Abstract
Iridoids are plant-derived terpenoids with a rich array of bioactivities. The key step in iridoid skeleton formation is the reduction of 8-oxogeranial by certain members of the progesterone 5β-reductase/iridoid synthase (PRISE) family of short-chain alcohol dehydrogenases. Other members of the PRISE family have previously been implicated in the biosynthesis of the triterpenoid class of cardenolides, which requires the reduction of progesterone. Here, we explore the occurrence and activity of PRISE across major lineages of plants. We observed trace activities toward either 8-oxogeranial or progesterone in all PRISEs, including those from nonseed plants and green algae. Phylogenetic analysis, coupled with enzymatic assays, show that these activities appear to have become specialized in specific angiosperm lineages. This broad analysis of the PRISE family provides insight into how these enzymes evolved in plants and also suggests that iridoid synthase activity is an ancestral trait in all land plants, which might have contributed to the rise of iridoid metabolites.
Introduction
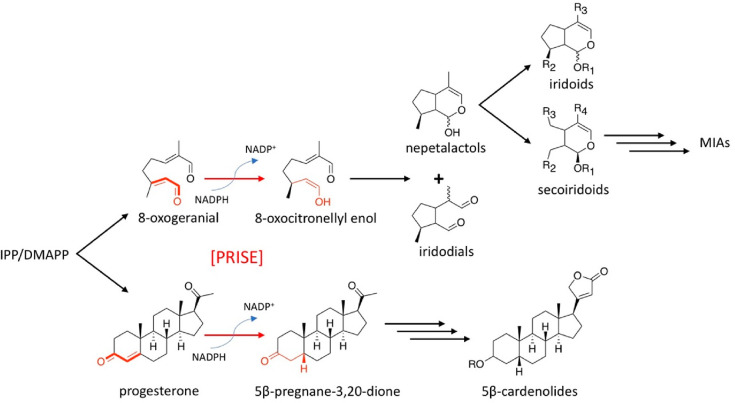
Iridoids constitute a noncanonical group of monoterpenoids with a distinctive cyclopentanopyran skeleton. Although members of this group were first isolated from insects (hence the name “iridoid”, from the rainbow ant genus Iridomyrmex), most known iridoids are synthesized by plants.1,2 Hundreds of naturally occurring iridoid structures act as defensive chemicals in 57 known families within the angiosperms.3,4 Iridoids also serve as precursors for a wide range of high-value monoterpenoid indole alkaloids, including the anticancer drugs vinblastine in Catharanthus roseus and campothecin in Camptotheca acuminata.5 Iridoid biosynthesis begins with the conversion of geranyl pyrophosphate, the general precursor of monoterpenoids, to geraniol via a typical monoterpene synthase, geraniol synthase. Geraniol is then hydroxylated and oxidized to form 8-oxogeranial, which, in turn, is subjected to reduction, catalyzed by a short-chain alcohol dehydrogenase called iridoid synthase (abbreviated hereafter as ISY), to form the reactive 8-oxocitronellyl enol. In the absence of additional enzymes that guide the stereoselective cyclization of this intermediate, it is converted nonenzymatically in aqueous solution to a combination of iriodial and nepetalactol stereoisomers (see Figure 1, as well as Figure S2 in the Supporting Information).6−9 The specific short-chain dehydrogenase family to which ISY belongs is known for its progesterone 5β-reductase activity, specifically, the stereoselective reduction of progesterone to 5β-pregnan-3,20-dione that occurs in the biosynthesis of cardenolide variety of triterpenoids.10,11 This enzyme family, named PRISE (for progesterone 5β-reductase/iridoid synthase activity), appears to play a critical ecophysiological role, since both iridoids and cardenolides are major groups of signaling molecules and semiochemicals.12 Intriguingly, however, members of the PRISE family have been reported in iridoid- and cardenolide-free species such as Arabidopsis.13−17 Earlier reports showed that both PRISEs from Catharanthus roseus, an iridoid-producing species, and PRISEs from the Brassicaceae, including those that do not produce iridoids and/or cardenolides, had different reductase activities toward 8-oxogeranial and progesterone.16,18,19 Promiscuous enzymatic activities are likely starting points for specialization under different conditions, as demonstrated by several examples in enzyme evolution.20−24 To determine whether such a process within the PRISE family played a role in iridoid metabolism evolution, we identified PRISE homologues from an encompassing range of plant lineages. We found that PRISEs appear to be ubiquitous in plants, and the substrate specificities of these enzymes for either 8-oxogeranial and progesterone, as measured within a phylogenetic framework, may provide clues regarding how iridoid pathways evolved.
Figure 1.
Progesterone 5β-reductase/iridoid synthase (PRISE) in the biosynthetic pathways of iridoids and monoterpenoid indole alkaloids (MIAs), and 5β-cardenolides. Bold red lines indicate the characteristic 4-en-3-one moeity of PRISE substrates. [IPP = isopentenyl pyprophosphate, DMAPP = dimethylallyl pyrophosphate.]
Results and Discussion
To probe the evolution of PRISE, we used C. roseus ISY (PRISE5) as a query to search for PRISE homologues in the publicly available nucleotide databases, including NCBI, the 1KP Project, and the bryophyte genome databases.25−27 BLAST searches revealed that PRISE homologues occur in all reported plant lineages. Outside angiosperms, we found full-length PRISE sequences in several families of gymnosperms (46%–56% sequence identity to C. roseus ISY), lycophytes (41%–56% sequence identity to C. roseus ISY), nonvascular plants (moss and liverwort) (46%–53% sequence identity to C. roseus ISY, and green algae (31%–47% sequence identity to C. roseus ISY).
We selected 113 sequences for phylogenetic reconstruction (see Table S1 in the Supporting Information). Although our sequence selection was biased toward plants that have genome or cDNA sequence data available, the available sequences cover major plant lineages including representative members of the core eudicots (orders Lamiales, Brassicales, Apiales, Caryophyllales, Cornales, Sapindales, Cucurbitales, Gentianales, Malvales, Asterales, Fabales, Malpighiales, and Vitales), monocots (orders Asparagales, Zingiberales, and Poales), basal angiosperm (order Amborellales), gymnosperms (order Pinales), lycophytes (orders Lycopodiales, Isoetales, and Selaginalles), and bryophytes (orders Marchantiales, Notothyladales, Metzgeriales, and Funariales). Importantly, the selected PRISE homologues include those from species that produce iridoids but not cardenolides (e.g., C. roseus), cardenolides but not iridoids (e.g., Digitalis lanata), neither iridoids nor cardenolides and with relatively limited observed secondary metabolites in general (e.g., the nonvascular bryophytes). Anywhere from 1–10 PRISE homologues were found in each of the species examined.
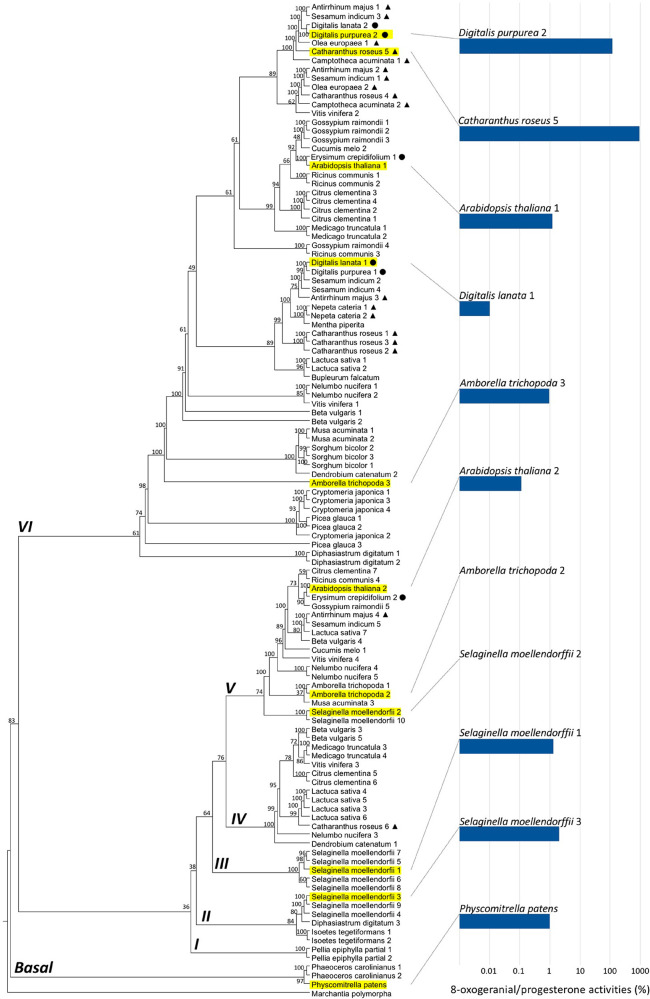
The phylogenetic analysis of this protein family indicates early duplications of PRISEs in land plants (Figure 2). All flowering plants examined here have at least one PRISE homologue in both clades IV and V (middle and top clades in Figure 2), while PRISEs from nonvascular plants (mosses) are found in the basal clade and clade I (bottom portion of Figure 2, which consists exclusively of homologues from nonvascular plants). Clade V (top) includes homologues from gymnosperms, and all functionally characterized ISYs involved in iridoid biosynthesis (e.g., Catharanthus), as well as PRISEs known to be involved in cardenolide biosynthesis (e.g., Digitalis).
Figure 2.
Phylogeny and 8-oxogeranial reduction activities of representative ISY/PRISE throughout land plants. Phylognetic tree inference was performed with W-IQ-Tree (http://iqtree.cibiv.univie.ac.at/) using maximum-likelihood method and JTT+I+G4 substitution model. Branch support values were calculated using ultrafast bootstraping with 1000 replicates. A tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Bars represent catalytic efficiencies (s–1 M–1) of 8-oxogeranial reduction activity, compared to those of progesterone reduction (as percentage) for selected ISY/PRISEs. [Circles (●) and triangles (▲) indicate the occurrence of cardenolides and iriroids, respectively, in the species to which the sequences belong.]
While a few PRISEs in clades IV and V from angiosperms have been assayed with the naturally occurring 4-en-3-ones, such as 8-oxogeranial and progesterone,15,16,19,28 we set out to biochemically characterize PRISE homologues from representative members of the angiosperms (clades IV and V), lycophytes (nonseed, vascular plants, clades II–IV), and nonvascular plants (basal clade). PRISEs from the model plant A. thaliana (clades IV and V), basal angiosperm Amborella trichopoda (clades IV and V), lycophyte Selaginella moellendorffii (clades II–IV), and the moss Physcomitrella patens (basal)—all species reported to lack iridoids and cardenolides—were compared to the reported PRISE enzymes in Catharanthus (iridoid producer) and Digitalis (cardenolide producer). In addition to members in land plants, a full-length PRISE homologue from green algae, Coccomyxa subellipsoidea, was also found and included in the analyses.
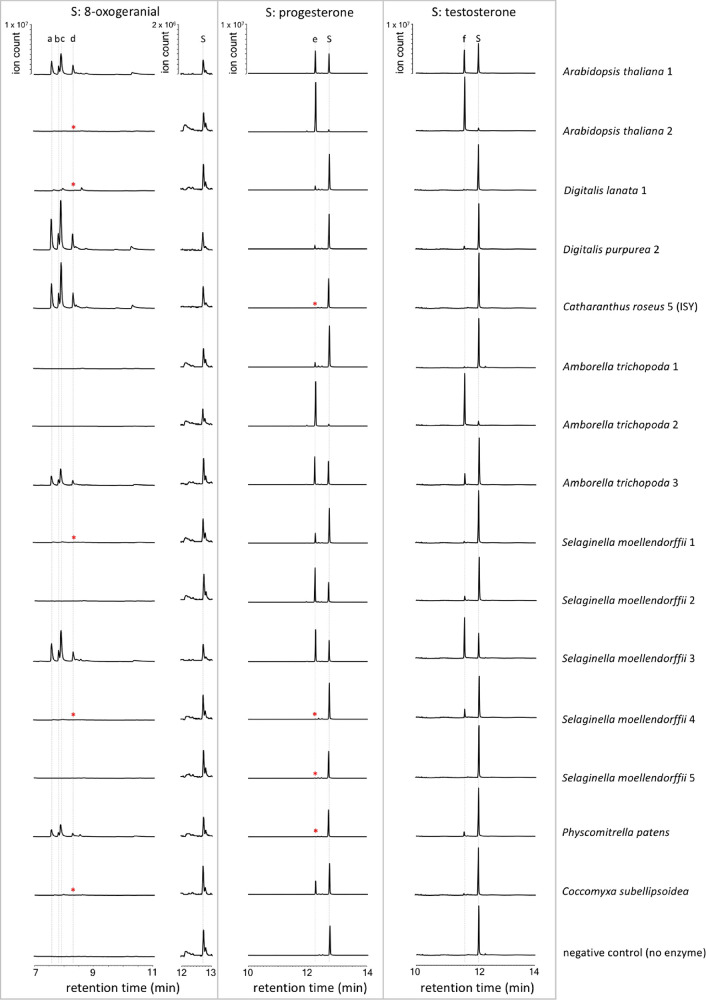
Given that PRISEs are found ubiquitously across the plant kingdom, it is likely that these enzymes have a more central role in plant metabolism beyond iridoid and cardenolide metabolism. Since sterols are one of the few plant central metabolites that contain the characteristic 4-en-3-one moiety characteristic of these enzyme substrates and are also found in all plant lineages, it is possible that sterols beyond progesterone may serve as substrates for these ubiquitous enzymes. Therefore, in addition to the two known, physiological substrates of the previously characterized PRISEs, 8-oxogeranial and progesterone, we tested each selected enzyme with two additional commercially available steroid substrates: testosterone, which is a steroid naturally occurring in some gymnosperm; and cholest-4-en-3-one, a close analogue of the precursor 24-methylcholest-4-en-3-one in brassinosteroid biosynthesis (see Figure S1 in the Supporting Information).29,30 Results showed that PRISEs displayed various activities against 8-oxogeranial, progesterone, and testosterone with nepetalactol/iridodial, 5β-pregnane-3,20-dione (or 5β-dihydroprogesterone), and possibly 5β-androstan-17β-ol-3-one (or 5β-dihydrotestosterone) as products, respectively (Figure 3 and Figures S2–S4 in the Supporting Information). No turnover of cholest-4-en-3-one could be observed in any assay, suggesting that an oxygenated group on the opposite end of the substrate (on the D-ring for steroid-like structures; see Figure S1) plays an essential role in substrate recognition. Steady-state kinetic parameters were characterized for 8-oxogeranial (the acyclic monoterpenoid iridoid precursor), and progesterone (a steroid substrate) in 12 homologues. These represent PRISEs from angiosperms, lycophytes, bryophytes, and green algae, and they include representatives of the same species in more than one clade where applicable.
Figure 3.
GC-MS analysis of activities of selected PRISEs. Total ion scan chromatograms are shown for each enzyme assayed with 8-oxogeranial (left), progesterone (middle), and testosterone (right). No activity on cholest-4-en-3-one was detected. [Legend: S, substrate; a–c, iridodials; d, nepetalactol; e, 5β-androstan-17β-ol-3-one (5β-dihydroprogesterone); and f, putative 5β-androstan-17β-ol-3-one (5β-dihydrotestosterone). Asterisk (*) denotes a detectable signal (see Figures S2 and S3).]
PRISEs in all clades displayed significant activities toward progesterone (see Figure 3, as well as Figure S3 in the Supporting Information). Interestingly, although optimized reduction/cyclization activities toward 8-oxogeranial are only found among clade V members as shown here and elsewhere,6,7 trace levels of 8-oxogeranial reductase activity were observed in PRISEs of the basal clade (moss, kcat = 0.066 s–1), and the vascular plants in clades II and III (Selaginella, kcat as high as 2 s–1) and of the basal angiosperms in clade V (Amborella, kcat = 0.321 s–1) (see Figure 3, as well as Figure S2). These activities seem to be maintained and further optimized in clade V (kcat > 5 s–1) while significantly reduced or completely lost (kcat/KM < 1000 s–1 M–1) in members of clade IV. This significant reduction and loss of ancestral 8-oxogeranial reduction activity is accompanied by the specialized activity on progesterone as seen in A. thaliana PRISE2 and A. trichopoda PRISE2 in clade V (see Table 1 and Figure 2).
Table 1. Comparison of Activities on 8-Oxogeranial and Progesterone in Selected PRISE Homologues (Mean ± SD, n = 4).
| 8-Oxogeranial |
Progesterone |
|||||
|---|---|---|---|---|---|---|
| KM (μM) | kcat (s–1) | kcat/KM (s–1 M–1) | KM (μM) | kcat (s–1) | kcat/KM (s–1 M–1) | |
| Coccomyxa subellipsoidea | 7.0 ± 1.6 | 0.025 ± 0.001 | 3578.5 | 23.6 ± 5.8 | 1.611 ± 0.148 | 68 360.9 |
| Physcomitrella patens | 31.3 ± 8.6 | 0.066 ± 0.005 | 2103.2 | 9.5 ± 3.1 | 2.101 ± 0.204 | 220 392.3 |
| Selaginella moellendorffii 1 | 4.6 ± 1.7 | 0.006 ± < 0.001 | 1273.0 | 11.7 ± 4.6 | 1.148 ± 0.143 | 98 119.7 |
| Selaginella moellendorffii 2 | n.d. | 11.5 ± 3.0 | 1.832 ± 0.157 | 159 443.0 | ||
| Selaginella moellendorffii 3 | 409.9 ± 74.2 | 2.029 ± 0.096 | 4950.0 | 8.5 ± 3.5 | 2.124 ± 0.254 | 249 032.7 |
| Amborella trichopoda 2 | n.d. | 3.1 ± 0.8 | 1.412 ± 0.089 | 458 441.6 | ||
| Amborella trichopoda 3 | 227.8 ± 29.7 | 0.321 ± 0.018 | 1410.0 | 4.5 ± 1.7 | 0.678 ± 0.067 | 149 958.8 |
| Arabidopsis thaliana 1 | 41.8 ± 2.75 | 0.155 ± 0.004 | 3706.6 | 6.0 ± 2.5 | 1.865 ± 0.216 | 312 081.7 |
| Arabidopsis thaliana 2 | 7.4 ± 3.0 | 0.002 ± < 0.001 | 308.1 | 5.7 ± 2.3 | 1.566 ± 0.148 | 275 026.3 |
| Digitalis lanata 1 | 3421.0 ± 1102.0 | 0.064 ± 0.015 | 18.6 | 6.5 ± 2.0 | 1.230 ± 0.111 | 189 376.4 |
| Digitalis purpurea 2 | 17.7 ± 5.9 | 6.317 ± 0.615 | 356 489.8 | 6.3 ± 1.9 | 1.881 ± 0.170 | 298 571.4 |
| Catharanthus roseus 5 | 8.5 ± 1.5 | 5.835 ± 0.302 | 688 739.4 | 28.8 ± 11.5 | 2.094 ± 0.332 | 72 784.2 |
From the universal occurrence of PRISE members in all plants, it is reasonable to speculate that PRISE members are involved in a reaction in primary metabolism that is central to plants, and this might be why they appeared early in plant evolution. Substrate tolerance for both progesterone and 8-oxogeranial has been reported in an PRISE homologue from various plant species, including A. thaliana and a gymnosperm,17,19 but our studies for the first time demonstrate that this ancestral promiscuity was retained throughout land plants from moss to angiosperms. We also cloned, for the first time, a PRISE homologue from green alga C. subellipsoidea, which showed broad substrate specificity toward steroidlike 4-en-3-ones (e.g., progesterone, testosterone) and acyclic allylic aldehydes (8-oxogeranial), although these activities were minimal.
To the best of our knowledge, iridoids and/or cardenolides are not reported in nonvascular plants and Selaginella. BLAST searches using enzymes in the iridoid pathway from C. roseus, as queries only produced significant hits for PRISE (∼45% amino acid identity) and no significant hits for geraniol synthase and geraniol-8-hydroxylase (<40% amino acid identity). This analysis suggests that, although low levels of iridoid synthase activity can be found in these plants, the precursor enzymes of the iridoid pathways do not occur in the basal plant lineages. Therefore, the ISY activity may not be the determining factor in the emergence of iridoids. Instead, other factors such as availability of 8-oxogeranial substrate may have played a greater role. In this regard, the emergence of ISYs/iridoids could be considered a type of “exaptation”, where the ancestral enzymes are able to catalyze 8-oxogeranial reduction but have no substrate. Once the substrate is available, these enzymes are primed for co-option into a new pathway.22
The capacity to reduce a variety of substrates could have been a detoxification mechanism for highly reactive α–β-unsaturated carbonyl compounds, as previously proposed by Kreis and coworkers.19 As land plants evolved, PRISE members appear to have been recruited for different pathways, and we see that certain groups seem to be optimized for steroidlike 4-en-3-ones while others accept acyclic allylic aldehydes almost exclusively. Examples for the latter group include geranial in the biosynthesis of the semiochemical (S)-β-citronellol in orchids, in addition to 8-oxogeranial.31
With its promiscuity feature, the PRISE family appears to be another example that supports the general hypothesis of evolution of specific enzyme activity from ancestral promiscuity.20,21 Our phylogenetic and biochemical analyses indicate that the promiscuity of the PRISE family is widespread and ancestral, and that the ability to reduce acyclic unsaturated carbonyl substrates such as C. roseus ISY’s reduction of 8-oxogeranial to nepetalactol and iridodials has not been recently “invented” but, instead, is very ancient (see Figure S2). Notably, this activity has been lost or reduced to negligible levels multiple times over the course of evolution of plants, as shown in in clade IV and certain subgroups of clade V in this study. This finding is also supported by a study on Antirrhinum majus PRISEs, in which A. majus PRISE1 (clade V) displayed comparable 8-oxogeranial reduction activity to that of C. roseus PRISE5 (ISY), while other A. majus PRISEs in clades IV and V only showed trace activities.8 Perhaps when 8-oxogeranial-like substrates were lost and/or new metabolic pathways that involved steroid-like enones emerged, certain ancient PRISEs were selected for alternative functions. From our data, it is also important to note that there exist PRISE homologues with nonexistent activity toward 8-oxogeranial and high activity toward steroidlike structures including progesterone in the iridoids- and cardenolides-free species A. thaliana, A. trichopoda, and S. moellendorffii. In contrast, progesterone 5β-reductase activity was observed at various degrees in all PRISE homologues, including C. roseus ISY. In PRISEs that accept such linear enones, the molecular flexibility of the substrate, as shown in at least one study on Plantago major PRISE,32 might render some promiscuity toward other structures. In PRISEs that only accept steroidlike enones, the rigidity of these four-ring structures could affort higher specificity at the expense of activities toward linear enones. This specialization, as evident in PRISEs in clade V here, suggests a committed role in yet-to-be-identified pathways, as opposed to general detoxification activities. Furthermore, PRISEs could also serve as another example of a plant’s “silent metabolism”, in which enzymes with broad-substrate specificity is retained and readily allow plants to chemically adapt to new conditions.33,34 To the best of our knowledge, except nonvascular plants (such as Physcomitrella and Marchantia), all plant species have more than one PRISE, allowing the optimization of specific activities in at least one homologue while retaining some promiscuity in others. Finally, the promiscuity of PRISE members is remarkable as progesterone and 8-oxogeranial reductions are catalyzed by two different types of enzymes in animals’ cardenolide and iridoid biosynthesis, respectively.2,28 This evolvable promiscuity provides not only an advantageous starting point in the establishment of novel metabolic pathways in plants, but also materials for many potential biochemical applications.
Methods
Phylogenetic Analysis
PRISE homologues across land plant lineages and green algae were identified by BLAST search using C. roseus ISY/PRISE5 as query. From available sequences on NCBI, 1KP Project, the Marchantia genome database, we selected 113 sequences, representing major orders covering angiosperms, gymnosperms, lycophytes, and bryophytes. These sequences were aligned with the multiple sequence alignment tool PRANK, and their maximum-likelihood phylogeny were reconstructed using the W-IQ-TREE server.35,36
Cloning
PRISE homologues from Physcomitrella patens, Selaginella moellendorffii, Amborella trichopoda, Digitalis purpurea, and the green alga Coccomyxa subellipsoidea were synthesized by ThermoFisher, in accordance with their published sequences without the start and stop codons, and with the additional sequences of AAGTTCTGTTTCAGGGCCCG and TAAAGCTTTCTAGACCAT at the 5′- and 3′-end, respectively (see Table S1 in the Supporting Information). PRISE1 (At4g24220) and PRISE2 (At5g58750) from Arabidopsis thaliana were cloned from cDNA using Phusion High-Fidelity DNA Polymerase (NEB) and the primer pairs of 5′-AAGTTCTGTTTCAGGGCCCGAGTTGGTGGTGGGCTGG-3′ (forward) and 5′-ATGGTCTAGAAAGCTTTAAGGTACGATCTTGAACGCC-3′ (reverse), and 5′-AAGTTCTGTTTCAGGGCCCGGGGTCTGAAAATGGCAGC-3′ (forward) and 5′-ATGGTCTAGAAAGCTTTACAAAGGAATGAGTTTTTCATCTCTCATC-3′ (reverse), respectively. C. roseus PRISE5 (ISY) and D. lanata PRISE1 (progesterone 5β-reductase) sequences codon-optimized for expression in Escherichia coli were obtained from a previous study.18 All PRISEs were cloned using In-Fusion HD Cloning Kit (Clontech Laboratories) into pOPINF expression vector as described in earlier reports, allowing expression of proteins with N-terminal fusion of a hexa-histidine tag.18,37 The constructs were confirmed by sequencing.
Expression and Protein Isolation
The pOPINF vectors harboring PRISE homologues were transformed to E. coli soluBL21 (DE3) cells (Genlantis). Transformed cells were inoculated overnight in LB medium supplemented with 100 μg/mL carbenicillin at 37 °C. The inoculates were then transferred to 1–2 L culture in 2xYT medium supplemented with 100 μg/mL carbenicillin in Erlenmeyer flasks with an inoculate:culture ratio of 1:100 and a culture:flask volume ratio of 1:2. When OD600 of the cultures reached ∼0.6 after 4–6 h at 37 °C, they were continued at 18 °C for 1 h, followed by addition of IPTG to a final concentration of 100 μM for induction. The induced cultures were continued for ∼16 h. Cells were collected by centrifugation and resuspended in buffer A (50 mM Tris-HCl buffer (pH 7.0) containing 50 mM glycine, 5% (v/v) glycerol, 500 mM NaCl, and one EDTA-free protease inhibitor tablet (Roche) per 50 mM buffer) containing 20 mM imidazole. Cells were lysed using a cell disruptor (Constant Systems, Ltd.) at 25 000 psi, followed by centrifugation at 35 000g for 20 min at 4 °C. All subsequent steps were performed at 4 °C. The supernatant was collected and mixed with Ni-NTA slurry (Qiagen) and incubated gently on rocking platform for 1 h. The slurry was subsequently collected by centrifugation at 2000g and washed three times with excessive amount of buffer A (15 mL of buffer for 1 mL of slurry). Target proteins were eluted by washing Ni-NTA slurry with buffer A containing 500 mM imidazole. Buffer was exchanged using PD 10 desalting columns (GE Healthcare) to 50 mM HEPES/NaOH (pH 7.0) buffer containing 100 mM NaCl.
Enzyme Assays and GC-MS Analysis
To test PRISE activities, each assay of 100 μL was set up with 2 μM enzyme, 500 μM NADPH, and 200 μM substrate in 50 mM HEPES/NaOH pH 7.0) buffer containing 100 mM NaCl. The steroid substrates (progesterone, testosterone, and cholest-4-en-3-one) were purchased from Sigma–Aldrich, while 8-oxogeranial was synthesized as previously described (4). The same reaction was set up without adding enzyme as a negative control. The reaction was allowed at room temperature (RT) with gentle agitation (60 rpm). After 3 h, 200 μL of ethyl acetate was added to the reaction and mixed vigorously for 30 s. The mixture was then centrifuged at 20 000g for 2 min using a benchtop centrifuge, and the ethyl acetate fraction was used for gas chromatography–mass spectrometry (GC-MS) analysis. The sample was injected in splitless mode using a Gerstel MPS autosampler on an Agilent 7890 GC system coupled with a Model 5973 mass-selective detector. The inlet temperatures were 100 and 250 °C for assays with 8-oxogeranial and with other substrates, respectively. GC separation was performed on an Agilent HP-5MS column (30 m × 320 μm) with helium at 1 mL/min as the mobile phase. The GC oven program was set to 80 °C for 1 min, followed by a linear gradient of 20 °C/min to 310 °C and held for 3 min.
Kinetic Analysis
Kinetics of PRISEs’ activities on 8-oxogeranial and progesterone were measured based on NADPH consumption. Reactions were set on a 96-well plate with each well containing 5–1000 nM enzyme, 250 μM NADPH, 1–100 μM substrate in 50 mM HEPES/NaOH (pH 7.0) buffer containing 100 mM NaCl to a total volume of 200 μL. Tetrahydrofuran at 0.1% and ethanol at 1.5% were used as cosolvent to ensure solubility of 8-oxogeranial and progesterone, respectively. Reactions were allowed at 25 °C, and NADPH consumption was monitored in a 96-well plate reader at 340 nm. Initial velocity was calculated based on NADPH standard and nonlinearly fit to the Michaelis–Menten curve using GraphPad Prism (GraphPad Software, Inc.).
Acknowledgments
We would like to thank B. Lichman, P. Soltis, and D. Soltis for helpful discussions. We also thank M. Kamileen for assistance with GC-MS analysis. The plant drawing in the Table of Contents graphic is a gift from K. Davis.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00220.
Table S1 and Figures S1–S4, with additional mass spectrometry chromatograms (PDF)
We gratefully acknowledge NSF Genome (Grant No.1444499).
The authors declare no competing financial interest.
Supplementary Material
References
- El-Naggar L. J.; Beal J. L. (1980) Iridoids: A review. J. Nat. Prod. 43, 649–707. 10.1021/np50012a001. [DOI] [PubMed] [Google Scholar]
- Beran F.; Köllner T. G.; Gershenzon J.; Tholl D. (2019) Chemical convergence between plants and insects: Biosynthetic origins and functions of common secondary metabolites. New Phytol. 223, 52–67. 10.1111/nph.15718. [DOI] [PubMed] [Google Scholar]
- Tundis R.; Loizzo M.; Menichini F.; Statti G.; Menichini F. (2008) Biological and pharmacological activities of iridoids: Recent developments. Mini-Rev. Med. Chem. 8, 399–420. 10.2174/138955708783955926. [DOI] [PubMed] [Google Scholar]
- Yamane H.; Konno K.; Sabelis M.; Takabayashi J.; Sassa T.; Oikawa H. (2010) Chemical defence and toxins of plants. Compr. Nat. Prod. II Chem. Biol. 4, 339–385. 10.1016/B978-008045382-8.00099-X. [DOI] [Google Scholar]
- Miettinen K.; Dong L.; Navrot N.; Schneider T.; Burlat V.; Pollier J.; Woittiez L.; Van Der Krol S.; Lugan R.; Ilc T.; Verpoorte R.; Oksman-Caldentey K. M.; Martinoia E.; Bouwmeester H.; Goossens A.; Memelink J.; Werck-Reichhart D. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5, 3606. 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F.; Sherden N. H.; Courdavault V.; Burlat V.; Glenn W. S.; Wu C.; Nims E.; Cui Y.; O’Connor S. E. (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492, 138–142. 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Alagna F.; Geu-Flores F.; Kries H.; Panara F.; Baldoni L.; O’Connor S. E.; Osbourn A. (2016) Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) fruits. J. Biol. Chem. 291, 5542–5554. 10.1074/jbc.M115.701276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kries H.; Kellner F.; Kamileen M. O.; O’Connor S. E. (2017) Inverted stereocontrol of iridoid synthase in snapdragon. J. Biol. Chem. 292, 14659–14667. 10.1074/jbc.M117.800979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichman B. R.; Kamileen M. O.; Titchiner G. R.; Saalbach G.; Stevenson C. E. M.; Lawson D. M.; O’Connor S. E. (2019) Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis. Nat. Chem. Biol. 15, 71–79. 10.1038/s41589-018-0185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner D. E.; Keilholz W.; Seitz H. U. (1994) Purification, characterization and partial peptide microsequencing of progesterone 5β-reductase from shoot cultures of Digitalis purpurea. Eur. J. Biochem. 225, 1125–1132. 10.1111/j.1432-1033.1994.1125b.x. [DOI] [PubMed] [Google Scholar]
- Agrawal A. A.; Petschenka G.; Bingham R. A.; Weber M. G.; Rasmann S. (2012) Toxic cardenolides: Chemical ecology and coevolution of specialized plant–herbivore interactions. New Phytol. 194, 28–45. 10.1111/j.1469-8137.2011.04049.x. [DOI] [PubMed] [Google Scholar]
- Dobler S.; Petschenka G.; Pankoke H. (2011) Coping with toxic plant compounds: The insect’s perspective on iridoid glycosides and cardenolides. Phytochemistry 72, 1593–1604. 10.1016/j.phytochem.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Petersen J.; Lanig H.; Munkert J.; Bauer P.; Müller-Uri F.; Kreis W. (2016) Progesterone 5β-reductases/iridoid synthases (PRISE): Gatekeeper role of highly conserved phenylalanines in substrate preference and trapping is supported by molecular dynamics simulations. J. Biomol. Struct. Dyn. 34, 1667–1680. 10.1080/07391102.2015.1088797. [DOI] [PubMed] [Google Scholar]
- Jun J. H.; Ha C. M.; Nam H. G. (2002) Involvement of the VEP1 gene in vascular strand development in Arabidopsis thaliana. Plant Cell Physiol. 43, 323–330. 10.1093/pcp/pcf042. [DOI] [PubMed] [Google Scholar]
- Bauer P.; Munkert J.; Brydziun M.; Burda E.; Müller-Uri F.; Gröger H.; Muller Y. A.; Kreis W. (2010) Highly conserved progesterone 5β-reductase genes (P5βR) from 5β-cardenolide-free and 5β-cardenolide-producing angiosperms. Phytochemistry 71, 1495–1505. 10.1016/j.phytochem.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Munkert J.; Pollier J.; Miettinen K.; Van Moerkercke A.; Payne R.; Müller-Uri F.; Burlat V.; O’Connor S. E.; Memelink J.; Kreis W.; Goossens A. (2015) Iridoid synthase activity is common among the plant progesterone 5β-reductase family. Mol. Plant 8, 136–152. 10.1016/j.molp.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Rudolph K.; Wiegert T.; Schubert R.; Müller-Uri F. (2016) The occurrence of progesterone 5β-reductase is not limited to the angiosperms: A functional gene was identified in Picea sitchensis and expressed in Escherichia coli. New Zeal. J. For. Sci. 46, 1–9. 10.1186/s40490-016-0063-1. [DOI] [Google Scholar]
- Kries H.; Caputi L.; Stevenson C. E. M.; Kamileen M. O.; Sherden N. H.; Geu-Flores F.; Lawson D. M.; O’Connor S. E. (2016) Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 12, 6–8. 10.1038/nchembio.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.; Petersen J.; Munkert J.; Egerer-Sieber C.; Hornig M.; Muller Y. A.; Kreis W. (2018) PRISEs (progesterone 5β-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism. Phytochemistry 156, 9–19. 10.1016/j.phytochem.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Jensen R. A. (1976) Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425. 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Tawfik O. K.; Tawfik D. S. (2010) Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505. 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Huang R.; O’Donnell A. J.; Barboline J. J.; Barkman T. J. (2016) Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. U. S. A. 113, 10613–10618. 10.1073/pnas.1602575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. S.; Facchini P. J. (2019) Molecular origins of functional diversity in benzylisoquinoline alkaloid methyltransferases. Front. Plant Sci. 10, 1–22. 10.3389/fpls.2019.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. D.; Kwon M.; Kim S. U.; Fischer C.; Ro D. K. (2019) Catalytic plasticity of germacrene A oxidase underlies sesquiterpene lactone diversification. Plant Physiol. 181, 945–960. 10.1104/pp.19.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N.; Hung L. H.; Yan Z.; Carpenter E. J.; Wickett N. J.; Mirarab S.; Nguyen N.; Warnow T.; Ayyampalayam S.; Barker M.; Burleigh J. G.; Gitzendanner M. A.; Wafula E.; Der J. P.; dePamphilis C. W.; Roure B.; Philippe H.; Ruhfel B. R.; Miles N. W.; Graham S. W.; Mathews S.; Surek B.; Melkonian M.; Soltis D. E.; Soltis P. S.; Rothfels C.; Pokorny L.; Shaw J. A.; DeGironimo L.; Stevenson D. W.; Villarreal J. C.; Chen T.; Kutchan T. M.; Rolf M.; Baucom R. S.; Deyholos M. K.; Samudrala R.; Tian Z.; Wu X.; Sun X.; Zhang Y.; Wang J.; Leebens-Mack J.; Wong G. K. S. (2014) Data access for the 1,000 Plants (1KP) Project. GigaScience 3, 1–10. 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S. A.; Lang D.; Zimmer A. D.; Terry A.; Salamov A.; Shapiro H.; Nishiyama T.; Perroud P. F.; Lindquist E. A.; Kamisugi Y.; Tanahashi T.; Sakakibara K.; Fujita T.; Oishi K.; Shin-I T.; Kuroki Y.; Toyoda A.; Suzuki Y.; Hashimoto S. I.; Yamaguchi K.; Sugano S.; Kohara Y.; Fujiyama A.; Anterola A.; Aoki S.; Ashton N.; Barbazuk W. B.; Barker E.; Bennetzen J. L.; Blankenship R.; Cho S. H.; Dutcher S. K.; Estelle M.; Fawcett J. A.; Gundlach H.; Hanada K.; Heyl A.; Hicks K. A.; Hughes J.; Lohr M.; Mayer K.; Melkozernov A.; Murata T.; Nelson D. R.; Pils B.; Prigge M.; Reiss B.; Renner T.; Rombauts S.; Rushton P. J.; Sanderfoot A.; Schween G.; Shiu S. H.; Stueber K.; Theodoulou F. L.; Tu H.; Van de Peer Y.; Verrier P. J.; Waters E.; Wood A.; Yang L.; Cove D.; Cuming A. C.; Hasebe M.; Lucas S.; Mishler B. D.; Reski R.; Grigoriev I. V.; Quatrano R. S.; Boore J. L. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Bowman J. L.; Kohchi T.; Yamato K. T.; Jenkins J.; Shu S.; Ishizaki K.; Yamaoka S.; Nishihama R.; Nakamura Y.; Berger F.; Adam C.; Aki S. S.; Althoff F.; Araki T.; Arteaga-Vazquez M. A.; Balasubrmanian S.; Barry K.; Bauer D.; Boehm C. R.; Briginshaw L.; Caballero-Perez J.; Catarino B.; Chen F.; Chiyoda S.; Chovatia M.; Davies K. M.; Delmans M.; Demura T.; Dierschke T.; Dolan L.; Dorantes-Acosta A. E.; Eklund D. M.; Florent S. N.; Flores-Sandoval E.; Fujiyama A.; Fukuzawa H.; Galik B.; Grimanelli D.; Grimwood J.; Grossniklaus U.; Hamada T.; Haseloff J.; Hetherington A. J.; Higo A.; Hirakawa Y.; Hundley H. N.; Ikeda Y.; Inoue K.; Inoue S. I.; Ishida S.; Jia Q.; Kakita M.; Kanazawa T.; Kawai Y.; Kawashima T.; Kennedy M.; Kinose K.; Kinoshita T.; Kohara Y.; Koide E.; Komatsu K.; Kopischke S.; Kubo M.; Kyozuka J.; Lagercrantz U.; Lin S. S.; Lindquist E.; Lipzen A. M.; Lu C. W.; De Luna E.; Martienssen R. A.; Minamino N.; Mizutani M.; Mizutani M.; Mochizuki N.; Monte I.; Mosher R.; Nagasaki H.; Nakagami H.; Naramoto S.; Nishitani K.; Ohtani M.; Okamoto T.; Okumura M.; Phillips J.; Pollak B.; Reinders A.; Rövekamp M.; Sano R.; Sawa S.; Schmid M. W.; Shirakawa M.; Solano R.; Spunde A.; Suetsugu N.; Sugano S.; Sugiyama A.; Sun R.; Suzuki Y.; Takenaka M.; Takezawa D.; Tomogane H.; Tsuzuki M.; Ueda T.; Umeda M.; Ward J. M.; Watanabe Y.; Yazaki K.; Yokoyama R.; Yoshitake Y.; Yotsui I.; Zachgo S.; Schmutz J. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- Gavidia I.; Tarrío R.; Rodríguez-Trelles F.; Pérez-Bermúdez P.; Ulrich Seitz H. (2007) Plant progesterone 5β-reductase is not homologous to the animal enzyme: Molecular evolutionary characterization of P5βR from Digitalis purpurea. Phytochemistry 68, 853–864. 10.1016/j.phytochem.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Šaden-Krehula M.; Tajić M.; Kolbah D. (1979) Sex hormones and corticosteroids in pollen of Pinus nigra. Phytochemistry 18, 345–346. 10.1016/0031-9422(79)80098-9. [DOI] [Google Scholar]
- Noguchi T.; Fujioka S.; Takatsuto S.; Sakurai A.; Yoshida S.; Li J.; Chory J. (1999) Arabidopsis Det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120, 833–839. 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Bohman B.; Wong D. C. J.; Rodriguez-Delgado C.; Scaffidi A.; Flematti G. R.; Phillips R. D.; Pichersky E.; Peakall R. (2017) Complex sexual deception in an orchid is achieved by co-opting two independent biosynthetic pathways for pollinator attraction. Curr. Biol. 27, 1867–1877.e5. 10.1016/j.cub.2017.05.065. [DOI] [PubMed] [Google Scholar]
- Fellows R.; Russo C. M.; Silva C. S.; Lee S. G.; Jez J. M.; Chisholm J. D.; Zubieta C.; Nanao M. H. (2018) A multisubstrate reductase from Plantago major: Structure-function in the short chain reductase superfamily. Sci. Rep. 8, 1–13. 10.1038/s41598-018-32967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. J.; Herschlag D. (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105. 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- Lewinsohn E.; Gijzen M. (2009) Phytochemical diversity: The sounds of silent metabolism. Plant Sci. 176, 161–169. 10.1016/j.plantsci.2008.09.018. [DOI] [Google Scholar]
- Chojnacki S.; Cowley A.; Lee J.; Foix A.; Lopez R. (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553. 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J.; Nguyen L. T.; von Haeseler A.; Minh B. Q. (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrow N. S.; Alderton D.; Sainsbury S.; Nettleship J.; Assenberg R.; Rahman N.; Stuart D. I.; Owens R. J. A. (2007) Versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45–e57. 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.