Abstract
The new SARS-CoV-2 poses a significant threat to human health but many aspects of its basic biology remain unknown. Its genome encodes accessory genes that differ significantly within coronaviruses and contribute to the virus pathogenicity. Among accessory genes, open reading frame 8 (ORF8) stands out by being highly variable and showing structural changes suspected to be related with the virus ability to spread. However, the function of ORF8 remains to be elucidated, making it less studied than other SARS-CoV-2 genes. Here I show that ORF8 is poorly conserved among related coronaviruses. The ORF8 phylogeny built using 11,113 SARS-CoV-2 sequences revealed traces of a typical expanding population with a small number of highly frequent lineages. Interestingly, I detected several nonsense mutations and three main deletions in the ORF8 gene that either remove or significantly change the ORF8 protein. These findings suggest that SARS-CoV-2 can persist without a functional ORF8 protein. Deletion breakpoints were found located in predicted hairpins suggesting a possible involvement of these elements in the rearrangement process. Although the function of ORF8 remains to be elucidated, its structural plasticity and high diversity suggest an important role in SARS-CoV-2 pathogenicity.
Keywords: COVID-19, Coronaviruses, ORF8 deletions, SARS-CoV-2 phylogeny, RNA structures
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the coronavirus disease 2019 (COVID-19), a large global outbreak with severe public health consequences (Wu et al., 2020; Zhou et al., 2020). SARS-CoV-2 belongs to the Betacoronavirus genus, which is divided into four lineages (A–D). Lineage B (subgenus Sarbecovirus) includes SARS-CoV-2 and SARS-CoV, the virus responsible for the 2002–2003 SARS outbreak in Asia (Cui et al., 2019; Forni et al., 2017). The Middle East respiratory syndrome coronavirus (MERS-CoV), responsible for several outbreaks since 2012, belongs to lineage C. The novel SARS-CoV-2 shares nearly 96% similarity to the bat coronavirus isolate RaTG13, suggesting these animals are the likely natural reservoir of the virus (Boni et al., 2020; Lu et al., 2020; Zhou et al., 2020).
The SARS-CoV-2 genome consists of a single, positive-stranded RNA with approximately 30,000 nucleotides (Fig. 1A). The genomic RNA serves as a mRNA upon entering the cell to produce several proteins from different open reading frames (ORFs) (Kim et al., 2020). Two overlapping ORF genes, ORF1a and ORF1b, encompass approximately two-thirds of the genome. The remaining 3′ part of the genome encodes four structural proteins (S, E, M and N) and several ‘accessory genes’ that encode additional non-structural proteins (3a, 6, 7a, 7b, 8, and 10). Despite the relevance for human health, it remains to be determined if all accessory ORFs are actually expressed and what is the function of the resulting proteins.
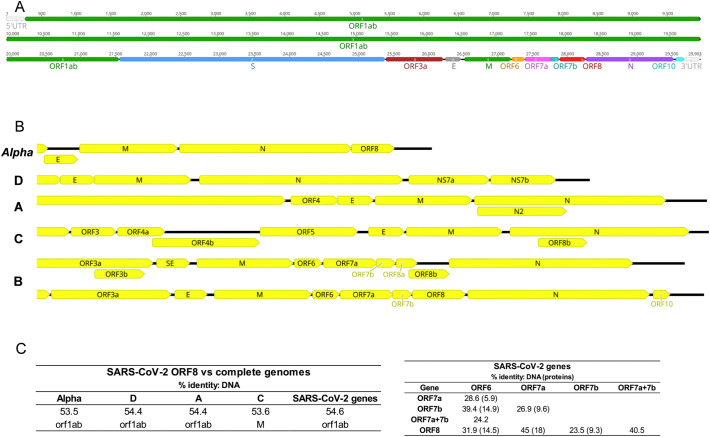
Fig. 1.
SARS-CoV-2 ORF8 genomic location. A) Genome structure of SARS-CoV-2. The ORF1ab (overlapping ORF1a and ORF1b) occupy approximately two-thirds of the genome. Genes for four structural proteins (S, E, M and N) and six non-structural proteins (3a, 6, 7a, 7b, 8, and 10) encompass the remaining genome. B) Genomic organization of betacoronaviruses lineages (A to D) and Aphacoronaviruses. The terminal 3′ section of the genomes is represented. The SARS-CoV-2 ORF8 is located in a cluster of ORFs between the M and N genes, flanked by ORF7b and N. A different gene organization was observed in the previous SARS epidemic, whereas ORF8 was splitted into two functional overlapping ORFs (ORF8a and ORF8b). C) Percentage of pairwise identity among ORF8 and other genes in SARS-CoV-2 and other viruses. The percentage of identity for protein comparisons is indicated in brackets next to the DNA percentage of identity.
Among accessory genes, ORF8 stands out by showing a series of intriguing features identified in SARS-CoV during the 2002–2003 SARS outbreak. ORF8 has only been detected in betacoronaviruses from lineage B (Cui et al., 2019; Forni et al., 2017; Wu et al., 2016). Most lineage B viruses retain a single continuous ORF8. In the reference SARS-CoV-2 genome (NC_045512.2), ORF8 has 366 nucleotides (positions 27,894–28,259) and encodes a protein with 121 amino acids. During the early phases of the SARS epidemic, human isolates were found to possess a unique continuous ORF8 with 366 nucleotides and a predicted protein with 122 amino acids. However, the middle and late phases of the SARS epidemic where characterized by the emergence and spread of strains with a 29-nucleotide deletion that created two functional ORFs (ORF8a and ORF8b), predicted to encode two small proteins, 8a with 39 amino acids and 8b with 84 amino acids (Consortium, 2004; Guan et al., 2003; Lau et al., 2005). Additionally, SARS-CoV lineages with a 83-nucleotide deletion within ORF8 and large deletions removing the complete gene were also detected during the SARS epidemic (Consortium, 2004). Interestingly, a large 382-nucleotide deletion, removing almost completely the ORF8 gene, has already been detected in COVID-19 patients from Singapore (45 samples) and Taiwan (one patient). The SARS-CoV-2 genomes with the 382-nucleotide deletion diverge by two or less mutations and most likely originated in Wuhan and spread to Singapore and Taiwan subsequently (Gong et al., 2020; Su et al., 2020). This deletion has been associated with a milder infection with less systemic release of proinflammatory cytokines and a more effective immune response to SARS-CoV-2 (Young et al., 2020). The propensity of the ORF8 region to suffer deletions has raised the hypothesis that RNA secondary structures could play a role in their formation (Consortium, 2004). Several hairpin structures have been already detected in SARS-CoV-2 (Andrews et al., 2020; Bartas et al., 2020; Lan et al., 2020; Rangan et al., 2020; Simmonds, 2020).
The precise functions of the ORF8 and the truncated ORF8a and ORFb genes remain unclear, although some studies on SARS-CoV have provided important clues about their putative cellular roles. Protein 8ab was found associated with the endoplasmic reticulum membrane at luminal surface, able to activate ATF6 and facilitate protein folding and processing (Oostra et al., 2007; Sung et al., 2009). Protein 8a was found localized in mitochondria where it could induce apoptosis via a caspase 3–dependent pathway (Chen et al., 2007). Protein 8b is involved in cellular degradation of the viral envelope protein and is also capable of inducing apoptosis (Chen et al., 2007; Keng et al., 2006; Law et al., 2006). Both proteins 8b and 8ab were found to inhibit the induction of interferon (IFN) via degradation of IRF3, allowing a high viral replication efficiency in cells (Wong et al., 2018). SARS-CoV-2 ORF8 was shown to inhibit type I interferon (IFN-β) activation and NF-κB pathway (Li et al., 2020). Muth et al. using viral reverse genetics and advanced cell culture models showed that the absence of 8ab (via the 29-nucleotide deletion) reduced significantly the replication capacity of SARS-CoV. The complete deletion of the ORF8 caused an even greater reduction in replicative capability. The study proved that this effect was independent of the type I interferon response. The spread of the deleted variants was hypothesized to be the result of a founder effect in the initial phase of the SARS epidemic (Muth et al., 2018). All these findings from studying SARS-CoV proteins remain to be tested in the new SARS-CoV-2.
The ORF8 locus is also peculiar by being one of the most variable genomic regions among betacoronaviruses (Cui et al., 2019; Wu et al., 2016). Comparison among betacoronavirures lineage B allowed researchers to identify three main lineages in phylogenetic analyses, named types I, II and III (Wu et al., 2016). Similarly, the first comparisons among SARS-CoV-2 and other SARS genomes placed ORF8 among the most divergent genes (Wu et al., 2020). After the S gene, ORF8 stands out as being the most divergent gene when SARS-CoV-2 is compared with bat and pangolin coronaviruses (Boni et al., 2020).
Here I provide a deep analysis of the evolutionary and structural features of ORF8 in SARS-CoV-2 and related betacoronaviruses. Predicted RNA structures are analyzed in the light of the observed ORF8 genetic diversity and genomic rearrangements. This new data clarifies the phylogenetic position of SARS-CoV-2 ORF8 and its mutational pattern in the ongoing pandemic.
2. Materials and methods
2.1. Sequence features
SARS-CoV-2 and other coronaviruses ORF8 gene sequences were obtained from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/) and the GISAID Initiative (https://www.gisaid.org/). The filters “complete”, “high coverage” and “low coverage excl” (all together) were used in the GISAID database. These filters excluded sequences with >1% Ns and with insertions/deletions not verified by the submitter. A total of 11,289 genomes were initially downloaded. I further removed from the dataset all sequences with any ambiguous nucleotide, resulting in a total of 11,113 ORF8 sequences. Data on the distribution of deleted lineages was obtained from the CoV-GLUE database (Singer et al., 2020) and China National Center for Bioinformation (CNCB) (Zhao et al., 2020), both accessed at 17 July 2020.
I also searched for homologous ORF8 gene and protein sequences using the SARS-CoV-2 reference genome (NC_045512.2) as a query in blastn (discontiguous megablast), blastx and blastp searched against the Nucleotide collection (nt) database (Altschul et al., 1990). I removed sequences significantly shorter than the query and/or with ambiguities. The sequences were aligned using the default parameters of the MAFFT version 7 online service (Katoh et al., 2019). Standard measures of genetic diversity were obtained using DNAsp 6.12.03 (Rozas et al., 2017). Gene and protein sequence features were obtained using the Geneious Prime 2019.0.4 (https://www.geneious.com). The CG content was calculated with a sliding window size of 15 nucleotides. Only repeats with 100% similarity (i.e., perfect repeats) and more than 8 nucleotides were identified.
2.2. Structural analyses
RNA secondary structures were predicted using the RNA Folding Form of the Mfold web server (Zuker, 2003) at a temperature of 37 °C. The RNA folding was limited to a maximum distance between paired bases of 75 nucleotides. The structures were predicted for the ORF8 gene region and the 50-nucleotide flanking regions with a total length of 466 nucleotides.
2.3. Phylogenetic analyses
Median-joining network (Bandelt et al., 1999) and mismatch distribution were calculated using the Network V10.1.0.0 software (http://www.fluxus-engineering.com) using default parameters. Bayesian analyses were performed with MrBayes on XSEDE (3.2.7a) software (Ronquist and Huelsenbeck, 2003) running on the CIPRES Science Gateway (Miller et al., 2010). The Metropolis-coupled Markov chain Monte Carlo process was set with two runs of four independent chains running simultaneously for 4,000,000 generations using the GTR + I + G mutation model. The average standard deviation of split frequencies of the final DNA tree was 0.007801 and the protein tree was 0.007183. A burn-in value of 0.25 was applied. Trees were edited in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. SARS-CoV-2 ORF8 has neither paralogues nor orthologues outside Betacoronavirus lineage B (subgenus Sarbecovirus)
SARS-CoV-2 open reading frame 8 (ORF8) gene only has homology in genomes of other lineage B betacoronaviruses (Fig. 1B). The attempts to align SARS-CoV-2 ORF8 with the full genome of other viruses failed (Fig. 1C). Similarly, blast searches with SARS-CoV-2 ORF8 DNA and protein sequences did not retrieve any match besides lineage B. Studies on SARS-CoV ORF8 also fail to identify homologies outside lineage B (Lau et al., 2015; Wu et al., 2016), a scenario that did not change with the new SARS-CoV-2 ORF8.
ORF8 is located between the M and N genes, in a cluster that also includes ORF6, ORF7a and ORF7b (Fig. 1A). It has been shown that gene duplication is rare in RNA viruses, but a few cases have been observed (Simon-Loriere and Holmes, 2013). Therefore, I tested the hypothesis that the cluster ORF6-ORF7a-ORF7b-ORF8 in SARS-CoV-2 may have evolved via past gene duplication events, resulting in paralogues. The highest similarity was found between ORF8 and ORF7a within SARS-CoV-2, with a percentage of identity of 45% for DNA and 18% for protein comparisons (Fig. 1C). The observed low similarity does not support the existence of paralogues resulting from a previous duplication event within this gene cluster.
3.2. SARS-CoV-2 ORF8 gene has several mutated positions at low frequencies, some causing premature stop codons
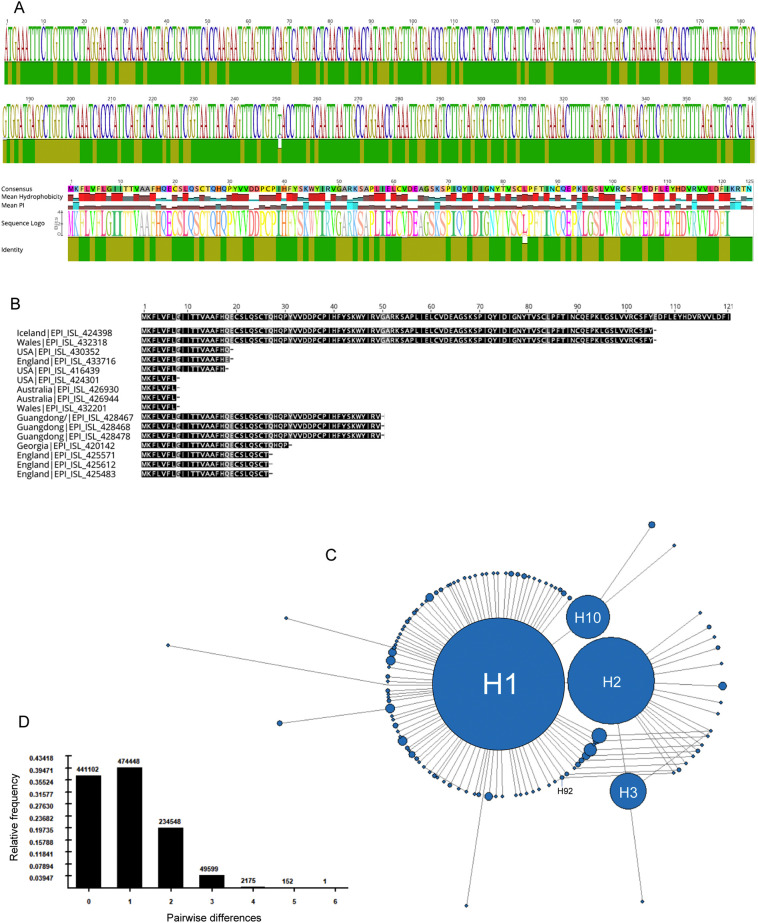
Alignment of the ORF8 gene from 11,113 SARS-CoV-2 genomes revealed 264 (72.1%) fully conserved sites out of 366 nucleotide positions. The 102 variable positions were found scattered along the ORF8 gene (Fig. 2A). Seven nucleotide positions were found with insertion/deletions in the alignment. The average number of pairwise matches across all the positions of the alignment (pairwise identity) was 99.9%, suggesting that most variable positions are defined by mutations in a small number of genomes. In fact, I identified 58 sites with singleton mutations in the 11,113 ORF8 sequences analyzed. The most variable site within ORF8 was position 28,144, presenting either a C (15%) or a U (85%) variant and a pairwise identity of 74.5%. This mutation changed the amino acid at position 84 of the protein from a leucine (observed in 85% of the sequences) to serine (in 15% of the sequences). I also identified 16 sequences with premature stop codons caused by seven different nonsense mutations (Fig. 2B). Five of the seven mutations causing a premature stop codon were found in more than one sequence reducing the likelihood of being sequencing errors. Moreover, several mutations were observed in viruses sequenced in different world regions by different laboratories. For example, the same stop codon was observed in the protein position 8 in sequences obtained from patients in USA, Australia and Wales. The nonsense mutation at position 106 was observed in viruses from patients in Iceland and Wales (Fig. 2B).
Fig. 2.
SARS-CoV-2 ORF8 genetic diversity and phylogeny. A) Identity plot and sequence logo for the alignments of 11,113 ORF8 gene and protein sequences. The most conserved positions are indicated by green bars. B) Location of the 16 stop codons (indicated by a dash) identified in the 11,113 ORF8 protein sequences. C) Median-joining network of ORF8 haplotypes. The area of the circles is proportional to the frequency of sequences, except H1 and H2 which are limited in size for visualization purposes. D) Mismatch distribution graph with the observed number of pairwise differences (y-axis) among ORF8 haplotypes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. SARS-CoV-2 ORF8 gene phylogeny is typical of an expanding population
Alignment of 11,113 ORF8 gene sequences yielded 118 different haplotypes. The phylogenetic network reveled a star-like pattern, typical of an expanding population (Fig. 2C). A similar pattern was observed using complete SARS-CoV-2 genomes (Forster et al., 2020). The central large haplotype (H1) was observed in 9086 sequences, including the reference NC_045512.2. The network also revealed three frequent haplotypes: H2 (1537 sequences) differing from the reference H1 at position 28,144; H10 (152 sequences) diverging from H1 at position 27,964 and H3 (108 sequences) diverging from H2 at position 28,077. The network shows reticulation at several points suggesting parallel mutations. The mismatch distribution shows that most ORF8 sequences diverge by a single mutation, representing ~40% of all pairwise comparisons (Fig. 2D). There are only two haplotypes that diverge by 6 mutations. All other haplotypes diverge by less than 6 mutations considering the ORF8 locus alone. The smooth unimodal distribution is in line with the recent population growth from a single ancestor.
3.4. Three different types of deletions were detected in the ORF8 gene region in patients sampled from different world regions
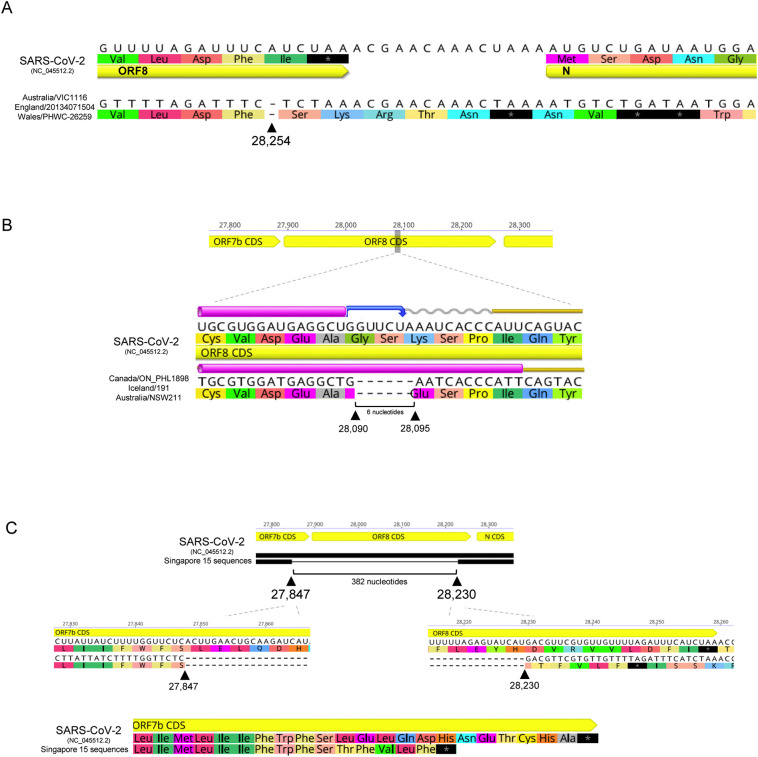
The ORF8 protein varies from 119 to 125 amino acids when considering the dataset with 11,113 sequences. The difference in length results from deletions in the ORF8 gene. First, three sequences had a deletion of a single base (loss of an adenine) at position 28,254 (Fig. 3A). The deletion occurs seven bases upstream the end of the gene, introducing a frameshift mutation in the second last amino acid. The resulting protein lacks the last isoleucine common to all other variants, and instead ends with a Ser-Lys-Arg-Thr-Asn sequence, which introduces an additional three amino acids. This change in the stop codon explains the ORF8 proteins with 125 amino acids. It is interesting to notice that a stop codon exists considering the new frame in the downstream region between ORF8 and N genes, allowing a protein that is only four amino acids longer than the average length. It remains to be determined if there is any selective pressure to maintain extra stop codons downstream of the gene to cope with the loss of the original stop codon. The three sequences belong to two different haplotypes: the samples from Australia and England cluster in the central H1 haplotype, while the sequence from Wales belongs to haplotype H92 (shared with another samples from England). The CNCB database reports 12 samples with this deletion when accessed at 17 July 2020.
Fig. 3.
SARS-CoV-2 ORF8 gene deletions. A) Deletion of a single base at position 28,254, upstream the end of the gene, introduces a frameshift mutation originating a longer protein with four more amino acids. B) Deletion of six nucleotide from positions 28,090 to 28,095 reduces the length of the protein in two amino acids. C) Deletion of 382 nucleotides reported in sequences from Singapore and Taiwan. The deletion removes the terminal part of the ORF7b gene and 336 nucleotides of the ORF8 gene. The predicted ORF7b protein is 7 amino acids shorter and no ORF8 protein is expected to be produced.
Second, a deletion of six nucleotide from positions 28,090 to 28,095 was observed in three sequences (Fig. 3B). The deletion occurs near the middle of the gene, 196 nucleotides downstream the beginning of the ORF8. The deletion does not match the reading frame, reducing the length of the protein in two amino acids by changing Gly-Ser-Lys (canonical protein) to Glu (deleted variant). This deletion explains the ORF8 proteins with only 119 amino acids. The deletion occurs in three sequences from Canada, Iceland and Australia. The Canadian sequence belongs to H1, while those from Iceland and Australia to H2 (Fig. 2C). According to the CoV-GLUE database, this deletion was detected in 24 samples from different lineages when accessed at 17 July 2020.
Finally, a large 382-nucleotide deletion has been reported in sequences from Singapore and Taiwan spanning most of the ORF8 gene (Gong et al., 2020; Su et al., 2020). The deletion removes the terminal part of the ORF7b gene and 336 nucleotides (91.8%) of the ORF8 gene (Fig. 3C). The 5′ breakpoint (27,847) is located 37 nucleotides upstream the ORF7b stop codon, while the 3′ breakpoint (28,230) is located 27 nucleotides upstream the ORF8 stop codon. Because the original ORF7b stop codon is removed, the predicted ORF7b protein is 7 amino acids shorter than the normal variant and ends with Thr-Phe-Val-Leu-Phe (Fig. 3C). The 382-nucleotide deletion also removes the region encoding the ORF8 protein, which is therefore missing in these SARS-CoV-2 viruses.
3.5. Several large hairpins are predicted for the SARS-CoV-2 ORF8 gene region
RNA genomes are known to form stable secondary structures implicated in a variety of regulatory functions (Brierley et al., 1989; Chen and Olsthoorn, 2010; Williams et al., 1999). The formation of secondary structures is often related with repeated sequences (Shapiro and von Sternberg, 2005; Zhao et al., 2012). I identified six perfect repeats within the ORF8 gene with more than 8 nucleotides (Supplementary Fig. S1). The largest is an overlapped inverted repeat (Repeat 1) with 10 nucleotides (CAAUUAAUUG) at positions 28,153–28,162. All other repeats have 8 nucleotides, with three direct repeats and two overlapped inverted repeats (Supplementary Fig. S1). Additionally, RNA regions with a high GC content may have more stable secondary structures. ORF8 has an overall GC content of 35.8%, slightly lower than the 38% observed in the complete genome (Supplementary Fig. S2). There are only four regions in the ORF8 gene with a GC content over 50% when considering 15 nucleotide windows.
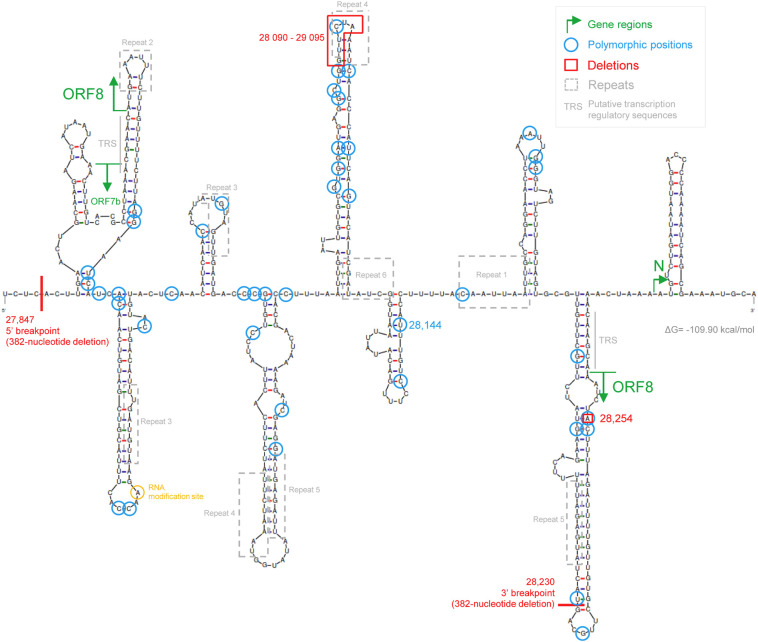
Using a standard nucleic acid folding method, I determined the secondary structure of the ORF8 gene region (Fig. 4 ). The folding was done considering only short distance interactions excluding possible pairing between distant bases in the genome. However, it was recently shown that long-distance interactions across the SARS-CoV-2 RNA have a small effect on the identity of local structures (Lan et al., 2020). The ORF8 start and stop codons are located in two hairpins. The 28,254 frameshift deletion is located on a paired base, which was also identified as polymorphic in a different haplotype. The six-nucleotide deletion (28,090–29,095) occurs at the tip of a large hairpin, where one of the identified perfect repeats (Repeat 4) is also located. The perfect repeats (Supplementary Fig. S1) are often associated with hairpin arms in this predicted structure. The 3′ breakpoint of the 382-nucleotide deletion is also located at the tip of a large hairpin.
Fig. 4.
SARS-CoV-2 ORF8 RNA secondary structure. The ORF8 start and stop codons are located in two hairpins (green arrows). Polymorphic sites (blue circles) are distributed equally by paired and unpaired nucleotides. The 28,090–29,095 deletion (red box) and the 28,230 3′ breakpoint (382-nucleotide deletion) are located at the tip of a hairpin. Most perfect repeats occur in hairpins. The RNA modification site identified by Kim, D. et al. (Cell, 2020, 181(4): 914–921) is located in an unpaired base in the terminal loop of a hairpin. The putative transcription-regulatory sequences (TRS) are indicated in grey. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The polymorphic positions identified in the alignment with 11,113 sequences that occur in more than one sequence (i.e., excluding singletons) were found slightly more in paired (59.5%) than in unpaired (40.5%) nucleotides (Fig. 4). The opposite trend was observed when considering the complete SARS-CoV-2 genome (Simmonds, 2020). It is possible that mutations on ORF8 are not dependent on the disruption of secondary structures as in other genomic regions. The position 28,144 is located in a paired region of a small hairpin. Finally, a RNA modification site identified on viral transcripts (Kim et al., 2020) is located in an unpaired base in the terminal loop of an hairpin.
3.6. The phylogenetic classification of ORF8 lineages are redefined by the new SARS-CoV-2 sequences
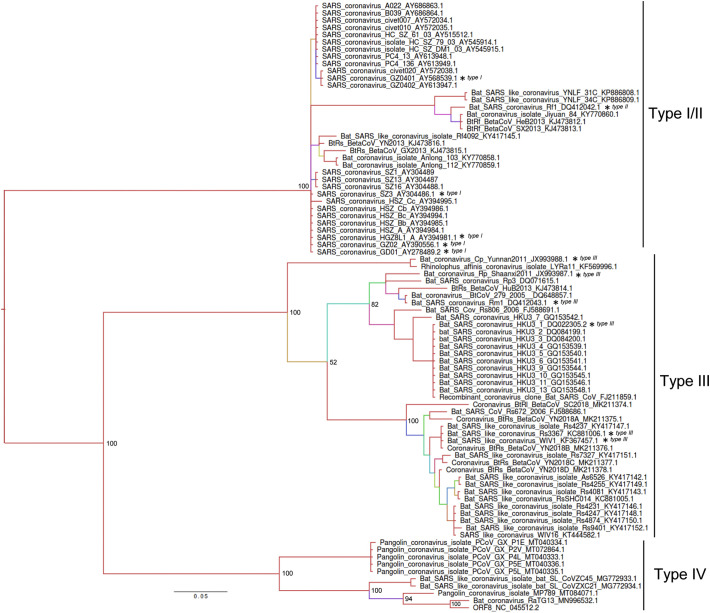
The ORF8 is located in a non-recombining region of sarbecoviruses allowing a reliable phylogenetic reconstruction (Boni et al., 2020). The Bayesian phylogenetic inference described in Fig. 5 placed SARS-CoV-2 ORF8 close to the RaTG13 previously detected in bats from the Yunnan province (Zhou et al., 2020), with a 97% sequence identity (Fig. 5). Close to SARS-CoV-2 are the pangolin coronaviruses, in particular sequence MT084071.1 (92.1% identity), and two SARS-like coronaviruses with 88.5% of identity (ZXC21 and ZC45), sampled in bats from the Zhejiang province of China (Hu et al., 2018). These ten sequences were now classified here as type IV. I followed the nomenclature for ORF8 lineages previously proposed (Wu et al., 2016), which identified three ORF8 types (I, II and III). All the sequences for the new type IV were not available at the time of the original classification. The remaining phylogenetic tree includes two main clades with the sequences previously clustered in type I, II and III. However, I found no support for a clear separation of types I and II, while sequences identified as Type III all belong to a monophyletic branch (Fig. 5).
Fig. 5.
Phylogeny of Betacoronavirus lineage B based on ORF8 gene sequences. The Bayesian phylogenetic tree was built with the reference SARS-CoV-2 and all related coronavirus sequences identified in blast searches (n = 84). Bayesian posterior probabilities are shown on basal nodes and as colours in branches (from high values in red to low values in violet). The scale bar indicates substitutions per site. The * indicates the sequences and classification used previously by Wu, Z. et al. The Journal of infectious diseases, 2016, 213(4): 579–583. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
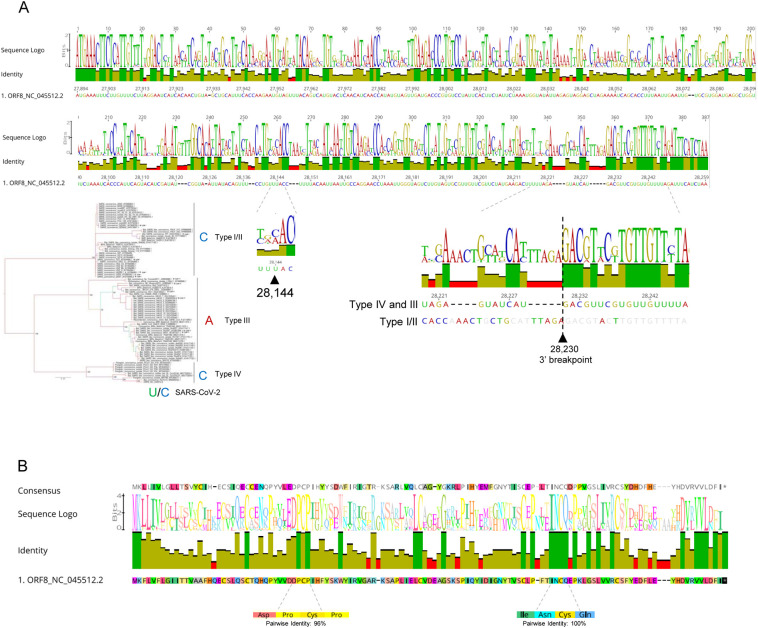
Position 28,144 has C in all type IV and type I/II Betacoronaviruses, as observed in 15% of human SARS-CoV-2 (Fig. 6A). Type III sequences have an A, while the most frequent U (85%) variant in human SARS-CoV-2 does not occur in any other coronavirus. The C variant seems to be the ancestral state in SARS-CoV-2 since it occurs in all other Type IV sequences. However, the early occurrence of both variants (U/C) in SARS-CoV-2 samples from Wuhan in early January 2020 and possible biases in early sampling does not allow a definitive conclusion to be drawn.
Fig. 6.
ORF8 genetic diversity in Betacoronavirus. A) Identity plot and sequence logo for the DNA alignments of reference SARS-CoV-2 and all related coronavirus sequences identified in blast searches (n = 84). The location of position 28,144 is indicated, as well as the nucleotide in all betacoronaviruses lineage B types (I to IV). The location of the 3′ breakpoint (28,230) of the 382-nucleotide deletion is also highlighted for the two main lineage B types. B) Identity plot and sequence logo for the protein alignments of the same viruses used in A).
Interestingly, the 3′ breakpoint 28,230 of the 382-nucleotide deletion occurs in a transition between a variable region (with 4 and 5 nucleotide insertion/deletions) and a conserved domain of the coronaviruses alignment (Fig. 6A). The phylogeny built with protein sequences resembles the one obtained with DNA (Supplementary Fig. S3). The reference SARS-CoV-2 ORF8 protein diverges only in six out of 122 amino acids in relation to the bat coronavirus RaTG13. The region at the C-terminus of the protein was found conserved, with a Val-Val-Leu stretch of amino acids equal in all available sequences. Additionally, two clusters of four amino acids are found conserved (Fig. 6B and Supplementary Fig. S4): Ile-Asn-Cys-Gln and Asp-Pro-Cys-Pro.
4. Discussion
The analyses of the newly discovered SARS-CoV-2 ORF8 confirmed the highly dynamic nature of this accessory gene in evolutionary and structural terms, as previously noted for the homologous SARS-CoV ORF8 locus during the SARS epidemic (Consortium, 2004; Cui et al., 2019; Forni et al., 2017; Stadler et al., 2003). Unfortunately, the origin of SARS-CoV-2 ORF8 remains unknown. I found no ORF8 orthologues in betacoronaviruses outside lineage B, supporting previous observations (Lau et al., 2015; Wu et al., 2016). The accumulation of mutations may have erased any trace of past homologies with other gene regions, either with viral or host genomes. The future identification of intermediates states among betacoronaviruses from lineage B may provide some clues about their origin, although the high diversity observed on these accessory genes makes such task very difficult. The phylogenies built using SARS-CoV-2 sequences allowed the redefinition of the classification of ORF8 lineages (Wu et al., 2016), with the inclusion of a new type (IV) defined by SARS-CoV-2 and the bat and pangolin related sequences. The new phylogeny did not support the separation of types ORF8 I and II, either in DNA or protein phylogenies, suggesting a revision of the nomenclature. Position 28,144 had the same state in all type I/II sequences, supporting their close phylogenetic relationship. In fact, the C seems to be the ancestral state according to the phylogeny, while the presence of an A defines type III ORF8.
Three different types of ORF8 deletions were detected in patients sampled from different world regions and often from different lineages. It has been suggested that the SARS-CoV 8a and 8b truncated products resulting from a large deletion in ORF8 might have affected the virus replication capacity and pathogenicity in a way that helped in the adaptation of SARS-CoV to humans during the 2002–2003 epidemic (Chen et al., 2007; Consortium, 2004; Guan et al., 2003; Lau et al., 2005). However, strains with a 83-nucleotide deletion within ORF8 and even the complete loss of the ORF8 region have been reported in SARS-CoV (Consortium, 2004), raising doubts regarding the functional relevance of the protein for the virus fitness in human hosts (Forni et al., 2017). Moreover, different studies showed that the 29-nucleotide deletion decreased the replication of the virus in different cell systems (Muth et al., 2018). The deleted ORF8 lineages detected in Singapore and Taiwan (Gong et al., 2020; Su et al., 2020) could be used to test the functional role of ORF8, by comparing transmission rates in deleted and non-deleted lineages. Su et al. found that SARS-CoV-2 with the 382-nucleotide deletion showed a higher replicative fitness in vitro than the wild type, with no differences in patient viral load. The clinical effect of the 382-nucleotide deletion appears to be a milder infection with less systemic release of proinflammatory cytokines (Young et al., 2020). The deletion was found associated with clinically significant illness, but infections tended to be milder compared with those caused by the non-deleted variants.
I identified six perfect repeats within the ORF8. Perfect direct repeats have been found distributed throughout the genome of several viruses (Zhao et al., 2012). Repetitive DNA sequence elements can be either random genomic features or be associated with the regulation of viral packaging, replication or transcription, directly or via the formation of secondary structures (Chew et al., 2004; Shapiro and von Sternberg, 2005). In fact, repeats 2 and 4 define the complementary nucleotides of the terminal loop of two large hairpins predicted for the ORF8 region. The six-nucleotide deletion (28,090–29,095) detected in SARS-CoV-2 lineages occurs at the tip of an hairpin, as observed for some of the previous SARS ORF8 deletions (Consortium, 2004). Most deletion breakpoint were found associated with hairpins, a phenomenon observed in other types of genomes (Bacolla et al., 2004; Damas et al., 2012). The formation of RNA structures has been shown to regulate ribosomal frameshifting, replication, translation and packaging in coronaviruses (Brierley et al., 1989; Chen and Olsthoorn, 2010; Williams et al., 1999). It has also been shown that RNA secondary structures can influence the generation of deletions by pausing the polymerase activity and increasing the chance of template slippage in RNA viruses (Pathak and Temin, 1992; Pita et al., 2007). Therefore, it is worth testing the possibility that some of these SARS-CoV-2 ORF8 deletions are related with the formation of secondary structures, as they are known to be sites of genomic instability (Kidd-Ljunggren et al., 2000; Romero et al., 2006).
Overall, SARS-CoV-2 ORF8 revealed many intriguing features that deserve further investigation. The occurrence of premature stop codons and large deletions described here reinforce the idea that ORF8 is dispensable for SARS-CoV-2 replication. However, the persistence of ORF8 in different lineages suggest it is playing an important yet uncovered role. It will be important to monitor the progression of variants with ORF8 deletions and nonsense mutations worldwide, as several were already detected. Hopefully, the deep study of accessory genes from an evolutionary and molecular perspective will help us to better understand the biology of these important viruses and learn how to deal with them in the future.
Funding sources
This work was supported by the Fundação para a Ciência e a Tecnologia [RESEARCH 4 COVID-19 project n. 029] and the EOSCsecretariat.eu COVID-19 Fast Track Funding.
Declaration of Competing Interest
None to declare.
Acknowledgments
I would like to thank all the researches who have kindly shared genomes on public databases.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104525.
Appendix A. Supplementary data
Supplementary material.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews R.J., Peterson J.M., Haniff H.F., Chen J., Williams C., Greffe M., Disney M.D., Moss W.N. An in silico map of the SARS-CoV-2 RNA structurome. BioRxiv. 2020 doi: 10.1101/2020.04.17.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bartas M., Brázda V., Bohálová N., Cantara A., Volná A., Stachurová T., Malachová K., Jagelská E.B., Porubiaková O., Červeň J. In-depth bioinformatic analyses of nidovirales including human SARS-CoV-2, SARS-CoV, MERS-CoV viruses suggest important roles of non-canonical nucleic acid structures in their lifecycles. Front. Microbiol. 2020;11:1583. doi: 10.3389/fmicb.2020.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0771-4. ahead of print. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y., Ping Y.-H., Lee H.-C., Chen K.-H., Lee Y.-M., Chan Y.-J., Lien T.-C., Jap T.-S., Lin C.-H., Kao L.-S. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew D.S., Choi K.P., Heidner H., Leung M.-Y. Palindromes in SARS and other coronaviruses. INFORMS J. Comput. 2004;16:331–340. doi: 10.1287/ijoc.1040.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium C.S.M.E. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Carneiro J., Goncalves J., Stewart J.B., Samuels D.C., Amorim A., Pereira F. Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 2012;40:7606–7621. doi: 10.1093/nar/gks500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.-N., Tsao K.-C., Hsiao M.-J., Huang C.-G., Huang P.-N., Huang P.-W., Lee K.-M., Liu Y.-C., Yang S.-L., Kuo R.-L. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microb. Infect. 2020:1–37. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., Luo S., Li P., Zhang L., Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., Yang L., Ding C., Zhu X., Lv R. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microb. Infect. 2018;7:1–10. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.-T., Choi Y.-W., Welkers M.R., Chan D.Z., Shen S., Lim S.G., Hong W., Tan Y.-J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd-Ljunggren K., Zuker M., Hofacker I.L., Kidd A.H. The hepatitis B virus pregenome: prediction of RNA structure and implications for the emergence of deletions. Intervirology. 2000;43:154–164. doi: 10.1159/000025041. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T.C., Allan M.F., Malsick L., Khandwala S., Nyeo S.S., Bathe M., Griffiths A., Rouskin S. Structure of the full SARS-CoV-2 RNA genome in infected cells. bioRxiv. 2020 [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.-W., Wong B.H., Wong S.S., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Feng Y., Chen H., Luk H.K., Yang W.-H., Li K.S., Zhang Y.-Z., Huang Y., Song Z.-Z., Chow W.-N. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.Y.P., Liu Y.-M., Geng H., Kwan K.H., Waye M.M.-Y., Ho Y.-Y. Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus. FEBS Lett. 2006;580:3643–3648. doi: 10.1016/j.febslet.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;198074 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. 2010 Gateway Computing Environments Workshop (GCE) IEEE; 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., Rottier P.J. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V.K., Temin H.M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita J.S., De Miranda J.R., Schneider W.L., Roossinck M.J. Environment determines fidelity for an RNA virus replicase. J. Virol. 2007;81:9072–9077. doi: 10.1128/JVI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero T.A., Tumban E., Jun J., Lott W.B., Hanley K.A. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 2006;87:3291–3296. doi: 10.1099/vir.0.82182-0. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Shapiro J.A., von Sternberg R. Why repetitive DNA is essential to genome function. Biol. Rev. 2005;80:227–250. doi: 10.1017/s1464793104006657. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses–an endeavour to understand its biological purpose. bioRxiv. 2020 doi: 10.1101/2020.06.17.155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E.C. Gene duplication is infrequent in the recent evolutionary history of RNA viruses. Mol. Biol. Evol. 2013;30:1263–1269. doi: 10.1093/molbev/mst044. [DOI] [PubMed] [Google Scholar]
- Singer J., Gifford R., Cotten M., Robertson D. 2020. CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation. [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.-D., Rappuoli R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G., Tan C.W. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11 doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S.-C., Chao C.-Y., Jeng K.-S., Yang J.-Y., Lai M.M. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Chang R.-Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., Zhang J., Yang F., Zhang S., Jin Q. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., Anderson D.E., Lee C.Y.-P., Amrun S.N., Lee B., Goh Y.S. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Tian Y., Yang R., Feng H., Ouyang Q., Tian Y., Tan Z., Li M., Niu Y., Jiang J. Coevolution between simple sequence repeats (SSRs) and virus genome size. BMC Genomics. 2012;13:435. doi: 10.1186/1471-2164-13-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.-M., Song S.-H., Chen M.-L., Zou D., Ma L.-N., Ma Y.-K., Li R.-J., Hao L.-L., Li C.-P., Tian D.-M. The 2019 novel coronavirus resource. Yi chuan=Hereditas. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.