Abstract
α-Linolenic acid (ALA, 18:3n-3) and γ-gamma linolenic acid (GLA, 18:3n-6) are polyunsaturated fatty acids (PUFA) that improve the human health. The present study focused on testing the in vitro antitumor actions of pure ALA and GLA on the HT-29 human colorectal cancer cell line. Cell viability was checked by MTT ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) test, cell membrane damage by the lactate dehydrogenase assay, apoptosis was tested by both caspase-3 activity trial and transmission electron microscopy images, and protein composition was analyzed by quantitative proteomics analysis. MTT test revealed IC50 values of 230 and 255 μM for ALA and GLA, respectively, at 72 h. After 24 h of incubation, both ALA and GLA induced apoptosis on HT-29 colorectal cancer cells according to the caspase-3 assay and microscopy images. SWATH/MS analysis evidenced that ALA significantly affected the mitochondrial protein import pathway and the citric acid cycle pathway, while GLA did not significantly affect any particular pathway. In summary, both ALA and GLA showed concentration-dependent inhibitory effects on HT-29 cells viability and induced cell death by apoptosis. ALA significantly affected cellular pathways, while GLA does not have specific actions on either pathway. Both n-3 and n-6 C18 PUFA are bioactive food components useful in the colorectal cancer prevention.
Keywords: α-linolenic acid, γ-linolenic acid, colorectal cancer, HT-29 cells, SWATH-MS, caspase-3
Graphical Abstract
Graphical Abstract.
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the fourth leading cause of death, with mortality close to 50% in Western countries, followed by lung cancer [1]. Also, several studies suggest that it is a disease strongly influenced by diet [2, 3], which can modulate inflammatory processes and also affect gene expression in the body and, hence, is linked to the incidence of several diseases [4]. Polyunsaturated fatty acids (PUFA) play a positive role against CRC cells [5, 6] and glioblastoma [7], among others. There are two important PUFA families in nature: the n-3 series, whose precursor is α-linolenic acid (ALA, 18:3n-3) and the n-6 series, which derives from linoleic acid (LA, 18:2n-6). These FA cannot be synthesized de novo in the body, so they must be obtained through diet to support normal growth and development. They are known as essential FA (EFA) [8]. ALA is a long-chain n-3 PUFA found in some seeds (chia and flaxseed) and nuts, and in some animal fats, which can be elongated and desaturated into eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n3), which develops a wide range of beneficial functions in the body [9, 10]. The available literature indicates that ALA has therapeutic potential in cervical cancer by modulating the expression of cyclooxygenase-2 (COX-2), which is a protein complex that controls DNA transcription called NF-κB, MMPs, vascular endothelial growth factor (VEGF) and c-Jun proteins. In addition, ALA affects key members of the mitogen-activated protein kinase signalling pathway such as ERK1/2 and p38 proteins in cervical [11] and breast cancer cells [12]. In breast cancer cells, Wiggins et al. [12] showed that ALA reduces cell growth and increases apoptosis. As for γ-linolenic acid (GLA, 18:3n-6), it is produced from LA by the action of the enzyme ∆6-desaturase, and is found in small amounts in some seed oils such as those of borage, black currant and evening primrose [13]. GLA has gained importance due to its anti-inflammatory and anti-cancer actions mediated by apoptosis and lipid peroxidation [14], and exerts a cytotoxic action by the release of lactate dehydrogenase (LDH) [15]. Also, GLA and its metabolites have been reported to modify the expression of important molecules that play a crucial role in the induction of apoptosis, such as COX-2 and MKP-1 [4, 16]. However, Awad et al. [17] concluded that ALA and GLA supplementation had no consistent effect on tumour growth on HT-29 CRC cells, while phospholipase C activity was not influenced by membrane FA modification.
According to De Roose and Romagnolo [18], proteomic technologies may be useful in determining the effects of FA on cancer growth. In this regard, a recently emerging technique, the sequential windowed acquisition of all theoretical fragment ion mass spectra (SWATH) mass spectrometry (MS) has been reported to provide an accurate quantitation of proteins compared to the traditional techniques [19].
This study was designed to elucidate the antitumor activity of ALA and GLA on the HT-29 CRC cell line and specifically, to identify the mechanisms involved in cell death and the pathways affected by in vitro exposition of cells to these PUFA.
Materials and Methods
Oil samples
Linseed (Linum usitatissimum L.) oil was used to obtain pure ALA, while GLA was purified from evening primrose (Oenothera biennis L.) oil. Both oils were obtained in the local markets of Almeria (Spain).
Purification process
GLA and ALA purification was carried out after the direct saponification and further fractionation of the oils with urea according to Spurvey and Shahidi [20]. The urea:FA ratio was 6:1 (w/w), and the urea:methanol ratio was 1:3 (w/v). PUFA were recovered with n-hexane to be analyzed by gas chromatography (GC).
GC analyses
FA determination was carried out by transesterification of FA to FA methyl esters (FAME) according to previous works [21, 22]. Briefly, 10 mg of sample and 50 μl of an internal standard (98% purity heptadecanoic acid, 17:0), H3500 from Sigma, St. Louis, USA) were introduced in test tubes and then 1 ml of n-hexane (10 mg/ml) and 1 ml of freshly prepared transesterification reagent (methanol and acetyl chloride 20:1 v/v) were subsequently added at 100°C for 30 min. FAME were analyzed in a Focus GC (Thermo Electron, Cambridge, UK) equipped with flame ionization detector (FID) and an Omegawax 250 capillary column (30 m × 0.25 mm ID × 0.25-μm film thickness) (Supelco, Bellefonte, PA, USA) The temperature of the oven was 90°C (1 min), 10°C/min–100°C (3 min), 6°C/min–260°C (5 min), and the injector temperature was 250°C with a split ratio of 50:1 and a volume of 4 μl. The detector temperature was 260°C, and the flow of carrier gas was 1 ml/min [23]. The peak area of the internal standard was used as a reference to compute the mass of every FA in the resulting chromatograms, and results were computed as FA% of total FA.
Cell assays
We used the well-established HT-29 CRC cell line, which was supplied by the Technical Instrumentation Service of University of Granada (Granada, Spain). This cell line is considered a pluripotent intestinal cell line, which can be used for the study of several structural and molecular events involved in cancer cell differentiation [24, 25]. The tests were the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) at 48 and 72 h, and caspase-3 at 24 h. Both were performed according to the manufacturer’s instructions and according to Ortea et al. [6]. LDH assay (Cytotoxicity Detection KitPLUS (LDH), Kit 04 744 926 001, Sigma-Aldrich, St. Louis, MO, USA) was conducted the same as for the MTT assay, but in this case, the cell density was 0.8 × 104 cells/well, and measurements were performed following the instructions of the manufacturer. The results were obtained by determining the absorbance at 490 nm with a reference filter at 690 nm at 24 h.
Transmission electron microscopy
The images of transmission electron microscopy (TEM) were obtained according to Ramos-Bueno et al. [26] in a Zeiss Libra 120 TEM (Carl Zeiss AG, Jena, Germany).
Quantitative proteomics analysis
The 24-h-cultured HT-29 cells in media supplemented with 470 μM ALA (n = 6) or 360 μM GLA (n = 6), and a control group of no-supplemented cells (n = 6), were recovered and lyzed to obtain protein extracts. These were cleaned by TCA/acetone precipitation, resuspended in 0.2% of RapiGest SF (Waters, Milford, MA, USA) and total protein was quantified using the Qubit Protein Assay kit (Thermo Scientific, Waltham, MA, USA). For each sample, 50 μg of protein were trypsin digested as previously described [27]. RePliCal iRT peptides (PolyQuant GmbH, Bad Abbach, Germany) were spiked into each sample to calibrate the peptide retention times later in the SWATH runs. The proteomes from the three cell groups (ALA, GLA and control) were quantitatively compared using a SWATH-MS approach as previously described [28]. Briefly, this approached consisted of identifying all detectable proteins in the samples using a data-dependent acquisition (DDA) LC-MS/MS method, building a peptidic MS/MS spectral library, analyzing the samples using a SWATH LC-MS data-independent acquisition (DIA) and using the peptide library to extract the peptide quantitative information from the SWATH chromatographic traces. For both DDA and DIA LC-MS analysis, a hybrid Quadrupole time-of-flight (Q-TOF) mass spectrometer (Triple TOF 5600+, Sciex, Redwood City, CA, USA) coupled to a nano-HPLC system (Ekspert nLC415, Eksigent, Dublin, CA, USA) was used. Chromatography gradient was 5–30% B (A: 0.1% FA in water; B: 0.1% in ACN) in 120 min, at a flow rate of 300 nl/min, using a 25-cm long × 75-μm internal diameter column (Acclaim PepMap100, Thermo Scientific) and a 2-cm × 100-μm trap column (Acclaim PepMap100, Thermo Scientific). For DDA runs, each group of six samples were pooled and run twice with a top 65 method, consisting on a TOF MS survey scan (250 ms acquisition time, 350–1250 m/z range) followed by the MS/MS scan (60 ms, 230–1700 m/z) of the highest ions as found in the survey scan, making a total cycle time of 4.2 s. The six runs were searched all together against a SwissProt human protein database (containing 20 200 protein entries, appended with the RePliCal iRT peptides and downloaded from UniProt on March 2017), using Protein Pilot v5.0.1 (Sciex), setting iodoacetamide as Cys alkylation, trypsin as enzyme and Triple TOF 5600 as instrument. The false discovery rate (FDR) was set to 1% for both peptides and proteins. For the SWATH DIA runs, each sample was analyzed using a variable window SWATH acquisition method consisting on a TOF MS (50 ms, 350–1200 m/z) followed by 60 MS/MS (90 ms) of precursor windows of variable m/z width in the whole range 350–1200 m/z. The size of the MS/MS windows was optimized according to the ion density found in the previous DDA runs. SWATH Acquisition MicroApp (v2.0, Sciex) was used for building the peptide spectral library and for extracting the fragment ion chromatographic traces from the SWATH runs. Peptide retention times were calibrated in all the SWATH runs using the RePliCal iRT peptides, spiked into each sample according to manufacturer’s instructions. To be confident on the proteins being quantified, only those showing confidence scores above 99% and FDR below 1% were included in the analysis. Protein abundances were normalized for inter-run variability using Marker View v1.3.1 (Sciex).
Pathway analysis
The significantly affected pathways were studied using the Reactome tool (https://reactome.org/), which is designed to give the user a graphical map of known biological processes and pathways with detailed information on components and their relations. We considered a restrictive scenario, namely a differential expression threshold of log (fold change) > 2.0 and adjusted P-value < 0.01, in order to have more confidence in selecting the proteins that presented real expression changes.
Statistical analysis
For cell assays (MTT, LDH and caspase-3 assays), statistical significance (P < 0.05) was determined by generalized linear models (GZLMs) and analysis of variance (ANOVA) using STATGRAPHICS Plus, version 5 (Statistical Graphic Corp., Warranton, VA, USA). Figure 1 shows mean data for three independent experiments ± standard deviation (SD). Calculations and data plotting for Fig. 1 were performed using Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA). For SWATH-MS protein quantitation, differences in protein abundance between groups were assessed by applying a Mann–Whitney nonparametric test, checking for multiple testing underestimation of P-values by obtaining a q-value estimation for FDR using the qvalue R package (accessed at http://github.com/jdstorey/qvalue on 29 March 2019).
Figure 1.
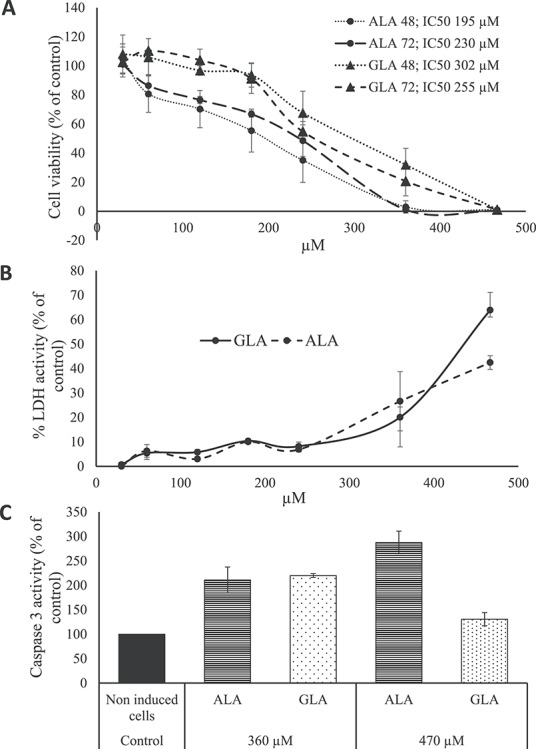
Plots showing results of cell assays. (A) Dose-dependent viability of HT-29 cells after ALA and GLA exposure for 48 and 72 h. (B) Dose-dependent LDH release from HT-29 colon cancer cells after ALA and GLA exposure. (C) Dose-dependent caspase-3 activity from HT-29 colon cancer cells in comparison with untreated cells (control). Data represent the mean of three complete independent experiments ± SD (error bars). Data were analyzed using GZLMs. There are no significant differences (P < 0.05) among series sharing the same letter.
Results and Discussion
The purity of the assayed PUFA was 97.8 and 94.6% for ALA and GLA.
ALA and GLA effects on HT-29 cells viability measured by the MTT test
To check the anti-proliferative activity of ALA and GLA on the HT-29 CRC cells, the MTT cell viability assay in both concentration and time-dependent manner was performed. Figure 1A shows the effects of ALA and GLA on HT-29 cells viability at different concentrations (30–470 μM) for 48 and 72 h. As shown in Fig. 1A, both ALA and GLA significantly decreased HT-29 cells viability in a dose-dependent manner, although both PUFA lacked a significant time response. The half-maximal inhibitory concentrations (IC50) at 48 and 72 h for ALA were 195 and 230 μM, while for GLA, values were 302 and 255 μM. In this regard, Zhang et al. [29] indicated 140–150 μM (ALA) and 200–300 μM (GLA) as the most effective concentrations for reducing LoVo and RKO CRC cells viability. For ALA, Chamberland and Moon [10], by the MTT test, showed a decrease of cell viability between 20 and 50% in HT-29 cells after a 1–5 mM treatment. Also, dose- and time-dependent growth inhibitory actions on breast cancer cells were reported for ALA in a [30], whereas Koralek et al. [31] found that ALA inhibited the production of urokinase, a protease enzyme that enhance carcinogenesis. Concerning GLA, in K562/ADM leukaemia cells, IC50 values were 113 and 101.59 μM at 48 and 72 h, respectively, [32, 14]. Taken together, the concentrations of ALA and GLA used in this study to inhibit the growth of HT-29 cells were higher than those reported for other cancer cells, although lower than concentrations determined as effective by Chamberland and Moon [10] in the same cell line. In this regard, Jiang et al. [33] and Habermann et al. [9] found that the growth inhibitory actions exerted by various PUFA on HT115 and LT97 CRC cells were significantly higher than that exercised on HT-29 cells. This fact may explain the high concentrations of ALA and GLA needed in this work to inhibit HT-29 cells growth. In previous works, we described experimentation on in vitro exposition of CCD-18 (normal colon cell line) to different PUFA (including both ALA and GLA) and no actions on cells viability were observed [26]. Thus, we conclude that the effects showed here are specific against cancer cells.
LDH assay
The LDH assay measures the activity of the LDH enzyme in the extracellular medium. The present work is the first study on the effects of GLA and ALA on cell membrane integrity of CRC cells. The release of LDH into the cellular medium is an indicator of irreversible cell death due to cell membrane damage [34]. To test whether cell number reduction was due to ALA- and GLA-mediated cell membrane damage, the LDH activity was measured (Fig. 1B). Note that LDH release increased in parallel with GLA and ALA concentrations: ALA and GLA at 30–240 μM showed no more than a 10% increase in LDH activity (% of control), whereas at 360 and 470 μM showed 63.9 and 42.4% LDH activity (% of control), respectively. In this regard, Kong et al. [32] studied the antitumor actions of GLA on human leukemic K562/ADM cells at 144 μM and concluded that significant LDH release indicated losses of plasma membrane integrity and the presence of necrosis, while Kim et al. [30] pointed out that ALA-treated breast cancer cells (0–100 μM) showed a significant increase in LDH release on a dose-dependent manner.
Caspase-3 assay
Apoptosis involves the sequential activation of a group of enzymes in the cysteine protease family named caspases. Caspase-3, which is activated in the apoptotic cell both by extrinsically (death ligand) and intrinsically (mitochondrial) pathways, is a caspase protein that interacts with caspase-8 and caspase-9 [35]. Thus, it is reliable indicator of cell death. According to Kapoor and Huang [4], the antitumor actions of GLA are mediated by apoptosis, inhibition of angiogenesis and gene activation, and also by production of eicosanoids via di-homo-gamma linolenic acid (DGLA, 20:3n-6), which competes with arachidonic acid (ARA, 20:4n-6) for several enzymes to produce prostaglandins and thromboxane’s. This conversion from GLA to DGLA is due to the ∆6-desaturase enzyme, which is very active in breast and CRC cells, according to Pender-Cudlip et al. [36]. In this work, it was found that both ALA- and GLA-induced significant caspase-3 activity as compared to untreated samples: ALA- and GLA-activated caspase-3 at 360 and 470 μM (Fig. 1C). Unlike expected, caspase-3 activity decreased significantly after 24 h incubation for 470 μM GLA-treated cells, as compared to 360 μM. Overall, these results agree well with recent results of Roy et al. [37] and Moravcová et al. [38], which showed a decrease of caspase-3 activity at high ALA and OA concentrations in breast cancer cells and hepatocytes. This effect could be related to the differential rate of incorporation of ALA and GLA into the cells due to differences in spatial configuration between n-3 PUFA (ALA) and n-6 PUFA (GLA). The latter could modify the membranes’ physical properties, bonds to receptors as well as transport [39]. In this regard, Tranchant et al. [40] concluded the existence of a carrier-mediated transport system for ALA and probably other long-chain PUFA, which could have low efficiency for GLA. Thus, high doses of GLA would not result in increased caspase-3 activity, and lingering-free GLA would be more prone to oxidation under the applied experimental conditions. Then, the generated H2O2 by the peroxidation process could inhibit apoptosis by depleting cells of ATP [41]. In other cancer cell lines, induction of apoptosis was found using lower ALA and GLA concentrations than those used here: Kong et al. [32] and Gillis et al. [42] reported that leukemic cells survival was reduced using doses of 144 and 100 μM GLA by inducing of apoptosis; while Deshpande et al. [43] indicated that 28–80 μM ALA treatment induced apoptosis in both breast and cervical cancer cell lines by activating of caspase-3. Taken together both LDH and caspase-3 tests, antitumor activities reached optimal values at 360 (GLA) and 470 (ALA) μM, especially for the caspase-3 trial. Thus, this was the concentration selected to conduct proteomic assays.
TEM
The goal of this study was to obtain TEM images of HT-29 CRC cells with and without the addition of antitumor-inducing agent (Fig. 2). Cells were supplemented with either with 470 μM ALA or 360 μM GLA. This tool helped to elucidate the various effects occurring in HT-29 CRC cells due to the presence either of GLA or ALA. Both GLA and ALA induced the appearance of amorphous mitochondria, apoptotic bodies and ‘ghost cells’, which are typical images of cells that have completed apoptosis and move erratically as granular debris. Note the presence of lipid droplets (LD), which in all cases are surrounded by endoplasmic reticulum. Previous studies [26, 29] on antitumor PUFA also indicated the presence of LD in PUFA-treated cancer cells. Although the role of LD in the development of cancer is still under investigation [44], it is assumed that these are involved in inflammatory and neoplastic processes, and it has been documented that CRC cells accumulate LD, with a documented PGE2 synthase localization and focal PGE2 synthesis [44, 45]. Bouzza and Viola [44] concluded that the inhibition of LD formation involves the inhibition of PGE2 synthesis, which is closely related to the pathogenesis of CRC. Another function of LD is to store FA, which will be used by the cancer cell in case of nutritional stress. In this sense, Jarc et al. [46] concluded that low concentrations (50 μM) of PUFA such as LA, EPA, DHA and ARA in breast cells were associated with protection from cell death due to starvation-induced degradation of LD, while higher concentrations (100 μM) induced cell death by oxidative stress of breast cancer cells. Gustin et al. [47] reported that 100 μM PUFA treatment stimulated LD accumulation in cervical and breast cancer cells, while 10 μM doses resulted in a slight decrease in LD content. Therefore, LD appearance depends on cell type and PUFA concentration, and it is not a conclusive sign of apoptosis caused by PUFA treatment.
Figure 2.
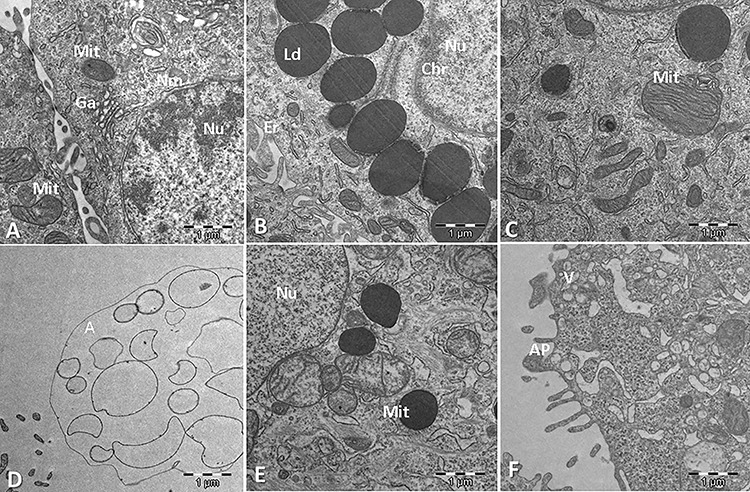
Morphological changes induced by ALA and GLA in HT-29 cells. A. Control cell, without cytotoxic agents added. Mit: mitochondria; Nu: nucleus; Nm: nuclear membrane; Ga: Golgi apparatus. B. Cells treated with GLA. Ld: lipid droplets. Note the intense chromatin (Chr) condensation. C. Cells treated with GLA. Notice broken amorphous Mit. D. Cells treated with ALA. Note the image of a cell that has completed apoptosis and is dispersing as granular debris (the ghost cell stage). E. Cells treated with ALA. Notice LD and broken amorphous Mit. F. Cells treated with ALA. Notice an apoptotic body (AP) budding off and heterogenous vesicles (V).
Quantitative proteomics analysis
For the massive identification of the proteins detectable in the samples, the DDA LC-MS/MS runs were searched against a human protein database using the search engine Protein Pilot. A total of 18 469 peptides and 2188 protein groups were identified with a 1% FDR at both peptide and protein levels. The list of identified proteins is shown in Supplementary Table S1. The file resulting from the database search was used to build a peptide spectral library, which contained 2090 proteins. This library, matching the MS/MS fragment ions to an identified peptide and a protein, was used to extract fragment ions chromatographic traces from the SWATH runs, obtaining quantitative data for a total of 1972 proteins in all the analyzed samples (Supplementary Table S2). Heat maps for each of the comparisons are shown for the fast visualization of the global quantitative result (Fig. 3). It can be seen that while control and ALA samples are completely separated from each other, meaning ALA addition is affecting globally the proteome, GLA samples could not be clearly separated from control samples, this meaning the effect of adding GLA is not so large. Supplementary Tables S3 and S4 show the results for the differential abundance tests for GLA vs. control and for ALA vs. control group comparisons, respectively, together with the corresponding fold changes found for each of the 1972 quantified proteins. For downstream analysis, a restrictive scenario of P-value<0.01 and a fold-change above 2.0 were considered, in order to be more confident in being selecting the proteins with real expression changes. According to these thresholds, 226 proteins were found to present expression changes as an effect of ALA supplementation (148 proteins up-regulated and 78 proteins down-regulated) (Supplementary Table S4). Interestingly, for GLA-treated samples, only eight proteins showed up as presenting differential abundance: six were up-regulated by GLA (TMM33, UD110, 5NTC, ATLA3, ODPX and HACD3) and two were down-regulated (PTMS and SEC13) (Supplementary File Table S3). These figures indicate a deeper effect on the HT-29 cell proteome for ALA than for GLA.
Figure 3.
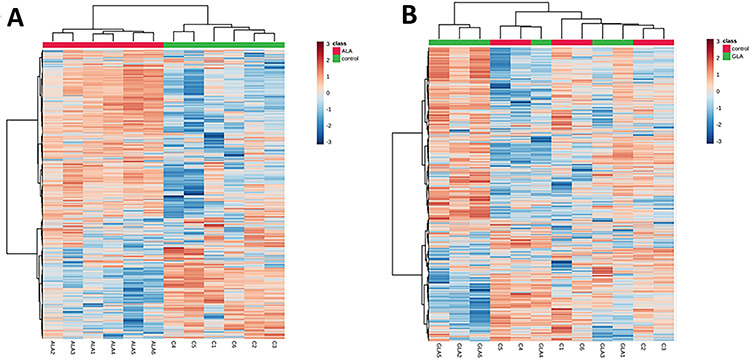
Heat maps including the quantitation results for 1972 proteins in all samples, for (a) ALA vs. control comparison and (b) GLA vs. control comparison.
Reactome pathway analysis
Significantly affected pathways were analyzed using Reactome software, which is a curated database of pathways and reactions in human biology. Reactome defines a ‘reaction’ as any event in biology that changes the state of a biological molecule. Pathways are represented by simple diagrams following an SBGN-like format. A binomial test is used to calculate the probability shown for each result, and P-values are corrected for the multiple testing (Benjamini–Hochberg procedure) that emerges from the evaluation of the submitted list of identifiers against each pathway. ALA showed more significant pathways in our analysis. A reactome pathway visualization is shown in Fig. 4. In our results, we found 77 and 41 pathways affected by ALA and GLA, respectively (P-value < 0.05), while FDR correction reduced the number of affected pathways to eight and zero, respectively (Supplementary Tables S5 and S6). With these data, we can conclude that GLA is not having a specific effect on any particular pathway. As far as ALA is concerned, the most significantly affected pathways were the mitochondrial protein import and the citric acid (TCA) cycle. In the mitochondrial protein import pathway, we found 11 proteins with significant differential abundance, being four of them up-regulated. Two of these up-regulated proteins are TOM40 and SAM50. While Ott et al. [48] concluded that TOM40 acts with the Bax translocation, Asmarinah et al. [49] suggested that the activities of the TOM complexes do not play any essential role in the development of prostate cancer. Regarding SAM, Rondepierre et al. [50] found three mitochondrial proteins in melanoma cells that had higher expression and were involved in the import protein machinery (SAM50). Mitochondrial homeostasis and function are dependent on the activity of the mitochondrial protein import machinery. For instance, recent findings imply a role of import machinery components in cancer development: numerous constituents of mitochondrial protein translocases are often up-regulated in cancer tissues, and this phenomenon is indicative of a poor survival prognosis [51]. TIMM17A, a mitochondrial inner membrane transport protein was much higher in breast cancer cell lines than in normal breast tissue and was directly correlated with breast cancer progression, so Xu et al. [52] indicated that TIMM17A may serve as a breast cancer marker in the clinic. The TCA cycle is a central route for oxidative phosphorylation in cells, macromolecule synthesis and redox balance occurring in the mitochondrial matrix. In the TCA pathway, we found 15 proteins with differential abundance, eight of them up-regulated and seven down-regulated. Among the up-regulated proteins, the cytochrome c oxidase (COX) should be highlighted. COX is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Also, Wu et al. [53] among others demonstrated an up-regulation of COX Va and Vb in CRC. Another interesting up-regulated protein found is PDHX, which is located in the mitochondrial matrix and is critical for mitochondrial fuel metabolism. Anderson et al. [54] concluded that different cancers are affected by changes in the TCA cycle enzymes, which result in characteristic metabolic changes that are correlated with disease transformation and progression. Montal et al. [55] showed that enzyme phosphoenolpyruvate carboxykinase (PEPCK) has the ability to regulate the TCA cycle and to metabolize lactate via the TCA cycle, so inhibition of PEPCK decreases lactate utilization and subsequently tumour cell growth. Another pathway affected by ALA is protein folding. The endoplasmic reticulum (ER) is the organelle responsible for intracellular Ca2+ homeostasis, lipid biosynthesis and protein folding and transport. Protein folding in the ER is highly sensitive to changes in the environment, which lead to the disruption of protein folding to cause accumulation of unfolded or misfolded proteins causing ER stress. The unfolded protein response (UPR) is a collection of pathways that evolved to maintain a productive ER protein-folding environment [56]. UPR activation contributes to both enhanced survival and cancer-cell-induced apoptosis.
Figure 4.
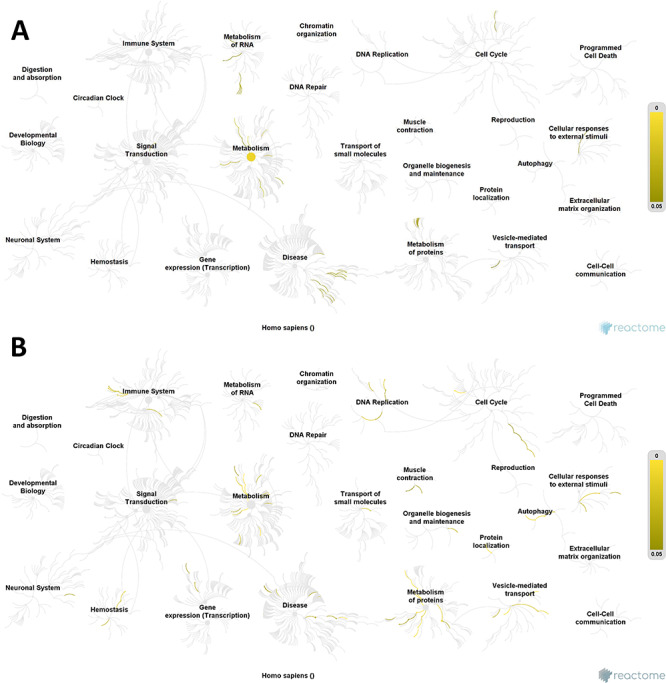
Reactome pathway visualization. Pathways affected by (A) GLA supplementation; (B) ALA supplementation. Yellow lines indicate number of pathways affected in the different groups, according to the differentially expressed proteins found. Note that ALA has a greatest effect than GLA due to the greater number of affected pathways such as metabolism of proteins and DNA replication pathways.
Conclusions
In summary, this work evidences that both ALA and GLA induced concentration-dependent inhibitory effects on HT-29 cells viability, evidenced by the MTT assay and cell membrane damage detected by the LDH test. The activation of caspase-3 suggests that ALA and GLA induce cell death by the mechanism of apoptosis. This fact is supported by the transmission electron microscopic images, which show deformation of certain organelles such as mitochondria and ghost cell formation. Analysis of pathways affected by ALA and GLA treatments has demonstrated that ALA significantly affects some cellular pathways, with the cycle of citric acid being most affected. However, GLA has no specific actions on any particular pathway. Future in vivo experimentation should be focused on the possibility of using ALA- and GLA-rich oils as bioactive food supplements for CRC prevention.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifierPXD017029.
Supplementary Material
Funding
MS analyses were performed at the IMIBIC Proteomics Unit. Ignacio Ortea is funded by the Miguel Servet Programme (grant CP19/00164) from the Instituto de Salud Carlos III, co-funded by the European Social Fund.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest
There are is conflict of interest to declare.
Abbreviations
ANOVA, analysis of variance; ALA, α-linolenic acid (18:3n-3); ARA, arachidonic acid (20:4n-6) CRC, colorectal cancer; COX-2, cyclooxygenase-2; DDA, data-dependent acquisition; DHA, docosahexaenoic acid (22:6n-3); EFA, essential FA; EPA, eicosapentaenoic acid (20:5n-3); ER, endoplasmic reticulum; FA, fatty acid ; FAME, FA methyl ester; GC, gas chromatography; GLA, γ-linolenic acid (18:3n-6); LA, linoleic acid (18:2n-6); LD, lipid droplet; LDH, lactate dehydrogenase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PEPCK, phosphoenolpyruvate carboxykinase; PUFA, polyunsaturated fatty acid; Q-TOF, Quadrupole time-of-flight; SWATH-MS, sequential windowed acquisition of all theoretical MS ; TCA, citric acid cycle; UPR, unfolded protein response; VEFG, Vascular Endothelial Growth Factor.
References
- 1. Vasaikar S, Huang C, Wang X et al. . Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 2019;177:1035 –49. doi: 10.1016/j.cell.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michalak A, Mosinska P, Fichna J. Polyunsaturated fatty acids and their derivatives: therapeutic value for inflammatory, functional gastrointestinal disorders, and colorectal cancer. Front Pharmacol 2016;7:1 –16. doi: 10.3389/fphar.2016.0045910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pietrzyk Ł. Food properties and dietary habits in colorectal cancer prevention and development. Int J Food Prop 2017;20:2323 –43. doi: 10.1080/10942912.2016.1236813. [DOI] [Google Scholar]
- 4. Kapoor R, Huang YS. Gamma Linolenic acid: an antiinflammatory omega-6 fatty acid. Curr Pharm Biotechnol 2006;7:531 –4. doi: 10.2174/138920106779116874. [DOI] [PubMed] [Google Scholar]
- 5. Colquhoun A, Schumacher RI. γ-Linolenic acid and eicosapentaenoic acid induce modifications in mitochondrial metabolism, reactive oxygen species generation, lipid peroxidation and apoptosis in Walker 256 rat carcinosarcoma cells. Biochim Biophys Acta 2001;1533:207 –19. doi: 10.1016/S1388-1981(01)00136-6. [DOI] [PubMed] [Google Scholar]
- 6. Ortea I, Gonzalez-Fernandez MJ, Ramos-Bueno RP et al. . Proteomics study reveals that docosahexaenoic and arachidonic acids exert different in vitro anticancer activities in colorectal cancer cells. J Agric Food Chem 2018;66:6003 –12. doi: 10.1021/acs.jafc.8b00915. [DOI] [PubMed] [Google Scholar]
- 7. Antal O Jr, Hackler L, Shen J et al. . Combination of unsaturated fatty acids and ionizing radiation on human glioma cells: cellular, biochemical and gene expression analysis. Lipids Health Dis 2014;13:142. doi: 10.1186/1476-511X-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Comba A, Lin YH, Renato-Eynard A et al. . Basic aspects of tumor cell fatty acid-regulated signaling and transcription factors. Cancer Metastasis Rev 2011;30:325 –42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Habermann N, Christian B, Luckas B et al. . Effects of fatty acids on metabolism and cell growth of human colon cell lines of different transformation state. Biofactors 2009;35:460 –7. doi: 10.1002/biof.60. [DOI] [PubMed] [Google Scholar]
- 10. Chamberland JP, Moon HS. Down-regulation of malignant potential by alpha linolenic acid in human and mouse colon cancer cells. Familial Cancer 2015;14:25 –30. doi: 10.1007/s10689-014-9762-z. [DOI] [PubMed] [Google Scholar]
- 11. Deshpande R, Mansara P, Kaul-Ghanekar R. Alpha-linolenic acid regulates Cox2/VEGF/MAP kinase pathway and decreases the expression of HPV oncoproteins E6/E7 through restoration of p53 and Rb expression in human cervical cancer cell lines. Tumor Biol 2016;37:3295 –305. doi: 10.1007/s13277-015-4170-z. [DOI] [PubMed] [Google Scholar]
- 12. Wiggins A, Mason J, Thompson L. Growth and gene expression differ overtime in alpha-linolenic acid treated breast cancer cells. Exp Cell Res 2015;333:147 –54. doi: 10.1016/j.yexcr.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 13. Rincón-Cervera MA, Galleguillos-Fernández R, González-Barriga V et al. . Concentration of gamma-linolenic and stearidonic acids as free fatty acids and ethyl esters from Viper’s bugloss seed oil by urea Complexation. Eur J Lipid Sci Technol 2018; 120: 1800208. doi: 10.1002/ejlt.201800208. [DOI] [Google Scholar]
- 14. Ge H, Kong X, Shi L et al. . Gamma-linolenic acid induces apoptosis and lipid peroxidation in human chronic myelogenous leukemia K562 cells. Cell Biol Int 2009;33:402 –10. doi: 10.1016/j.cellbi.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 15. Das UN. γ-Linolenic acid therapy of human glioma-a review of in vitro, in vivo, and clinical studies. Med Sci Monit 2007;13:RA119 –31. [PubMed] [Google Scholar]
- 16. Serini S, Piccioni E, Merendino N et al. . Dietary polyunsaturated fatty acids as inducers of apoptosis: implications for cancer. Apoptosis 2009;14:135 –52. doi: 10.1007/s10495-008-0298-2. [DOI] [PubMed] [Google Scholar]
- 17. Awad AB, Young AL, Fink CS. The effect of unsaturated fatty acids on membrane composition and signal transduction in HT-29 human colon cancer cells. Cancer Lett 1996;108:25 –33. [DOI] [PubMed] [Google Scholar]
- 18. De Roos B, Romagnolo DF. Proteomic approaches to predict bioavailability of fatty acids and their influence on cancer and chronic disease prevention. J Nutr 2012;142:1370S –6S. doi: 10.3945/jn.111.157206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu J, Tang J, Wang Y et al. . Discovery of the consistently well-performed analysis chain for SWATH-MS based pharmacoproteomic quantification. Front Pharmacol 2018;9:681. doi: 10.3389/fphar.2018.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spurvey S, Shahidi F. Concentration of gamma linolenic acid (GLA) from borage oil by urea complexation: optimization of reaction conditions. J Food Lipids 2000;7:163 –74. doi: 10.1111/j.1745-4522.2000.tb00169.x. [DOI] [Google Scholar]
- 21. Lepage G, Roy C. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J Lipid Res 1984;25:1391 –6. [PubMed] [Google Scholar]
- 22. Rodríguez-Ruiz J, Belarbi EH, Sánchez JLG et al. . Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol Tech 1998;12:689 –91. doi: 10.1023/A:1008812904017. [DOI] [Google Scholar]
- 23. González-Fernández MJ, Ramos-Bueno RP, Rodríguez-García I et al. . Purification process for MUFA- and PUFA-based monoacylglycerols from edible oils. Biochimie 2017;139:107 –14. doi: 10.1016/j.biochi.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 24. Fogh J. Human Tumor Cells In Vitro. New York and London: Plenum Press, 1975, 115–159. doi: 10.1007/978-1-4757-1647-4_5 [DOI] [Google Scholar]
- 25. Martínez-Maqueda D, Miralles B and Recio I. HT29 cell line In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, and Mackie A (eds), The Impact of Food Bioactives on Health. Cham: Springer International Publishing, 2015, 113–24. doi: 10.1007/978-3-319-16104-4_1 [DOI] [Google Scholar]
- 26. Ramos-Bueno RP, González-Fernández MJ, Guil-Guerrero JL. Various acylglycerols from common oils exert different antitumor activities on colorectal cancer cells. Nutr Cancer 2016;68:518 –29. doi: 10.1080/01635581.2016.1152382. [DOI] [PubMed] [Google Scholar]
- 27. Ortea I, Ruiz-Sánchez I, Cañete R et al. . Identification of candidate serum biomarkers of childhood-onset growth hormone deficiency using SWATH-MS and feature selection. J Proteome 2018;175:105 –13. doi: 10.1016/j.jprot.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 28. González-Fernández MJ, Fabrikov D, Ramos-Bueno RP et al. . SWATH Diferential abundance proteomics and cellular assays show in vitro anticancer activity of arachidonic acid- and docosahexaenoic acid-based monoacylglycerols in HT-29 colorectal cancer cells. Nutrients 2019;11:2984. doi: 10.3390/nu11122984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang C, Yu H, Shen Y et al. . Polyunsaturated fatty acids trigger apoptosis of colon cancer cells through a mitochondrial pathway. Arch Med Sci 2015;11:1081 –94. doi: 10.5114/aoms.2015.54865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim JY, Park HD, Park E et al. . Growth-inhibitory and proapoptotic effects of alpha-linolenic acid on estrogen-positive breast cancer cells. Second look at n-3 fatty acid. Natural compounds and their role in apoptotic cell signaling pathways. Ann N Y Acad Sci 2009;1171:190 –5. doi: 10.1111/j.1749-6632.2009.04897.x. [DOI] [PubMed] [Google Scholar]
- 31. Koralek DO, Peters U, Andriole G et al. . A prospective study of dietary alpha-linolenic acid and the risk of prostate cancer (United States). Cancer Causes Control 2006;17:783 –91. doi: 10.1007/s10552-006-0014-x. [DOI] [PubMed] [Google Scholar]
- 32. Kong X, Ge H, Hou L et al. . Induction of apoptosis in K562/ADM cells by gamma-linolenic acid involves lipid peroxidation and activation of caspase-3. Chem Biol Interact 2006;162:140 –8. doi: 10.1016/j.cbi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 33. Jiang WG, Hiscoxl S, Hallett MB et al. . Inhibition of hepatocyte growth factor-induced motility and in vitro invasion of human colon cancer cells by gamma-linolenic acid. Brit J Cancer 1995;71:744 –52. doi: 10.1038/bjc.1995.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fotakis G, Timbrell JA. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett 2006;160:171 –7. doi: 10.1016/j.toxlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 35. Ediriweera MK, Tennekoon KH, Samarakoon SR. In vitro assays and techniques utilized in anticancer drug discovery. J Appl Toxicol 2018;39:38 –71. doi: 10.1002/jat.3658. [DOI] [PubMed] [Google Scholar]
- 36. Pender-Cudlip MC, Krag KJ, Martini D et al. . Delta-6-desaturase activity and arachidonic acid synthesis are increased in human breast cancer tissue. Cancer Sci 2013;104:760 –4. doi: 10.1111/cas.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roy S, Singh M, Rawat A et al. . GLA supplementation regulates PHD2 mediated hypoxia and mitochondrial apoptosis in DMBA induced mammary gland carcinoma. Int J Biochem Cell Biol 2018;96:51 –62. doi: 10.1016/j.biocel.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 38. Moravcová A, Červinková Z, Kučera O et al. . The effect of oleic and palmitic acid on induction of steatosis and cytotoxicity on rat hepatocytes in primary culture. Physiol Res 2015;64:S627 –36. [DOI] [PubMed] [Google Scholar]
- 39. Mengeaud V, Nano JL, Fournel S et al. . Effects of eicosapentaenoic acid, gamma-linolenic acid and prostaglandin E1 on three human colon carcinoma cell lines. Prostag, Leukotr ESS 1992;47:313 –9. doi: 10.1016/0952-3278(92)90204-v. [DOI] [PubMed] [Google Scholar]
- 40. Tranchant T, Besson P, Hoinard C et al. . Mechanisms and kinetics of α-linolenic acid uptake in caco-2 clone TC7. BBA 1997;1345:151 –61. doi: 10.1016/s0005-2760(96)00171-3. [DOI] [PubMed] [Google Scholar]
- 41. Lee YJ, Shacter E. Oxidative stress inhibits apoptosis in human lymphoma cells. J Biol Chem 1999;274:19792 –8. doi: 10.1074/jbc.274.28.19792. [DOI] [PubMed] [Google Scholar]
- 42. Gillis RC, Daley BJ, Enderson BL et al. . Eicosapentaenoic acid and γ-linolenic acid induce apoptosis in HL-60 cells. J Surg Res 2002;107:145–53. doi: 10.1006/jsre.2002.6496. [DOI] [PubMed] [Google Scholar]
- 43. Deshpande R, Mansara P, Suryavanshi S et al. . Alpha-linolenic acid regulates the growth of breast and cervical cancer cell lines through regulation of NO release and induction of lipid peroxidation. J Mol Biochem 2013;2:6 –17. [Google Scholar]
- 44. Bouzza PT, Viola JPB. Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fatty Acids 2010;82:243 –50. doi: 10.1016/j.plefa.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 45. Tirinato L, Pagliari F, Limongi T et al. . An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int 2017;2017:1 –17. doi: 10.1155/2017/1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jarc E, Kump A, Malavašič P et al. . Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. BBA Mol Cell 2018;1863:247 –65. doi: 10.1016/j.bbalip.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 47. Gustin E, Jarc E, Kump A et al. . Lipid droplet formation in HeLa cervical cancer cells depends on cell density and the concentration of exogenous unsaturated fatty acids. Acta Chim Slov 2017;64:549 –54. doi: 10.17344/acsi.2016.2908. [DOI] [PubMed] [Google Scholar]
- 48. Ott M, Norberg E, Zhivotovsky B et al. . Mitochondrial targeting of tBid/Bax: a role for the TOM complex? Cell Death Differ 2009;16:1075 –82. doi: 10.1038/cdd.2009.61. [DOI] [PubMed] [Google Scholar]
- 49. Asmarinah A, Paradowska-Dogan A, Kodariah R et al. . Expression of the Bcl-2 family genes and complexes involved in the mitochondrial transport in prostate cancer cells. Int J Oncol 2014;45:1489 –96. doi: 10.3892/ijo.2014.2576. [DOI] [PubMed] [Google Scholar]
- 50. Rondepierre F, Bouchon B, Papon J et al. . Proteomic studies of B16 lines: involvement of annexin A1 in melanoma dissemination. Biochim Biophys Acta 2009;1794:61 –9. doi: 10.1016/j.bbapap.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 51. Opalinska M, Meisinger C. Metabolic control via the mitochondrial protein import machinery. Curr Opin Cell Biol 2015;33:42 –8. doi: 10.1016/j.ceb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 52. Xu X, Qiao M, Zhang Y et al. . Quantitative proteomics study of breast cancer cell lines isolated from a single patient: discovery of TIMM17A as a marker for breast cancer. Proteomics 2010;10:1374 –90. doi: 10.1002/pmic.200900380. [DOI] [PubMed] [Google Scholar]
- 53. Wu H, Rao GN, Dai B et al. . Autocrine Gastrins in colon cancer cells up-regulate cytochromec oxidase Vb and down-regulate efflux of cytochromec and activation of caspase-3. J Biol Chem 2000;275:32491 –8. doi: 10.1074/jbc.M002458200. [DOI] [PubMed] [Google Scholar]
- 54. Anderson NM, Mucka P, Kern JG et al. . The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018;9:216 –37. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Montal ED, Bhalla K, Dewi RE et al. . Inhibition of phosphoenolpyruvate carboxykinase blocks lactate utilization and impairs tumor growth in colorectal cancer. Cancer Metabolism 2019;7:8. doi: 10.1186/s40170-019-0199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nature 2014;14:581 –97. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


