Abstract
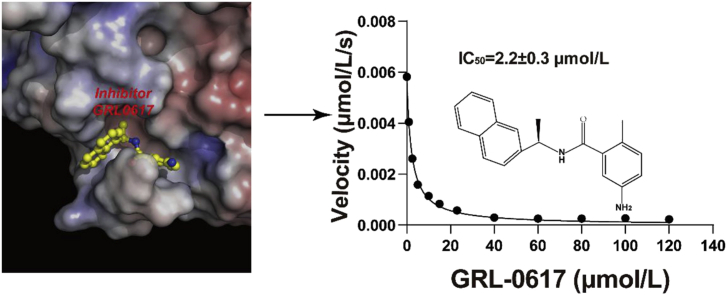
The pandemic of coronavirus disease 2019 (COVID-19) is changing the world like never before. This crisis is unlikely contained in the absence of effective therapeutics or vaccine. The papain-like protease (PLpro) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) plays essential roles in virus replication and immune evasion, presenting a charming drug target. Given the PLpro proteases of SARS-CoV-2 and SARS-CoV share significant homology, inhibitor developed for SARS-CoV PLpro is a promising starting point of therapeutic development. In this study, we sought to provide structural frameworks for PLpro inhibitor design. We determined the unliganded structure of SARS-CoV-2 PLpro mutant C111S, which shares many structural features of SARS-CoV PLpro. This crystal form has unique packing, high solvent content and reasonable resolution 2.5 Å, hence provides a good possibility for fragment-based screening using crystallographic approach. We characterized the protease activity of PLpro in cleaving synthetic peptide harboring nsp2/nsp3 juncture. We demonstrate that a potent SARS-CoV PLpro inhibitor GRL0617 is highly effective in inhibiting protease activity of SARS-CoV-2 with the IC50 of 2.2 ± 0.3 μmol/L. We then determined the structure of SARS-CoV-2 PLpro complexed by GRL0617 to 2.6 Å, showing the inhibitor accommodates the S3–S4 pockets of the substrate binding cleft. The binding of GRL0617 induces closure of the BL2 loop and narrows the substrate binding cleft, whereas the binding of a tetrapeptide substrate enlarges the cleft. Hence, our results suggest a mechanism of GRL0617 inhibition, that GRL0617 not only occupies the substrate pockets, but also seals the entrance to the substrate binding cleft hence prevents the binding of the LXGG motif of the substrate.
KEY WORDS: SARS-CoV-2, PLpro, Proteinase inhibitor, Crystal structure, Antiviral drug, Drug design
Graphical abstract
This study indicates a mechanism of GRL0617 inhibition and provides a framework for further inhibitor development targeting SARS-CoV-2 PLpro.
1. Introduction
The 2019 novel coronavirus was officially named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV)1. This virus is the causative agent of the current pandemic of coronavirus disease 2019 (COVID-19)2, 3, 4, 5. To date, approximately 15 million COVID-19 cases have been reported globally and more than 610,000 lives have lost. In the absence of either effective vaccine or drugs, the peak of COVID-19 pandemic is yet to come.
Comparing to SARS-CoV, while the structural and accessory proteins are variable in SARS-CoV-2, the virally-encoded replicases are highly conserved. One of the best-characterized CoV replicases is the papain-like protease domain (PLpro)6,7 from the membrane anchored multi-domain protein nsp3. PLpro and the main protease (Mpro) are together responsible for the processing of viral polyproteins (pp1a and pp1ab) yielding mature viral proteins6,8,9. In addition, PLpro suppresses innate immunity through reversing the ubiquitination and ISGylation events10, 11, 12, 13, 14, 15. In this regard, SARS-CoV-2 PLpro prefers the ISGylated proteins, whereas SARS-CoV PLpro prefers the ubiquitinated substrates16,17. The dual functionality of PLpro makes it an attractive antiviral drug target18, 19, 20. In the past few months, several studies have reported the structures of the SARS-CoV-2 PLpro complexed by ubiquitin, ISG15 or inhibitors; thus provided the basis of substrate specificity10,16.
Here, we determined the crystal structure of the unliganded SARS-CoV-2 PLpro exhibiting unique crystal packing, high solvent content and reasonable resolution, suggesting this crystal form is useful for fragment-based screening using crystallographic approach. We characterized the protease activity of PLpro in cleaving synthetic peptide harboring nsp2/nsp3 juncture and established an in vitro assay to assess inhibitor efficacy. We evaluated the efficacy of a series of inhibitors and found that the GRL0617 was the most potent. We finally determined the crystal structure of SARS-CoV-2 PLpro complexed by GRL0617, and gained novel insights into the mechanism of inhibition. These results provide a framework for further inhibitor development targeting SARS-CoV-2 PLpro.
2. Materials and methods
2.1. Reagents
All chemicals used in this study were purchased from Sigma–Aldrich (Shanghai, China) unless otherwise specified.
2.2. Construction plasmids, protein expression and purification
The gene encoding PLpro was synthesized from Sangon company (Shanghai, China). The coding sequence of SARS-CoV-2 PLpro was amplified by polymerase chain reaction (PCR). The PCR product was then inserted into pET-28a and pET-28a-SUMO vectors as previously described21, expressing PLpro with a C-terminal His-tag (pET-28a-PLpro-C-His) and N-terminal 6× His-SUMO tag (pET-28a-PLpro-N-His-SUMO) respectively. The sequence of the vectors was verified by DNA sequencing. The plasmid expressing C111S mutant was constructed using site-directed mutagenesis (QuickChange). For protein expression and production, the plasmids were transformed in to Escherichia coli BL21 (DE3) competent cells. The bacteria colony was picked from a LB plate and inoculated in 10 mL LB medium at 37 °C overnight. The protein expression was induced by applying 0.5 mmol/L isopropyl β-d-1-thiogalactopyranoside (IPTG) when the culture reached an OD600 ∼0.8.
To purified recombinant PLpro and its variants, bacterial cells were pelleted by centrifugation and resuspended in lysis buffer (20 mmol/L Tris-HCl, pH 8.0, 300 mmol/L NaCl, 10 mmol/L imidazole, 10 mmol/L β-mercaptoethanol and 1 mmol/L PMSF). The cells were disrupted by ultrasonication. The cell debris was pelleted by centrifugation at 18,000×g for 50 min at 4 °C and the clarified supernatant was filtered through a 0.45 μm syringe filter and applied to Ni-NTA resin (QIAGEN, Shenzhen, China). The resin was washed three times with 10 times column volume with the lysis buffer, and the target protein was then eluted with the elution buffer containing 300 mmol/L imidazole. The 6× His-SUMO tag was cleaved in the dialysis buffer (10 mmol/L HEPES pH = 7.4, 100 mmol/L NaCl, and 10 mmol/L DTT) containing Ulp1 peptidase at 4 °C overnight. The resulting sample was loaded to Ni-NTA resin and the non-tagged PLpro was collected in flow through. The final purification was size-exclusion chromatography using the Superdex 200 10/300 column (GE Healthcare, Little Chalfont, Buckinghamshire, UK) pre-equilibrated with buffer containing 10 mmol/L HEPES pH = 7.4, 100 mmol/L NaCl, and 10 mmol/L DTT. The C-terminal His-tagged PLpro was purified with the same gel filtration column equilibrated with a buffer containing 20 mmol/L Tris-HCl pH = 8.0, 100 mmol/L NaCl, and 2 mmol/L DTT.
2.3. Crystallization and structure determination
Crystallization trials of C-terminal His-tagged C111S PLpro mutant was performed in a hanging drop vapor diffusion system at 18 °C. C111S mutant was concentrated to 10 mg/mL before crystallization. The crystallization was conducted by mixing 1 μL sample and 1 μL reservoir buffer containing 3% dextran sulfate sodium salt, 0.1 mol/L Bicine pH = 8.5, and 15% PEG 20,000. To crystalize SARS-CoV-2 PLpro GRL0617 (MedChemExpress, Monmouth Junction, NJ, USA) complex, wild-type PLpro was concentrated to 1 mg/mL before adding GRL0617. The molar ratio of PLpro:GRL0617 was 1:10. After incubating at 4 °C overnight, the mixture was concentrated to approximately 10 mg/mL. The crystallization was achieved by mixing 1 μL sample with 1 μL reservoir buffer containing 0.1 mol/L sodium citrate pH = 5.6, 0.24 mol/L ammonium acetate and 24% PEG 4000. Subsequently, the crystals were soaked in reservoir buffer supplemented with 20% ethylene glycol (or 15% glycerol for GRL0617 complex crystals) and the crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K at the Shanghai synchrotron radiation facility (SSRF) beamline BL17U (Shanghai, China). Datasets were processed and scaled using XDS Package (Max Planck Institute for Medical Research, Heidelberg Baden-Württemberg, Germany)22. The structures were solved by molecular replacement using Phaser MR23 (University of Cambridge, Cambridge Cambridgeshire, UK), yielding interpretable initial electron density map. Manual model building and refinement were performed using software Coot (University of Oxford, Oxford Oxfordshire, UK) and PHENIX (Lawrence Berkeley National Laboratory, Berkeley, CA, USA)24,25. The refined structures were validated by Molprobity (Duke University, Durham, NC, USA)26. Statistics of data collection and refinement are summerized in Supporting Information Table S1. Structural figures were generated by software Pymol (Schrödinger, New York, NY, USA) and Coot.
2.4. In vitro proteinase assay
Fluorogenic peptide substrate containing fluorescence quenching pair Dabcyl and Edans, Dabcyl-FTLKGG↓APTKVTE-Edans, was derived from the proteolytic processing site between nsp2/nsp3 of pp1a polyprotein. The Fluorogenic peptide was synthesized from GL Biochem (Shanghai) Ltd., Shanghai, China. Fluorescence signal of the cleaved product, APTKVTE-Edans, was measured with SpectraMax® M5 Microplate reader (Molecular Devices, San Jose, CA, USA). Each reaction mixture (100 μL) contains 10 mmol/L HEPES pH = 7.4, 100 mmol/L NaCl, 10 mmol/L DTT, 1 μmol/L SARS-CoV-2 PLpro and the indicated amount of peptide substrate. The reactions were performed in a 96-well plate at 30 °C. The fluorescent signals were recorded with the excitation at 340 nm and emission at 500 nm. The relationship between relative fluorescence unit (RFU) and peptide concentration was calibrated with Edans standard. The reactions were monitored every 30 s, and the initial hydrolysis rate was plotted as the function of substrate concentration. The plot was then fitted with the Michaelis–Menten equation using software GraphPad (La Jolla, CA, USA) to yield kinetic parameters.
2.5. Determination of IC50
Each reaction mixture contains 10 mmol/L HEPES pH = 7.4, 100 mmol/L NaCl, 10 mmol/L DTT, 1 μmol/L SARS-CoV-2 PLpro and increasing amount of inhibitor. The reaction was initiated by adding 11 μmol/L fluorogenic peptide and the mixtures were incubated at 37 °C for 10 min. The initial hydrolysis rate was plotted as the function of inhibitor concentration and the plot was fitted by Eq. (1) using software GraphPad.
| (1) |
3. Results and discussion
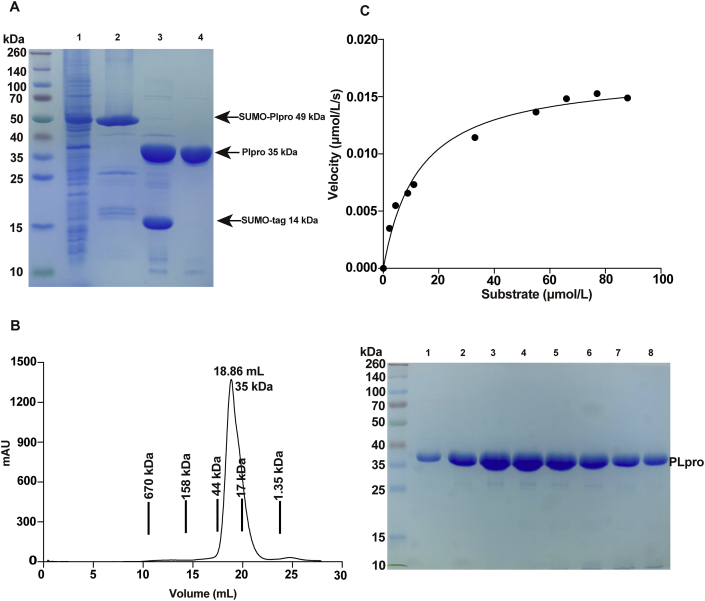
To gain structural and biochemical insight into SARS-CoV-2 PLpro domain (nsp3 746-1063aa, Supporting Information Fig. S1), we expressed two PLpro variants. One contains a C-terminal His-tag, the other harbors an N-terminal His-SUMO tag and the tag was subsequently removed by Ulp1 proteinase (Fig. 1A and B), thus yielded the non-tagged PLpro. The activity of both variants was evaluated using a fluorogenic peptide based in vitro protease assay as previously described27. The synthetic peptide substrate (13-mer) was derived from the proteolytic processing site between nsp2/nsp3 of pp1a polyprotein, which contains substrate recognition LKGG motif. Both variants exhibited proteolytic activity (Fig. 1C and Supporting Information Fig. S2). The catalytic efficiency (kcat/Km) of the C-terminal His-tagged and the non-tagged PLpro proteins were 1074.6 ± 261.9 and 1339.1 ± 362.3 L/mol·s, which is similar to that of SARS-CoV PLpro (Supporting Information Table S2). The catalytically null mutant C111S served as the negative control. The results demonstrate that the C-terminal His-tag has subtle effect on PLpro activity, therefore we used the non-tagged protein for further biochemical characterization and inhibitor evaluation in this study.
Figure 1.
Production and biochemical characterization of the non-tagged SARS-CoV-2 PLpro. (A) Expression of the non-tagged SARS-CoV PLpro. Lane 1, supernatant of the bacteria lysis overexpressing His-SUMO tagged PLpro; lane 2, eluate from the Ni-NTA resin; lane 3, digestion by Ulp1 overnight; lane 4, flow through from the Ni–NTA column. While the cleaved His-SUMO tag and undigested PLpro species were trapped on the column, non-tagged PLpro proteins were collected. (B) Final purification of PLpro. Left, PLpro was loaded to Superdex 200 10/300GL column, the peak fractions were pooled and concentrated for follow up experiments. Right, SDS-PAGE analysis of the peak fractions. (C) Protease activity of PLpro in cleaving a short synthetic peptide harboring nsp2/nsp3 juncture. The velocity of substrate hydrolysis is plotted as the function of the substrate concentration. The kinetic parameters Km and Vmax were calculated via nonlinear fitting to the Michaelis–Menten equation. The curves were fitted by single measurements, and errors were estimated by curve fitting.
Comparing to the activity of SARS-CoV-2 PLpro in processing ISGylated or ubiquitinated substrates (kcat/Km for deISGylating 30,210 L/mol·s)10, the activity in cleaving short peptide is much less efficient. This substrate specificity is in line with SARS-CoV PLpro; deubiquitination activity of SARS-CoV PLpro is ∼220 folds more efficient than cleaving small peptide substrate14. Recent structural investigations revealed the distinct binding sites for ISG15 outside the substrate binding cleft (S1–S4 pockets)10, which offered the structural basis for PLpro preference towards the ISGylated substrate.
Next, we characterized SARS-CoV-2 PLpro using crystallographic methods. In the absence of ligand, only the C-terminal His-tagged PLpro C111S mutant yielded measurable crystals. The crystals belonged to the space group of C2 and diffracted the X ray to 2.5 Å. Comparing to several SARS-CoV PLpro structures with C2 space group (PDB ID: 2FE8, 3MJ5 and 4OW0), the current crystal form of SARS-CoV-2 PLpro has larger cell dimensions (Table S1), higher solvent content (56%) and unique crystal packing. Instead of containing a crystallographic trimer in the asymmetry unit (ASU) in apo SARS-CoV PLpro structure, our structure has two SARS-CoV-2 PLpro dimers in the ASU (Supporting Information Fig. S3A–S3C). Intriguingly, the dimer interface involves an intermolecular disulfate-bond bridging two C270 residues of adjacent PLpro copies (Fig. S3A). The unique packing interaction, large solvent space and reasonable resolution (2.5 Å) of the current crystal form offer several advantages for fragment-based screening using crystallographic approach: (1) Although the active site cysteine mutation C111S of this crystal form prevents the screening of cysteine reactive inhibitor/fragment, it is still useful for identifying non-covalent inhibitor targeting substrate binding pockets S1–S4 and allosteric sites. For example, several crystal structures of SARS-CoV-2 PLpro C111S mutant complexed by various non-covalent inhibitors have been recently deposited in Protein Data Bank (PDB ID: 7JIT, 7JIR and 7JIV), which support our assertion. (2) PLpro is challenging to crystalize in the absence of ligand/inhibitor. The frequent observed crystal forms containing crystallographic trimer (2FE8 for SARS-CoV PLpro and 6W9C for SARS-CoV-2 PLpro) are difficult for compound soaking, probably due to the tight packing between PLpro monomers. Our crystal form has relatively higher solvent content and looser packing (Fig. S3B and S3C), thus may facilitate rapid diffusion of compound into crystals.
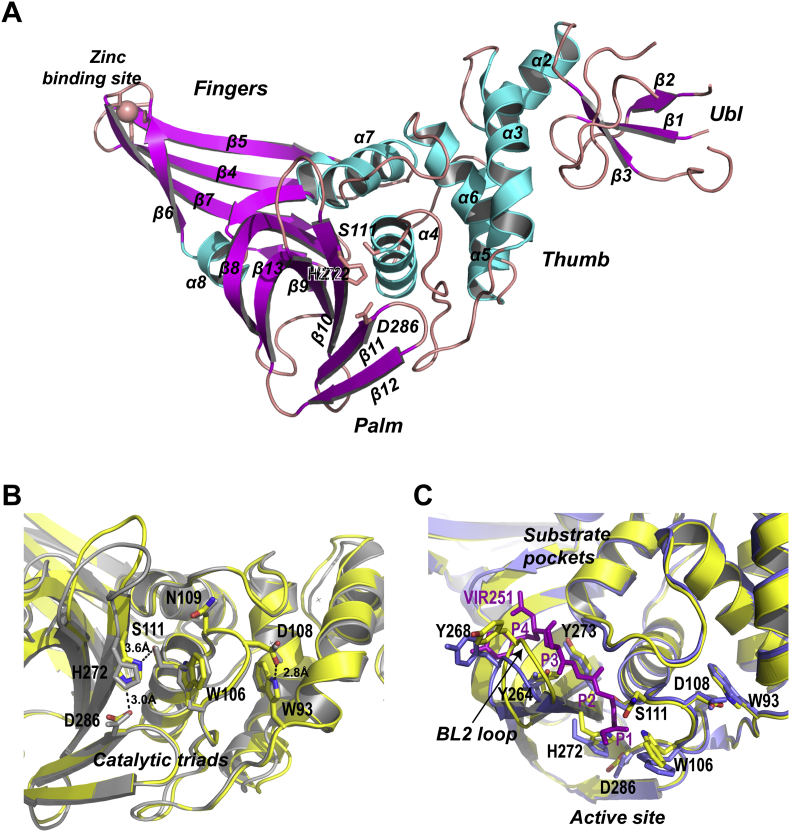
Given that SARS-CoV-2 and SARS-CoV PLpro proteases share ∼82% amino acid sequence identity, most structural features of the orthologs are conserved (Fig. S1). We searched the unliganded SARS-CoV-2 PLpro structure against all entries in the Protein Data Bank using DALI server. The top hit was a structure of SARS-CoV PLpro inhibitor complex (PDB: 3E9S, Z-score = 41.2, RMSD = 1.0 Å). To facilitate structural comparison, we labeled the secondary structural elements of SARS-CoV-2 PLpro following the same scheme for SARS-CoV PLpro (Fig. S1). SARS-CoV-2 PLpro is divided into four sub-domains, the N-terminal ubiquitin-like domain (Ubl, β1–3), the α-helical Thumb domain (α2–7), the β-stranded Finger domain (β4–7) and the Palm domain (β8–13) (Fig. 2A). In the Finger sub-domain, four conserved cysteine (C189 and C192 on the loop between β4–5, C224 and C226 on the loop between β6–7) form a zinc finger belonging to the “zinc ribbon” fold group28.
Figure 2.
Crystal structure of the unliganded SARS-CoV-2 PLpro. (A) Ribbon model of the unliganded SARS-CoV-2 PLpro containing C111S mutation. The structure is colored by secondary structural elements, β-strands magenta, α-helices cyan. PLpro subdomains Ub1, Thumb, Finger and Palm are indicated. Four cysteine residues forming the zine finger on the Finger subdomain are shown with stick model. (B) The view of the active site of SARS-CoV-2 PLpro (yellow) superimposed with SARS-CoV PLpro (gray). The catalytic triads C111S-H272-D286, and other catalytically important residues W93 W106, D108 and N109 are shown with stick model. Their counterparts in SARS-CoV PLpro are shown with gray stick model. Key hydrogen bonds are indicated with the dashed lines. (C) Superimposition of the unliganded SARS-CoV-2 PLpro (yellow, PDB ID: 7CJD, C111S mutant) with SARS-CoV-2 PLpro (blue) complexed by peptide inhibitor VIR251 (magenta). The BL2 loop undergoes marked conformational changes upon substrate binding.
The geometry of SARS-CoV-2 PLpro active site resembles that of SARS-CoV PLpro. All catalytically important residues are invariant (Fig. 2B and Fig. S1). The catalytic triads include residues C111, H272 and D286. Residue C111 (mutated to serine in apo structure) is located 3.6 Å away from the catalytic histidine H272; the corresponding distance SARS-CoV PLpro is 3.7 Å. Residue H272 donates a hydrogen bond to D286 with the length of 3.0 Å; the corresponding distance in SARS-CoV PLpro is 2.7 Å. The hydrogen bond between D108 and W93 (2.8 Å) strengthens the conformation of oxygen anion hole; the corresponding hydrogen bond in SARS-CoV PLpro is 3.0 Å in length.
To gain insight of substrate binding mechanism, we superimposed the unliganded SARS-CoV-2 PLpro structure to an inhibitor bound SARS-CoV-2 PLpro structure (PDB ID: 6WX4). The conformational change of the BL2 loop is remarkable. The BL2 loop is located between β11–12 strands, spanning residues 267–271. This is a flexible loop that recognizes P2–P4 of the LXGG motif of substrate. In the unliganded PLpro (Fig. 2C), the BL2 loop adopts a relatively close confirmation. Whereas in the presence of the tetrapeptide inhibitor VIR251, the loop moves outward by ∼3.2 Å to provide enough room for the tetrapeptide. Similar conformational changes were observed for SARS-CoV PLpro20, suggesting that the substrate recognition mechanism is well preserved.
To expedite the development of COVID-19 therapeutics, drug repurposing is a welcomed strategy. We evaluated the potency of several U.S. Food and Drug Administration (FDA)-approved or preclinical drugs, including disulfiram, 6-TG (6-thioguanine), famotidine, ebselen and 4′-O-methylbavachalcone on SARS-CoV-2 PLpro activity using the fluorogenic peptide-based protease assay. Disulfiram is an alcohol antagonist for treating chronic alcoholism. The drug also has anti-HIV and anti-HCV activities29, 30, 31. 6-TG is an FDA approved drug treating leukemia and Crohn's disease32. Famotidine is a histamine receptor antagonist which was proven to mitigate COVID-19 disease in clinics33,34. Famotidine was predicted to target the catalytic site of PLpro in a in silico screening of the licensed compound libraries33,35; however, in the following experimental study, famotidine neither binds nor inhibits the PLpro36,37. Ebselen has therapeutic activity in neurological disorders, bipolar disorders and hearing loss38, 39, 40, and it was recently shown that the drug is potent against PLpro and has anti-SARS-CoV-2 activity41,42. 4′-O-Methylbavachalcone is a nature metabolites from Psoralea corylifolia, which is high effective against the SARS-CoV PLpro43. To our surprises, we found that only 6-TG inhibited PLpro weakly (IC50 = 103.7 ± 49.4 μmol/L, Supporting Information Fig. S4), but all other drugs were ineffective. These results contradict with several published studies which demonstrated that disulfiram44, 6-TG45,46 and ebselen43 inhibited the protease activity of MERS-CoV and SARS-CoV PLpro in vitro, and 6-TG inhibited SARS-CoV-2 replication at submicromolar levels47. The discrepancy may stem from different substrates used for assessing inhibitor efficacy. For example, while Węglarz-Tomczak et al.41 used ubiquitin conjugated fluorophore substrate (Ub–AMC) for assessing protease activity of PLpro, we used a 13-mer synthetic peptide substrate harboring the P1–P4 residue lacking the ubiquitin portion. Given Ub–AMC is cleaved more efficiently by PLpro, the inhibitory effect of ebselen might be more pronounced when using Ub–AMC as substrate. Our result on famotidine is consistent with a recently study showing that famotidine neither inhibit the activity of PLpro nor the replication of SARS-CoV-2 in cells36,37.
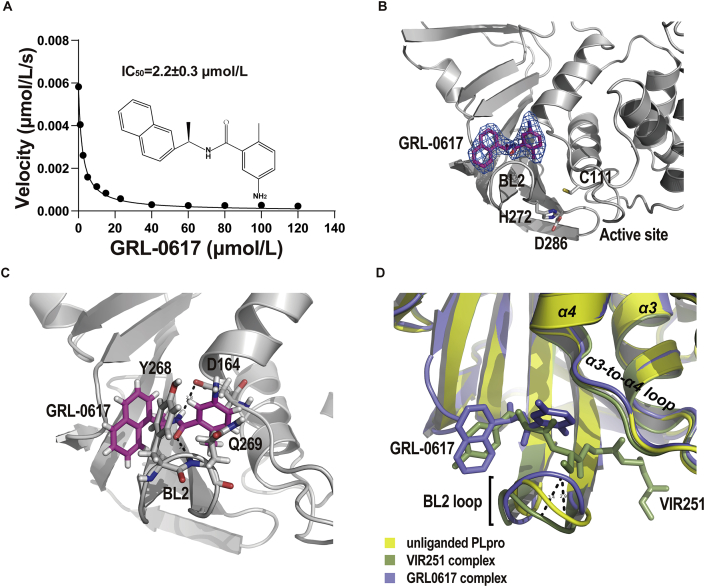
Given SARS-CoV and SARS-CoV-2 PLpro domains share the most structural features, inhibitors developed for SARS-CoV PLpro has a good chance to be effective on SARS-CoV-2 PLpro. GRL0617 is one of the most potent inhibitor for SARS-CoV PLpro7. Several recent studies demonstrated that GRL0617 inhibits the activity of SARS-CoV-2 PLpro17. In revising our manuscript, two independent group also reported the crystal structure of SARS-CoV-2 PLpro in complex with GRL0617 in preprints48,49. We first confirmed that GRL0617 inhibits the activity of SARS-CoV-2 PLpro with an IC50 of 2.2 ± 0.3 μmol/L (Fig. 3A), which is similar to its efficacy against SARS-CoV PLpro (IC50 = 0.6 ± 0.1 μmol/L, Supporting Information Table S3). To reveal the structural basis for GRL0617 inhibition, we next determined the crystal structure of SARS-CoV-2 PLpro complexed by GRL0617. The non-tagged wild-type protein was used in co-crystallization. The crystals of the GRL0617 complex diffracted the X ray to 2.6 Å. It belonged to the space group of P21 and contained 4 PLpro copies in ASU. GRL0617 binds to all PLpro copies in ASU and the inhibitor is associated with well-defined electron density (Fig. 3B). Using software phenix.polder, we calculated a polder map with GRL0617 itself omitted. Positive electron density clearly delineates the shape of the inhibitor, which confirmed the presence of GRL0617 in crystals (Supporting Information Fig. S5). The binding mode of GRL0617 with SARS-CoV-2 PLpro is nearly identical to that of SARS-CoV PLpro (Supporting Information Fig. S6A). Briefly, GRL0617 accommodates in the substrate cleft formed between the BL2 loop and the loop connecting α3 and α4 (α3-to-α4 loop), where it occupies the S3 and S4 pockets. Two highly specific hydrogen bonds are critical to PLpro–GRL0617 interaction (Fig. 3C). The carboxylate side chain of residue D164 accepts one hydrogen bond (angle = 145°, length = 2.8 Å) from N2 nitrogen of GRL0617, and the O7 oxygen of the inhibitor accepts another hydrogen bond (angle = 158°, length = 2.4 Å) from the NH group of Q269. It is worth noting that comparing to apo structure, the peptide backbone of Q269 is flipped out by ∼180° (Fig. S6B) in GRL0617–PLpro complex, so that the NH group of Q269 is oriented in an optimal conformation hydrogen bonding with the inhibitor. Importantly, these hydrogen bonds essentially anchor the flexible BL2 loop to the body of PLpro and fix the loop in a closed conformation.
Figure 3.
The potency of GRL0617 against the protease activity of SARS-CoV-2 PLpro and the inhibitor accommodates the S3–S4 pockets of the protease. (A) Assessment of IC50 of GRL0617 in inhibiting PLpro protease activity in vitro. The data are shown as mean ± SEM (n = 3). (B) Crystal structure of SARS-CoV-2 PLpro complexed by GRL0617. PLpro is shown with ribbon model. The inhibitor is shown with stick model (magenta); the final 2Fo-Fc electron density map (blue mesh, contour level, 1.5 σ) is superimposed. The catalytic triads of PLpro are highlighted with stick model. (C) Two specific hydrogen bonds recognizing GRL0617 are indicated with the dashed lines. The carboxylate side chain of D164 donates a hydrogen bond (angle = 145°, length = 2.8 Å) to N2 nitrogen of GRL0617; the O7 oxygen accepts a hydrogen bond (angle = 158°, length = 2.4 Å) from the NH group of Q269. (D) The unliganded SARS-CoV-2 PLpro (yellow, PDB ID: 7CJD, C111S mutant) is superimposed with the structures of PLpro complexed by VIR251 (light blue, PDB ID: 6WX4) and by GRL0617 (green, PDB ID: 7CMD, complexed by GRL0617).
The closure of the BL2 loop induced by GRL0617 is remarkable. Superimposing apo structure to the tetrapeptide and GRL0617 bound structures clearly show that while the binding of GRL0617 narrows the substrate cleft between the BL2 loop and the α3-to-α4 loop, the binding of the tetrapeptide VIR251 widen the cleft (Fig. 3D). The BL2 loop in the closed conformation would clash with the tetrapeptide. Multiple conformations of the BL2 loop were observed in the unliganded and inhibitor-bound SARS-CoV PLpro structures7,19,50,51. The intrinsic plasticity of the BL2 loop suggests an induced-fit mechanism adopted by PLpro in substrate recognition as well as its ability to accommodate structurally different substrates. Depending on the size of inhibitor, the BL2 loop can adopt either more close or more open conformations. GRL0617 presents one of the smaller inhibitors that trap the BL2 loop in more close conformation and induces backbone flipping of the loop. Thus, the closed BL2 loop may restrict the access of substrate hence inhibits the protease activity. VIR251 presents one of the larger inhibitors that stabilizes the BL2 loop in open conformation. The opened BL2 loop shares the similar backbone conformation with the unliganded BL2 loop. The mechanism of the larger inhibitor might be binding competition for substrate pockets, hence inhibits the protease activity. Taken together, our results suggest a possible mechanism for PLpro inhibition: GRL0617 not only occupies S3–S4 pockets, but also seals the substrate binding cleft in the narrowest conformation hence prevents the entering of the LXGG motif of the substrate.
More than 15-year CoV PLpro inhibitor development, mainly targeting SARS-CoV PLpro, has identified numerous compounds. Given high similarity SARS-CoV-2 PLpro shares with the PLpro of SARS and other CoVs, prior knowledge is certainly useful to COVID-19 drug development. Known PLpro inhibitors include not only the catalytic C111 reactive compounds but also a large collection of non-covalent compounds. Those range from the inhibitors discovered from the yeast-based screening, thiopurine compounds, natural compounds to the naphthalene-based compounds. Of those, the naphthalene-based compounds are of particular interests for its potent efficacy in proteinase inhibition and low molecular mass. GRL0617 and GRL0667 are two of the most potent naphthalene-based inhibitors despite they occupy the S4/S3 pockets that is ∼8 Å away from the catalytic triad. As demonstrated by our structural characterizations, while the naphthalene portion of GRL0617 accommodates the S4 pocket of SARS-CoV-2 PLpro, the benzene ring fits the S3 pocket. Because the S4 pocket determines the specificity of the LXGG motif via recognizing the P4 leucine side chain, the naphthalene occupying the S4 pocket is good candidate for inhibitor design. By contrast, the S3 pocket recognizes the backbone of the P3, thus accepts any residues; therefore, the 1-naphthyl substitution and position of the hydrogen donating nitrogen should be fixed and the S3 is variable in future drug design and modification. Very recently, Osipiuk and colleagues48 synthesized six novel compounds, of which five are benzamine functionalized derivatives of GRL0617 and one is a GRL0617 variant lacking the chirality center, and all six retained the naphthalene portion. Using crystallographic approaches, they revealed that compounds 1–3 bind to the same site as GRL0617. However, the inhibitory effects of these compounds on the proteinase activity in vitro are not connected to their efficacy in suppressing SARS-CoV-2 replication in cells. The weakest proteinase inhibitor compound 5 performed unexpectedly well in SARS-CoV-2 replication assays, whereas the potent proteinase inhibitors compounds 2 and 3 failed to show any antiviral activity. Their results suggest that improving cell permeability, solubility as well as minor modification of the compounds are important for the next-stage optimization.
4. Conclusions
Two important structures of SARS-CoV-2 PLpro protease were determined. The unliganded structure has novel crystal packing, high solvent content and reasonable resolution; thus, it offers a good foundation for fragment-based screening targeting the enzyme. The GRL0617 bound structure provides valuable insight into the inhibition mechanism at atomic level. Given that GRL0617 is one of the most promising inhibitors of CoV PLpro, our findings will aid further optimization of the inhibitor, which may contribute to speed up therapeutic development of COVID-19.
Accession codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes:7CJD and 7CMD.
Acknowledgments
The authors thank the staff of PX III beamline at the Swiss Light Source, Paul Scherrer Institute (Villigen, Switzerland) for assistance in data collection. The authors thank the staff of BL17B/BL18U1/BL19U1/BL19U2/BL01B beamlines at National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility (Shanghai, China) for the help with data collection. This work was supported by the National Key Research and Development Program of China (2016YFD0500300); National Science and Technology Major Project (2018ZX10101001, China); National Natural Science Foundation of China (Grant Nos. 81572005, 81772207, 81971985, 11775308 and 81802057); Beijing Municipal Natural Science Foundation (Grant Nos.7182117 and 7174288, China); Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (Grant Nos. 2017-I2M-1-014 and 2016-I2M-1-013, China); and Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (Grant Nos. 2018PT51009 and 2017PT31049, China).
Author contributions
Sheng Cui and Xiaopan Gao designed the study. Sheng Cui and Xiaopan Gao solved the structure and wrote the paper. Xiaopan Gao, Bo Qin, Pu Chen, Kaixiang Zhu and Pengjiao Hou performed experiments. Xiaopan Gao, Bo Qin, Meitian Wang, Justyna Aleksandra Wojdyla and Sheng Cui analyzed the data and revised the paper. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.08.014.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim K.P., Ng L.F., Liu D.X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P. Mechanism and inhibition of SARS-CoV-2 PLpro. bioRxiv. 2020 doi: 10.1101/2020.06.18.160614. Available from: [DOI] [Google Scholar]
- 11.Bekes M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol Cell. 2016;62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bekes M., Rut W., Kasperkiewicz P., Mulder M.P., Ovaa H., Drag M. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem J. 2015;468:215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1101/2020.04.29.068890. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 18.Lee H., Lei H., Santarsiero B.D., Gatuz J.L., Cao S., Rice A.J. Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem Biol. 2015;10:1456–1465. doi: 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X., Zhu K., Wojdyla J.A., Chen P., Qin B., Li Z. Crystal structure of the NS3-like helicase from Alongshan virus. IUCrJ. 2020;7:375–382. doi: 10.1107/S2052252520003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Echols N., Grosse-Kunstleve R.W., Afonine P.V., Bunkoczi G., Chen V.B., Headd J.J. Graphical tools for macromolecular crystallography in PHENIX. J Appl Crystallogr. 2012;45:581–586. doi: 10.1107/S0021889812017293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui S., Wang J., Fan T., Qin B., Guo L., Lei X. Crystal structure of human enterovirus 71 3C protease. J Mol Biol. 2011;408:449–461. doi: 10.1016/j.jmb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishna S.S., Majumdar I., Grishin N.V. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krampe H., Ehrenreich H. Supervised disulfiram as adjunct to psychotherapy in alcoholism treatment. Curr Pharmaceut Des. 2010;16:2076–2090. doi: 10.2174/138161210791516431. [DOI] [PubMed] [Google Scholar]
- 30.Elliott J.H., McMahon J.H., Chang C.C., Lee S.A., Hartogensis W., Bumpus N. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV. 2015;2:e520–e529. doi: 10.1016/S2352-3018(15)00226-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y.M., Duh Y., Wang S.T., Lai M.M., Yuan H.S., Lim C. Using an old drug to target a new drug site: application of disulfiram to target the Zn-site in HCV NS5A protein. J Am Chem Soc. 2016;138:3856–3862. doi: 10.1021/jacs.6b00299. [DOI] [PubMed] [Google Scholar]
- 32.Bayoumy A.B., Simsek M., Seinen M.L., Mulder C.J.J., Ansari A., Peters G.J. The continuous rediscovery and the benefit-risk ratio of thioguanine, a comprehensive review. Expet Opin Drug Metabol Toxicol. 2020;16:111–123. doi: 10.1080/17425255.2020.1719996. [DOI] [PubMed] [Google Scholar]
- 33.Shaffer L. 15 drugs being tested to treat COVID-19 and how they would work. Nat Med. 2020 doi: 10.1038/d41591-020-00019-9. Available from: [DOI] [PubMed] [Google Scholar]
- 34.Janowitz T., Gablenz E., Pattinson D., Wang T.C., Conigliaro J., Tracey K. Famotidine use and quantitative symptom tracking for COVID-19 in non-hospitalised patients: a case series. Gut. 2020;69:1592–1597. doi: 10.1136/gutjnl-2020-321852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malone R.W., Tisdall P., Fremont-Smith P., Liu Y., Huang X.-P., White K.M. Famotidine, histamine, mast cells, and mechanisms. Research Square. 2020 doi: 10.21203/rs.3.rs-30934/v1. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loffredo M., Lucero H., Chen D.Y., O'Connell A., Bergqvist S., Munawar A. The Effect of famotidine on SARS-CoV-2 proteases and virus replication. bioRxiv. 2020 doi: 10.1101/2020.07.15.203059. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chantadul V., Wright G.S.A., Amporndanai K., Shahid M., Antonyuk S.V., Washbourn G. Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy. Commun Biol. 2020;3:97. doi: 10.1038/s42003-020-0826-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh N., Halliday A.C., Thomas J.M., Kuznetsova O.V., Baldwin R., Woon E.C. A safe lithium mimetic for bipolar disorder. Nat Commun. 2013;4:1332. doi: 10.1038/ncomms2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kil J., Pierce C., Tran H., Gu R., Lynch E.D. Ebselen treatment reduces noise induced hearing loss via the mimicry and induction of glutathione peroxidase. Hear Res. 2007;226:44–51. doi: 10.1016/j.heares.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Węglarz-Tomczak E., Tomczak J.M., Talma M., Brul S. Ebselen as a highly active inhibitor of PLpro CoV2. bioRxiv. 2020 doi: 10.1101/2020.05.17.100768. Available from: [DOI] [Google Scholar]
- 42.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 43.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzym Inhib Med Chem. 2014;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 44.Lin M.H., Moses D.C., Hsieh C.H., Cheng S.C., Chen Y.H., Sun C.Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou C.Y., Chien C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng K.W., Cheng S.C., Chen W.Y., Lin M.H., Chuang S.J., Cheng I.H. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swaim C.D., Perng Y.C., Zhao X., Canadeo L.A., Harastani H.H., Darling T.L. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro protease activities. bioRxiv. 2020 doi: 10.1101/2020.07.01.183020. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osipiuk J., Azizi S.-A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. bioRxiv. 2020 doi: 10.1101/2020.08.06.240192. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y. Structural basis for the inhibition of the papain-like protease of SARS-CoV-2 by small molecules. bioRxiv. 2020 doi: 10.1101/2020.07.17.208959. Available from: [DOI] [Google Scholar]
- 50.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh A.K., Takayama J., Rao K.V., Ratia K., Chaudhuri R., Mulhearn D.C. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J Med Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.