Abstract
Matrix-assisted laser desorption/ionization (MALDI)-MS imaging has been utilized to image a variety of biomolecules, including neuropeptides. Washing a tissue section is an effective way to eliminate interfering background and improve detection of low concentration target analyte molecules; however, many previous methods have not been tested for neuropeptide analysis via MALDI-MS imaging. Using crustacean as a neurological model organism, we developed a new, simple washing procedure and applied this method to characterize neuropeptide changes due to hypoxia stress. By using a 10 second 50:50 EtOH:H2O wash, neuropeptide coverage was improved by 1.15-fold, while normalized signal intensities were increased by 5.28-fold.Specifically, hypoxia and hypercapnia stress conditions were investigated due to their environmental relevance to marine invertebrates. Many neuropeptides, including RFamides, pyrokinin, and cardioactive peptides, showed distinct up- and down-regulation for specific neuropeptide isoforms. Since crustacean neuropeptides are homologous to those found in humans, results from these studies can be applied to understand potential roles of neuropeptides involved in medical hypoxia and hypercapnia.
Graphical Abstract
Introduction
Mass spectrometry (MS) imaging has found popularity because of its ability to provide relative molecular abundance and localization information simultaneously within a single tissue section. Unlike immuno-based assays, which require prior knowledge about the molecules of interest, thousands of species can be imaged in a single sample run, including unknown molecules.1 Originally developed in 1997, matrix-assisted laser desorption/ionization (MALDI)-MS imaging has been applied to several molecular species, including metabolites, peptides, and proteins.1–2 In general, MS imaging has the power to map the spatial localization of a molecule, which could provide important clue to possible function of this molecule.
As with all analytical techniques, the success of MALDI-MS imaging is dependent upon proper sample preparation. For MALDI-MS imaging, the basic workflow requires an optimized matrix application step after proper sectioning of the biological tissue. While this has been successful in the past, innovative techniques are being incorporated to this workflow to increase molecular depth and image quality.1 In particular, researchers are focusing on both (a) properly removing contaminants and (b) modifying molecules of interest prior to matrix application. Some examples include enzymatic digestion, chemical derivatization, and even incubation vapor chambers.1, 3–5 Washing is one of the simplest methods to remove contaminants and enrich molecules. By immersing the tissue section in a solvent of choice for a desired amount of time, one can fix proteins (e.g., Carnoy’s solution) or release molecules from formalin-fixed paraffin-embedded tissue section.1, 6–7 The most common and effective wash solvents seen in the literature are alcohol-based.8
Compared to proteins, very little effort has been put towards developing washing protocols for other molecular species, including neuropeptides.9 Dysregulation of neuropeptides, which are diverse signaling molecules in the brain, can have long-lasting physiological effects, suggesting they are of high interest in many social and biochemical behavioral studies.10 Extensive washing tends to remove peptides due to their solubility in water, thus these washes need to be short and precisely timed. One group used a 70% ethanol (EtOH) (10 seconds) followed by two 90% EtOH washes (10 seconds each) for rat neuropeptide analysis, although this sequential ethanol wash procedure had been primarily used for protein MS imaging analysis and unlikely was optimized for MS imaging of neuropeptides.11
Compared to mammals, crustaceans have been utilized as a model system for neuropeptide-based studies due to their simple, well-characterized networks.12 This is especially useful for comprehensive global analysis, which lends well to untargeted MS imaging-based studies.13 Crustacean neuropeptides are also known to be homologous to many of those found in humans, thus findings from crustacean-based studies can be applied to human studies in the future.14–17 Specifically, crustaceans have been used to study the role of neuropeptides in the stress response, including temperature and salinity stress.13, 18–19 Other stressors, including hypoxia (e.g., low oxygen (O2) levels) and hypercapnia (e.g., low O2 and high carbon dioxide (CO2) levels), are interesting due to their relevance to human respiratory distress (e.g., asthma) or disease (e.g., cancer).20–24 In particular, there is a lack of understanding of the molecular changes that occur due to hypoxia and hypercapnia stress.Hypoxia and related pH stress (i.e., hypercapnia) are well documented as environmental barriers for crustacean species.25–32 Cellular respiration is a key process in the body, so it is imperative to further our knowledge of hypoxia and hypercapnia’s effect on the nervous system.20
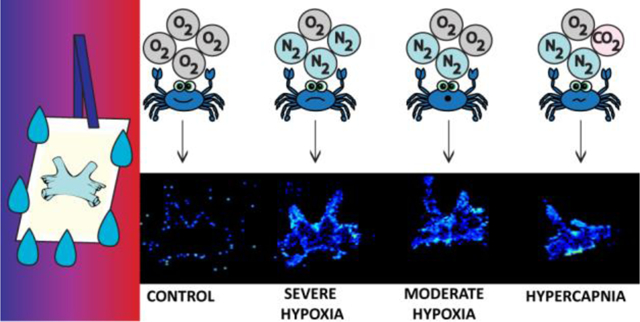
Here, we present a new washing method to enhance the detection of crustacean neuropeptides in brain tissue. Upon selection of a 10 second 50:50 EtOH:H2O wash, the new sample preparation method has been applied to characterize and quantify the localization changes of crustacean neuropeptides in the brain from four different stress conditions: (a) control (pH = 8.3, 100% O2 water saturation), (b) severe hypoxia (pH = 8.3, 10% O2 water saturation), (c) mild hypoxia (pH = 8.3, 50% O2 water saturation), and (d) hypercapnia (pH = 7.6–7.8, 50% O2 water saturation) for 2 hours. This study highlights (a) the first washing system that is optimized for neuropeptides and (b) a novel study of the neuropeptide localization and abundance changes due to hypoxia and hypercapnia stress in the crustacean model system.
Methods
Materials
All water (H2O) used in this study was doubly distilled on a Millipore filtration system (Burlington, MA) or Fisher HPLC grade. Difco™ gelatin, plain glass microscope slides, all crab saline components (see below), and methanol (MeOH) were obtained from Fisher Scientific (Pittsburgh, PA). EtOH utilized in this study was from Pharmco-Aaper (Chicago, IL). Formic acid (FA) was purchased from Fluka (Mexico City, Mexico), and the 2, 5-dihydroxybenzoic acid (DHB) was obtained from Acros Organics (Morris, New Jersey).
Animals and Stress Experiments
All female blue crabs, Callinectes sapidus, were either purchased from Midway Asian Market (Madison, WI) or LA Crawfish Company (Natchitoches, LA). After transport, crabs were allowed to recover in seawater made to be 35 parts per thousand salinity (i.e., salt concentration), 17–18 °C, 8–10 parts per million (ppm) O2 (80–100% O2 saturation), and pH = 8.3 for several (>5) days prior to being exposed to stressful conditions. For hypoxia experiments, a crab-less 10- gallon tank was sparged with N2 gas for 30–40 minutes to bring the dissolved O2 down to the desired level (i.e., 1 ppm or 5 ppm O2) (10% and 50% O2 saturation, respectively). For hypercapnia experiments, a crab-less 10-gallon tank was sparged with CO2 gas for 5 minutes to lower the pH to 7.6–7.8, which also lowered the dissolved O2 levels (i.e., 5 ppm O2) (50% O2 saturation). A plastic tarp is placed on top of the water’s surface to minimize H2O-air O2 exchange during the course of the experiment. A crab was then placed in the tank for the desired amount of time (i.e., 2 hours) before being anesthetized on ice and obtaining the brain as previously described.33 All dissections were performed in chilled (~ 10 °C) physiological saline (composition:440 mM NaCl; 11 mM KCl; 13 mM CaCl2; 26 mM MgCl2; 10 mM Trizma acid; pH 7.4 (adjusted with NaOH)).
Sample Preparation
Freshly dissected brains were embedded in gelatin (in small plastic cups) and sectioned using a Micro HM525 cryostat (Thermo Scientific). The 12 micron thick sections were thaw mounted onto plain glass microscope slides and stored at −80 °C until needed. Prior to use, samples were dried in a vacuum chamber for 10–20 minutes. Samples were then submerged into varying ratios of EtOH:H2O (i.e., 100:0, 85:15, 70:30, 50:50, 30:70, 15:85, and 0:100 (volume (v):v)) for either 10 or 30 seconds using a slide staining system (Tissue-Tek). The optimal wash for crustacean neuropeptides was found to be 50:50 EtOH:H2O for 10 seconds and applied to all samples post-optimization.
After drying under vacuum again (i.e., 15–20 minutes), matrix (i.e., 40 mg/mL DHB in 50:50 MeOH:H2O and 0.1% FA) was applied with a commercial TM-Sprayer (HTX Technologies, LLC) with the following optimized parameters: 0.1 mL/min syringe flow rate; 12 passes, 0.5 min dry time, 1250 mm/min nozzle velocity, 80 °C,and 3 mm track spacing. Samples were then analyzed immediately after matrix application. Control brains were used for all method optimization.
For pericardial organ (PO) imaging, freshly dissected pairs of POs were either submerged in water for 2 seconds (i.e., control) or 50:50 EtOH:H2O for 10 seconds (i.e., experimental) prior to being positioned in the slide. All slides were dried under vacuum and sprayed with matrix following the protocol above. Due to the thinness of the PO, no sectioning was required.
To determine if salts was being removed due to washing, the osmolarity (i.e., salt content) of the wash solution before and after the brain tissue section was submerged in it was determined by using an Advanced Instruments Model 325 Single-Sample Osmometer. To characterize the wash off, samples were spotted 1:1 with 150 mg/mL DHB (in 50:50 MeOH:H2O and 0.1% FA).
Data Collection and Analysis
Samples were analyzed on a MALDI-LTQ-Orbitrap XL (Thermo Fisher). A mass range of m/z 500–2000 was used along with the following parameters: 30,000 mass resolution (at m/z 400) and 75 micron spatial resolution. Raw data files were exported into an imZML format (using Thermo Fisher ImageQuest; Version 1.1.0 Build 54) to be imported into MSiReader (Version 1.00) or SCiLs lab software (Bremen, Germany).34 Files were loaded into MSiReader together and normalized to the TIC to generate images. Accurate mass matching (AMM) (±10 parts per million (ppm)) to a homebuilt database was used to manually identify neuropeptides and their distributions. Lipids were identified by AMM (± 5 ppm) to the online LIPID MAPS database.35 When distributions were compared, all images were normalized to the same intensity scale.
Co-localization analysis was performed using the Coloc2 FIJI plugin on FIJI (FIJI Is Just ImageJ).36 Images are overlaid in ImageJ using a linear least square regression for technical replicates of this analysis, using at least 5 points as coordinates for each registration. For intensity analysis, Pearson’s Correlation Coefficient (r) is used to measure the correlation between intensities of two images for each pixel. This analysis uses a scale from −1 to 1, where 1 is perfect correlation, 0 is no correlation, and −1 is anti-correlation. To evaluate spatial distribution conservation, we also use a Manders’ split coefficient analysis. The Manders’ coefficient is a value between 0 to 1, which provides information on the colocalization of two images, in that it measures how well the distributions of the images match based on the spatial distribution of the analyte. Each of the images was scaled using the same scale bar, and the color scale used in the analysis was greyscale. For comprehensive statistical analysis, we also perform a Costes analysis, which will tell us if the Pearson Correlation Coefficient and the Manders’ Coefficients are better than pure chance or not. This is done by shuffling the pixels in one of the images, and then reperforming the Manders’ analysis and Pearson Correlation analysis. A p-value of 1.0 means that none of the randomized images had better correlation.
To determine statistically significant changes between control, hypoxia, and hypercapnia samples, samples were identified by MMA (±5 ppm) and analyzed by a student’s t-test using SCiLs software (Bruker; Bremen, Germany). In order to correct for the multi-comparison analysis, a Bonferroni correction was applied for the four comparisons, decreased the desired p-value from 0.05 to 0.0125.37
Results and Discussion
Wash Optimization and Characterization
In order to increase neuropeptide signals in the crustacean brain tissue, several different washes with varying amount of EtOH:H2O were investigated: 100:0, 85:15, 70:30, 50:50, 30:70, 15:85, and 0:100 (v:v). All washes were investigated for either 10 seconds or 30 seconds. A basic workflow is shown in Figure 1.
Figure 1.
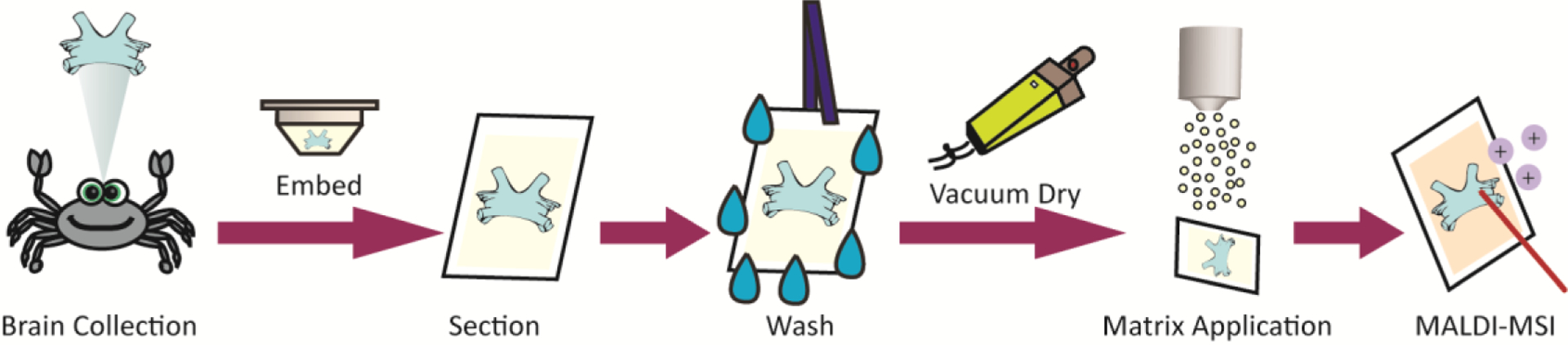
A pictorial schematic of the proposed wash method for crustacean neuropeptides. After collecting the brain from the control or exposed crab, the tissue is embedded in gelatin for sectioning using a cryostat. Next, sections are washed in the optimal wash solution and dried under vacuum. Matrix is then applied prior to be analyzed by MALDI-MS imaging.
As the amount of H2O increased, the number of neuropeptide identifications increased, with 50:50 to 15:85 (EtOH:H2O) showing the highest number of neuropeptides (Avg. ~30 neuropeptides), although 100% water showed a major decrease in neuropeptides when washed for 30 seconds compared to 10 seconds (18 versus 25 neuropeptides, respectively). The top 3 wash solutions were 50:50, 30:70, and 15:85 EtOH:H2O, and each time duration was investigated further with intensity analysis. Average normalized intensity showed an increase for all washes compared to a control by 3–5 fold, although the 50:50 wash, both 10 seconds and 30 seconds, provided the highest overall signal intensity (~5x), as shown in Figure 2.
Figure 2.
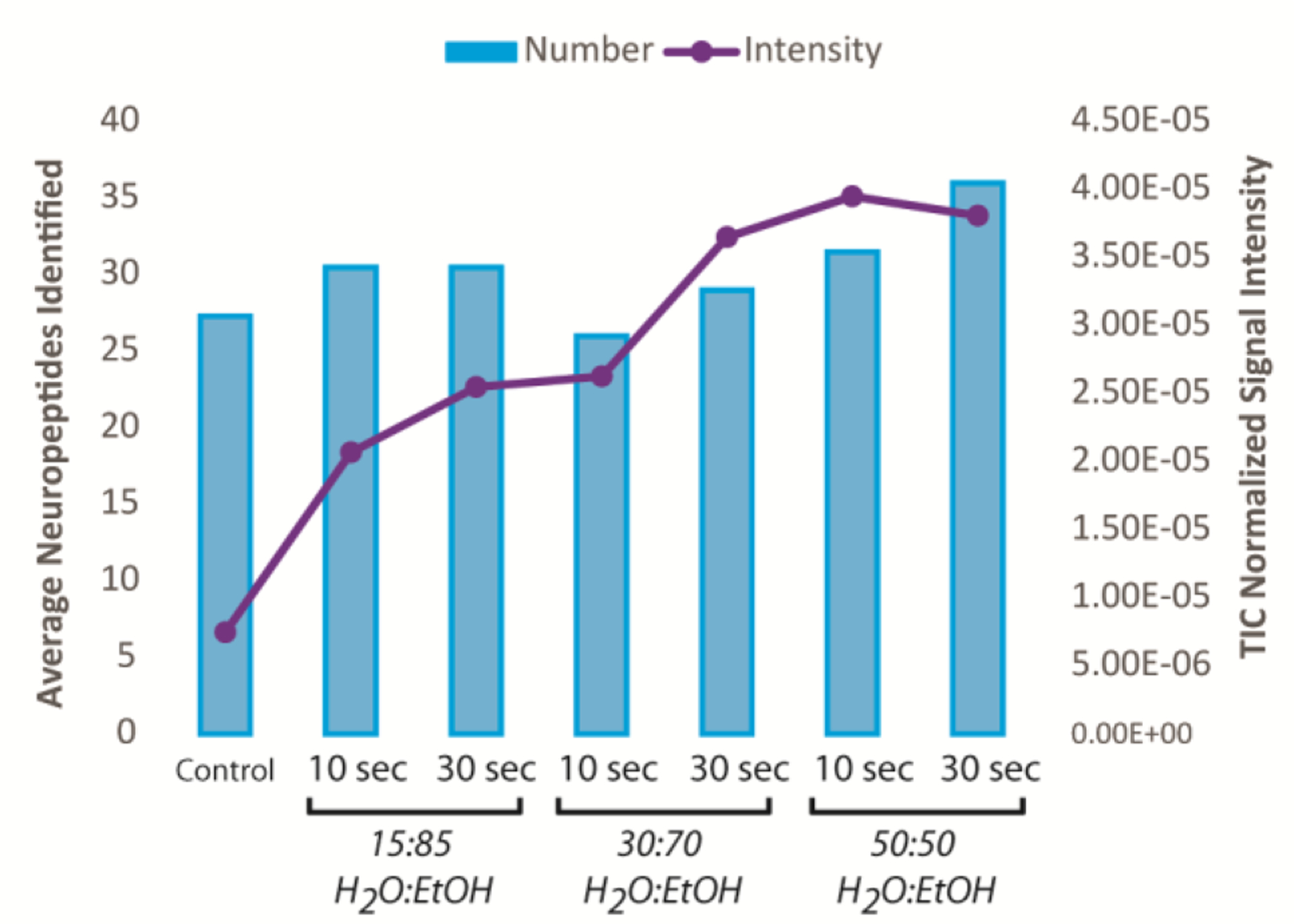
Comparison of the top three washing protocols with number of neuropeptides identified (blue bars) and the normalized signal intensity (purple dots) (n=3 technical replicates). The number of neuropeptides varied slightly between these conditions and a control, but the intensity spiked for the 50:50 H2O:EtOH washes.
In order to pick the most effective wash, co-localization analysis was performed on the 10 and 30 second 50:50 EtOH:H2O washes. For m/z 865.516 (RFamide RQFLRFamide), we obtained a Pearson’s correlation coefficient of 0.71 and a Mander’s split coefficients 1 and 2 values of 0.896 and 0.897, respectively (Table S1).36 The Pearson’s correlation coefficient, which ranges from −1 to 1, relates the intensity colocalization between two images, in which values closer to one are considered more colocalized.36 The Mander’s coefficient, which ranges from 0 to 1, determines spatial correlation between two images, where values closer to 1 are considered to be more correlated36. Thus, for m/z 865.516, it appears that the washes show identical changes to the localization of the neuropeptide. For m/z 1071.562 (TNYGGFLRFamide), 1119.646 (SMPTLRLRFamide), 1124.632 (GLSRNYLRFamide), and 1150.648 (ALDRNFLRFamide), high values for intensity and spatial distribution colocalization were found for both 10 second and 30 second washes (Table S1). Due to the lack of differences between these two wash conditions, the shorter, 10 second 50:50 EtOH:H2O was chosen as the optimal wash for neuropeptides in this study. It should be noted that these colocalization tests here were automatically thresholded in FIJI to account for non-zero pixel overlap, however new statistical colocalization algorithms are currently in development that are not dependent upon the background to minimize the accounted effect of non-zero pixel overlap, which can artificially increase colocalization coefficient values.36, 38
For this wash system (i.e., 50:50 EtOH:H2O for 10 seconds), several neuropeptides were enriched. In fact, compared to a control (n=4), 34 neuropeptides showed an increase in signal after washing. It should be noted that three neuropeptides did not show any signal change, and six neuropeptides showed a signal decrease after washing. Several examples are shown in Figure 3.
Figure 3.
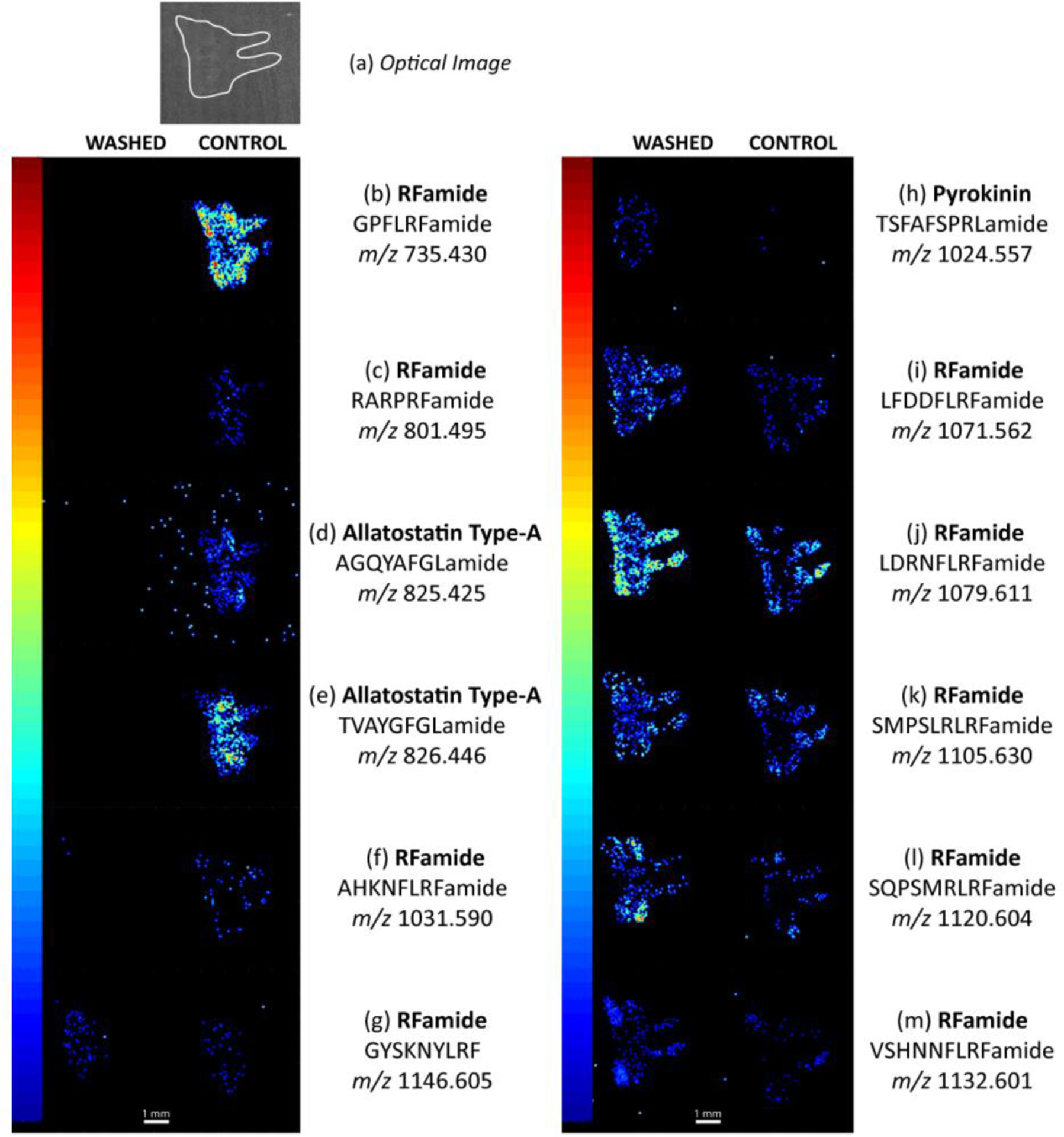
Examples of neuropeptide image changes due to the wash step in serial sections of a crustacean brain. (a) An optical image of the crustacean brain, outlined in white. The elongated tubes point towards the commissural ganglion of the crustacean nervous system. (b)-(f) Crustacean neuropeptides that were removed due to the washing step. (g) A neuropeptide image example that did not have a change in intensity or localization due to the washing step. (h)-(m) Neuropeptides that had a clear signal increase due to the washing step. All washed-control comparisons are generated at the same TIC normalized signal intensity level (0 to (b) 1.82×10−3, (c) 2.40×10−3, (d) 3.00×10−3, (e) 2.07×10−3, (f) 1.15×10−3, (g) 2.38×10−3, (h) 1.96×10−3, (i) 2.55×10−3, (j) 1.27×10−3, (k) 2.05×10−3, (l) 1.18×10−3, (m) 3.34×10−3). The white line represents a 1 mm scale bar.
These trends were seen consistently across bioreplicates and technical replicates, and an example of both is shown in Figures S1 and S2, respectively. While we lost some neuropeptides in the washing process, it was clear that there was an overall positive effect with enhanced detection of majority of neuropeptides due to the wash step. Compared to the brain, neuropeptide identifications (or the neuropeptides’ normalized intensities) in the PO tissue did not improve using the optimized wash system (i.e., 50:50 EtOH:H2O for 10 seconds) (n=4). This observation is likely due to the thinness of the tissue and its unique preparation and different molecular composition, as it does not require any sectioning and the tissue is less lipid-rich compared to the brain tissue.
Hydrophobicity analysis was also performed to find trends in which neuropeptides were removed while others increased in signal.Based upon the number of residues that are characterized as “hydrophobic” divided by the total number of residues, each peptide’s hydrophobicity was calculated and compared. Other online calculators, including Peptide 2.0, SSR Calculator, and Biosyn, were used to confirm our calculations and observations.39 Overall, it appears that there is no clear trend, but, interestingly, most of the peptides that are more hydrophobic show a signal decrease after the wash (Table S2). It should also be noted that most of these peptides were smaller, including m/z 808.435 (allatostatin A-type AAPYAFGLamide) and m/z 826.446 (allatostatin A-type TVAYGFGLamide), likely due to the fact that amino acids with hydrophobic side chains have a bigger effect on smaller peptides. There were obvious outliers to this trend though, including m/z 1019.590 (RFamide APRNFLRFamide), m/z 1061.574 (RFamide LPGVNFLRFamide), and m/z 1106.611 (RFamide LNPSNFLRFamide), which all decreased but showed low or no hydrophobic nature (Table S2). In general, smaller neuropeptides appeared to decrease in signal after being washed, while larger neuropeptides appeared to increase in signal, which could be related to hydrophobicity. Unfortunately, these results appear counterintuitive when considering how water-based washes tend to remove hydrophilic species, including salt. Thus, our results may be due to other, unknown molecular characteristics of the neuropeptides.
Several different avenues were investigated to determine factors that contributed to enhanced detection of neuropeptides after washing. Other biologically relevant species, such as lipids and interfering background such as salts, appeared to show signal increase or minor decrease, respectively, compared to control. Thus, while the washes were increasing neuropeptide signals, they were also increasing lipid signals, likely due to the removal of salts. Preliminary measurements of the osmolality of the pre- and post-wash solution (e.g., 1 and 4 mOsm before and after washing, respectively (n=1)) do indicate some salts are being removed. Although, the measured values were near zero and could be considered experimental variation when considering the rated standard deviation of osmolality measurements in this range (i.e., 0–400 mOsm/kg H2O is ±2 mOsm/kg H2O). When analyzing the post-tissue wash solution, it did contain a few neuropeptides, such as m/z 1061.574 (orcokinin TRPDIANLYamide), that were observed to be removed from the tissue, confirming that the wash itself was removing these neuropeptides. One trend to note was that many RFamides and other arginine containing neuropeptides were preferentially enriched. Arginine contains a guanidinium group, which has a high pKa, meaning it tends to maintain a positive change. This feature and other properties of arginine along with high proton affinity of arginine in the gas phase would help to explain enhanced detection of these neuropeptides after the washing procedure (i.e., 50:50 EtOH:H2O for 10 seconds) employed in this study.
Hypoxia and Hypercapnia Comparison
The optimized wash system was utilized to characterize the neuropeptidomic changes between four different conditions: a) control (pH = 8.3, 100% O2 water saturation), (b) severe hypoxia (pH = 8.3, 10% O2 water saturation), (c) mild hypoxia (pH = 8.3, 50% O2 water saturation), and (d) hypercapnia (pH = 7.6–7.8, 50% O2 water saturation), all exposed for 2 hours. A representative group of neuropeptides that were calculated to have statistically significant changes are shown in Table S3.
Figure 4 illustrates several neuropeptides that are dysregulated due to hypoxia or hypercapnia stress. In some cases, there were clear changes in intensity and localization, such as Figures 4d–f, while Figures 4b–c require more sophisticated computational analysis to discriminate changes. Using the student’s t-test for determining statistical significance (Table S3), many of the conditions were different from the control, including m/z 975.541 (cardioacceletory peptide (CAP) pELYAFPRVamide), m/z 1031.590 (RFamide AHKNFLRFamdie), m/z 1061.626 (RFamide LPGVNFLRFamide), and m/z 1079.611 (RFamide LDRNFLRFamide). In particular, Figure 4d shows that the neuropeptide (RFamide AHKNFLRFamide, m/z 1031.590) was absent in the control but was readily detected when the crustacean was exposed to hypoxia or related stress. This was also the case in Figures 4f and 4g. The opposite trend was seen in Figure 4e, where the neuropeptide appears to be released into the hemolymph (i.e., crustacean hemolymph) or degraded due to exposure to stress since there was only localization around the edge of the tissue. Figure 4b and 4c are difficult to infer what was occurring in the system, although it was clear that the neuropeptides exhibited substantial changes in relative abundance throughout the entire tissue (Figure 4b) or in specific ganglia (Figure 4c).
Figure 4.

Examples of neuropeptides and their changes due to hypoxia and hypercapnia stress compared to a control. (a) Optical image of crab brain under each washed condition, outlined in white. The elongated tubes point towards the commissural ganglion of the crustacean nervous system. (b)-(g) Statistically significant (p<0.0125) neuropeptides between either severe hypoxia vs. control, moderate hypoxia vs. control, hypercapnia vs. control, severe hypoxia vs. moderate hypoxia, and/or moderate hypoxia vs. hypercapnia, with a table of these results provided in Table S2. All control-hypoxia-hypercapnia comparisons are generated at the same TIC normalized signal intensity level (0 to (b) 2.22×10−4, (c) 5.31×10−4, (d) 5.65×10−4, (e) 6.03×10−4, (f) 6.73×10−4, (g) 8.29×10−4). The white line represented a 1 mm scale bar. All biological replicates of each condition for the m/z values shown are available in Figures S3–8.
Besides comparing in situ peptide expression patterns only between the stress condition and a control, differences were also tested between both the moderate and severe hypoxia (i.e., was there difference in localization and intensity due to the hypoxic severity?) and moderate hypoxia and hypercapnia (i.e., were the changes due to the hypoxia or also the inclusion of CO2?). This can be aligned with statistically significant differences from the control (m/z 1065.595, 1119.646, and 1120.604) as well as not being significantly different from the control (m/z 1007.579). In the case of m/z 1007.579 (RFamide PKSNFLRFamide), both the two hypoxia severities were different from each other, suggesting that dysregulation of this neuropeptide was related to the hypoxic severity. More biological replicates are required to distinguish these minute differences. Another interesting trend was that several neuropeptides only exhibited significant changes due to pH stress, such as m/z 1024.557 (RFamide GLSRNYLRFamide), which suggested that the changes were directly related to the inclusion of CO2, not just hypoxia.
Interestingly, most of the neuropeptides exhibiting statistically significant changes were RFamides, a crustacean neuropeptide family that is homologous to opioids and neuropeptide Y (NPY).15–17 In crustaceans, RFamides, also known as FMRFamide-like peptides (FLPs), have various well documented functions, including modulation of the cardiac and stomatogastric neuromuscular system along with exoskeleton muscles.40 They also work as autocrine/paracrine modulators while also circulating as hormones. Overall, it is obvious that RFamides can play various roles due to its diverse targets and functional roles. NPY has already been implicated in hypoxia stress (e.g., in rats), demonstrating that crustacean neuropeptides function similarly to those in higher order organisms.41–42 Another crustacean neuropeptide family (i.e., tachykinin) that is homologous to human substance P, which has previously been implicated in hypoxia stress were also identified in our experiments.14, 41 Like RFamides, tachykinin and related peptides work both locally and as long-distance circulating hormones as well as being implicated in decapod neuromuscular modulation. Unlike RFamides, most tachykinin-related neuropeptides did not exhibit statistically significant changes except one (m/z 605.304; APSFGQamide). For this neuropeptide, only the two hypoxia severities (i.e., 10% vs. 50% O2) and the hypoxia and hypercapnia (i.e., 50% O2 vs. pH stress) conditions were significantly different, meaning none of the 3 stress conditions were significantly different from a control. This could mean that, since RFamides seem to play a more comprehensive role in how crustaceans handle hypoxia and hypercapnia stress, NPY could also be a more dominant player how humans handle respiratory distress and other hypoxia/hypercapnia prominent conditions.
Conclusions
Crustacean neuropeptides are important signaling molecules due to their role in the stress response. Using MALDI-MS imaging, a new wash-based sample preparation protocol was developed in order to improve detection of neuropeptides and better understand their role in both hypoxia and hypercapnia stress. It would be of interest to further expand our ability to image other crustacean neuropeptides, specifically by utilizing different matrices and exploring other enrichment strategies. Comparison to quantitative analysis of tissue and hemolymph samples is also needed to fully understand the dynamic changes due to stress, such as the distinction between degradation and release into the crustacean hemolymph (i.e., blood). Furthermore, this wash method can be applied to future imaging studies including mapping their localization in other crustacean tissues (e.g., pericardial organ) as well as other biological systems.
Supplementary Material
Acknowledgements
This work was supported by a National Science Foundation grant (CHE-1710140) and the National Institutes of Health (NIH) through grants 1R01DK071801 and R56 MH110215. The MALDI-LTQ-Orbitrap instrument was purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531). J.J. and L.L. acknowledge the University of Wisconsin Carbone Cancer Center Pancreas Cancer Task Force for the funds to complete this project. We would also like to thank the Zeeh Station in the University of Wisconsin-Madison School of Pharmacy for use of the Osmometer.A.R.B. acknowledges the NIH General Medical Sciences F31 Fellowship (1F31GM119365) for funding. N.Q.V. would like to thank a UW-Madison Biotechnology Training Program Traineeship sponsored by the National Institute of General Medical Sciences of the NIH under Award Number T32GM008349.K.D. acknowledges a predoctoral fellowship supported by the National Institutes of Health, under Ruth L. Kirschstein National Research Service Award T32 HL 007936 from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center and the National Institutes of Health-General Medical Sciences F31 National Research Service Award (1F31GM126870) for funding. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Supporting Information: Supporting Figures S1–S8 and Supporting Tables S1–S3 (.docx)
References
- 1.Buchberger AR; DeLaney K; Johnson J; Li LJ, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Analytical Chemistry 2018, 90 (1), 240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caprioli RM; Farmer TB; Gile J, Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Analytical Chemistry 1997, 69 (23), 4751–4760. [DOI] [PubMed] [Google Scholar]
- 3.Heijs B; Holst S; Briaire-de Bruijn IH; van Pelt GW; de Ru AH; van Veelen PA; Drake RR; Mehta AS; Mesker WE; Tollenaar RA; Bovee J; Wuhrer M; McDonnell LA, Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Analytical Chemistry 2016, 88 (15), 7745–7753. [DOI] [PubMed] [Google Scholar]
- 4.Barre FPY; Flinders B; Garcia JP; Jansen I; Huizing LRS; Porta T; Creemers LB; Heeren RMA; Cillero-Pastor B, Derivatization Strategies for the Detection of Triamcinolone Acetonide in Cartilage by Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Analytical Chemistry 2016, 88 (24), 12051–12059. [DOI] [PubMed] [Google Scholar]
- 5.Angerer TB; Magnusson Y; Landberg G; Fletcher JS, Lipid Heterogeneity Resulting from Fatty Acid Processing in the Human Breast Cancer Microenvironment Identified by GCIB-ToF-SIMS Imaging. Analytical Chemistry 2016, 88 (23), 11946–11954. [DOI] [PubMed] [Google Scholar]
- 6.O’Rourke MB; Raymond BBA; Djordjevic SP; Padula MP, A versatile cost-effective method for the analysis of fresh frozen tissue sections via matrix-assisted laser desorption/ionisation imaging mass spectrometry. Rapid Communications in Mass Spectrometry 2015, 29 (7), 637–644. [DOI] [PubMed] [Google Scholar]
- 7.O’Rourke MB; Padula MP, Analysis of formalin-fixed, paraffin-embedded (FFPE) tissue via proteomic techniques and misconceptions of antigen retrieval. Biotechniques 2016, 60 (5), 229-+. [DOI] [PubMed] [Google Scholar]
- 8.Seeley EH; Oppenheimer SR; Mi D; Chaurand P; Caprioli RM, Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. Journal of the American Society for Mass Spectrometry 2008, 19 (8), 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchberger A; Yu Q; Li L, Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annu Rev Anal Chem (Palo Alto Calif) 2015, 8 (1), 485–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hokfelt T; Broberger C; Xu ZQD; Sergeyev V; Ubink R; Diez M, Neuropeptides - an overview. Neuropharmacology 2000, 39 (8), 1337–1356. [DOI] [PubMed] [Google Scholar]
- 11.Hanrieder J; Ljungdahl A; Andersson M, MALDI imaging mass spectrometry of neuropeptides in Parkinson’s disease. Journal of visualized experiments : JoVE 2012,(60). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins PM, The eyes have it: A brief history of crustacean neuroendocrinology. General and Comparative Endocrinology 2012, 175 (3), 357–366. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y; Buchberger A; Muthuvel G; Li L, Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satake H; Kawada T; Nomoto K; Minakata H, Insight into tachykinin-related peptides, their receptors, and invertebrate tachykinins: a review. Zoolog Sci 2003, 20 (5), 533–49. [DOI] [PubMed] [Google Scholar]
- 15.Husson SJ; Mertens I; Janssen T; Lindemans M; Schoofs L, Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog Neurobiol 2007, 82 (1), 33–55. [DOI] [PubMed] [Google Scholar]
- 16.Dockray GJ, The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol 2004, 89 (3), 229–35. [DOI] [PubMed] [Google Scholar]
- 17.Coast GM; Schooley DA, Toward a consensus nomenclature for insect neuropeptides and peptide hormones. Peptides 2011, 32 (3), 620–31. [DOI] [PubMed] [Google Scholar]
- 18.Chen RB; Xiao MM; Buchberger A; Li LJ, Quantitative Neuropeptidomics Study of the Effects of Temperature Change in the Crab Cancer borealis. Journal of Proteome Research 2014, 13 (12), 5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JH; Zhang YZ; Xiang F; Zhang ZC; Li LJ, Combining capillary electrophoresis matrix-assisted laser desorption/ionization mass spectrometry and stable isotopic labeling techniques for comparative crustacean peptidomics. Journal of Chromatography A 2010, 1217 (26), 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolfe DFS; Brown GC, Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological Reviews 1997, 77 (3), 731–758. [DOI] [PubMed] [Google Scholar]
- 21.Harris AL, Hypoxia - A key regulatory factor in tumour growth. Nature Reviews Cancer 2002, 2 (1), 38–47. [DOI] [PubMed] [Google Scholar]
- 22.Kelsen SG; Fleegler B; Altose MD, Respiratory Neuromuscular Response to Hypoxia, Hypercapnia, and Obstruction to Air-Flow in Asthma. American Review of Respiratory Disease 1979, 120 (3), 517–527. [DOI] [PubMed] [Google Scholar]
- 23.Semenza GL, HIF-1: mediator of physiological and pathophysiological responses to hypoxia. Journal of Applied Physiology 2000, 88 (4), 1474–1480. [DOI] [PubMed] [Google Scholar]
- 24.Sud S; Friedrich JO; Taccone P; Polli F; Adhikari NKJ; Latini R; Pesenti A; Guerin C; Mancebo J; Curley MAQ; Fernandez R; Chan MC; Beuret P; Voggenreiter G; Sud M; Tognoni G; Gattinoni L, Prone ventilation reduces mortality in patients with acute respiratory failure and severe hypoxemia: systematic review and meta-analysis. Intensive Care Medicine 2010, 36 (4), 585–599. [DOI] [PubMed] [Google Scholar]
- 25.Bell GW; Eggleston DB; Noga EJ, Environmental and Physiological Controls of Blue Crab Avoidance Behavior During Exposure to Hypoxia. Biological Bulletin 2009, 217 (2), 161–172. [DOI] [PubMed] [Google Scholar]
- 26.Bell GW; Eggleston DB; Noga EJ, Molecular keys unlock the mysteries of variable survival responses of blue crabs to hypoxia. Oecologia 2010, 163 (1), 57–68. [DOI] [PubMed] [Google Scholar]
- 27.Brouwer M; Larkin P; Brown-Peterson N; King C; Manning S; Denslow N, Effects of hypoxia on gene and protein expression in the blue crab, Callinectes sapidus. Mar Environ Res 2004, 58 (2–5), 787–92. [DOI] [PubMed] [Google Scholar]
- 28.Stover KK; Burnett KG; McElroy EJ; Burnett LE, Locomotory Fatigue During Moderate and Severe Hypoxia and Hypercapnia in the Atlantic Blue Crab, Callinectes sapidus. Biological Bulletin 2013, 224 (2), 68–78. [DOI] [PubMed] [Google Scholar]
- 29.Tanner CA; Burnett LE; Burnett KG, The effects of hypoxia and pH on phenoloxidase activity in the Atlantic blue crab, Callinectes sapidus. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology 2006, 144 (2), 218–223. [DOI] [PubMed] [Google Scholar]
- 30.Chung JS; Zmora N, Functional studies of crustacean hyperglycemic hormones (CHHs) of the blue crab, Callinectes sapidus - the expression and release of CHH in eyestalk and pericardial organ in response to environmental stress. Febs Journal 2008, 275 (4), 693–704. [DOI] [PubMed] [Google Scholar]
- 31.Hardy KM; Burnett KG; Burnett LE, Effect of hypercapnic hypoxia and bacterial infection (Vibrio campbellii) on protein synthesis rates in the Pacific whiteleg shrimp, Litopenaeus vannamei. American Journal of Physiology-Regulatory Integrative and Comparative Physiology 2013, 305 (11), R1356–R1366. [DOI] [PubMed] [Google Scholar]
- 32.Hardy KM; Follett CR; Burnett LE; Lema SC, Gene transcripts encoding hypoxia-inducible factor (HIF) exhibit tissue- and muscle fiber type-dependent responses to hypoxia and hypercapnic hypoxia in the Atlantic blue crab, Callinectes sapidus. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology 2012, 163 (1), 137–146. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez GJ; Grashow RG, Cancer borealis stomatogastric nervous system dissection. J Vis Exp 2009,(25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bokhart MT; Nazari M; Garrard KP; Muddiman DC, MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. Journal of the American Society for Mass Spectrometry 2018, 29 (1), 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sud M; Fahy E; Cotter D; Brown A; Dennis EA; Glass CK; Merrill AH; Murphy RC; Raetz CRH; Russell DW; Subramaniam S, LMSD: LIPID MAPS structure database. Nucleic Acids Research 2007, 35, D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aaron JS; Taylor AB; Chew TL, Image co-localization - co-occurrence versus correlation. Journal of Cell Science 2018, 131 (3), 10. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y; Hochberg Y, CONTROLLING THE FALSE DISCOVERY RATE - A PRACTICAL AND POWERFUL APPROACH TO MULTIPLE TESTING. Journal of the Royal Statistical Society Series B-Methodological 1995, 57 (1), 289–300. [Google Scholar]
- 38.Wang SL; Arena ET; Eliceiri KW; Yuan M, Automated and Robust Quantification of Colocalization in Dual-Color Fluorescence Microscopy: A Nonparametric Statistical Approach. Ieee Transactions on Image Processing 2018, 27 (2), 622–636. [DOI] [PubMed] [Google Scholar]
- 39.Wang S; Xiao CS; Jiang LY; Ling L; Chen XS; Guo XH, A high sensitive and contaminant tolerant matrix for facile detection of membrane proteins by matrix-assisted laser desorption/ionization mass spectrometry. Analytica Chimica Acta 2018, 999, 114–122. [DOI] [PubMed] [Google Scholar]
- 40.Christie AE; Stemmler EA; Dickinson PS, Crustacean neuropeptides. Cell Mol Life Sci 2010, 67 (24), 4135–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poncet L; Denoroy L; Dalmaz Y; Pequignot JM; Jouvet M, Alteration in central and peripheral substance P- and neuropeptide Y-like immunoreactivity after chronic hypoxia in the rat. Brain Research 1996, 733 (1), 64–72. [DOI] [PubMed] [Google Scholar]
- 42.Lee EW; Michalkiewicz M; Kitlinska J; Kalezic I; Switalska H; Yoo P; Sangkharat A; Ji H; Li LJ; Michalkiewicz T; Ljubisavljevic M; Johansson H; Grant DS; Zukowska Z, Neuropeptide Y induces ischemic angiogenesis and restores fimction of ischemic skeletal muscles. Journal of Clinical Investigation 2003, 111 (12), 1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


