Abstract
Nanopores have become an important tool for the detection and analysis of molecules at the single-molecule level. Surface modification of solid-state nanopores can improve their durability and efficiency. Peptides are ideal for surface modifications as they allow tailoring of multiple properties by a rational design of their sequence. Here, silicon nitride nanopores were coated by a dipeptide layer where a l-3,4-dihydroxyphenylalanine (DOPA) residue is the anchoring element and the other amino acid moiety is the functional element. DOPA binds tightly to many types of surfaces and allows a one-step functionalization of surfaces by simple immersion. As a result, the lifetime of coated nanopores increased from hours to months and the current-stability has significantly improved with respect to uncoated pores. This improvement is achieved by controlling the surface wettability and charge. Peptide-coated nanopores can be utilized as sensitive sensors that can be adjusted based on the choice of the functional moiety of the coated peptide. In addition, the coating slows down dsDNA translocation because of the DNA interaction with the pore coating.
Keywords: Nanopores, peptides, DOPA, self-assembly, dipeptides
Introduction
Nanopores are selective and sensitive sensors at the single-molecule level. They are used to detect various analytes, like single ions, organic molecules, and more complex molecules such as proteins and nucleic acid polymers. They are mainly used for rapid and low-cost DNA sequencing.1
A nanopore is a nanometer-sized hole, embedded in an insulating membrane. Electric potential is applied across the insulating membrane that separates two chambers filled with ionic solution, resulting in ions current through the nanopore. Molecule detection and characterization with nanopores is based on a constant flow of ions through the nanopore. When an analyte is translocated through the pore, the flow of ions is normally blocked and results in a change in the monitored current.
Solid-state nanopores are created by drilling a nanoscale hole in a synthetic membrane such as silicon nitride, silicon oxide, or graphene.2 Silicon-based materials have been popular for nanopores, because of their low mechanical stress, high chemical resistance, and processability.3 The pore is typically formed in the membrane either by a focused ion beam4 or by a transmission electron microscope (TEM) beam.5
It has been shown that interaction of analytes, such as DNA, with the walls of the solid state nanopore can affect their translocation dynamics.6 One of the ways to improve sequencing with solid-state nanopores is to slow down the translocation speed by chemical modification of solid-state nanopores’ surface, thus improving the nanopore efficiency in detecting and analyzing analytes.7 Various approaches for nanopore modification have been reported, including metal8 and oxides9 deposition, various organic modifications,9,10 assembly of fluidic lipid-bilayer on the surface of a solid state nanopore,11 and coating the pore surface with DNA hairpins.12 Nanopore modification with peptides is attractive for cooperative control over the translocation,13 and broadens the functionality of solid-state nanopores.14 Peptides characteristics and configurations depend on their primary sequence. Therefore, they allow tailoring of multiple properties by rational design of their sequence. Peptides have been studied thoroughly for their solid-surface coating ability, because of the presence of specific bond formation between the peptide and the solid surface.15−17 In addition, peptides are environmentally friendly and biocompatible material, and can provide a required functionality by combining different amino acids with several properties.18 However, these modifications have several limitations, such as the need of treating the membranes with silane before peptide coating.19
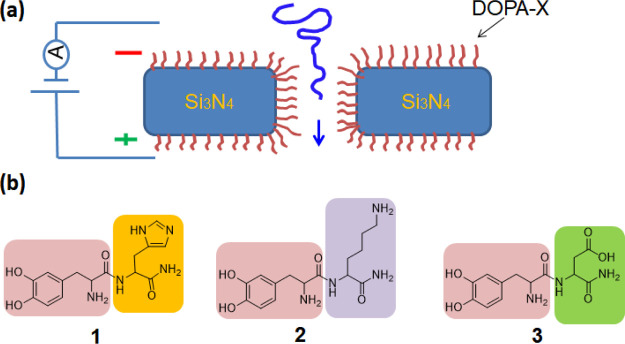
Here, we present coating of Si3N4 nanopore surfaces with various self-assembled dipeptide monolayers. These monolayers enable manipulation of the nanopore properties and affect its interaction with analytes such as DNA and proteins. We report coating by dipeptides comprising the amino l-3,4-dihydroxyphenylalanine (DOPA) (Figure 1). DOPA is known for its surface-adhesive properties.18,20−23 It binds to different materials including metals, oxides, and polymers as it can form various types of bonds with the surface.21,24 The interaction of the catechol group of DOPA with the Si3N4 surface is pH-dependent. At pH 7.2, which was used here during the coating, two coordination bonds per catechol are formed with the Si3N4 surface.25 We used DOPA as an anchoring group to functionalize the pore with different amino acids (positively or negatively charged) that determine the surface chemistry of the pore. We investigated coating with histidine, lysine, and glutamate having pKa values of 6.02, 10.52, and 4.25,26 respectively (Figure 1b). As we show below, the coating modified the contact angle of the surface as well as the nanopore behavior, indicating that coating indeed took place. These modifications allow nanopore usage for at least 7 months without further treatment, whereas uncoated nanopores need cleaning and functionalization after each use (hours). Furthermore, they enable the manipulation of the nanopore surface charge and influence the interaction with translocated objects such as DNA.
Figure 1.
(a) Schematic diagram of the experimental setup. SiN nanopore is coated with DOPA-X dipeptides. (b) Chemical structure of the studied dipeptides, DOPA-His (1), DOPA-Lys (2), and DOPA-Glu (3).
Materials and Methods
Nanopore Fabrication
Nanopores were fabricated by drilling in a 30 nm thick, low-stress SiN membrane (50 × 50 μm2) supported by a silicon chip (Protochips, Inc.) using a focused electron beam of 200 keV high-resolution TEM (FEI Tecnai G2-F20). The drilling process involves alignment of the electron beam and adjustment of the condenser astigmatism. The condenser lens was then used to direct the intense beam to a target point.27 After the alignment procedure, nanopores with the diameter in a range from 8 to 12 nm were directly drilled. The time for pore formation in a 30 nm thick Si3N4 membrane was in the range of 30–60 s. Once the pores were drilled, they were stored in ethanol/triple distilled water (TDW) (1:1, v/v) immediately to avoid contamination.28 Following drilling, the nanopore was imaged and characterized by the TEM imaging mode.
Dipeptide Synthesis
The dipeptides DOPA-His, DOPA-Lys, and DOPA-Glu (Figure 1) were synthesized using 9-fluorenylmethoxycarbonyl (Fmoc)-based solid-phase peptide chemistry manually. Standard coupling conditions using AA/HATU/DIPEA were employed to obtain the desired peptides. The peptides were synthesized on Fmoc-Rink amide resin, which was subjected to Fmoc removal before coupling the AA residues to yield C-terminus amides. Amino acids were coupled in fivefold excess in the synthesis and all residues were coupled once for 1 h. The coupling reactions were monitored by the Kaiser ninhydrin test. Removal of the Fmoc group was performed using 20% Piperidine in DMF for 15 min twice and the residual piperidine was removed by three consecutive washes with DMF. Peptide cleavage from the resin support was performed using 95% trifluoroacetic acid, 2.5% water, and 2.5% triisopropylsilane (5 mL/183 mg of resin) for 2 h at room temperature around 25 °C. Crude peptides were purified by preparative high-performance liquid chromatography.
Peptide identity was confirmed using electrospray ionization mass spectrometry (LC(UV)MS/MS, Agilent 6520 QTOF analyzer for DOPA-His and ESI-MS, Waters ZQ4000 for the rest) (Figure S1). Pure peptides were stored at −20 °C.
Nanopore Modification with a Dipeptide
Nanopore membranes were treated in a Plasma Cleaner for 30 s before modification with the dipeptide to improve binding. The Nanopore membrane was immersed in the dipeptide solution (0.5 mg/mL dipeptide dissolved in Tris-HCl/ethanol (1:1, v/v), pH 7.2) overnight at room temperature and then washed with 3 mL of ethanol. The SiN surface modification was characterized by contact angle measurements. Membranes coated with the dipeptides were stored in ethanol/TDW (1:1, v/v).
To examine the chemical identity of the surfaces after coating, X-ray photoelectron spectroscopy analysis was performed (Figure S2).
Nanopore Activity Recording
A coated chip with a drilled pore is mounted in an electrophoresis flow cell. Two reservoirs on each side with a volume of 100 μL (trans and cis) were filled with filtered and degassed buffer of 0.02–1 M KCl, 10 nM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA) (pH 6,7.5, 9), or 0.02–1 M KCl, 10 nM succinic acid, 1 mM EDTA (pH 4.5). Linear double-stranded DNA (2 kb NoLimits DNA Fragment and 48 kb Lambda DNA (Thermo Fisher Scientific)) translocation experiments were done with 1 M KCl, 10 mM Tris-HCl, 1 mM EDTA, 10% glycerol (pH 7.5). A pair of Ag/AgCl pellet electrodes were immersed in the two reservoirs and connected to an Axopatch 200B amplifier (Molecular Devices, Inc.) to record ionic current flow through the nanopore. The whole setup was placed in a double Faraday cage to lower the external electrostatic interference. Signals were collected at 10 kHz sampling rate using a Digi-data 1440A (Molecular Devices, Inc.) and filtered at 1 kHz using the built-in low pass Bessel filter of the Axopatch.
After each measurement, the chips were washed with TDW to remove salt residues, and then installed back into the flow cell without any further cleaning.
Results and Discussion
Effect of Dipeptide Modification on Nanopore Usability and Stability
Nanopores were coated with a layer of dipeptides (DOPA-amino acid) by immersion overnight after 30 s of cleaning with plasma. This was done without any chemical treatment of the silicon surface as in other peptides binding procedures reported for solid state nanopores.29,30 In a typical experiment, uncoated Si3N4 nanopores are treated with plasma or piranha solution right before every usage.31,32 However, in these nanopores a hydrophilic surface is not maintained along the experiments. As a result, the current through the nanopore varies for the same pore from experiment to experiment.
We studied the effect of peptide coating of Si3N4 surfaces by contact angle measurements (Figure 2). For the three types of coatings, DOPA-His, DOPA-Glu, and DOPA-Lys, the contact angle was ∼50% smaller than for surfaces treated for 30 s with plasma, indicating improved wetting.
Figure 2.
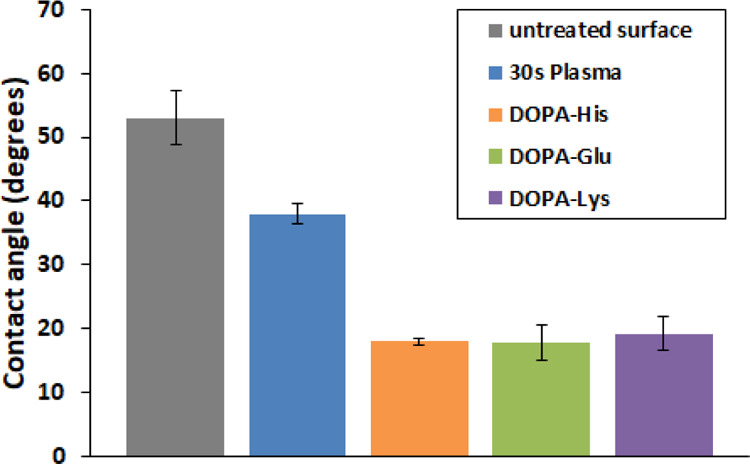
Contact angle measurements for different Si3N4 surfaces before and after 30 s plasma treatment, and DOPA-His, DOPA-Lys, and DOPA-Glu coating. For all the five columns, measurements were conducted for three different SiN chips, pH 7.5.
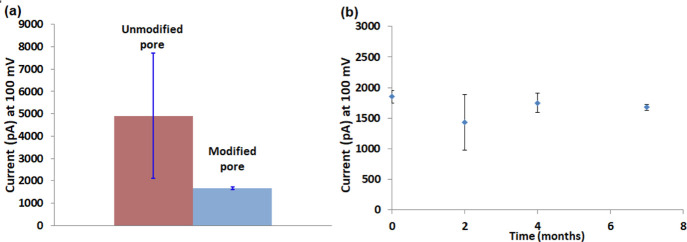
We tested the stability of the ionic current through the nanopore after peptide coating. The term stability is used here to describe the reproducibility of the results upon repeating the experiments with the same pore several times. To test the stability, currents through nanopores with similar diameter (∼12 nm) were measured repeatedly five times, all at the same day and under the same conditions. The current was reduced after coating because of a decrease in pore diameter. We assume that this reduction is in correlation with DOPA-His molecule length (∼1.5 nm) that may take some space in the pore. As conductance through the pore depends on pore size,33 the decrease in pore diameter reduces the current through the coated pore. The variability of the current through DOPA-His-coated nanopores was much smaller compared with unmodified nanopores, which were treated with only plasma (see error bar in Figure 3a). Thus, after modifying the pore with DOPA-His, the ionic current is much more stable. In our system, a nanopore is considered ready for measurements only after becoming hydrophilic, as all the measurements are in aqueous medium. When the hydrophilicity of the pore is constant, the current through the pore remains stable, including in repeated measurements through the same pore. As mentioned above, the typical wetting of Si3N4 surfaces can be achieved by plasma or piranha solution. These treatments cause only a temporal oxidation of the surface. The dipeptides that were used in this study contain, in addition to DOPA, a charged amino acid. Therefore, this peptide coating allows covering the nanopores with a durable (spatial and temporal) charged layer that keeps the pore constantly wet. Importantly, the peptide coating considerably reduces the current background noise level (Figure S3).
Figure 3.
(a) Currents through representative uncoated (red) and DOPA-His dipeptide-coated (blue) 12 nm diameter nanopores at 100 mV. Currents through each nanopore were measured repeatedly five times in 1 M KCl, 10 mM Tris-HCl, 1 mM EDTA (pH 7.5). After each measurement, the chambers were cleaned with water and the solution was refilled. The graph presents the average of these five measurements. Note the much larger error bar for the uncoated nanopore. (b) Similar currents were measured [under the same conditions as in (a)] through a single nanopore along several months without any further treatment.
To check the durability of the coated pore, we measured currents in the coated pore from time to time. Similar ionic currents were measured through peptide-coated nanopores for at least 7 months without any additional treatment (Figure 3b). These results indicate that the peptide coating improves the nanopore durability, stability, and usability, both for short and long terms.
Effect of pH on the Conductance through the Dipeptide-Treated Nanopores
Ionic transport through nanoscale pores is affected by their surface charge, which influences the flow of counterions near the pore walls.7 The effect of pH (in the range 4.5—9) that governs the amino acids charge, on the conductance through three types of dipeptide-coated nanopores, was investigated and is shown in Figure 4. The results are in good correlation with the nature of the amino acid residue at the C-terminus of the dipeptide. Glutamic acid, which is an acidic amino acid with pKa of 4.25, becomes more negatively charged as the pH rises, whereas histidine, which is a basic amino acid with pKa of 6.02, become more positively charged as the pH is reduced. Therefore, when the pH is raised, the charge of DOPA-Glu peptides is increased, whereas the charge of DOPA-His peptides is decreased. These changes affect the conductivity of the peptide-treated nanopore (Figure 4). On the other hand, the current through the DOPA-Lys-coated nanopore was relatively high and constant along the tested pH range because Lys is practically fully charged within this pH range as its pKa is 10.5.
Figure 4.
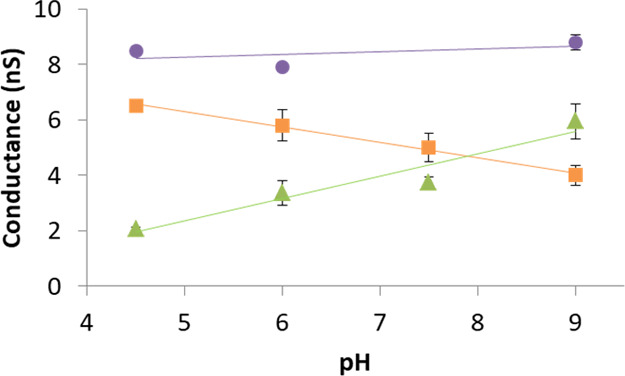
Conductance measurements through dipeptide-treated nanopores as a function of pH change (DOPA-Lys: purple, DOPA-His: orange, and DOPA-Glu: green). Measurements were performed in 0.14 M KCl, 10 mM Tris-HCl/succinic acid, 1 mM EDTA (pH 4.5, 6, 7.5, and 9) at 100 mV. In some of the points, the error bars are smaller than the marking.
The pH effect on the conductivity through peptide-treated nanopores is reduced as the salt concentration is increased and is abolished around 0.5 M KCl (Figures 5, S4). The ratio between the conductance at pH 4.5 and pH 9 at various salt concentrations is shown in Figure 5. For nanopores treated with DOPA-His and DOPA-Glu, this ratio becomes closer to 1 as the salt concentration is increased. This indicates a loss of pH effect at high salt concentrations. The conductivity through nanopores treated with DOPA-Lys is pH-independent at any salt concertation, as expected.
Figure 5.
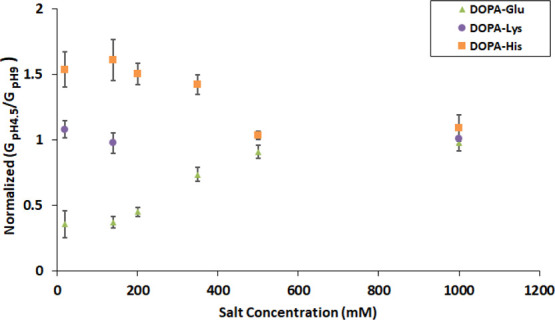
Ratio of conductance at pH 4.5 to conductance at pH 9 at different salt concentrations, for nanopores coated with DOPA-His, DOPA-Glu, and DOPA-Lys.
These results are not surprising as the pH effect on the amino acids charge is dependent on their Debye length (how far a charge carrier’s electrostatic effect persists)34 and Debye length becomes shorter as the salt concentration is increased. For instance, at 0.14 M KCl, the Debye length is ∼0.8 nm and at 1.0 M KCl the Debye length is only ∼0.3 nm. Thus, the surface charge of DOPA-His- and DOPA-Glu-treated nanopores, which is pH-dependent, has more effect on the ion conductivity through the nanopore when the salt concentration is low.
The peptide-treated nanopores can thus be utilized as small and sensitive pH and salt concentration sensors and can be adjusted based on the coated peptide identity.
DNA Translocation through Dipeptide-Treated Nanopores
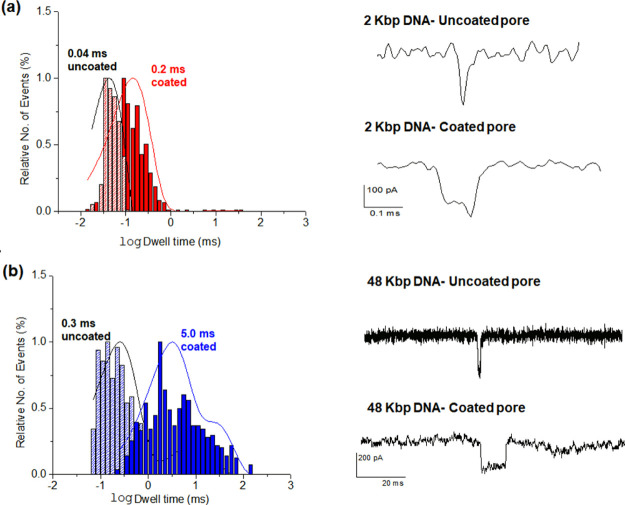
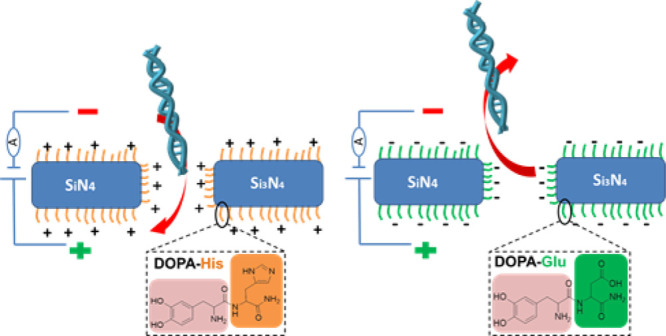
dsDNA translocation through dipeptide-treated nanopores was also investigated. The change in ionic conductance when dsDNA was translocated through peptide-coated nanopores was measured with a buffer of 1 M KCl, pH 7.5. Blockage events were observed when dsDNA was added to a cell with DOPA-His-coated nanopores (Figure 6). The results demonstrate that the dsDNA was translocated through the nanopore. The translocation of shorter DNA (2 kbp) had a shorter dwell time, 0.1 ms (Figure 6a), than that of the long dsDNA (48 kbp), 5.0 ms (Figure 6b), confirming that dsDNA is indeed the source of the blockage events. Figure S5a shows the dwell time and amplitude for dsDNA blocking events measured at various voltages for the nanopore coated with DOPA-His, further strengthening the conclusion that dsDNA is indeed translocated. An exponential dependence of the DNA translocation dwell time (Figure S5b,c) on the voltage was observed. This fits to electrophoretic dragging of the DNA through a pore.34 All the above results indicate that dsDNA was indeed translocated through the coated nanopore.
Figure 6.
dsDNA translocation time through 10 nm untreated and DOPA-His-coated nanopores. The measurements were done at 1 M KCl, 10 mM Tris-HCl, 1 mM EDTA, and pH 7.5, 200 mV, pore size 10 nm. (a) Translocation dwell time histogram for 2 kbp DNA (left) and an example of 2 kbp DNA translocation events (right), through DOPA-His-coated and uncoated nanopores. (b) Translocation dwell time histogram for 48 kbp DNA (left) and an example of 48 kbp DNA translocation events (right) through DOPA-His-coated and uncoated nanopore.
No conductance change, indicating translocation, was measured for nanopores treated with DOPA-Glu (Figure S6). As Glu (pKa 4.25) is negatively charged at pH 7.5, it repels the DNA from the pore. Currents measured through nanopores treated with DOPA-Lys fluctuate between two levels (Figure S7). The source of these fluctuations is unclear. After DNA addition to the cis chamber, no new current changes that indicate translocation were observed. It is possible that either the blockage level of DNA translocation is of the same magnitude as that of the current fluctuations and cannot be distinguished or that a large portion of the DNA binds tightly to the positively charged DOPA-Lys coating, and therefore cannot be translocated through the nanopore and blocks further translocation.
The DOPA-His coating reduces the DNA translocation by about an order of magnitude with respect to the uncoated nanopore (Figure 6a,b). After the coating, the dwell time of 2 kbp dsDNA increased from 0.040 ± 0.025 to 0.20 ± 0.10 ms (n = 3) on average and the dwell time of 48 kbp dsDNA increased from 0.50 ± 0.20 to 5.0 ± 1.4 ms (n = 3) on average. The slowdown may result from either the positive charge residing in the coated pore (His is positively charged at pH 7.5) and/or from a possible higher friction. A further slowdown of the DNA translocation might be achieved by optimization of the used peptides based on their charge and size, thus addressing a central challenge for DNA sequencing.
Conclusions
Our results show that DOPA allows a simple one-step peptide functionalization of surfaces. We show that Si3N4 nanopores can be easily modified by a simple immersion in DOPA-based peptides solutions. This modification increases the nanopores’ durability and allows using them for many months without special cleaning or necessity for any other treatment. The coating enables to use them for a range of sensing applications (such as pH and salt) and to control the translocation time of analytes such as DNA. As peptides allow tailoring of multiple properties by rational design of their sequence, solid-state nanopores can be easily adapted to numerous functions by coating with predesigned DOPA-based peptides.
Acknowledgments
This work was supported by the Israel Science Foundation (ISF grant 1589/14), the ISF-NSFC (grant 2556/17), the Minerva Centre for bio-hybrid complex systems. D.P. thanks the Etta and Paul Schankerman Chair of Molecular Biomedicine.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.0c00062.
Conductance measurements through dipeptide-treated nanopores as a function of pH change, under various KCl concentrations; 2 kbp DNA translocation through 10 nm nanopores coated with DOPA-His under various potentials; current through DOPA-Glu-modified nanopore with or without 2 kbp DNA; and current trace through the DOPA-Lys-modified nanopore (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Li J.; Gershow M.; Stein D.; Brandin E.; Golovchenko J. A. DNA Molecules and Configurations in a Solid-State Nanopore Microscope. Nat. Mater. 2003, 2, 611–615. 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- Fischbein M. D.; Drndić M. Electron Beam Nanosculpting of Suspended Graphene Sheets. Appl. Phys. Lett. 2008, 93, 113107–113109. 10.1063/1.2980518. [DOI] [Google Scholar]
- Venkatesan B. M.; Bashir R. Nanopore Sensors for Nucleic Acid Analysis. Nat. Nanotechnol. 2011, 6, 615–624. 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- Li J.; Stein D.; McMullan C.; Branton D.; Aziz M. J.; Golovchenko J. A. Ion-Beam Sculpting at Nanometre Length Scales. Nature 2001, 412, 166–169. 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- Branton D.; Deamer D. W.; Marziali A.; Bayley H.; Benner S. A.; Butler T.; Di Ventra M.; Garaj S.; Hibbs A.; Huang X.; Jovanovich S. B.; Krstic P. S.; Lindsay S.; Ling X. S.; Mastrangelo C. H.; Meller A.; Oliver J. S.; Pershin Y. V.; Ramsey J. M.; Riehn R.; Soni G. V.; Tabard-Cossa V.; Wanunu M.; Wiggin M.; Schloss J. A. The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M.; Sutin J.; McNally B.; Chow A.; Meller A. DNA Translocation Governed by Interactions with Solid-State Nanopores. Biophys. J. 2008, 95, 4716–4725. 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M.; Meller A. Chemically Modified Solid-State Nanopores. Nano Lett. 2007, 7, 1580–1585. 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- Harrell C. C.; Lee S. B.; Martin C. R. Synthetic Single-Nanopore and Nanotube Membranes. Anal. Chem. 2003, 75, 6861–6867. 10.1021/ac034602n. [DOI] [PubMed] [Google Scholar]
- Nilsson J.; Lee J. R. I.; Ratto T. V.; Létant S. E. Localized Functionalization of Single Nanopores. Adv. Mater. 2006, 18, 427–431. 10.1002/adma.200501991. [DOI] [Google Scholar]
- Wang G.; Zhang B.; Wayment J. R.; Harris J. M.; White H. S. Electrostatic-Gated Transport in Chemically Modified Glass Nanopore Electrodes. J. Am. Chem. Soc. 2006, 128, 7679–7686. 10.1021/ja061357r. [DOI] [PubMed] [Google Scholar]
- Yusko E. C.; Johnson J. M.; Majd S.; Prangkio P.; Rollings R. C.; Li J.; Yang J.; Mayer M. Controlling Protein Translocation through Nanopores with Bio-Inspired Fluid Walls. Nat. Nanotechnol. 2011, 6, 253–260. 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S. M.; Akin D.; Bashir R. Solid-State Nanopore Channels with DNA Selectivity. Nat. Nanotechnol. 2007, 2, 243–248. 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- Hou X.; Liu Y.; Dong H.; Yang F.; Li L.; Jiang L. A pH-Gating Ionic Transport Nanodevice: Asymmetric Chemical Modification of Single Nanochannels. Adv. Mater. 2010, 22, 2440–2443. 10.1002/adma.200904268. [DOI] [PubMed] [Google Scholar]
- Kang Q.; Guo W.; Tagliazucchi M.; Szleifer I.. 4-Biomimetic Smart Nanopores and Nanochannels. In Chemically Modified Nanopores and Nanochannels; William Andrew Publishing: Boston, 2017; pp 85–102. [Google Scholar]
- Sarikaya M.; Tamerler C.; Jen A. K.-Y.; Schulten K.; Baneyx F. Molecular Biomimetics: Nanotechnology through Biology. Nat. Mater. 2003, 2, 577–585. 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- Yazici H.; O’Neill M. B.; Kacar T.; Wilson B. R.; Oren E. E.; Sarikaya M.; Tamerler C. Engineered Chimeric Peptides as Antimicrobial Surface Coating Agents toward Infection-Free Implants. ACS Appl. Mater. Interfaces 2016, 8, 5070–5081. 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerler C.; Khatayevich D.; Gungormus M.; Kacar T.; Oren E. E.; Hnilova M.; Sarikaya M. Molecular Biomimetics: Gepi-Based Biological Routes to Technology. Pept. Sci. 2010, 94, 78–94. 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- Maity S.; Nir S.; Zada T.; Reches M. Self-Assembly of a Tripeptide into a Functional Coating That Resists Fouling. Chem. Commun. 2014, 50, 11154–11157. 10.1039/c4cc03578j. [DOI] [PubMed] [Google Scholar]
- Kowalczyk S. W.; Kapinos L.; Blosser T. R.; Magalhães T.; van Nies P.; Lim R. Y. H.; Dekker C. Single-Molecule Transport across an Individual Biomimetic Nuclear Pore Complex. Nat. Nanotechnol. 2011, 6, 433–438. 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Wu Y.; Wang L.; Zhang M.; Chen X.; Liu M.; Fan J.; Liu J.; Zhou F.; Wang Z. Bio-Inspired Reversible Underwater Adhesive. Nat. Commun. 2017, 8, 2218–2225. 10.1038/s41467-017-02387-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Scherer N. F.; Messersmith P. B. Single-Molecule Mechanics of Mussel Adhesion. Proceedings of the National Academy of Sciences 2006, 103, 12999–13003. 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite J. H.; Tanzer M. L. Polyphenolic Substance of Mytilus Edulis: Novel Adhesive Containing L-Dopa and Hydroxyproline. Science 1981, 212, 1038–1040. 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- Lee B. P.; Messersmith P. B.; Israelachvili J. N.; Waite J. H. Mussel-Inspired Adhesives and Coatings. Annu. Rev. Mater. Res. 2011, 41, 99–132. 10.1146/annurev-matsci-062910-100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaw S. L.; Sakala G.; Nir S.; Saha A.; Xu Z. J.; Lee P. S.; Reches M. Rational Design of Amphiphilic Peptides and Its Effect on Antifouling Performance. Biomacromolecules 2018, 19, 3620–3627. 10.1021/acs.biomac.8b00587. [DOI] [PubMed] [Google Scholar]
- Waite J. H. Mussel adhesion - essential footwork. J. Exp. Biol. 2017, 220, 517–530. 10.1242/jeb.134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel U., Cravatt B., Graslund A., von Heijne N., Zorko M., Land T., Niessen S.. Introduction to Peptides and Proteins; CRC Press: Boca Raton, 2010. [Google Scholar]
- Kim M. J.; Wanunu M.; Bell D. C.; Meller A. Rapid Fabrication of Uniformly Sized Nanopores and Nanopore Arrays for Parallel DNA Analysis. Adv. Mater. 2006, 18, 3149–3153. 10.1002/adma.200601191. [DOI] [Google Scholar]
- Storm A. J.; Chen J. H.; Ling X. S.; Zandbergen H. W.; Dekker C. Fabrication of Solid-State Nanopores with Single-Nanometre Precision. Nat. Mater. 2003, 2, 537–540. 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- Liebes-Peer Y.; Rapaport H.; Ashkenasy N. Amplification of Single Molecule Translocation Signal Using β-Strand Peptide Functionalized Nanopores. ACS Nano 2014, 8, 6822–6832. 10.1021/nn501331u. [DOI] [PubMed] [Google Scholar]
- Smeets R. M. M.; Kowalczyk S. W.; Hall A. R.; Dekker N. H.; Dekker C. Translocation of Reca-Coated Double-Stranded DNA through Solid-State Nanopores. Nano Lett. 2009, 9, 3089–3095. 10.1021/nl803189k. [DOI] [PubMed] [Google Scholar]
- Tabard-Cossa V.; Dhruti T.; Matthew W.; Nahid N. J.; Andre M. Noise Analysis and Reduction in Solid-State Nanopores. Nanotechnology 2007, 18, 305505–305510. 10.1088/0957-4484/18/30/305505. [DOI] [Google Scholar]
- Beamish E.; Harold K.; Vincent T.-C.; Michel G. Precise Control of the Size and Noise of Solid-State Nanopores Using High Electric Fields. Nanotechnology 2012, 23, 405301–405307. 10.1088/0957-4484/23/40/405301. [DOI] [PubMed] [Google Scholar]
- Kowalczyk S. W.; Grosberg A. Y.; Rabin Y.; Dekker C. Modeling the Conductance and DNA Blockade of Solid-State Nanopores. Nanotechnology 2011, 22, 315101–315105. 10.1088/0957-4484/22/31/315101. [DOI] [PubMed] [Google Scholar]
- Anderson B. N.; Muthukumar M.; Meller A. pH Tuning of DNA Translocation Time through Organically Functionalized Nanopores. ACS Nano 2013, 7, 1408–1414. 10.1021/nn3051677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.