Abstract
Recently, a new coronavirus (SARS-CoV-2) was discovered in China. Due to its high level of contagion, it has already reached most countries, quickly becoming a pandemic. Although the most common symptoms are related to breathing problems, SARS-CoV-2 infections also affect the gastrointestinal tract culminating in inflammation and diarrhea. However, the mechanisms related to these enteric manifestations are still not well understood. Evidence shows that the SARS-CoV-2 binds to the angiotensin-converting enzyme receptor 2 (ACE2) in host cells as a viral invasion mechanism and can infect the lungs and the gut. Other viruses have already been linked to intestinal symptoms through binding to ACE2. In turn, this medical hypothesis article conjectures that the ACE2 downregulation caused by the SARS-CoV-2 internalization could lead to decreased activation of the mechanistic target of mTOR with increased autophagy and lead to intestinal dysbiosis, resulting in diarrhea. Besides that, dysbiosis can directly affect the respiratory system through the lungs. Although there are clues to other viruses that modulate the ACE2/gut/lungs axis, including the participation of autophagy and dysbiosis in the development of gastrointestinal symptoms, there is still no evidence of the ACE2/mTOR/autophagy pathway in SARS-CoV-2 infections. Thus, we propose that the new coronavirus causes a change in the intestinal microbiota, which culminates in a diarrheal process through the ACE2/mTOR/autophagy pathway into enterocytes. Our assumption is supported by premises that unregulated intestinal microbiota increases the susceptibility to other diseases and extra-intestinal manifestations, which can even cause remote damage in lungs. These putative connections lead us to suggest and encourage future studies aiming at assessing the aforementioned hypothesis and regulating dysbiosis caused by SARS-CoV-2 infection, in order to confirm the decrease in lung injuries and the improvement in the prognosis of the disease.
Keywords: Coronavirus, SARS-CoV-2, Diarrhea, Dysbiosis
Introduction
Coronavirus is a virus belonging to the Coronaviridaea family that has simple and enveloped RNA-type genetic material [1], [2]. It is related to the development of severe acute respiratory syndrome (SARS), the most severe form of the disease. According to WHO data, some outbreaks of coronavirus infection have raised concerns from the world population, such as SARS-CoV in 2002 (close to 8,000 cases and 800 deaths) and the Middle East Respiratory Syndrome, MERS-CoV, which infected approximately 2,500 people, causing 858 deaths [3], [4].
Recently, a new coronavirus was discovered in Wuhan, China. SARS-CoV-2 has a lower lethality rate when compared to the previous ones, but the contagion level is accentuated [5] reaching most of the countries worldwide, quickly becoming a pandemic [6]. Although the most common symptoms are related to respiratory problems, coronavirus infections also affect the gastrointestinal tract. Recent studies with SARS-CoV2 demonstrate a high concentration of viral genetic material in the anal swab of infected patients (approximately 40%) [7] and in wastewater [8], [9], thus characterizing intestinal infection. Intestinal infection by the virus can lead to a range of complications, from inflammation and intestinal cramps to diarrhea and although variable, can reach 30% of cases [10], [11].
Conceptually, diarrhea is characterized by an increase in intestinal transit causing in an intensified number of daily gut movements, 3 times or more and may be accompanied by mucus or blood depending on its origin, causing difficulty in the absorption of nutrients, loss of electrolytes and water [12], [13]. Among various etiological forms, there is viral diarrhea, which can be resulting from adenovirus, human calicivirus, astrovirus, rotavirus and norovirus, as common examples [14].
Several factors may be included in the diarrheal process, such as a decrease in resident microbiota due to the use of antibiotics (dysbiosis) causing imbalance of intestinal homeostasis [15] through the ACE2, which allows absorption of amino acids by enterocytes and consequently the production of antimicrobial peptides [16]. The signaling of intracellular pathways as the mechanistic target of rapamycin (mTOR) is a set of kinase-type proteins [17] that are directly associated to cellular autophagy, aiding in the flow of intestinal Na+/H+ [18] and channel openings through the ATPase pump (Na+/K+), which are also involved in this disorder [19], [20].
Dysbiosis has a fundamental role during the development of diarrheal conditions. However, it also plays an important role in the homeostasis of other systems and organs. Ahluwalia and colleagues [21] showed that changes in dysbiosis can cause disturbances in the intestine-liver-brain axis, associated with a pro-inflammatory systemic condition, altered brain function and liver cirrhosis. The imbalance of intestinal microbiota can affect the respiratory system through the lungs, since it contributes to pulmonary immunity and defense of the host [22]. Therefore, studies of the intestinal microbiota have been strongly related to the development of respiratory diseases [23], [24].
The hypothesis
SARS-CoV-2 is known to bind to ACE2, which is also present in enterocytes. The binding of the virus to the enzyme leads to its deregulation both in its ancestor SARS-CoV and in SARS-CoV-2. In other viruses, a decrease in mTOR was observed, triggered by virus internalization, which has already been associated with increased autophagy and dysbiosis. However, the mechanism of dysbiosis in infections by SARS-CoV-2 remains unknown up to this date. We speculate that the new coronavirus causes an alteration of the intestinal microbiota through the ACE2/mTOR/autophagy pathway in the enterocytes, which culminates in gastrointestinal symptoms in patients with COVID-19.
Evaluation of the hypothesis
Due to the urgency to find new therapeutic targets against SARS-CoV-2, several studies focus on understanding the coronavirus infection and replication tactics. Literature reports evidences that the viral surface spike glycoprotein (S protein) of SARS-CoV-2 binds to ACE2 and also requires proteolytic cleavage of S protein by host proteases, such as membrane-associated type II transmembrane serine protease (TMPRSS2) as a viral invasion mechanism [25], [26], [27], [28].
The virus is still known to have the capacity to infect and replicate in intestinal cells, being the viral genetic material directly identified in the intestine, since they also express ACE2 and TMPRSS2 [29], [30], [31]. That is believed because these patients can manifest enteric symptoms such as diarrhea, vomiting and abdominal pain in earlier COVID-19 course [32], [33], [34], [35]. On account of that, the implications of SARS-CoV-2 on the gastrointestinal tract must be taken into account in infection treatment and control [33], [34], [36].
Other coronaviruses caused by SARS-CoV and MERS-CoV have the ability to survive in the gastrointestinal environment, infect intestinal cells and cause diarrhea [32], [37]. However, an important observation is that while spike proteins in SARS-CoV and SARS-CoV-2 viruses bind to ACE2 in enterocytes to promote their gastrointestinal effects, while MERS-CoV binds to dipeptidyl peptidase 4 (DPP4), and can act by a different mechanism [28], [38].
Studies suggest that one of the reasons for the new coronavirus to cause the diarrheal process is probably due to a change in mucosal permeability of infected intestinal cells, resulting in malabsorption in the enterocytes [39]. Moreover, some other viruses cause intestinal disturbances through the ACE2 dysregulation [40], [41], [42]. That is probably because a putative ACE2 present in these cells is involved in amino acids uptake, such as tryptophan, and that results in mTOR activation, a regulator related to cell proliferation, synthesis and production of proteins, which is activated directly by the detection of nutrients or tryptophan/nicotinamide pathway [16]. mTOR acts on the expression of antimicrobial agents by Paneth cells in the small intestine, which regulates the intestinal microbiota. Studies suggest that when ACE2 is downregulated, there is a tryptophan reduction, which causes a decrease in mTOR activation and intestinal dysbiosis, resulting in diarrhea and increased susceptibility to intestinal inflammation from other diseases, such as colitis [16], [43]. In addition, scientific studies show that other proteins and substrates correlated with ACE2 deregulation, such as depletion of angiotensin 1–7 and neutral amino acid transporter B0AT1 deficiency, are correlated with decreased amino acid uptake, such as tryptophan, and gastrointestinal manifestations [43], [44], [45].
Still, an important fact to be discussed and investigated is that the mTOR present in the mTORC1 signaling complex is also related to autophagy, an endocytosis and degradation process of damaged intracellular components in a double-membrane structure, which fuses with a lysosome for the autophagolysome formation [46], [47]. Studies show that this process is involved with α and β-coronavirus replication, where the co-localization of proteins that participate in viral replication with endogenous LC3 (a protein marker for autophagosome) has been found [48]. The new coronavirus is believed to possibly use this same replication mechanism in human hosts [47].
The mTOR inhibition causes the activation of intestinal autophagy, demonstrated by the expression of the autophagosome marker Atg5. This process causes the autophagic degradation of NHE3, the abundant intestinal epithelial brush-border Na+/H+ exchanger protein located in enterocytes, related with the absorption of NaCl [18]. According to Yang and coworkers [18], the administration of rapamycin, an inhibitor of the mTORC1 complex, reduces NHE3 levels, with a consequent reduction in Na+/H+ exchange activity, with water absorption in the humans and rodents gut, and increase in intestinal fluids, featuring secretory-type diarrhea. Thus, we speculate that the role of mTOR would be to limit the formation of the autophagosome. The reduction of its activation, which is a consequent step followed by ACE2 downregulation during the SARS-CoV-2 infection into the intestinal cells, may lead to autophagy triggered and a subsequent viral replication accompanied by diarrhea.
Interestingly, studies have displayed that the autophagy process also regulates the intestinal microbiota, where its increase is related to diarrhea [49], [50]. Atg5, a protein that is required for the autophagosome formation, also regulates Paneth cells, which end up preventing microorganisms from coming into contact with the intestinal epithelial surface [49], [50]. These evidences lead us to infer that the autophagy process may be related to changes in the intestinal microbiota in COVID-19. Still, the microbiota by itself can also prompt the mTOR and directly regulate intestinal autophagy [51], [52], [53], [54]. As seen earlier, the deregulation of this pathway culminates in a secretory diarrheal process.
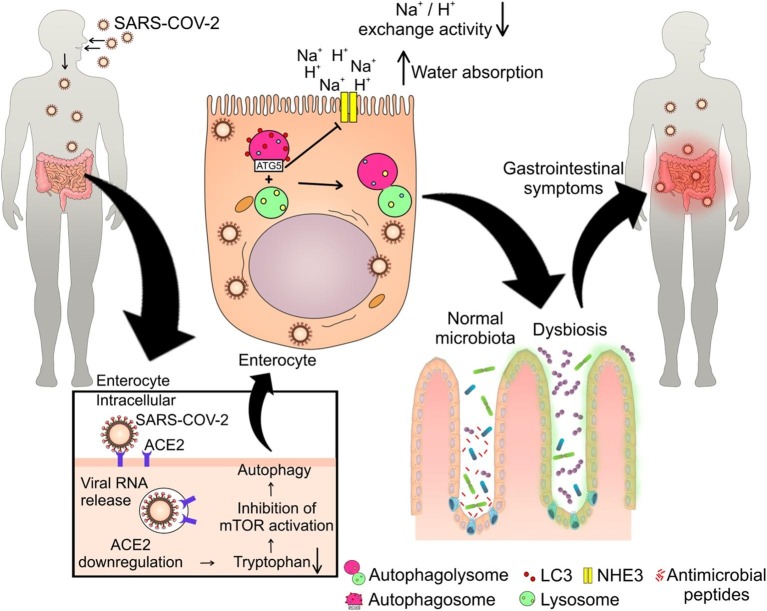
As previously reported, the mechanisms related to enteric manifestations caused by the new coronavirus are still not well understood. It is known that this virus alters the microbiota, but there is still no evidence involving the ACE2/mTOR/autophagy pathway in this process. The SARS-CoV-2 compilations show: i) interaction with ACE2 and followed by its downregulation; ii) there are evidences of the occurrence of intestinal dysbiosis; iii) mTORC1 is downregulated in cells infected with SARS-COV-2. Moreover, studies show that other coronaviruses are accompanied by an altered microbiota [33], [55], [56], [57], [58], [59], [60], [61]. Upon entering the cells via ACE2 [25], SARS-CoV-2 may be triggering the mTOR inhibition [57] that can culminate in the activation of autophagolysosomes and assembly into intestinal cells, causing dysbiosis, intestinal inflammation and diarrhea, as has been shown with the administration of rapamycin [18], [43]. Moreover, deregulations in the ACE2 pathway and substrates cause a reduction in the tryptophan uptake, a process identified as one of the causes of mTOR inhibition in other studies, which are related to intestinal disorders [16], [43], [44], [45]. Thus, we propose that SARS-CoV-2 causes alteration of the intestinal microbiota, which culminates in a diarrheal process through the ACE2/mTOR/autophagy pathway in enterocytes. Our hypothesis and the supporting evidences discussed above are shown in Fig. 1 .
Fig. 1.
Suggested mechanism of intestinal dysbiosis and diarrhea induced by SARS-COV-2. After entering the body, SARS-COV-2 manages to infect intestinal cells by binding with ACE2, which leads to the dysregulation of this human enzyme, which results in less uptake of tryptophan, which will cause less activation of mTOR. This can lead to autophagolysome formation that cause reduction in Na+/H+ exchange activity, increased water absorption and diarrhea. Moreover, the reduction of mTOR activity decrease the antimicrobial peptides production by Paneth cells, which will generate the intestinal dysbiosis and gastrointestinal problems related to intestinal microbiota imbalance.
Thus, the confirmation of the hypothesis tested may generate new therapeutic perspectives aimed at the microbiota focused on mitigate diarrheal symptoms in patients infected by SARS-CoV-2. Still, the literature reports that unregulated intestinal microbiota may increase susceptibility to other diseases and extra-intestinal manifestations, since the enteric immune system is important in the homeostasis of the immune system of other organs, such as the lung [62], [63].
Consequences of the hypothesis and discussion
Alterations in the composition and function of digestive tract microbiota may mutually affect the respiratory tract through immune regulation, through the so-called “intestine-lung axis” [62], [63]. Furthermore, a high level of pro-inflammatory cytokines caused by dysbiosis was previously associated with impairment of lung integrity in other diseases, such as tuberculosis [23]. Evidence shows that SARS-CoV-2 damages the digestive system through a chain of inflammatory responses [11].
However, up to the present date, no associations have yet been made between such gastrointestinal harm and lung injuries in SARS-CoV-2 infection through the orchestrated steps proposed in this hypothetical article. We believe that microbiota dysregulation caused by the possible increased autophagy from the ACE2/mTOR/autophagy pathway is leading to a positive feedback, where the increase in pro-inflammatory cytokines over intestinal dysbiosis may be associated with a worsening of lung injuries in positive patients for SARS-CoV-2. This evidence leads us to suggest and encourage future studies aimed at regulating dysbiosis in COVID-19 or underlying diseases in patients infected with the virus, as well as in patients undergoing antibiotic therapy, in order to confirm the decrease in lung injuries and the improvement in the prognosis of the disease.
It is noteworthy that gastrointestinal symptoms are part of the symptoms present in patients with COVID-19. Future studies are encouraged to investigate whether these patients previously had dysbiosis caused by a basic disease or poor diet that may have been amplified by the SARS-CoV-2 infection, using the path proposed in this medical hypothesis. These reports corroborate and support our hypothesis, demonstrating the importance of future studies elucidating mechanisms and treatments for diarrheal symptoms in SARS-CoV-2 infections in order to better understand the association between viral infection and symptoms on gastrointestinal tract, as well as the possible participation of dysbiosis in worsening pulmonary symptoms.
Author’s contributions
Ana Patrícia de Oliveira, André Luis Fernandes Lopes and Gabriella Pacheco developed and wrote the hypothesis. Isabela Noleto elaborated and developed the figure and format of the article. Jand Medeiros and Lucas Nicolau helped in the writing, review and scientific contribution.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the financial support from the National Council of Technological and Scientific Development—CNPq (Brazil), Concil for Advanced Professional Training (CAPES) and Research foundation for the state of Piauí—FAPEPI.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.110243.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Masters P.S., Perlman S. Coronaviridae. Fields Virology. 2013;6(1):825–858. [Google Scholar]
- 2.Song Z., Xu Y., Bao L. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):592019. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/emergencies/mers-cov/en/ (accessed April 14, 2020).
- 4.World Health Organization. Cumulative number of reported probable cases of Severe Acute Respiratory Syndrome (SARS) https://www.who.int/csr/sars/country/en/ (accessed April 14, 2020).
- 5.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N.W., Ke R. The novel coronavirus, 2019-nCoV, is highly contagious and more infectious than initially estimated. ArXiv.org. 2020:1–34. https://arxiv.org/abs/2002.03268. [Google Scholar]
- 6.Hick J.L., Biddinger P.D. Novel coronavirus and old lessons—preparing the health system for the pandemic. N Engl J Med. 2020:1–3. doi: 10.1056/NEJMp2005118. [DOI] [PubMed] [Google Scholar]
- 7.Chen W., Lan Y., Yuan X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodder W., Husman A.M.R. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F., Xiao A., Zhang J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MedRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W., Feng Z., Rao S. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020 doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 11.Pan L., Mu M., Yang P. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiller L.R. Definitions, pathophysiology, and evaluation of chronic diarrhoea. Best Pract Res Clin Gastroenterol. 2012;26(5):551–562. doi: 10.1016/j.bpg.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Whyte L.A., Jenkins H.R. Pathophysiology of diarrhoea. Paediatr Child Health (Oxford) 2012;22(10):443–447. doi: 10.1016/j.paed.2012.05.006. [DOI] [Google Scholar]
- 14.Musher D.M., Musher B.L. Contagious acute gastrointestinal infections. N Engl J Med. 2004;351(23):2417–2427. doi: 10.1056/NEJMra041837. [DOI] [PubMed] [Google Scholar]
- 15.Long C., Liu Y., He L. Bacterial lactase genes diversity in intestinal mucosa of mice with dysbacterial diarrhea induced by antibiotics. 3 Biotech. 2018;8(176):1–9. doi: 10.1007/s13205-018-1191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto T., Perlot T., Rehman A. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chagin A.S. Effectors of mTOR-autophagy pathway: targeting cancer, affecting the skeleton. Curr Opin Pharmacol. 2016;28:1–7. doi: 10.1016/j.coph.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Zhao X., Patel A. Rapamycin inhibition of mTOR reduces levels of the Na+/H+ exchanger 3 in intestines of mice and humans, leading to diarrhea. Gastroenterology. 2015;149(1):151–162. doi: 10.1053/j.gastro.2015.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araújo T.S.L., de Oliveira T.M., de Sousa N.A. Biopolymer extracted from anadenanthera colubrina (Red angico gum) exerts therapeutic potential in mice: antidiarrheal activity and safety assessment. Pharmaceuticals (Basel) 2020;13(1):1–27. doi: 10.3390/ph13010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachmilewitz D., Karmeli F., Sharon P. Decreased colonic Na-K-ATPase activity in active ulcerative colitis. Isr J Med Sci. 1984;20(8,):681–684. PMID: 6088428. [PubMed] [Google Scholar]
- 21.Ahluwalia V., Betrapally N.S., Hylemon P.B. Impaired gut-liver-brain axis in patients with cirrhosis. Sci Rep. 2016;6(1):1–11. doi: 10.1038/srep26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuelson D.R., Welsh D.A., Shellito J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front Immunol. 2015;1–14:1085. doi: 10.3389/fmicb.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan N., Vidyarthi A., Nadeem S., Negi S., Nair G., Agrewala J.N. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol. 2016;7:529. doi: 10.3389/fimmu.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuelson D.R., Siggins R.W., Ruan S. Alcohol consumption increases susceptibility to pneumococcal pneumonia in a humanized murine HIV model mediated by intestinal dysbiosis. Alcohol. 2019;80:33–43. doi: 10.1016/j.alcohol.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamers M.M., Beumer J., van der Vaart J. SARS-CoV-2 productively infects human gut enterocytes. bioRxiv. 2020 doi: 10.1101/2020.04.25.060350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang R., Castro M.F.G., McCune B.T. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5:1–10. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian Q., Fan L., Liu W. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan J.F.W., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Z., Wang Y., Qi W. The small intestine, an underestimated site of SARS-CoV-2 infection: from red queen effect to probiotics. Preprints. 2020 doi: 10.20944/preprints202003.0161.v1. [DOI] [Google Scholar]
- 34.Lee J.J., Kopetz S., Vilar E., Shen J.P., Chen K., Maitra A. Relative abundance of SARS-CoV-2 entry genes in the enterocytes of the lower gastrointestinal tract. bioRxivorg. 2020 doi: 10.1101/2020.04.08.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H., Kang Z., Gong H. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxivorg. 2020 doi: 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- 36.Wong S.H., Rashid N.S.L., Sung J.Y.J. Covid-19 and the digestive system. JGH Open. 2020 doi: 10.1111/jgh.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan P.K., To K.F., Lo A.W. Persistent infection of SARS coronavirus in colonic cells in vitro. J Med Virol. 2004;74(1):1–7. doi: 10.1002/jmv.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020:1–2. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer D.D., Kandasamy S., Paim F.C. Protein malnutrition alters tryptophan and angiotensin-converting enzyme 2 homeostasis and adaptive immune responses in human rotavirus-infected gnotobiotic pigs with human infant fecal microbiota transplant. Clin Vaccine Immunol. 2017;24(8):e00172–e217. doi: 10.1128/CVI.00172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shenoy V., Yang T., Rubiano A. ACE2 activator, diminazene, rebalances gut microbial dysbiosis and attenuates pulmonary hypertension. Hypertension. 2015;66(1) doi: 10.1161/hyp.66.suppl_1.028. A028–A028. [DOI] [Google Scholar]
- 42.Andrade J.M.O., Lelis D.F., Mafra V., Cota J. The angiotensin converting enzyme 2 (ACE2), gut microbiota, and cardiovascular health. Protein Pept Lett. 2017;24(9):827–832. doi: 10.2174/0929866524666170728145333. [DOI] [PubMed] [Google Scholar]
- 43.Perlot T., Penninger J.M. ACE2–From the renin–angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira L.P., Guimarães V.H.D., Oliveira J.R. Genetic deletion of the angiotensin-(1–7) receptor mas leads to alterations in gut villi length modulating TLR4/PI3K/AKT and produces microbiome dysbiosis. Neuropeptides. 2020;102056 doi: 10.1016/j.npep.2020.102056. [DOI] [PubMed] [Google Scholar]
- 45.Kleta R., Romeo E., Ristic Z. Mutations in SLC6A19, encoding B0AT1, cause Hartnup disorder. Nat Genet. 2004;36:999–1002. doi: 10.1038/ng1405. [DOI] [PubMed] [Google Scholar]
- 46.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci. 2020;16(10):1724. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cadwell K., Liu J.Y., Brown S.L. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008:456–1259. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cadwell K., Patel K.K., Komatsu M., Virgin I., Herbert W., Stappenbeck T.S. A common role for Atg16L1, Atg5, and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donohoe D.R., Garge N., Zhang X. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kar S.K., Jansman A.J.M., Benis N. Dietary protein sources differentially affect microbiota, mTOR activity and transcription of mTOR signaling pathways in the small intestine. PLoS ONE. 2017;12(11):1–19. doi: 10.1371/journal.pone.0188282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin P.K., Marchiando A., Xu R. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol. 2018;3(10):1131–1141. doi: 10.1038/s41564-018-0229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y., Chen F., Wu W. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11(3):752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson G., Reiter R.J. Melatonin: roles in influenza, covid-19 and other viral infections. Rev Med Virol. 2020;2109:1–10. doi: 10.1002/rmv.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao Q.Y., Chen Y.X., Fang J.Y. 2019 novel coronavirus infection and gastrointestinal tract. J Dig Dis. 2020;21(3):125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gassen N.C., Papies J., Bajaj T. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. BioRxivorg. 2020;1:13. doi: 10.1101/2020.04.15.997254. [DOI] [Google Scholar]
- 58.Gheblawi M., Wang K., Viveiros A. Angiotensin converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ Res. 2020:1–35. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Z., Xiao Y., Kang L. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin Infect Dis. 2019;2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meazzi S., Lauzi S., Stranieri A., Paltrinieri S., Giordano A. The gut microbiome and mucosal defenses in cats with coronaviruses: a pilot study. HAF. 2017;4(1s) doi: 10.13130/2283-3927/8410. [DOI] [Google Scholar]
- 61.Meazzi S., Stranieri A., Lauzi S. Feline gut microbiota composition in association with feline coronavirus infection: a pilot study. Res Vet Sci. 2019;125:272–278. doi: 10.1016/j.rvsc.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Budden K.F., Gellatly S.L., Wood D.L. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 63.Musa S. Hepatic and gastrointestinal involvement in coronavirus disease 2019 (COVID-19): what do we know till now? Arab J Gastroenterol. 2020;21(1):3–8. doi: 10.1016/j.ajg.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.