Abstract
Corona virus disease-2019 (COVID-19) is a pandemic caused by novel coronavirus. COVID-19 is spreading rapidly throughout the world. The gold standard for diagnosing COVID-19 is reverse transcription-polymerase chain reaction (RT-PCR) test. However, the facility for RT-PCR test is limited, which causes early diagnosis of the disease difficult. Easily available modalities like X-ray can be used to detect specific symptoms associated with COVID-19. Pre-trained convolutional neural networks are widely used for computer-aided detection of diseases from smaller datasets. This paper investigates the effectiveness of multi-CNN, a combination of several pre-trained CNNs, for the automated detection of COVID-19 from X-ray images. The method uses a combination of features extracted from multi-CNN with correlation based feature selection (CFS) technique and Bayesnet classifier for the prediction of COVID-19. The method was tested using two public datasets and achieved promising results on both the datasets. In the first dataset consisting of 453 COVID-19 images and 497 non-COVID images, the method achieved an AUC of 0.963 and an accuracy of 91.16%. In the second dataset consisting of 71 COVID-19 images and 7 non-COVID images, the method achieved an AUC of 0.911 and an accuracy of 97.44%. The experiments performed in this study proved the effectiveness of pre-trained multi-CNN over single CNN in the detection of COVID-19.
Keywords: COVID-19, X-ray, CNN, Bayesnet, Multi-CNN
1. Introduction
Coronavirus disease 2019 (COVID-19) is a kind of viral pneumonia which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It is one among the three respiratory disease outbreak caused by the coronavirus, other two being severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). As on 7th of May 2020, more than 3.5 million cases of COVID-19 and 250,000 deaths due to the disease have been reported by the World Health Organization (WHO) [1]. WHO has listed COVID-19 as a Public Health Emergency of International Concern (PHEIC) [2]. There is an urgent need for early diagnosis of the disease to prevent further spreading and control the death toll. The gold standard for diagnosing COVID-19 is reverse transcription-polymerase chain reaction (RT-PCR) test [2], [3]. However, the RT-PCR testing facility is inadequate in most of the areas hit by the COVID-19 outbreak [3]. COVID-19 is characterized by a lung infection in the post of the patients [4]. Easily available modalities like X-ray and CT can be used for detecting lung infections [4]. It is proven that X-ray and computed tomography (CT) scan can be used effectively for the diagnosis of COVID-19 [5]. However, manual reading of X-ray and CT scan of a large number of patients could be time-consuming. A computer-aided diagnosis method could assist the radiologists in predicting COVID-19 from X-ray and CT-scan images [3].
Convolutional neural network (CNN) has shown promising results in the area of computer-aided detection and diagnosis of various diseases. CNN requires a large amount of data for training from scratch. In the case of medical images, it is difficult to obtain a huge number of labelled images. In such cases pre-trained CNNs trained on a large number of natural images like ImageNet can be used [6]. Pre-trained CNNs were earlier used successfully in diagnosis of prostate cancer [7], [8], breast cancer [9], brain diseases [10], leukemia [11], etc. to name a few. Pre-trained CNN is also found successful in predicting COVID-19 [12], [13], [14]. This paper presents a method for the prediction of COVID-19 using features extracted from multiple pre-trained networks.
The paper is organized as follows. Section 1.1 discusses the related works in the area of computer-aided detection of COVID-19. Section 1.2 describes the contributions of the proposed method. Section 2 describes the proposed multi-CNN, feature selection technique and classifier. Section 3 discusses the results achieved using various combinations of multi-CNN. The section also analyzes the results achieved using various classifiers in comparison with the proposed classifier. Section 4 explains the conclusions reached based on the experimental analysis.
1.1. Related works
Recently, a number of papers were published in the area of computer-aided detection of COVID-19 using pre-trained CNNs from X-ray and CT images. Shi et al. [3] performed a detailed review of the state-of-the-art computer-aided techniques for the detection of COVID-19 from X-ray and CT scans. Narin et al. [12] used Resnet-50 for predicting COVID-19 from a balanced set of 50 COVID-19 and 50 non-COVID cases. Castiglioni et al. [13] used Resnet-50 for classification of COVID-19 and non-COVID instances using a balanced dataset of 250 COVID-19 cases and 250 non-COVID cases. Hemdan et al. [14] employed Densenet to predict COVID-19 from a balanced dataset of 25 COVID-19 and 25 non-COVID images. Panwar et al. [15] proposed a transfer learning based model, nCOVnet, which adds 5 custom layers to VGG 16 network. The method used 142 COVID-19 images and 142 normal images. Pereira et al. [16] used features extracted using InceptionV3 in combination with texture features extracted using local binary pattern (LBP), elongated quinary patterns (EQP), local directional number (LDN), locally encoded transform feature histogram (LETRIST), binarized statistical image features (BSIF), local phase quantization (LPQ) and oriented basic image features (OBIFs). Training data was resampled in order to solve the class imbalance problem. The method used multi-class classification to classify images into COVID-19, normal, MERS, SARS, Varicella, Streptococcus and Pneumocystis. The total samples used were 1144 out of which 90 images belong to COVID-19 category. Toraman et al. [17] used a capsule network with 4 convolution layers and a primary capsule layer. The method used 231 COVID-19 images, 1050 pneumonia images and 1050 images with no-findings.
The above-mentioned methods use X-ray images for the computer-aided detection of COVID-19. Few recent works also prove the effectiveness of CT scans in the detection of COVID-19. He et al. [18] performed transfer learning on a CT dataset, containing 349 COVID-19 CT scans and 397 normal CT scans. The method proposed a novel transfer learning technology called self-trans that learns features that are robust to overfitting. Mei et al. [19] used two CNN models (one for slice selection and another one for diagnosis) in combination with clinical data to predict COVID-19 using CT scans. Shan et al. [20] proposed a deep learning based scheme that uses 'VB-Net' , a modification of V-Net architecture for the segmentation of COVID-19 affected areas in chest CT scans. The method used CT scans of 249 subjects for training and 300 subjects for validation. Chen et al. [21] proposed a Residual Attention U-Net for the segmentation of COVID-19 affected areas in CT scans. The dataset used for the method contains 110 CT images. Chen et al. [22] further proposed a contrastive learning technique to train an encoder for the detection of COVID-19 from CT scans. Fan et al. [23] proposed a model that employs implicit reverse attention and explicit edge-attention for the segmentation of COVID-19 infected areas in CT scans.
In this work, we have chosen to use X-ray for COVID-19 detection as X-ray is cost-effective compared to CT scans.
1.2. Contribution of the proposed method
The aforementioned methods have used a single pre-trained CNN to predict COVID-19 from a balanced dataset. The discriminatory features extracted from each pre-trained network will be different. Combination of features extracted from pre-trained CNN is expected to improve the performance of computer-aided diagnosis systems. The proposed method explores a combination of features from multiple CNNs for the diagnosis of COVID-19.
The proposed method has the following contributions:
-
i.
The method uses a multi-CNN comprising of several pre-trained CNNs for the extraction of features from chest X-ray images for the diagnosis of COVID-19. Most of the existing methods have used a single pre-trained CNN.
-
ii.
The method uses a combination of multi-CNN with correlation-based feature selection (CFS) and Bayesnet classifier. No existing state-of-the-art methods have employed multi-CNN, CFS based feature selection and Bayesnet classifier for the diagnosis of COVID-19.
-
iii.
Most of the existing works have used a small dataset whereas the proposed method used a relatively large number of COVID-19 cases.
2. Materials and methods
2.1. Dataset
The method was implemented using two COVID-19 public datasets. The first dataset created by Cohen et al. [24] consists of 560 chest X-ray images. The 560 chest X-ray images were composed of 453 COVID-19 and 107 non-COVID images. 107 non-COVID images consists of either bacterial pneumonia or viral pneumonia. Images with no findings were excluded. 390 chest X-ray images of viral and bacterial pneumonia taken from a Kaggle dataset [25], [26] were added to the 107 non-COVID images to make the dataset a balanced one. The combined dataset (DATASET-1) consists of 950 images of which 453 are COVID-19 and 497 are non-COVID.
The second dataset consisting of 71 COVID-19 and 7 non-COVID chest X-ray images was taken from Kaggle [27]. The sub classification of non-COVID images are not available. For performing experimental analysis Dataset-1 was used due to its larger size. To confirm robustness of the method, the results of both Dataset-1 and Dataset-2 are considered. Fig. 1 show sample COVID-19 X-ray images.
Fig. 1.
The four X-ray images in first row corresponds to COVID-19 and the images in second row corresponds to non-COVID.
2.2. Feature extraction
The multi-CNN architecture used in the proposed method consists of a set of pre-trained CNN. Each CNN used in the method was pre-trained using Imagenet [28] which has more than a million natural images belonging to 1000 different classes. Different combinations of pre-trained CNNs are used for feature extraction. CNN consists of three basic layers: convolution, pooling and fully connected layers [29]. Convolution layers perform feature extraction by convolving the input image with a set of learned kernels. The layer typically consists of a combination of convolution operation and activation function. The convolution operation between an image I of dimension p × q and a kernel W of size x × y that produces a feature map s is defined by the following dot product:
| (1) |
The output of the convolution layer is then passed through a non-linear activation function. The most common non-linear activation function used is the rectified linear unit (ReLU) and its variant Leaky ReLu. ReLu is represented as:
| (2) |
where is the input at location (i, j) on the nth feature map at kth layer. Leaky ReLu is represented as:
| (3) |
where is the input at location (i, j) on the nth feature map at kth layer and β is the slope of negative linear function [30].
The pooling layer is used to reduce the spatial resolution of the activation map and thereby reducing the number of parameters. Pooling helps to decrease the computational cost and over-fitting. Max-pooling and average pooling are the most common methods of pooling.
Every neuron in the previous layer are connected to a fully connected (FC) layer. Features generated by the previous layer are flattened in a feature vector by the FC layer. It then performs weight updates to improve the predicting ability of feature vector.
Densenet201 [31], InceptionResnetV2 [32], Shufflenet [33], Resnet-101 [34], Darknet-53 [35], MobilenetV2 [36], NasnetLarge [37], Xception [38], VGG-19 [39] and Squeezenet [40] are the pre-trained networks used for experimental analysis in this work. The input X-ray images used in this study are of different formats. The dimension of input images also varies. The input size of Densenet-201, Shufflenet, MobilenetV2, VGG-19 and Resnet-101 are 224 × 224 whereas that of InceptionResnetV2 and Xception are 299 × 299. The input size of Nasnetlarge, Darknet-53 and Squeezenet are 331 × 331, 256 × 256 and 227 × 227, respectively. The size of the X-ray images present in the datasets vary. Before passing the images to the pre-trained networks a preprocessing is done to make the size of images uniform and also to replicate the colour channels of the grayscale images in the dataset. The images are scaled using bilinear interpolation to make them compatible with input size of the pre-trained network. Features are extracted from the last fully connected layer of the pre-trained CNNs with 1000 neurons. Each pre-trained CNN produces a feature matrix of size n × 1000, where n is the number of X-ray images. The feature matrices of the muti-CNN are combined together to form a feature matrix of dimension n × 1000m, where m is the number of pre-trained networks used in the multi-CNN. The best performing multi-CNN of this study used 5 pre-trained CNNs (Squeezenet, Darknet-53, MobilenetV2, Xception, Shufflenet) to produce a feature matrix of dimension 950 × 5000.
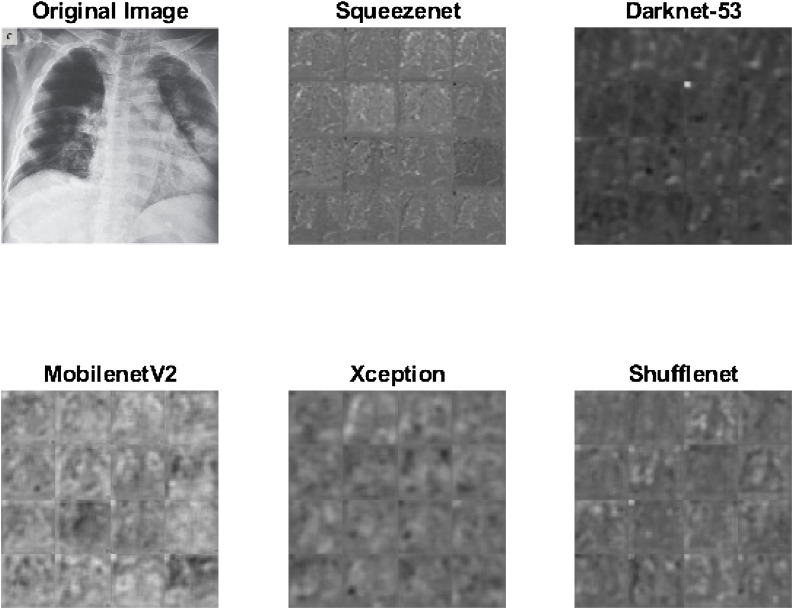
Sample activations using last convolutional layer of Squeezenet, Darknet-53, MobilenetV2, Xception and Shufflenet is shown in Fig. 2 .
Fig. 2.
Sample activations. Each image contains sixteen tiles corresponding to sample activations of the original image.
2.3. Feature selection
Feature matrix of size n × 1000m is passed to a feature selection unit for selecting the most distinguishing features. A correlation-based feature selection (CFS) algorithm [41] in combination with subset size forward selection (SSFS), a linear forward selection based search technique [42] was utilized to determine the optimal feature subset. CFS evaluates the merit of a subset of features by considering the individual predictive ability of each attribute along with the degree of redundancy among them. Merit of a subset of features is given by:
| (4) |
where n is the number of features present in the subset, r fc is the mean feature correlation and r ic is the average value of feature intercorrelation. The numerator of the equation represents ability of a set of features in predicting a class whereas the denominator indicates redundancy among them. After computing the merit of subset of features, SSFS based search is performed. SSFS performs an interior cross-validation to determine the effectiveness of feature subsets. A linear forward selection (LFS) is performed on each fold. To estimate the optimal subset size, scores achieved on the test data for each subset size are averaged and subset size with highest average is chosen. Search terminates at the optimal subset size. Finally, a linear forward selection up to the optimal size of subset is conducted on the whole data.
CFS in combination with SSFS reduces the dimensionality of features from n × 1000m to n × p, where p is the reduced number of features. The algorithm reduced the dimension of feature vector corresponding to the best performing multi-CNN of this study from 950 × 5000 to 950 × 45.
2.4. BayesNet classifier
Consider a set of variables, V = m 1, …, m n, where n ≥ 1. A Bayesian network G over a set of variables V represents a network structure G S, and a set of probability tables G P [43]. G S is a directed acyclic graph (DAG) over V.
| (5) |
where is the set of parents of in G S. A Bayesian network indicates the following probability distribution [43].
| (6) |
Let m = m 1, m 2, m 3, …, m n be a set of attribute variables. A classifier q : m → c is a function that maps an instance of m to a class c. For using Bayesian network as a classifier, argmax c P(c|m) is computed using the distribution P(V) [43].
The feature matrix of dimension n × p is passed to a Bayesnet classifier which classifies the images into COVID-19 and non-COVID categories.
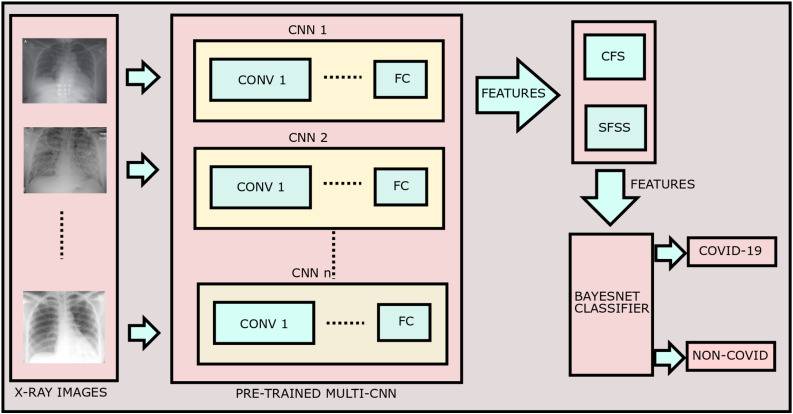
Architecture of the proposed method is shown in Fig. 3 .
Fig. 3.
Architecture of the proposed method.
3. Results and discussions
3.1. Experimental setup
The experiments were performed using a core i7, GTX 1060 6GB GPU. Feature extraction was performed using MATLAB 2020a and classification using Weka 3.6. Area under the receiver operating characteristic curve (AUC) along with accuracy are used as the major performance metrics. Precision, recall and F-measure in predicting COVID-19 class are used as the auxilliary performance measures.
3.2. Classification results
A ten-fold cross-validation was performed on both the datasets. The results achieved are shown in Table 1 . Confusion matrices corresponding to the results are given in Fig. 4 . ROC curves corresponding to results are shown in Fig. 5 . The method achieved considerable performance in both the datasets. 446 among the 453 instances of COVID-19 cases were classified correctly in Dataset-1, achieving a recall of 98.5%. One except all among the 71 COVID-19 cases were predicted correctly in Dataset-2 achieving a recall of 98.6%. The recall obtained in predicting non-COVID cases were 84.5% and 85.7% respectively, in Dataset-1 and Dataset-2. The method achieved a precision of 85.3% and 98.4% respectively, in predicting COVID-19 and non-COVID images of Dataset-1. It further achieved a precision of 98.6% and 85.7% in predicting COVID-19 and non-COVID images of Dataset-2. Feature extraction, feature selection and training the classifier using Dataset-1 required a computational time of 165.33 s. Testing using 10% of the images (96 images) required only 15.8136 s. Feature extraction, feature selection and training the classifier using Dataset-1 required a computational time of 18.91 s. Testing using 10% of the images of DATASET-2 required only 3.02 s.
Table 1.
Results achieved using various datasets.
Fig. 4.
Confusion matrices corresponding to Dataset-1 and Dataset2. The bottom-most diagonal elements indicated in yellow colour represent accuracy. Elements in right most columns represent recall and bottom most rows represent precision.
Fig. 5.
ROC curves corresponding to results achieved in Dataset-1 and Dataset-2.
3.3. Parameter setting
Default parameter settings were used for all the pre-trained networks used for creating multi-CNN.
Table 2, Table 3 displays the parameter settings of the feature selection method and Bayesnet classifier in WEKA. The algorithm used to determine the subset size was set to sequential minimal optimization (SMO). The number of cross-validation folds used for subset size determination was set to 5. The algorithm for determining the conditional probability tables of the Bayes network was set to simple estimator algorithm. Parameter alpha used for determining the conditional probability tables of the Bayes network was set to 0.3. The search algorithm for searching the network structures was set to hill-climbing algorithm, K2.
Table 2.
Parameter settings of CFS.
| Subset size evaluator | Kernel | Number of CV folds | Seed |
|---|---|---|---|
| SMO | Polynomial | 5 | 3 |
Table 3.
Parameter settings of Bayesnet.
| Estimator | Alpha | Search algorithm |
|---|---|---|
| Simple estimator | 0.3 | K2 hill climbing algorithm |
3.4. Selection of search algorithm associated with feature selection technique
Three algorithms, SSFS, best first and greedy stepwise were compared to select the best performing one. Best first and greedy stepwise algorithms achieved the same results in combination with CFS. Best performing algorithm among the three was SSFS. Even though SSFS achieved a slightly lower recall compared to other algorithms, it achieved better precision, F-measure, AUC and accuracy than the other two algorithms. The results achieved using the three algorithms are displayed in Table 4 .
Table 4.
Comparison of results achieved using various search algorithms in combination with proposed feature selection technique. Results obtained using the best performing algorithm is indicated in bold.
| Search algorithm | Precision | Recall | F-measure | AUC | Accuracy |
|---|---|---|---|---|---|
| SSFS | 0.853 | 0.985 | 0.914 | 0.963 | 91.1579 |
| Best first | 0.817 | 0.998 | 0.899 | 0.912 | 89.2632 |
| Greedy stepwise | 0.817 | 0.998 | 0.899 | 0.913 | 89.2632 |
3.5. Comparison of the proposed pre-trained multi-CNN with single pre-trained CNN
Experiments were conducted on features extracted using different combinations of pre-trained networks. The results of various combinations of pre-trained CNNs are shown in Table 5 . The experiments show that combinations of multiple pre-trained CNNs outperform that of single CNN. Among the combinations of pre-trained CNNs we experimented with, two multi-CNNs composed of 5 pre-trained networks and three multi-CNN composed of 4 pre-trained networks achieved an AUC above 95% and accuracy of 90% or above. A multi-CNN which uses a combination of features extracted from 5 different pre-trained networks (Squeezenet, Darknet-53, MobilenetV2, Xception, Shufflenet) achieved the best performance. The multi-CNN achieved an AUC of 96.3% and accuracy of 91.1579%. Results achieved using single pre-trained CNNs were less compared to most of the multi CNNs composed of 3 or more pre-trained CNNs. All single pre-trained networks could achieve an AUC of less than 95% and accuracy less than 90% only. The best performing single CNN was MobilenetV2. Even though MobilenetV2 achieved an AUC of 94.2%, its accuracy was less than 90% only. The experimental analysis proves the efficiency of pre-trained multi-CNN over single pre-trained CNN.
Table 5.
Comparison of results achieved using various pre-trained networks in combination with proposed classifier. Best results achieved using multi-CNN is indicated in bold.
| Network | Precision | Recall | F-measure | AUC | Accuracy |
|---|---|---|---|---|---|
| Squeezenet+Darknet-53+MobilenetV2+Xception+Shufflenet | 0.853 | 0.985 | 0.914 | 0.963 | 91.1579 |
| Darknet-53+MobilenetV2+Resnet-101+NasnetLarge+Xception | 0.839 | 0.969 | 0.9 | 0.959 | 89.6842 |
| Shufflenet+Darknet-53+Mobilenet+Resnet-101+NasnetLarge | 0.826 | 0.974 | 0.894 | 0.952 | 88.9474 |
| Resnet-101+NasnetLarge+Xception+VGG-19+Squeezenet | 0.833 | 0.98 | 0.901 | 0.952 | 89.6842 |
| Densenet-201+InceptionResnetV2+Shufflenet+Darknet-53+MobilenetV2 | 0.846 | 0.967 | 0.902 | 0.952 | 90 |
| Densenet+InceptionResnetV2+Shufflenet+Darknet-53 | 0.837 | 0.951 | 0.89 | 0.944 | 88.8421 |
| Squeezenet+Darknet-53+MobilenetV2+Xception | 0.842 | 0.985 | 0.907 | 0.962 | 90.4211 |
| Squeezenet+Darknet-53+Shufflenet+Xception | 0.837 | 0.987 | 0.906 | 0.955 | 90.2105 |
| InceptionResnetV2+Shufflenet+Darknet-53+MobilenetV2 | 0.846 | 0.969 | 0.903 | 0.951 | 90.1053 |
| Squeezenet+Darknet-53+Shufflenet | 0.815 | 0.971 | 0.886 | 0.943 | 88.1053 |
| Squeezenet+Darknet-53+MobilenetV2 | 0.826 | 0.974 | 0.894 | 0.948 | 88.9474 |
| Darknet+InceptionResnetV2+MobilenetV2 | 0.847 | 0.951 | 0.896 | 0.954 | 89.4737 |
| Densenet-201+Darknet-53 | 0.815 | 0.98 | 0.89 | 0.931 | 88.4211 |
| Darknet-53+InceptionResnetV2 | 0.84 | 0.929 | 0.883 | 0.94 | 88.2105 |
| Squeezenet+Shufflenet | 0.809 | 0.971 | 0.883 | 0.933 | 87.6842 |
| Densenet-201 | 0.809 | 0.947 | 0.873 | 0.924 | 86.8421 |
| Darknet-53 | 0.8 | 0.956 | 0.871 | 0.919 | 86.5263 |
| InceptionResnetV2 | 0.806 | 0.96 | 0.876 | 0.918 | 87.0526 |
| MobilenetV2 | 0.83 | 0.956 | 0.888 | 0.942 | 88.5263 |
| NasnetLarge | 0.819 | 0.936 | 0.873 | 0.906 | 87.0526 |
| Resnet-101 | 0.814 | 0.958 | 0.88 | 0.925 | 87.5789 |
| Shufflenet | 0.813 | 0.971 | 0.885 | 0.925 | 88 |
| Squeezenet | 0.803 | 0.934 | 0.863 | 0.908 | 85.8947 |
| VGG-19 | 0.816 | 0.949 | 0.878 | 0.914 | 87.3684 |
| Xception | 0.817 | 0.985 | 0.893 | 0.91 | 88.7368 |
3.6. Comparison of results achieved using various classifiers
Table 6 displays the results achieved using various classifiers in combination with the best performing multi-CNN composed of 5 pre-trained CNNs. Only Bayesnet achieved an accuracy above 90%. All other classifiers could achieve an accuracy below 90% only. Bayesnet, NaiveBayes, LogisticRegression, Random Forest, ADTree and NBTree achieved an AUC above 90%. SVM and AdaBoostM1 could achieve AUC below 90% only. Bayesnet achieved better precision and F-measure than the other classifiers. However, the recall achieved by Bayesnet was slightly lower than that achieved by NaiveBayes, SVM and AdaBoostM1. The experimental analysis proves the efficiency of Bayesnet classifier in combination with multi-CNN and correlation-based feature selection technique for the detection of COVID-19.
Table 6.
Comparison of results achieved using various classifiers in combination with the proposed network. Results achieved using proposed classifier is shown in bold.
| Classifier | Precision | Recall | F-measure | AUC | Accuracy |
|---|---|---|---|---|---|
| Bayesnet | 0.853 | 0.985 | 0.914 | 0.963 | 91.1579 |
| NaiveBayes | 0.83 | 0.989 | 0.902 | 0.947 | 89.7895 |
| SVM | 0.822 | 0.989 | 0.898 | 0.897 | 89.2632 |
| LogisticRegresion | 0.846 | 0.909 | 0.877 | 0.941 | 87.7895 |
| AdaBoostM1 | 0.81 | 0.998 | 0.894 | 0.898 | 88.7368 |
| Random Forest | 0.828 | 0.987 | 0.9 | 0.94 | 89.5789 |
| ADTree | 0.828 | 0.938 | 0.88 | 0.922 | 87.7895 |
| NBTree | 0.84 | 0.96 | 0.896 | 0.949 | 89.3684 |
3.7. Comparison with other state-of-the-art methods
Most of the existing methods used a small dataset for classification whereas the proposed method used a relatively large dataset. The method was further tested in a second dataset and achieved promising results in both the datasets. The number of images and validation techniques used by the various state-of-the-art methods are different. Table 7 shows the number of images and validation method used by the various authors. Table 8 displays the results of other major state-of-the-art methods along with the proposed best performing multi-CNN (Squeezenet+Darknet-53+Mobilenet+Xception+Shufflenet). A fair comparison of results is not possible due to the difference in the datasets, performance metrics and the validation techniques. However, it is noteworthy that the proposed method has proven its effectiveness in a relatively large dataset consisting of 453 COVID-19 images. The number of COVID-19 images used by the other methods is considerably less. The method by Panwar et al. [15] achieved a recall of 0.972 in a balanced dataset of 142 COVID-19 and 142 normal images. The result was achieved on a held-out test dataset consisting of 30% of the images. Toraman et al. [17] achieved a precision of 0.916, recall of 0.96, F-measure of 0.938 and an accuracy of 91.24% based on 10-fold cross-validation performed on 231 COVID-19 images and 500 images with no-findings. The method by Narin et al. [12] achieved a recall of 0.96, F-measure of 0.98 and an accuracy of 98%. However, the method was implemented on a small balanced dataset of 50 COVID-19 and 50 non-COVID images. [13] used a balanced dataset of 250 COVID-19 and 250 non-COVID images to achieve a recall of 0.78 and an AUC of 0.98. The proposed method achieved a better recall and AUC in both the datasets. Zhang et al. [44] achieved a recall of 0.96 and AUC of 0.95 using 100 COVID-19 images and 1431 non-COVID images. However, the result was based on splitting the dataset into training and testing data and not based on cross-validation. Hemdan et al. [14] achieved a precision of 0.83, recall of 1.00 and F-measure of 0.91 using a small balanced dataset of 25 COVID-19 and 25 non-COVID images. The result was based on a held-out test dataset obtained by partitioning the dataset into 80% training data and 20% test data. Unlike cross-validation, testing on a held-out test dataset does not ensure the robustness of the method.
Table 7.
Number of images and validation techniques used by the various state-of-the-art methods.
| Method | Number of images | Validation |
|---|---|---|
| Multi-CNN | 453 COVID-19 vs. 497 non-COVID (Dataset-1) | 10-fold CV |
| Multi-CNN | 71 COVID-19 vs. 7 non-COVID (Dataset-2) | 10-fold CV |
| [17] | 231 COVID-19 vs. 500 no-findings | 10-fold CV |
| [15] | 142 COVID-19 vs. 142 normal | 70% data for training and 30% testing |
| [12] | 50 COVID-19 vs. 50 normal | 5-fold CV |
| [13] | 250 COVID-19 vs. 250 non-COVID | 10-fold CV |
| [13] | Training: 250 COVID-19 vs. 250 non-COVID | Testing: 74 COVID-19 vs. 36 non-COVID |
| [44] | 100 COVID-19 vs. 1431 non-COVID | 2-fold CV |
| [14] | 25 COVID-19 vs. 25 non-COVID | 80% data for training and 20% testing |
Number of images and validation techniques used by the proposed method are indicated in bold.
Table 8.
Results reported by various state-of-the-art methods. Results achieved using the proposed method is indicated in bold.
| Method | Precision | Recall | F-measure | AUC | Accuracy |
|---|---|---|---|---|---|
| Multi-CNN (Dataset-1) | 0.853 | 0.985 | 0.914 | 0.963 | 91.1579 |
| Multi-CNN (Dataset-2) | 0.986 | 0.986 | 0.986 | 0.911 | 97.4359 |
| [17] | 0.916 | 0.96 | 0.938 | – | 91.24 |
| [15] | – | 0.972 | – | – | – |
| [12] | – | 0.96 | 0.98 | – | 98 |
| [13] (10-fold CV) | 0.81 | 0.78 | – | 0.89 | – |
| [13] (test dataset) | 0.89 | 0.80 | – | 0.81 | – |
| [44] | – | 0.96 | – | 0.95 | – |
| [14] | 0.83 | 1.00 | 0.91 | – | – |
3.8. Limitations of the proposed method
Even though the method produced good results in a considerably larger dataset, it has few limitations worth mentioning. The method performs classification between COVID-19 and non-COVID X-ray images only. It is not tested in a multi-class classification scenario where the images can be classified as COVID-19, normal and pneumonic. The method does not perform segmentation of the infected region. The combinations of all multi-CNNs are not explored in the paper. It is left to the readers and other researchers to explore more combinations of pre-trained CNNs for the prediction of COVID-19. Being more economical and easily available modality compared to CT scans, the proposed method focused on COVID-19 detection using X-ray images. As a future research direction we propose the use of multi-CNN to extract features from CT scans for the detection of COVID-19 and other lung infections.
4. Conclusion
In this paper, the effectiveness of pre-trained multi-CNN in predicting COVID-19 from X-ray images is investigated. A combination of features extracted from several pre-trained networks in combination with Correlation-based Feature Selection technique and Bayesnet classifier is employed in the method. The best performing multi-CNN used in this study employs a combination of 5 pre-trained CNNs: Squeezenet, Darknet-53, MobilenetV2, Xception and Shufflenet. Results prove the effectiveness of pre-trained multi-CNN over pre-trained single CNNs. Experimental analysis performed using two public datasets show that pre-trained multi-CNN in combination with CFS and Bayesnet is effective in the diagnosis of COVID-19.
Author contributions
Bejoy Abraham and Madhu S. Nair: Conception and design of study, acquisition of data, analysis and/or interpretation of data, drafting the manuscript, revising the manuscript critically for important intellectual content, approval of the version of the manuscript to be published.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.WHO . 2020. WHO situation report-108. Available from: http://www.who.int/292docs/default-source/coronaviruse/situation-reports/29320200507covid-19-sitrep-108.pdf?sfvrsn=44cc8ed8_2. [Accessed 9 May 2020] [Google Scholar]
- 2.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of covid-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev Biomed Eng. 2020 doi: 10.1109/rbme.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 4.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J. Radiological findings from 81 patients with covid-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (covid-19): a pictorial review. Clin Imaging. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham B., Nair M.S. Computer-aided grading of prostate cancer from MRI images using convolutional neural networks. J Intell Fuzzy Syst. 2019;36(3):2015–2024. [Google Scholar]
- 7.Abraham B., Nair M.S. Automated grading of prostate cancer using convolutional neural network and ordinal class classifier. Inform Med Unlocked. 2019;17:100256. [Google Scholar]
- 8.Abraham B., Nair M.S. Computer-aided diagnosis of clinically significant prostate cancer from MRI images using sparse autoencoder and random forest classifier. Biocybern Biomed Eng. 2018;38(3):733–744. [Google Scholar]
- 9.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit Lett. 2020;133:232–239. [Google Scholar]
- 10.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput Med Imaging Graph. 2019;78:101673. doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 11.Doan M., Case M., Masic D., Hennig H., McQuin C., Caicedo J. Label-free leukemia monitoring by computer vision. Cytometry A. 2020;97(4):407–414. doi: 10.1002/cyto.a.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castiglioni I., Ippolito D., Interlenghi M., Monti C.B., Salvatore C., Schiaffino S. Artificial intelligence applied on chest X-ray can aid in the diagnosis of covid-19 infection: a first experience from Lombardy, Italy. medRxiv. 2020 doi: 10.1186/s41747-020-00203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 15.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of covid-19 in X-rays using ncovnet. Chaos Solitons Fractals. 2020:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Jr, Costa Y.M. 2020. Covid-19 identification in chest X-ray images on flat and hierarchical classification scenarios. arXiv preprint arXiv:2004.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toraman S., Alakuş T.B., Türkoğlu İ. Convolutional capsnet: A novel artificial neural network approach to detect covid-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020:110122. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E. Sample-efficient deep learning for covid-19 diagnosis based on CT scans. medRxiv. 2020 [Google Scholar]
- 19.Mei X., Lee H.-C., Diao K.-y., Huang M., Lin B., Liu C. Artificial intelligence-enabled rapid diagnosis of patients with covid-19. Nat Med. 2020:1–5. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M. 2020. Lung infection quantification of covid-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. [Google Scholar]
- 21.Chen X., Yao L., Zhang Y. 2020. Residual attention u-net for automated multi-class segmentation of covid-19 chest CT images. arXiv preprint arXiv:2004.05645. [Google Scholar]
- 22.Chen X., Yao L., Zhou T., Dong J., Zhang Y. 2020. Momentum contrastive learning for few-shot covid-19 diagnosis from chest CT images. arXiv preprint arXiv:2006.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H. Inf-net: automatic covid-19 lung infection segmentation from CT images. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 24.Cohen J.P., Morrison P., Dao L. 2020. Covid-19 image data collection. arXiv 2003.11597. https://github.com/ieee8023/covid-chestxray-dataset. [Google Scholar]
- 25.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Mooney P. 2018. Pneumonia X rays. Available from: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Accessed 14 July 2020] [Google Scholar]
- 27.Dadario A.M.V. 2020. Covid-19 X rays. Available from: https://www.kaggle.com/dsv/1019469. [Accessed 26 April 2020] [DOI] [Google Scholar]
- 28.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 29.Yamashita R., Nishio M., Do R.K.G., Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernal J., Kushibar K., Asfaw D.S., Valverde S., Oliver A., Martí R. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med. 2019;95:64–81. doi: 10.1016/j.artmed.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 32.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Thirty-first AAAI Conference on Artificial Intelligence. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning. [Google Scholar]
- 33.Zhang X., Zhou X., Lin M., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Shufflenet: an extremely efficient convolutional neural network for mobile devices; pp. 6848–6856. [Google Scholar]
- 34.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 35.Redmon J., Farhadi A. 2018. Yolov3: an incremental improvement. arXiv preprint arXiv:1804.02767. [Google Scholar]
- 36.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.-C. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Mobilenetv2: inverted residuals and linear bottlenecks; pp. 4510–4520. [Google Scholar]
- 37.Zoph B., Vasudevan V., Shlens J., Le Q.V. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Learning transferable architectures for scalable image recognition; pp. 8697–8710. [Google Scholar]
- 38.Chollet F., Xception: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 39.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 40.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. 2016. Squeezenet: Alexnet-level accuracy with 50x fewer parameters and 0.5 MB model size. arXiv preprint arXiv:1602.07360. [Google Scholar]
- 41.Hall M.A. Thesis submitted in partial fulfillment of the requirements of the degree of Doctor of Philosophy at the University of Waikato; 1998. Correlation-based feature subset selection for machine learning. [Google Scholar]
- 42.Gutlein M., Frank E., Hall M., Karwath A. 2009 IEEE Symposium on Computational Intelligence and Data Mining. IEEE; 2009. Large-scale attribute selection using wrappers; pp. 332–339. [Google Scholar]
- 43.Bouckaert R.R. Bayesian network classifiers in weka for version 3-5-7. Artif Intell Tools. 2008;11(3):369–387. [Google Scholar]
- 44.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. Covid-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338. [Google Scholar]