, 4.10, 3.87 and 2.42% of the overall genetic diversity (Table S4). The PC1 vs. PC2 plot (total of 29.5% of explained diversity; Figure 2a) depicted the two major groups of Occidental and Oriental pears, with a number of interspecific hybrids in between. Additionally, the three slightly overlapping clusters of the groups P. communis (including P. communis, P. communis subsp. caucasica and P. communis subsp. pyraster), Group 1 (species that are considered wild relatives of P. communis) and Group 2 (Middle East/Central Asia arid-adapted species) could be identified within the Occidental cluster, and the two groups Group 3 (East Asian “pea” pear species) and Group 4 (East Asian large-fruited cultivars and wild relatives) were distinguishable within the Oriental cluster.
Table 2. Classification of Pyrus species into different groups as reported in Montanari et al. (2019).
| Occidental species | Oriental species |
|---|---|
| Group Communis | Group 3 (East Asian “pea” pears) |
| Pyrus communis | Pyrus betulaefolia |
| Pyrus communis subsp. caucasica | Pyrus calleryana |
| Pyrus communis subsp. pyraster | Pyrus calleryana f. graciliflora |
| Group 1 (Europe, North Africa – P. communis wild relatives) | Pyrus dimorphophylla |
| Pyrus cordata | Pyrus fauriei |
| Pyrus cossonii | Pyrus koehnei |
| Pyrus gharbiana | Group 4 (East Asian large-fruited cultivars and wild relatives) |
| Pyrus korshinskyi | Pyrus ×bretschneideri |
| Pyrus mamorensis | Pyrus ×sinkiangensis |
| Pyrus nivalis | Pyrus hondoensis |
| Group 2 (Middle East/Central Asia arid-adapted species) | Pyrus pashia |
| Pyrus ×canescens | Pyrus pseudopashia |
| Pyrus elaeagrifolia | Pyrus pyrifolia |
| Pyrus glabra | Pyrus ussuriensis |
| Pyrus regelii | Pyrus xerophila |
| Pyrus sachokiana | |
| Pyrus salicifolia | |
| Pyrus spinosa | |
| Pyrus syriaca |
Figure 2.
Principal component analysis plots. PC1 vs. PC2 plots are shown for a) all accessions; b) Occidental accessions; c) Admixed accessions; and d) Oriental accessions. Colors are assigned based on the known species assignment for a, and on the new species assignment proposed in this study for b, c and d. The percentages of variation accounted for by each PC1 and PC2 are displayed on the axes. In plot a the major groups of species are shown with circles on the chart, and with bars on the legend.
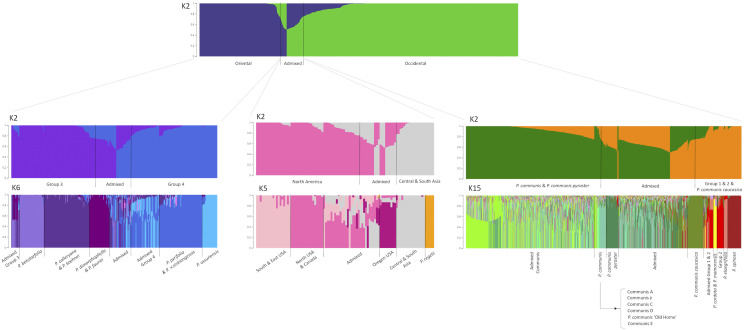
It was difficult to identify the optimal number of subpopulations on the overall population, therefore a hierarchical approach was applied. At K = 2, the two major groups of Occidental and Oriental pears were clearly identified, and the analysis was repeated for each of these subpopulations, as well as for the admixed group (Figure 3). Optimal values of K for the Occidental population were 12 to 15, according to the Evanno’s procedure (the fastSTRUCTURE algorithm for multiple choices of K gave uncertain results). At K = 2, one subpopulation included the pure P. communis and the P. communis subsp. pyraster samples, and the other one included P. communis subsp. caucasica and Group 1 and Group 2 accessions; a large number of admixed samples was found. At K = 15, the following subpopulations were identified: P. cordata with P. mamorensis; P. elaeagrifolia; P. spinosa; the rest of the Group 2 species (P. salicifolia, P. syriaca and P. sachokiana); P. communis subsp. caucasica; P. communis subsp. pyraster; six separate groups of pure P. communis; Group 1/Group2 hybrids; some more complex hybrids; and a large number of samples with admixture of different P. communis groups and subspecies. Within the Oriental population, at K = 2 the two subgroups Group 3 and Group 4 could be separated, and at the optimal number of K = 6 the following subpopulations were identified: P. betulaefolia; P. calleryana with P. koehnei; P. dimorphophylla with P. fauriei; P. ussuriensis; and P. pyrifolia with P. sinkiangensis. Similar to the Occidental group, samples with admixture of Group 3 species, samples with admixture of Group 4 species, and more complex hybrids admixed from the two groups were also found. Some subpopulations, apparently based on geographical origin, could be identified even among the Occidental/Oriental admixed. At K = 2, a group of North American and a group of Central and South Asian hybrids could be distinguished. At the optimal value of K = 5 a subpopulation for P. regelii could be separated from the Central and South Asian samples; a group of Northern USA and Canada hybrids and a group of South-Eastern USA hybrids could be identified within the North American subpopulation; and a group of accessions developed in Oregon, USA could be separated from the other admixed.
Figure 3.
Hierarchical population structure analysis plots. The first plot shows the structure of all samples at K = 2, and the plots below show the structure of the Oriental, the Admixed and the Occidental groups at K = 2 and at the respective optimal Ks.
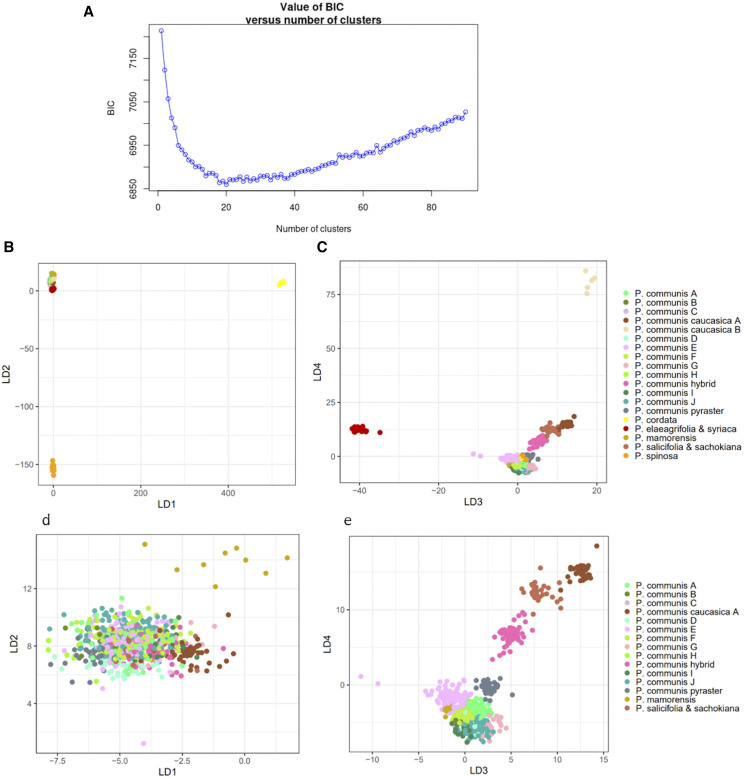
A DAPC was also run on the complex Occidental group, using 20 clusters, which was the number of clusters with the lowest BIC value (Figure 4a). Ten separate groups of P. communis were identified (P. communis A through J), two of P. communis subsp. caucasica (P. communis caucasica A and B) and then one each for P. communis hybrid; P. communis subsp. pyraster; P. cordata; P. elaeagrifolia with P. syriaca; P. mamorensis; P. salicifolia with P. sachokiana; and P. spinosa. The plot of discriminant functions 1 and 2 (LD1 vs. LD2) showed that P. cordata and P. spinosa were the most diverse groups (Figure 4b), while the LD3 vs. LD4 plot showed that the groups P. communis subsp. caucasica B and P. elaeagrifolia & syriaca were the most diverse (Figure 4c). Results were then plotted again after removal of these four groups, and while at the LD1 vs. LD2 plot the groups were indistinguishable, except for P. mamorensis (Figure 4d), at the LD3 vs. LD4 plot clusters for each group were more compact, although still largely overlapping, except for P. communis subsp. caucasica A, P. salicifolia & sachokiana, and P. communis hybrid.
Figure 4.
Discriminant analysis of principal components (DAPC) in the Occidental group. In a) the plot of BIC values vs. number of clusters; in b) the discriminant functions 1 vs. 2 (LD1 vs. LD2) plot and in c) the LD3 vs. LD4 plot for all groups identified with the DAPC; in d) the LD1 vs. LD2 plot and in e) the LD3 vs. LD4 plots for all groups identified with the DAPC excluding P. cordata, P. spinosa, P communis caucasica B and P. elaeagrifolia & syriaca.
Proposed new sample classification
Based on the results of the hierarchical structure and PC analysis, a new taxonomic classification was proposed for a number of accessions (Table S4). There were several accessions that resulted from hybridization between pure P. communis and its subspecies caucasica and pyraster, making their re-classification complicated. Several P. nivalis accessions appeared as mis-classified P. communis subsp. caucasica or P. communis hybrids. P. korshinskyi accessions appeared to be either P. communis subsp. caucasica or complex hybrids with various degrees of subsp. caucasica ancestry. Several accessions appeared mis-classified within Group 4. Here, two main subpopulations were identified, one for P. ussuriensis and one for P. pyrifolia; however, a number of samples that were assigned to P. ussuriensis, and a few assigned to P. pyrifolia, appeared to be hybrids between the two species. Additionally, approximately half of the P. bretschneideri and the P. hondoensis samples are likely hybrids between P. ussuriensis and P. pyrifolia, while the second half were reassigned to either one of the two species. Classification of P. sinkiangensis was rather difficult in this study. Of the eight samples analyzed, three were in the Occidental groups based on both analyses, and were reassigned to P. communis or P. communis subsp. pyraster; one was admixed between Oriental and Occidental; and four (of which two were PO related) formed a subpopulation with P. pyrifolia. The hierarchal structure analysis and PCA also allowed the inclusion in this species of two more samples that were mis-classified, bringing the number of putative P. sinkiangensis accessions to six. The small number of P. pashia, P. pseudopashia and P. xerophila samples appeared mis-classified.
Discussion
The high-density genotyping performed in this study gave relevant information for germplasm conservation and Pyrus taxonomic classification, and it enabled a large pedigree reconstruction for cultivars held at the NCGR. The Axiom Pear 70 K Genotyping Array (Montanari et al. 2019) was a highly useful and efficient tool for high-throughput genotyping in a diverse number of Pyrus species. To the best of our knowledge, this is the largest germplasm characterization study performed in pear, encompassing 1,331 unique genotypes across 36 species, interspecific and intergeneric hybrids, and one of the largest pedigree reconstruction efforts in perennial fruit species, being on the same scale of the recent work in apple by Muranty et al. (2020).
Genotyping tools are useful for optimization of conservation strategies at germplasm repositories
A large number of accessions that were collected in the wild or received from other germplasm repositories or donors from all over the world turned out to be identical to cultivars or accessions already present at the NCGR. All biological samples must undergo an expensive and time-consuming quarantine, pathogen testing and clean-up process before being released to the NCGR. Many of these efforts could have been avoided if synonymy or duplication with accessions already present at the NCGR collection was first determined by genotype comparison. Furthermore, potential labeling errors at the NCGR were flagged, and numerous previously unknown synonyms were discovered. Molecular markers could therefore provide a very useful tool to optimize material exchange between countries and avoid unnecessary expenses.
Faulty historical pedigrees and high degree of inbreeding
The number of errors in historical pedigree records of pear cultivars appeared to be very high, with approximately 80 trios and duos that showed inconsistencies compared to that reported in the literature or in passport data at the NCGR. This was somehow expected, as several cultivars analyzed in this study are ancient and documentation is vague. However, this also indicates the necessity of using molecular markers to confirm or elucidate the parentage of new cultivars to be released, as well as of accessions recurrently used in breeding programs. It is, however, important to underline that the method applied in this study for the pedigree reconstruction could be subject to a certain degree of error and, even if a stringent threshold was used in the final Mendelian test, certain FS and GPO relationships could have been mistakenly identified as PO, particularly in the case of inbreeding. For example, the two accessions OH×F 247 and OH×F 512 appeared offspring of ‘Old Home’ × ‘Anjou’, and therefore different from all other rootstock accession of the same series. Interestingly, ‘Anjou’ is the parent of ‘Farmingdale’, the previously claimed parent of the OH×F series (Postman et al. 2013), thus it is possible that Anjou has a GPO, instead of PO, relationship with OH×F 247 and 512. Additionally, when documented year and origin of cultivars were unreliable or unavailable, it was not possible to determine the direction of the duos with certainty. Notes about ambiguous results have been appropriately reported in Table S3.
We found a high degree of inbreeding among the P. communis cultivars analyzed, with a small number of old pear cultivars as the main founders, and the same scenario was observed for Oriental Pyrus species. Despite the recent inbreeding, however, pear species are still highly heterozygous, likely because of their history of self-incompatibility (Wu et al. 2013; Chagné et al. 2014; Volk and Cornille 2019).
A hierarchical population structure
The PC1 vs. PC2 plot for all the accessions (Figure 2a) showed a sample clustering very similar to what was observed in our previous work (Montanari et al. 2019) (where a smaller number of accessions was used), and mostly depicted the known groups of species identified by Challice and Westwood (1973). While the structure analysis reflected the results of the PCA, it also allowed a better representation of the genetic differentiation within some species groups (Figure 3). With both analysis, Occidental and Oriental accessions appeared genetically very different, reflecting their morphological diversity and their independent domestication events (Wu et al. 2018). These two major groups were themselves clusters of different subpopulations, which were depicted with the hierarchical approach.
The Occidental group included P. communis sensu lato and Groups 1 and 2, as described in Montanari et al. (2019) and in Table 2. Within the P. communis cluster, the two subspecies pyraster and caucasica formed two slightly distinct clusters closer to Group 1, although they appeared to largely overlap with several pure P. communis accessions (Figure 2b). In the structure analysis, these two subspecies formed their own subpopulations. While Challice and Westwood (1973) classified P. nivalis and P. cordata together with P. communis in the group “European species”, and P. cossonii, P. gharbiana and P. mamorensis together in the group “North African species” such classification was not confirmed, neither in the PCA nor from the structure analysis. In Montanari et al. (2019) all these species were assigned to Group 1 (Table 2), which formed a sparse cluster in between P. communis and Group 2 in the PC1 vs. PC2 plot (Figure 2b). In the structure analysis, the majority of P. cordata samples formed a subpopulation with P. mamorensis, while at the DAPC these two species could be clearly differentiated (Figure 4b). P. nivalis samples did not show a consistent organization, and only two samples each for P. cossonii and P. gharbiana were available and had complex hybrid structures, thus it was difficult to make any conclusion about these three species. Group 1 species P. korshinskyi also showed an unclear pattern. Group 2 formed a well-identifiable cluster in the PC1 vs. PC2 plot (Figure 2b), including the species P. elaeagrifolia, P. sachokiana, P. salicifolia, P. spinosa (syn. P. amygdaliformis) and P. syriaca. Challice and Westwood (1973) assigned all these species to the group “West Asian Species”, except for P. sachokiana. In the structure analysis, P. elaeagrifolia and P. spinosa stood out as two separate subpopulations, while P. sachokiana formed a subpopulation with P. salicifolia and P. syriaca. On the other hand, at the DAPC P. syriaca grouped with P. elaeagrifolia and not with P. salicifolia and P. sachokiana (Figure 4). Montanari et al. (2019) assigned to Group 2 also the species P. glabra and the hybrid P. canescens (Table 2). However, samples from P. glabra appeared admixed between Group 2 and P. communis subsp. caucasica, and the only one sample available for P. canescensand was likely a mis-classified P. communis accession. The species P. regelii, which was assigned to Group 2/“West Asian Species” (Table 2; Challice and Westwood (1973)), appeared to be quite distinct instead, forming its own cluster in between the Occidental and Oriental accessions (Figure 2a), as well as its own subpopulation among the Central and South Asian admixed group (Figure 2c and Figure 3).
The intergeneric hybrids that passed genotyping standards appeared to be admixed with a majority of Occidental ancestry. These included the Pyronia accession CIGC 9.001 (Pyronia veitchii), which was reported as a P. communis × Cydonia oblonga, and the Sorbopyrus accession CIGC 28.001 (Pollwiller Pear), which was reported as P. communis × Sorbus aria. However, the fact that these accessions easily passed the genotyping thresholds applied for the Pyrus species might be an indication that they either have very small proportions of Cydonia and Sorbus genomes, or that they were mis-classified and actually are interspecific hybrids of two (or more) Pyrus species.
Within the Oriental major group, Groups 3 and 4 formed two clearly distinguishable clusters and subpopulations (Figure 2d and Figure 3). According to both Challice and Westwood (1973) and Montanari et al. (2019), Group 3 included the species P. betulaefolia, P. calleryana, P. dimorphophylla, P. fauriei and P. koehnei, and Group 4 the species P. hondoensis, P. pashia, P. pyrifolia and P. ussuriensis (Table 2). P. betulaefolia appeared distinct from the other Group 3 species, and was located farther away from the domesticated Group 4 accessions in the PC1 vs. PC2 plot (Figure 2d), indicating a possible more ancestral origin for this species. P. calleryana and P. koehnei were genetically similar, and so were P. dimorphophylla and P. fauriei. While the close grouping of P. calleryana and P. koehnei is not surprising, as they are also morphologically very similar, P. dimorphophylla and P. fauriei have distinct phenotypic characters and originate in different countries (Japan vs. Korea). The structure of Group 4 was a little more unclear, which could however be attributed to mis-classification of several accessions. P. ussuriensis was distinguishable from the other species, although there were several samples grouping with P. pyrifolia or appearing as hybrids of Group 4 species. Most of the P. pyrifolia accessions formed a subpopulation with the few samples of P. sinkiangensis, one of the major cultivated species in Asia, which was not reported by Challice and Westwood (1973). P. hondoensis samples were spread across the Group 4 cluster in the PC1 vs. PC2 plot (Figure 2d), and in the structure analysis they either grouped with P. ussuriensis, or appeared admixed with a majority of Group 4 ancestry. Group 4 also included P. bretschneideri, P. pseudopashia and P. xerophila. P. bretschneideri accessions appeared either admixed between P. ussuriensis and P. pyrifolia, or they were part of the P. pyrifolia/ P. sinkiangensis subpopulation. The other two Group 4 species showed inconsistent structural organization, casting doubts on their taxonomic classification.
Finally, a large number of true interspecific hybrids between Occidental and Oriental species could be confirmed or newly identified. The structure analysis highlighted a certain genetic similarity among hybrids of common geographical origin (Central and Southern Asia, Northern USA and Canada, Southern and Eastern USA, and Oregon, USA), probably as a result of breeding programs based on interspecific crosses or targeting adaptation to specific environmental conditions (University of Tennessee Agricultural Experiment et al. 1954; Westwood and Lombard 1977; Peteršon and Waples 1988; Bassil et al. 2008; Bell and Itai 2011). The assignment of some interspecific hybrids to their own species, such as P. complexa, P. phaeocarpa and P. uyematsuana, is arguable.
Genetic diversity of the various species evaluated
Within the Occidental group, pure P. communis cultivars and accessions showed a wide diversity. Of all the 457 P. communis genotypes, the structure analysis separated only 39 of them into six distinguishable subpopulations (Communis A through E and Communis ‘Old Home’), which however could not be related to their geographic origin. On the other hand, the majority of the pure P. communis samples appeared to be admixed among these six subpopulations and were indicated as Admixed Communis in Figure 3. The P. communis accessions as a whole did not reveal a particular structure, as the attempt to identify subpopulations within them did not give any clear results (data not shown). The complexity of P. communis could not be resolved even with the DAPC, which returned ten different groups that, however, did not appear very diverse (Figure 4). There is confusion in the literature about subspecies caucasica and pyraster, which are considered by some as primary Pyrus species (Zheng et al. 2014; Wu et al. 2018), and by others as subspecies of P. communis (Challice and Westwood 1973; Asanidze et al. 2014). Our structure analysis suggested that they are genetically diverse from each other and from P. communis, enough to form their own subpopulations (Figure 3 and Figure 4), and they may therefore be considered as true species. P. communis subsps. caucasica and pyraster are believed to be the direct ancestors of the domesticated P. communis (Asanidze et al. 2014; Zheng et al. 2014), and the present study clearly showed that pyraster is more closely related to pure P. communis cultivars than caucasica (Figure 2b and Figure 4e). Challice and Westwood (1973) reported that several P. communis cultivars may also have originated from hybridization events between subspecies caucasica and pyraster with P. nivalis; however, it was not possible to confirm such hypothesis, since several P. nivalis accessions here evaluated appeared mis-classified, and the few remaining had an admixed ancestry between Group 2 species and P. communis, subsp. caucasica or subsp. pyraster (Table S4). It is worth noting, however, that the DAPC identified two separate clusters for P. communis subsp. caucasica (Figure 4), and one (P. communis caucasica B) was composed of accessions originally classified as P. nivalis.
Contradictory results were observed for the Group 1 species P. cordata and P. mamorensis, which appeared related to each other at the structure analysis (Figure 3), but very diverse at the DAPC (Figure 4b). Challice and Westwood (1973) believed that P. cordata had a central position in the evolution of Pyrus, being related to all Oriental and Occidental groups. However, such a unique connecting role could hardly be supported by the results of the present study. Analysis of more accessions from these two species will be necessary to better understand their relatedness and connection to other Pyrus species.
On the contrary to what Zheng et al. (2014) reported, we found P. elaeagrifolia was a well-defined species (Figure 3), although composed of two subgroups, one (CPYR 1482.001, 1483.001 and 1604.001) closer to P. communis and with a lower percentage of Oriental ancestry than the other one (Figure 2b). Also P. spinosa stood out as a subpopulation within the Occidental group, with its accessions being genetically very uniform, although they did not have any first-degree relationship with each other (Figure 1). P. korshinskyi accessions CPYR 2522.001 through 009 were re-assigned to P. communis subsp. caucasica by Volk et al. (2006). In our study, this classification was confirmed only for three of these accessions, while all other P. korshinskyi samples showed a more complex ancestry (although certainly involving subsp. caucasica), suggesting that it should not be considered as a true species. Challice and Westwood (1973) raised doubts about the classification of P. glabra as a true species as well, and its complex Group 2 and P. communis hybrid structure that resulted from the present study seems to confirm that hypothesis (Table S4). Finally, the species P. salicifolia and P. sachokiana were shown to be related, while P. syriaca might represent a connection between them and P. elaeagrifolia (Figure 2b, Figure 3 and Figure 4).
The classification of Group 3 was in accordance with what already reported by Challice and Westwood (1973), with P. betulaefolia the more clearly distinguishable species, P. calleryana related to P. koehnei, and P. dimorphophylla related to P. fauriei (which appeared as a true species, in disagreement with Wu et al. (2018)). However, it is possible that P. betulaefolia is the more ancient species within Group 3, as it is the most distant from the large-fruited Group 4 species (Figure 2d), which seems to be in disagreement with what was reported by Challice and Westwood (1973).
P. bretschneideri was long regarded as an interspecific hybrid (Challice and Westwood 1973; Zheng et al. 2014), and only recently had been reported as a true species (Wu et al. 2013, 2018). Results from the present study seems to reject the latter hypothesis, though, and in contrast support a P. ussuriensis P. pyrifolia origin of P. bretschneideri (Table S4). Similarly, P. hondoensis also appeared to be a P. ussuriensis P. pyrifolia hybrid. The conclusion that P. sinkiangensis is a hybrid between cultivated European and Asian pears (Wu et al. 2018) is not supported by this study, as only one out of eight analyzed samples had admixed ancestry. On the contrary, it appeared to be a Group 4 species, related to P. pyrifolia but distinct from it (Figure 2d and Figure 3). However, the number of P. sinkiangensis accessions was too low to make final conclusions about its origin and classification.
Very few samples were analyzed for P. pashia, P. pseudopashia and P. xerophila, and they all appeared either mis-classified or admixed, therefore preventing any further understanding of these species. All of the P. xerophila samples were seedlings from a single seedlot of uncertain provenance and tree phenotypes are consistent with that of P. pyrifolia hybrids.
The species P. regelii was probably the one that gave the most unexpected results. It was considered part of the “West Asian” (Group 2) species, although its morphology suggested it to be a divergent and more ancient species (Challice and Westwood 1973; Zheng et al. 2014). The structure analysis in the present study clearly showed P. regelii to have an admixed ancestry between Occidental and Oriental pears; however, it could be readily-separated from other hybrids and formed an unambiguous distinct subpopulation (Figure 2a, c and Figure 3). This is somehow in contrast with that reported by Wu et al. (2018), who suggested that the highly admixed ancestry of P. regelii was an indication of its re-classification as an “interspecies”, rather than a true species. In view of the structure analysis, it is more likely that P. regelii is a true species that resulted from hybridization of ancestral Oriental and Occidental pears and remained isolated, or a connecting link between the two major groups of species.
Conclusions
This is the first study that genetically characterized the entire Pyrus germplasm collection held at the NCGR, one of the largest Pyrus repository in the world. The in depth genotyping performed with the Axiom Pear 70 K Genotyping Array (Montanari et al. 2019) allowed the identification of several duplicated samples in the collection. Those that have been flagged as possible sampling errors will be verified by comparison of the morphology of the original trees at the NCGR collection and/or by SSR fingerprinting, as in Montanari et al. (2019). This information will be particularly useful for the optimization of the conservation strategy at the repository. Additionally, by analyzing a large number of samples, this study was able to reconstruct the parentage (or partial parentage) of 637 accessions, giving insights into the level of inbreeding in cultivated pear. Pear breeders across the world will be able to use this extended pedigree to make more informed decisions in their crossing schemes, while maximizing efforts to maintain diversity within their programs. The population structure analysis, made possible by the high quality of the SNPs included in the Axiom Pear 70 K Genotyping Array, enabled the re-classification of a large number of accessions and improved our understanding of the genetic diversity of Pyrus species. Further analysis of this dataset in conjunction with morphological and phenological data will be performed to better evaluate the genetic diversity of the different Pyrus species. Phylogeneticists and taxonomists can build on the information reported here to better elucidate the evolution and domestication of pear.
Additional Information
We found that the large and complex pedigree network built in this work was better represented with the software Helium (Shaw et al. 2014), which allows interactive visualization. The software is downloadable for free at https://github.com/cardinalb/helium-docs/wiki and the input files are given in Files S1-S5.
Acknowledgments
This project was funded by the California Pear Advisory Board and the Pear Pest Management Research Fund. The funding body had no role in the design of the study, collection, analysis, or interpretation of data, or in writing the manuscript. We wish to acknowledge Rachel Elkins (UC Cooperative Extension) for her help and support during the entire project. We also wish to thank Brian J. Allen (UC Davis) for his contribution to the leaf collection and DNA extractions and Jason Zurn (USDA-ARS) for his suggestions on the genetic diversity analysis. Finally, we would like to acknowledge the Natural Park of Paneveggio Pale di San Martino (TN, Italy) and Michela Troggio and Luca Bianco (Fondazione Edmund Mach), as well as Charles-Eric Durel and Caroline Denance (INRA, Angers) for providing leaf material of local landraces and cultivars.All authors contributed to the study conception and design. Material preparation was performed by Sara Montanari, Joseph Postman and Nahla Bassil. Data collection and analysis were performed by Sara Montanari. The first draft of the manuscript was written by Sara Montanari and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12186105.
Communicating editor: I. Parkin
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arab M. M., Marrano A., Abdollahi-Arpanahi R., Leslie C. A., Askari H. et al. , 2019. Genome-wide patterns of population structure and association mapping of nut-related traits in Persian walnut populations from Iran using the Axiom J. regia 700K SNP array. Sci. Rep. 9: 6376 10.1038/s41598-019-42940-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanidze Z., Akhalkatsi M., Henk A. D., Richards C. M., and Volk G. M., 2014. Genetic relationships between wild progenitor pear (Pyrus L.) species and local cultivars native to Georgia, South Caucasus. Flora 209: 504–512. 10.1016/j.flora.2014.06.013 [DOI] [Google Scholar]
- Bassil N., and Postman J. D., 2010. Identification of European and Asian pears using EST-SSRs from Pyrus. Genet. Resour. Crop Evol. 57: 357–370. 10.1007/s10722-009-9474-7 [DOI] [Google Scholar]
- Bassil N., Postman J., Hummer K., Dolan S., and Lawliss L., 2008. Molecular fingerprints identify historic pear trees in two U.S. National Parks. Acta Hortic. (800): 417–422. 10.17660/ActaHortic.2008.800.52 [DOI] [Google Scholar]
- Bassil N. V., Postman J. D., and Neou C., 2005. Pyrus microsatellite markers from GenBank sequences. Acta Hortic. (671): 289–292. 10.17660/ActaHortic.2005.671.41 [DOI] [Google Scholar]
- Bell R. L., and Itai A., 2011. Pyrus, pp. 147–179 in Wild Crop Relatives: Genomic and Breeding Resources: Temperate Fruits, edited by Kole C. Springer Science+Business Media, Berlin, Germany. [Google Scholar]
- Bell R. L., Van Der Zwet T., Castagnoli S., Einhorn T., Turner J. D. et al. , 2014. ‘Gem’ Pear. HortScience 49: 361–363. 10.21273/HORTSCI.49.3.361 [DOI] [Google Scholar]
- Bink M. C. A. M., Boer M. P., Braak C. J. F., Jansen J., Voorrips R. E. et al. , 2007. Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 161: 85–96. 10.1007/s10681-007-9516-1 [DOI] [Google Scholar]
- Butts C. T., 2008. network: a package for managing relational data in R. J. Stat. Softw. 24: 1–36. 10.18637/jss.v024.i0218612375 [DOI] [Google Scholar]
- Cellon C., Amadeu R. R., Olmstead J. W., Mattia M. R., Ferrao L. F. V. et al. , 2018. Estimation of genetic parameters and prediction of breeding values in an autotetraploid blueberry breeding population with extensive pedigree data. Euphytica 214: 87 10.1007/s10681-018-2165-8 [DOI] [Google Scholar]
- Chagné D., Crowhurst R. N., Pindo M., Thrimawithana A., Deng C. et al. , 2014. The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS One 9: e92644 10.1371/journal.pone.0092644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challice J. S., and Westwood M. N., 1973. Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot. J. Linn. Soc. 67: 121–148. 10.1111/j.1095-8339.1973.tb01734.x [DOI] [Google Scholar]
- Chevreau E., Evans K. M., Chagné D., and Montanari S., 2020. Pyrus spp. pear and Cydonia spp. quince, pp. 581–605 in Biotechnology of fruit and nut crops, edited by Litz R. E., Pliego-Alfaro F., and Hormaza J. I.. CABI Publishing, Oxfordshire, England: 10.1079/9781780648279.0581 [DOI] [Google Scholar]
- Drain, B. D., and G. A. Shuey, 1954 Breeding and testing fire blight-resistant pears. University of Tennessee Agricultural Experiment Station Bulletins 236: 1–20.
- Evanno G., Regnaut S., and Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14: 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Evans K. M., and Fernández-Fernández N., Bassil N., Nyberg A. and Postman J., 2015. Comparison of accessions from the UK and US National Pear Germplasm Collections with a standardized set of microsatellite markers. Acta Hortic. (1094): 41–46. 10.17660/ActaHortic.2015.1094.2 [DOI] [Google Scholar]
- Guzman D., 2018. Evaluation of pre-breeding resources for Pyrus spp. pp 1–129: Washington State University, Pullman, WA. [Google Scholar]
- Hedrick, U. P., G. H. Howe, and O. M. Taylor, F. E. H., and H. B. Tukey, 1921 The pears of New York. J.B. Lyon Company, Printers, Albany. [Google Scholar]
- Hinze L. L., Hulse-Kemp A. M., Wilson I. W., Zhu Q. H., Llewellyn D. J. et al. , 2017. Diversity analysis of cotton (Gossypium hirsutum L.) germplasm using the CottonSNP63K Array. BMC Plant Biol. 17: 37 10.1186/s12870-017-0981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob H. B., 1998. Pyrodwarf, a new clonal rootstock for high density pear orchards. Acta Hortic. (475): 169–178. 10.17660/ActaHortic.1998.475.20 [DOI] [Google Scholar]
- Jakobsson M., and Rosenberg N. A., 2007. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jombart T., 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jombart T., and Ahmed I., 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Devillard S., and Balloux F., 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11: 94 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Oh Y., Han H., Oh S., Lim H. et al. , 2019. Genetic relationships and population structure of pears (Pyrus spp.) assessed with genome-wide SNPs detected by genotyping-by-sequencing. Hortic. Environ. Biotechnol. 60: 945–953. 10.1007/s13580-019-00178-w [DOI] [Google Scholar]
- Kouassi A. B., Durel C. E., Costa F., Tartarini S., van de Weg E. et al. , 2009. Estimation of genetic parameters and prediction of breeding values for apple fruit-quality traits using pedigreed plant material in Europe. Tree Genet. Genomes 5: 659–672. 10.1007/s11295-009-0217-x [DOI] [Google Scholar]
- Kumar S., Garrick D. J., Bink M. C., Whitworth C., Chagné D. et al. , 2013. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genomics 14: 393 10.1186/1471-2164-14-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Kirk C., Deng C., Wiedow C., Knaebel M. et al. , 2017. Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hortic. Res. 4: 17015 10.1038/hortres.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemopoulos A., Prokkola J. M., Uusi-Heikkilä S., Vasemägi A., Huusko A. et al. , 2019. Comparing RADseq and microsatellites for estimating genetic diversity and relatedness — Implications for brown trout conservation. Ecol. Evol. 9: 2106–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmith G., Rombauts S., Montanari S., Deng C. H., Celton J. M. et al. , 2019. Pseudo-chromosome-length genome assembly of a double haploid “Bartlett” pear (Pyrus communis L.). Gigascience 8: 1–17. 10.1093/gigascience/giz138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sale M. et al. , 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke E. A., and Smith L., 2002. Evaluation of the Horner rootstocks. Acta Hortic. (596): 325–330. 10.17660/ActaHortic.2002.596.50 [DOI] [Google Scholar]
- Montanari S., Bianco L., Allen B. J., Martínez-garcía P. J., Bassil N. V. et al. , 2019. Development of a highly efficient Axiom 70 K SNP array for Pyrus and evaluation for high-density mapping and germplasm characterization. BMC Genomics 20: 331 10.1186/s12864-019-5712-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J., 2015. The Book of Pears: The Definitive History and Guide to Over 500 Varieties, edited by Dobell S. Chelsea Green Publishing, Vermont. [Google Scholar]
- Muranty H., Denancé C., Feugey L., Crépin J., Barbier Y. et al. , 2020. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 20: 2 10.1186/s12870-019-2171-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio S., Takada N., Saito T., Yamamoto T., and Iketani H., 2016. Estimation of loss of genetic diversity in modern Japanese cultivars by comparison of diverse genetic resources in Asian pear (Pyrus spp.). BMC Genet. 17: 81 10.1186/s12863-016-0380-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini E., Civolani S., Musacchi S., Ancarani V., Dondini L. et al. , 2006. Cacopsylla pyri behaviour on new pear selections for host resistance programs. Bull. Insectol. 59: 27–37. [Google Scholar]
- Peteršon R., and Waples J., 1988. ‘Gourmet’ Pear. HortScience 23: 633. [Google Scholar]
- Piaskowski J., Hardner C., Cai L., Zhao Y., Iezzoni A. et al. , 2018. Genomic heritability estimates in sweet cherry reveal non-additive genetic variance is relevant for industry-prioritized traits. BMC Genet. 19: 23 10.1186/s12863-018-0609-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postman J. D., 2008a The USDA quince and pear genebank in Oregon, a world source of fire blight resistance. Acta Hortic. (793): 357–362. 10.17660/ActaHortic.2008.793.53 [DOI] [Google Scholar]
- Postman J. D., 2008b World Pyrus collection at USDA genebank in Corvallis, Oregon. Acta Hortic. (800): 527–533. 10.17660/ActaHortic.2008.800.69 [DOI] [Google Scholar]
- Postman J., Kim D., and Bassil N., 2013. OH x F paternity perplexes pear producers. J. Am. Pomol. Soc. 67: 157–167. [Google Scholar]
- Rafalski A., 2002. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5: 94–100. 10.1016/S1369-5266(02)00240-6 [DOI] [PubMed] [Google Scholar]
- Raj A., Stephens M., and Pritchard J. K., 2014. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197: 573–589. 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy R. K., Ramasamy S., Bindroo B. B., and Naik V. G., 2014. STRUCTURE PLOT: A program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus 3: 431 10.1186/2193-1801-3-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufo R., Alvaro F., Royo C., and Soriano J. M., 2019. From landraces to improved cultivars: Assessment of genetic diversity and population structure of Mediterranean wheat using SNP markers. PLoS One 14: e0219867 10.1371/journal.pone.0219867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura Y., Takada N., Yamamoto T., Saito T., Kimura T. et al. , 2008. Identification of parent-offspring relationships in 55 Japanese pear cultivars using S-RNase allele and SSR markers. J. Jpn. Soc. Hortic. Sci. 77: 364–373. 10.2503/jjshs1.77.364 [DOI] [Google Scholar]
- Shaw P. D., Graham M., Kennedy J., Milne I., and Marshall D. F., 2014. Helium: visualization of large scale plant pedigrees. BMC Bioinformatics 15: 259 10.1186/1471-2105-15-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard M. H., and Michelesi J. C., 2002. “Pyriam”: a new pear rootstock. Acta Hortic. (596): 351–355. 10.17660/ActaHortic.2002.596.54 [DOI] [Google Scholar]
- Urrestarazu J., Royo J. B., Santesteban L. G., and Miranda C., 2015. Evaluating the influence of the microsatellite marker set on the genetic structure inferred in Pyrus communis L. PLoS One 10: e0138417 10.1371/journal.pone.0138417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk G. M., and Cornille A., 2019. Genetic diversity and domestication history in Pyrus, pp. 51–62 in The Pear Genome, edited by Korban S. S. Springer International Publishing, Cham: 10.1007/978-3-030-11048-2_3 [DOI] [Google Scholar]
- Volk G. M., Henk A. D., Richards C. M., Bassil N., and Postman J., 2019. Chloroplast sequence data differentiate Maleae, and specifically Pyrus, species in the USDA-ARS National Plant Germplasm System. Genet. Resour. Crop Evol. 66: 5–15. 10.1007/s10722-018-0691-9 [DOI] [Google Scholar]
- Volk G. M., Richards C. M., Henk A. D., and Rillery A. A., 2006. Diversity of wild Pyrus communis based on microsatellite analyses. J. Am. Soc. Hortic. Sci. 131: 408–417. 10.21273/JASHS.131.3.408 [DOI] [Google Scholar]
- Westwood M. N., and Lombard P. B., 1977. Pear rootstock and Pyrus research in Oregon. Acta Hortic. (69): 117–122. 10.17660/ActaHortic.1977.69.14 [DOI] [Google Scholar]
- Wu J., Wang Z., Shi Z., Zhang S., Ming R. et al. , 2013. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23: 396–408. 10.1101/gr.144311.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang Y., Xu J., Korban S. S., Fei Z. et al. , 2018. Diversification and independent domestication of Asian and European pears. Genome Biol. 19: 77 10.1186/s13059-018-1452-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W., Luo T., Zhang W., Mason A. S., Huang D. et al. , 2019. Development of high-density snp markers and their application in evaluating genetic diversity and population structure in Elaeis guineensis. Front. Plant Sci. 10: 130 10.3389/fpls.2019.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Cai D., Potter D., Postman J., Liu J. et al. , 2014. Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol. Phylogenet. Evol. 80: 54–65. 10.1016/j.ympev.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Zheng X., Levine D., Shen J., Gogarten S. M., Laurie C. et al. , 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurn J. D., Norelli J. L., Montanari S., Bell R., and Bassil N. V., 2020. Dissecting Genetic Resistance to Fire Blight in Three Pear Populations. Phytopathology 110: 1305–1311. 10.1094/PHYTO-02-20-0051-R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental data (Tables S1-S4; Figure S1; Files S1-S5) are provided through figshare. The genotyping data for the 1,749 samples that passed genotyping standards and 64,571 SNPs that had unique, high-quality alignment to the DH Bartlett Genome and that were classified as PHR are provided through the Genome Database for Rosaceae (GDR, https://www.rosaceae.org/, accession number tfGDR1042). Supplemental material available at figshare: https://doi.org/10.25387/g3.12186105.