Abstract
Background
Chronic kidney disease (CKD) patients under hemodialysis show a higher risk of cardiovascular (CV) mortality and morbidity than the general population. This study aims to identify genetic markers that could explain the increased CV risk in hemodialysis.
Methods
A total of 245 CKD patients under hemodialysis were recruited and followed up for 5 years to record CV events. Genetic analysis was performed using single-nucleotide polymorphisms (SNPs) genotyping by Infinium Expanded Multi-Ethnic Genotyping Array (Illumina, San Diego, CA, USA) comparing patients with and without a history of CV events [161 cardiovascular diseases (CVDs) and 84 no CVDs]. The fixation index (Fst) measure was used to identify the most differentiated SNPs, and gene ontology analysis [Protein Analysis THrough Evolutionary Relationships (PANTHER) and Ingenuity Pathway Analysis (IPA)] was applied to define the biological/pathological roles of the associated SNPs. Partitioning tree analysis interrogated the genotype–phenotype relationship between discovered genetic variants and CV phenotypes. Cox regression analysis measured the effect of these SNPs on new CV events during the follow-up (FU).
Results
Fst analysis identified 3218 SNPs that were significantly different between CVD and no CVD. Gene ontology analysis identified two of these SNPs as involved in cardiovascular disease pathways (Ingenuity Pathway) and heart development (Panther) and belonging to 2 different genes: Glucagon-like peptide-1 receptor (GLP1R) and Sarcoglycan delta (SGCD). The phenotype–genotype analysis found a higher percentage of CVD patients carrying the GLP1R rs10305445 allele A (P = 0.03) and lower percentages of CVD patients carrying the SGCD rs145292439 allele A (P = 0.038). Moreover, SGCD rs145292439 was associated with higher levels of high-density lipoprotein (P = 0.015). Cox analysis confirmed the increased frequency of CV events during the 5-year FU in patients carrying GLP1R rs1035445 allele A but it did not show any significant association with SGCD rs145292439.
Conclusions
This study identified GLP1R rs10305445 and SCGD rs145292439 as potential genetic markers that may explain the higher risk of CVD in hemodialysis patients.
Keywords: cardiovascular disease, chronic kidney disease, genetics, gene polymorphism, hemodialysis
INTRODUCTION
Cardiovascular disease (CVD) is the main cause of death and morbidity in patients with chronic kidney disease (CKD). The incidence of cardiovascular (CV) mortality is 10–20 times higher in CKD patients compared with the general population and it increases with the severity of CKD. Consequently, ∼20% of hemodialysis patients die for CV events each year [1–5]. Traditional predisposing factors have been demonstrated to not to play a role in predicting CV risk in CKD patients; nevertheless, CV risk in CKD has been associated to mineral metabolism alterations, anemia and uremic toxin levels that may lead to vascular and coronary stiffness and left ventricular hypertrophy [6–9].
Twin and family studies have documented the heritability of susceptibility to CV events in patients with normal renal function. Genetic studies and genome-wide association studies (GWASs) have identified multiple loci associated with potential causal factors as well as pathophysiological pathways and therapeutic targets implicated in CVD [10, 11].
Studies in CKD patients investigated single candidate genes potentially involved in CV risk, and found potentially genetic risk factors, such as Kelch Like ECH Associated Protein 1 (Keap1), Vascular Endothelial Growth Factor (VEGF), Monocyte Chemotactic Protein-1 (MCP-1), Interleukin 6 (IL-6), Nuclear Erythroid Factor 2 (Nrf2), Vitamin D Receptor (VDR), Heme Oxigenase 1 (HMOX1) and Receptor of Advanced Glycosylation End-Product (RAGE). [12–19]. No GWASs were conducted in CKD patients to explore genes and their variants associated with CVD. A recent study investigated the genetic risk of CVD in CKD [20] using a multilocus risk factor score, as the sum of the number of risk alleles across 9 variants identified from a GWAS in patients with normal kidney function. This score was able to predict an increase in coronary heart disease in CKD Stages 4 and 5 patients more accurately than traditional risk factors [21].
The aim of this study was to interrogate the whole-genome variants to identify genetic markers for CVD in CKD patients undergoing hemodialysis. We genotyped 245 hemodialysis patients using Infinium Expanded Multi-Ethnic Genotyping Array (MEGAEX; Illumina, San Diego, CA, USA) and correlated those variants to the patients’ history of CV events and to new events occurring during a 5-year follow-up (FU).
MATERIALS AND METHODS
Patients, clinical data and samples
A total of 245 hemodialysis patients were enrolled in a prospective study from three dialysis centers in Milan (Italy): San Raffaele Hospital, San Paolo Hospital and Cernusco sul Naviglio Uboldo Hospital. CKD causes are summarized in Supplementary data, Table S1 and the patients were treated according to the clinical guidelines and patient needs. Exclusion criteria were a 5-year history of neoplasia, acute kidney disorders, liver insufficiency or cirrhosis, malabsorption, dementia and motorial inability and autoimmune disorders.
Incident CV events were recorded during a 5-year FU; previous CV events were documented from clinical records (the included CVD are listed in the Supplementary data file). All-cause mortality was also considered. Patients were grouped into two subpopulations: individuals with or without a history of CV events (CVD and no CVD, respectively). Secondarily, patients after the 5-year FU were reclassified for the absence or presence of new CV events (CVD at FU and no CVD at FU, respectively).
Serum concentrations of phosphate, total calcium, creatinine, C-reactive protein (CRP), 25-hydroxyvitamin D [25(OH)D], 1,25-dihydroxy-vitamin D [1,25(OH)2D], intact parathyroid hormone (PTH) and fibroblast growth factor 23 (FGF23) were measured at the study baseline. In particular, PTH and 1,25(OH)2D were measured by chemiluminescence immunoassay (CLIA); 25(OH)D by radioimmunoassay CLIA (DiaSorin, Stillwater, MN, USA); and full-length FGF23 by a two-step enzyme-linked immunoassay method (Kainos Laboratories, Tokyo, Japan) [22]. Peripheral blood samples were collected at the baseline in ethylenediaminetetraacetic acid (EDTA) tubes and stored for genotyping analysis.
The study was approved by the San Raffaele Scientific Institute Ethical Committee. All participants signed informed consent to participate in this study.
Genotyping
Genomic DNA was extracted from EDTA blood samples using the NucleoSpin Blood DNA Isolation kit (Macherey-Nagel, Düren, Germany). DNA was checked for quantity and quality using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and FlexStation3 (Molecular Devices, San Jose, CA, USA) and then processed for genotyping using the MEGAEX. See the detailed method in the Supplementary data.
Statistics
Phenotypic data were expressed as mean ± standard deviation. Differences between mean values were assessed using Student’s t-test. A two-sided P-value < 0.05 (after Bonferroni correction) was considered indicative of statistical significance.
Pairwise identity was calculated by state between individuals using PLINK version 1.07 [23], and all close relatives were removed up to the second cousin using a ρ^ cut-off of 0.125. The initial dataset composed of 245 genotyped individuals was reduced to 208 unrelated individuals.
For analyses of markers with a steep difference in allele frequency between no CVD and CVD, the following filters were applied: minor allele frequency > 0.05, Hardy–Weinberg equilibrium exact test P > 1−7, marker in linkage disequilibrium (r2 > 0.4) and genotyping rate > 0.99.
Subsequently the fixation index (Fst) was estimated [24] as a measure of genetic differentiation for the same marker between two subpopulations in order to find the most differentiated single-nucleotide polymorphisms (SNPs) among no CVD and CVD patients taking only the variants that fall over the 99th percentile of the genome-wide Fst distribution. Variants were annotated using the variant effect predictor (VEP) [25]. Gene ontology analyses were performed using Protein Analysis THrough Evolutionary Relationships (PANTHER) [26]. Pathways analyses were performed using Ingenuity Pathway Analysis (IPA) [27].
Conditional inference-based recursive partitioning (hereafter ‘tree regression analysis’) implemented in the R ‘party’ package [28] was used to describe the genotype–phenotype relationship between genetic variants discovered, covariates and CV phenotype. The procedure utilizes the variable with the lowest P-value (after Bonferroni correction) as the first node of the decision tree; subsequently, subgroups are created and the variable with the lowest P-value (if there is one) is taken as the second or third node.
SNPs associated with the previous history of CV events were then tested for the association with new CV events during the 5-year FU using two Cox regression models. The first model included variables associated with CV events in the univariate analysis and the second considered variables potentially implicated in CV events. The odds ratio (OR) and 95% confidence interval (CI) were calculated to express the risk of CV events related to the patient genotype.
Statistical analyses were conducted using SPSS Statistics (IBM, Armonk, NY, USA) and R (R Foundation, Vienna, Austria) [28].
Predictive functional analysis
Prediction tools were used to test any function effect of SNPs associated with CV events on the respective genes. The RegulomeDB database annotates SNPs with known and predicted regulatory DNA elements in the human genome, including regions of DNAase hypersensitivity, binding sites of transcription factors and promoter regions [29]. HaploReg version 4.1 explores annotations of SNPs for chromatin state, protein-binding annotation, sequence conservation across mammals and the effect on regulatory motifs and on expression from expression quantitative trait loci studies [30]. The SNP identification (rs# from the dbSNP database) was used as data for entry into both RegulomeDB and HaploReg. Match 1.0 Public is a weight matrix–based program for predicting transcription factor binding sites in DNA sequences that uses a library of positional weight matrices from TRANSFAC 6.0 [31]. Sequences (100 nt) for each variant were imputed as FASTA format.
RESULTS
Patients’ characteristics
Among 245 participants, 161 patients (65.7%) had suffered from previous CV events (CVD). Compared with 84 patients without a CV history (no CVD), these patients were older, spent more time on dialysis, had a higher incidence of hypertension and had higher CRP values. They also developed more CV events during the 5-year FU (Table 1).
Table 1.
Characteristics of patients divided according to CV history
| Variables | Total patients | Patients CVD | Patients no CVD | P-value |
|---|---|---|---|---|
| Patients, n (M/F) | 245 (168/77) | 161(112/49) | 84(56/28) | 0.64 |
| Age (years) | 67 ± 14 | 70 ± 12 | 62 ± 15 | 0.0001 |
| Body weight (kg) | 66 ± 14 | 66 ± 14 | 66 ± 13 | 0.87 |
| Dialysis vintage (months) | 62 ± 64 | 71 ± 67 | 45 ± 55 | 0.002 |
| Serum creatinine (mg/dL) | 8.34 ± 2.36 | 8.25 ± 2.25 | 8.51 ± 2.54 | 0.41 |
| Serum calcium (mmol/L) | 2.22 ± 0.18 | 2.21 ± 0.18 | 2.24 ± 0.18 | 0.15 |
| Serum phosphate (mmol/L) | 1.48 ± 0.54 | 1.44 ± 0.55 | 1.55 ± 0.53 | 0.15 |
| Serum protein (g/dL) | 6.54 ± 0.67 | 6.55 ± 0.68 | 6.53 ± 0.69 | 0.81 |
| Serum PTH (pg/mL) | 235 ± 197 | 228 ± 197 | 248 ± 197 | 0.44 |
| Serum 1,25(OH)2D (pg/mL) | 10.3 ± 10.1 | 9.5 ± 6.2 | 11.7 ± 14.5 | 0.23 |
| Serum 25(OH)D (ng/mL) | 13.1 ± 9.8 | 12.4 ± 10.5 | 14.3 ± 8.5 | 0.32 |
| Serum FGF23 (pg/mL) | 2158 ± 3543 | 1874 ± 3181 | 2654 ± 4078 | 0.13 |
| Serum CRP (mg/L) | 9.1 ± 16.1 | 11.1 ± 18.8 | 5.1 ± 7.8 | 0.001 |
| CV events in a 5-year FU, n (%) | 79 (32.2) | 73 (45.3) | 6 (7.1) | 0.0001 |
| CV deaths in a 5-year FU, n (%) | 34 (13.9) | 34 (21.1) | 0 | 0.0001 |
| All-cause deaths in FU, n (%) | 62 (25.3) | 49 (30.4) | 13 (15.5) | 0.011 |
| Diabetes mellitus, n (%) | 76 (31) | 54 (33.5) | 22 (26.2) | 0.24 |
| Arterial hypertension, n (%) | 235 (95.9) | 159 (98.8) | 76 (90.5) | 0.002 |
Values are presented as mean ± standard deviation unless stated otherwise.
Seventy-nine patients suffered from CV events during the 5-year FU and they were characterized by a higher percentage of past CV events, increased body weight and lower serum PTH. The effect of age, dialysis vintage and hypertension was confirmed with the CV events recorded during the FU (Table 2).
Table 2.
Characteristics of patients who suffered from CV events during the 5-year FU
| Variables | Patients CVD at FU | Patients no CVD at FU | P-value |
|---|---|---|---|
| n (M/F) | 79 (56/23) | 166 (112/54) | 0.59 |
| Age (years) | 70 ± 12 | 62 ± 15 | 0.031 |
| Body weight (kg) | 63 ± 14 | 67 ± 13 | 0.038 |
| Dialysis vintage (months) | 70 ± 68 | 58 ± 62 | 0.002 |
| Serum creatinine (mg/dL) | 8.31 ± 2.31 | 8.36 ± 2.39 | 0.88 |
| Serum calcium (mmol/L) | 2.20 ± 0.16 | 2.22 ± 0.19 | 0.39 |
| Serum phosphate (mmol/L) | 1.47 ± 0.48 | 1.48 ± 0.57 | 0.84 |
| Serum protein (g/dL) | 6.59 ± 0.59 | 6.52 ± 0.71 | 0.81 |
| Serum PTH (pg/mL) | 197 ± 155 | 253 ± 211 | 0.039 |
| Serum 1,25(OH)2D (pg/mL) | 9.2 ± 7.1 | 10.8 ± 11.2 | 0.31 |
| Serum 25(OH)D (ng/mL) | 13.4 ± 13.8 | 13 ± 7.4 | 0.85 |
| Serum FGF23 (pg/mL) | 2092 ± 3482 | 2193 ± 3588 | 0.85 |
| Serum CRP (mg/L) | 11.7 ± 18.7 | 7.8 ± 14.6 | 0.11 |
| Diabetes mellitus, n (%) | 23 (29.1) | 59 (31.9) | 0.66 |
| Arterial hypertension, n (%) | 78 (98.7) | 157 (94.6) | 0.038 |
| Past CV events, n (%) | 73 (92.4) | 88 (50) | 0.0001 |
Values are presented as mean ± standard deviation unless stated otherwise.
Genetic analysis
A SNP genotyping array was used to investigate potential genetic markers predisposing to CVD in hemodialysis patients.
From an initial dataset of 245 genotyped patients, the genetically close relatives were removed and 208 unrelated individuals were used for the analysis (Supplementary data, Figure S1). At the baseline, from 208 subjects, we defined a group of 135 CVD subjects and a group of 73 no CVD subjects that shared similar clinical characteristics of the entire population at baseline and at FU (Supplementary data, Tables S2 and S3).
After applying genetic analysis filters, 321 788 polymorphic variants were obtained from the 2 million interrogated SNPs. The Fst was estimated as a measure of the genetic differentiation between CVD and no CVD and resulted in 3218 markers with Fst >99th percentile of the genome-wide distribution (which represents the most differentiated SNPs between CVD and no CVD; Supplementary data, Figure S2).
Gene ontology analysis
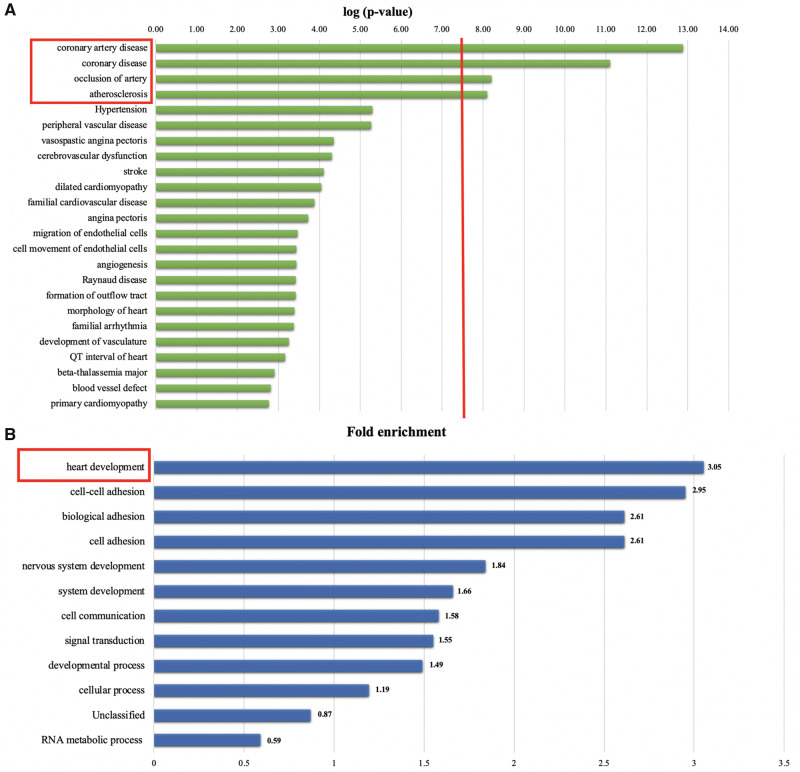
IPA was performed to identify possible functional roles of the 3218 selected variants. Twelve disease pathways were identified with –log(P-value) > 7.5, among which CVD scored in the top 10 (Supplementary data, Figure S3). We focused on the disease categories from the CVD pathway and we identified four enriched categories with a –log(P-value) > 7.5 (Figure 1A): coronary artery disease, coronary disease, arterial occlusion and atherosclerosis.
FIGURE 1.
Gene Ontology Analysis. (A) IPA shows the enriched categories among CVD in CVD versus no CVD. The red line represents the P-value cut-off of 7.5 –log(P-value): 12 disease pathways were identified with a log(P-value) > 7.5, among which CVD scored in the top 10. In the ‘CVD pathway’, four enriched categories with a –log(P-value) > 7.5 were identified: coronary artery disease, coronary disease, arterial occlusion and atherosclerosis. (B) PANTHER analysis resulted in the heart development category as most significant comparing CVD versus no CVD (3.05-fold enrichment, P = 2.65 × 10−2).
Gene ontology analyses using PANTHER identified the heart development pathway as the one with the highest value of gene enrichment (3.05-fold, P = 2.65 × 10−2) (Figure 1B and Supplementary data, Figure S4).
Selecting the only significant results from both the IPA and PANTHER analyses resulted in the identification of two genes: Glucagon-like peptide 1 receptor (GLP1R) and sarcoglycan delta (SGCD) (Supplementary data, Table S4).
Tree regression analysis and CV history
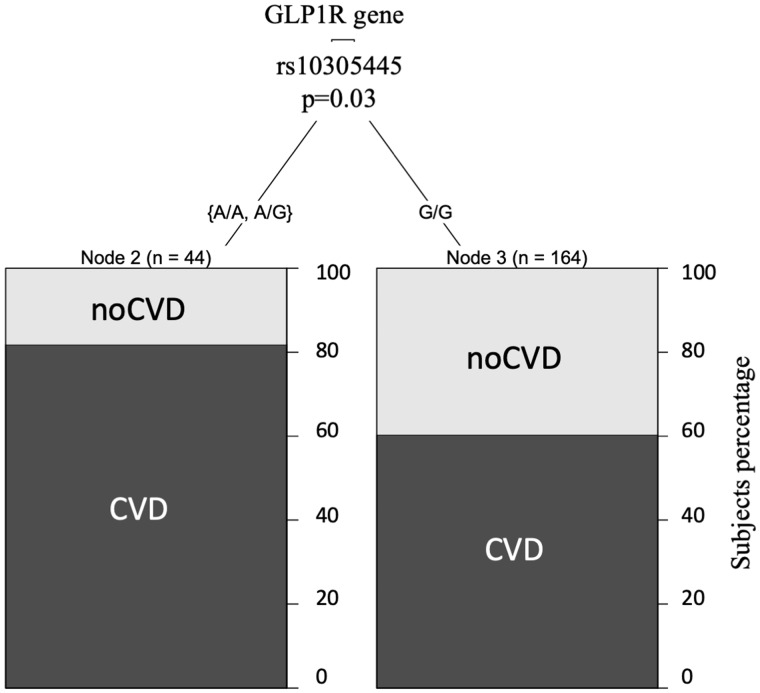
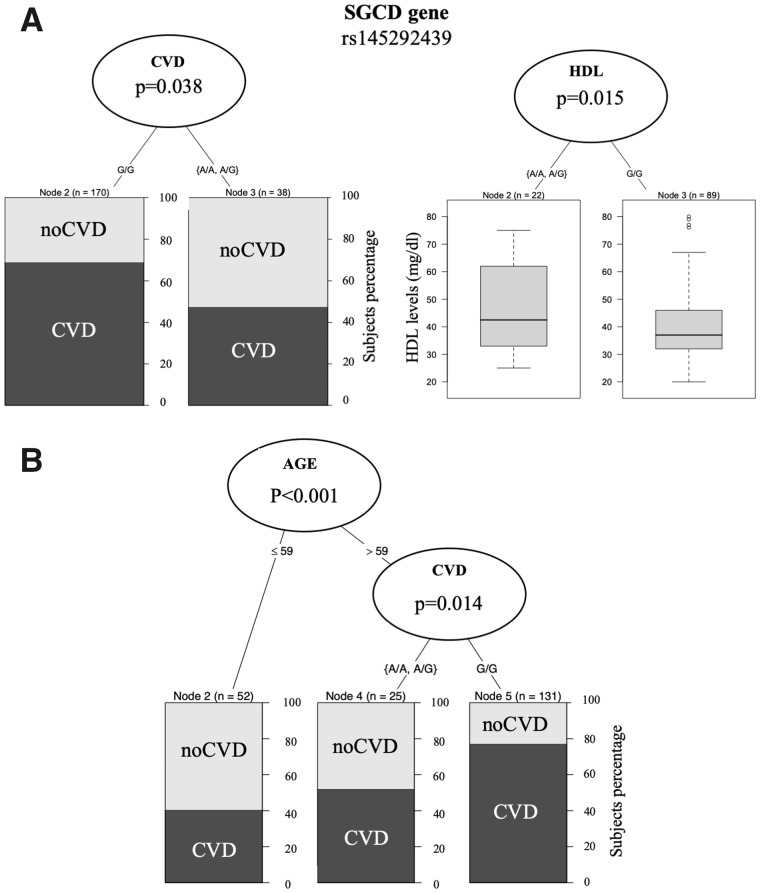
The tree regression analysis was used to investigate any correlation between SGCD and GLP1R genetic variants annotated in the SNP array and any CV events or CV risk factors at the baseline. Among the subjects homozygous for the GLP1R rs10305445 allele A, we found a higher number of subjects with previous CV events (81%) compared with heterozygous subjects (60% CVD) (Figure 2). In contrast, SCGD rs145292439 allele A associated with a lower number of subjects with previous CV events and a higher level of high-density lipoprotein (HDL; Figure 3A), with 68% CVD among the homozygous subjects for allele G versus 45% CVD among the heterozygous subjects. HDL levels were lower among subjects carrying only allele G (41.83 ± 13.84 mg/dL) with respect to the ones carrying allele A in both the homozygous and heterozygous state (45.96 ± 14.98 mg/dL). Furthermore, regression tree analysis using age as a covariate found that individuals >59 years of age and homozygous carriers of the common allele G in SCGD have a higher risk with respect to individuals that carry the variant allele A (78% in homozygous G versus 50% in heterozygous) (Figure 3B), confirming the age effect on CVD.
FIGURE 2.
The tree regression analysis investigated any correlation between SNP GLP1R rs10305445 variant allele A and CV events and risk factors. SNP GLP1R rs10305445 statistically associated with CVD in HD patients, with 81% CVD in subjects among those carrying the A allele compared with 60% CVD in subjects carrying the G allele.
FIGURE 3.
The tree regression analysis investigated any correlation between the SNP SCGD rs145292439 allele A and CV events and CVD risk factors. SNP SCGD rs145292439 significantly associated with a lower percentage of CVD (45% CVD among the heterozygous versus 68% CVD among those homozygous for the allele G) and a higher HDL level in heterozygous HD patients (A). When we included age as covariate, the regression analyses confirmed that age (>9 years) and SCGD allele G increases CVD risk (B), with a higher percentage of CVD subjects among those >59 years of age and carrying allele G.
Cox regression analysis of CV events during the FU
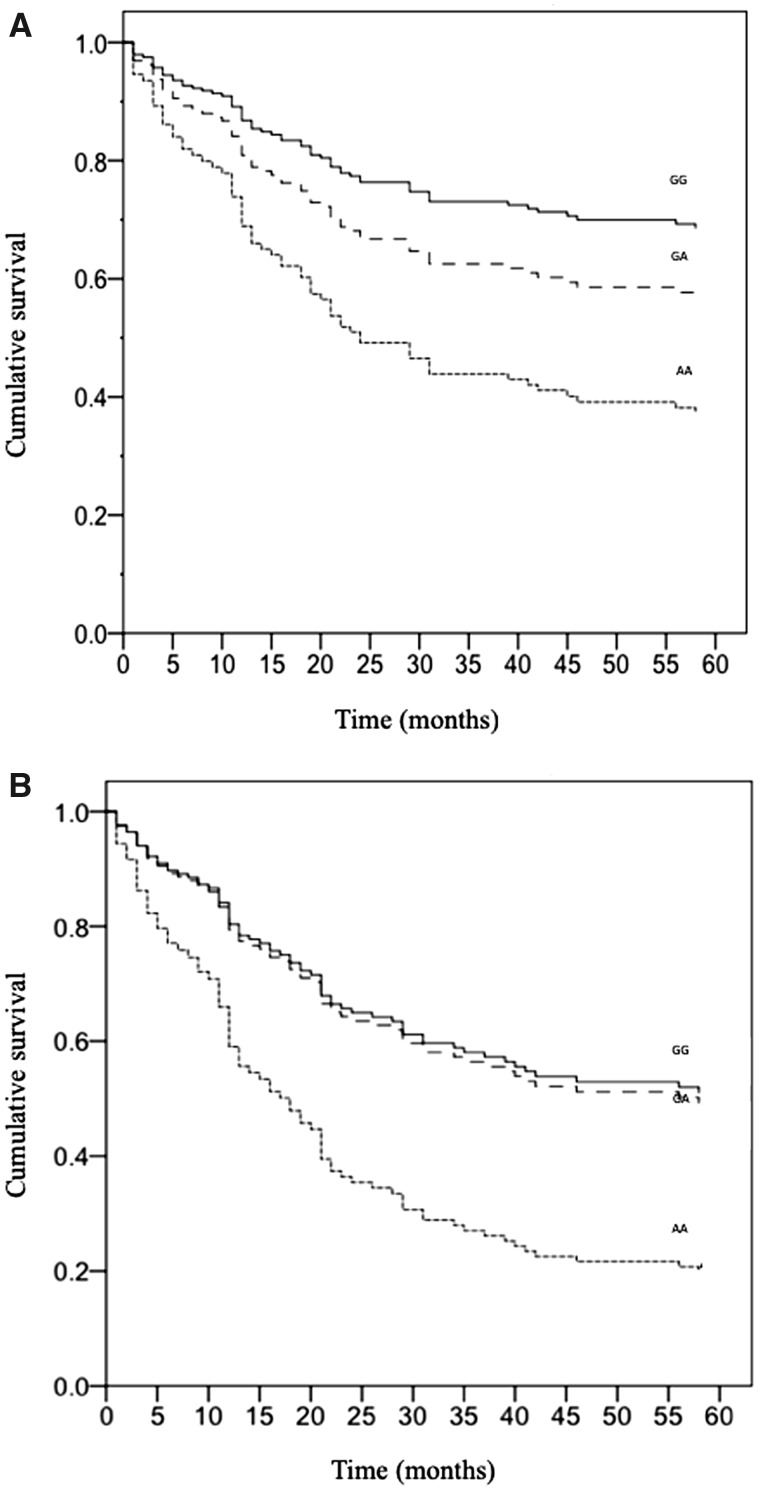
The occurrence of CV events was analyzed with Cox regression in patients (n = 245) with different genotypes at rs10305445 and rs145292439 during the 5-year FU. The OR of CV events resulted in an increase in homozygous patients for the GLP1R rs10305445 an allele A (n = 9) compared with homozygous patients for allele G [n = 185; OR = 3.3 (95% CI 1.4–7.7), P = 0.006; Figure 4A]. This finding was confirmed when we considered variables associated to CV events in the univariate analysis: gender and tertiles of age, serum PTH and body weight [AA patients: OR 2.7 (95% CI 1.1–6.5), P = 0.025; GA patients: OR 1.4 (95% CI 0.8–2.5), P = 0.21; GG as the reference group]. The association was still significant after having included in the analysis variables potentially correlated to CV events: gender, diabetes, arterial hypertension, tertiles of age, body weight, serum phosphate, PTH and dialysis vintage (AA patients: OR = 3, 95% CI 1.2–7.4, P = 0.019; GA patients, OR = 1.4, 95% CI 0.8–2.4, P = 0.25; GG as the reference group).
FIGURE 4.
Cox regression analysis of CV events during the 5-year FU in (A) the entire population of 245 patients [OR 3.3 (95% CI 1.4–7.7), P = 0.006] and in (B) 161 hemodialysis patients with a previous history of CV events divided according to their genotype at GLP1R rs10305445 (G > A) [AA patients: OR 2.4 (95% CI 1–5.6), P = 0.043; GA patients: OR 1.1 (95% CI 0.6–1.8), P = 0.86; GG as the reference group].
The Cox regression including only the patients with a clinical history of CV events at the baseline (n = 161) confirmed that CV events during the FU were more frequent in AA patients for GLP1R rs10305445 [AA patients: OR 2.4 (95% CI 1–5.6), P = 0.043; GA patients: OR 1.1 (95% CI 0.6–1.8), P = 0.86; GG as the reference group; Figure 4B]. There was no association in patients without previous events.
When we repeated the Cox regression in genetically unrelated patients (n = 208), again homozygous patients for GLP1R rs10305445 allele A (n = 6) had a higher OR for CV events compared with the homozygous patients for allele G (n = 164) in the unadjusted analysis [OR 3.6 (95% CI 1.3–10), P = 0.015] and in the analysis adjusted for the above-mentioned variables potentially implicated in CV events [OR 4.1 (95% CI 1.4–12.2), P = 0.012].
The distribution of CV events during the FU was not related to genotypes at SCGD rs145292439 [AA OR 1.2 (95% CI 0.4–4.1), P = 0.72; GG as the reference group; data not shown].
In the Supplementary data, Tables S5 and S6 show the genotypes distribution in our population at the two SNPs that demonstrated a greater number of subjects carrying GLP1R rs10305445 allele A (n carriers = 44) compared with SCGD rs145292439 allele A (n carriers = 38), with allele frequencies of ∼0.11 and ∼0.09, respectively, in our sample.
To complete the genetic analysis, we interrogated our genotyping array for the nine SNPs previously associated with the risk of coronary heart diseases in CKD [20]. Only to SNPs (WDR12 and LPA markers) were present in our genotyping array and none were significantly associated with CV events in our sample set (Supplementary data, Table S7).
Prediction analysis of the SNP functional activity
RegulomeDB, HaploReg v4.1 and Match 1.0 Public were interrogated for any potential function effect of the two SNPs associated with CV events. RegulomeDB analysis of GLP1R rs10305445 allele A resulted in a weak binding site for the transcription factor Runx-1, which was not confirmed in HaploReg, where both alleles resulted as a potential binding site for Runx-1. SCGS rs145292439 allele A creates a low-score binding site for the Oct-1 and POU3F2 transcription sites, according to RegulomeDB. The data were not confirmed by HaploReg analysis, which produced changes in 11 different motifs not including Oct-1 and POUF3F2 (see Supplementary data, Figure S5).
Match 1.0 did not identify any potential transcription binding sites around the rs10305445 and rs145292439 regions.
DISCUSSION
Our aim was to interrogate the whole-genome variants to identify any genetic marker indicative of CVD in hemodialysis patients. We genotyped 245 hemodialysis patients using the Infinium MEGAEX and correlated those variants to the history of CV events. Previous GWASs investigated genes associated with CKD and dialysis [32–34] and, to our knowledge, no whole-genome variant array was performed to analyze CVD in hemodialysis. Considering the small sample size, we shifted from the GWAS analysis to a population-based approach analyzing the Fst distance between the subjects with previous CV events (CVD) and without events (no CVD). With this approach, we identified 3218 SNPs differentiating the CVD group from the no CVD group, and among these we found two gene variants related to CVD and heart development using the Gene Ontology Analysis.
We identified an association of GLP1R rs10305445 with previous CV events and SCGD rs145292439 with no previous CV events and a higher level of HDL, suggesting a high-risk association of GLP1R rs10305445 with CV events, while SCGD rs145292439 can be associated with a low risk for CVD. When age was added as a covariate, the results for SCGD rs145292439 were significant only in subjects >59 years of age, confirming the role of the age in the risk of CVD.
Survival analysis over the 5-year FU confirmed the increase of new CV events for patients carrying the allele A of GLP1R rs1035445 but it did not show any protective effect of SCGD. This result showed that the GLP1R variant has a strong predisposition towards increasing the risk of CV events in hemodialysis patients both at baseline and at FU.
Previously GLP1R and SGCD were independently associated with CVD and CV risk factors. GLP1R, expressed in the pancreas, is the GLP1 receptor that is involved in blood sugar homeostasis. GLP1R genetic variants are associated with diabetes and obesity [35]. The GLP1R rs6923761 variant allele was demonstrated to improve weight loss after bariatric surgery and to reduce CV comorbidity [36]. Another study found similar results with GLP1R rs10305492, which is associated with a lower risk of diabetes and increased protection for coronary heart disease [37]. Recently GLP1R has been targeted for therapeutic treatment in CKD patients affected by type 2 diabetes. Although the clinical experience is still limited, GLP1R agonists are promising alternatives to the use of glucose-lowering agents in CKD diabetic patients [38]. Our study suggests that the genetic background should be considered as a factor affecting the response to the treatment and that a pharmacogenomic approach could produce stronger results.
SGCD was associated with the heart and muscle development in Drosophila models and confirmed in a GWAS human study that identified rs6877118 as associated with plasma HDL [39, 40]. Interestingly the SGCD variant associated with lower risk of CV events is practically absent in Africa (allele frequency = 0.002) and East Asia subjects, instead is present in Europeans (allele frequency = 0.07) (1000 genome database) [41].
A previous study identified a genetic risk of coronary heart diseases in CKD using a multilocus genetic risk score from nine genetic variants [20]. None of these variants include the two SNPs that we identified in our study, probably due to selection of the nine variants from a healthy-population GWAS. We then analyzed the frequency of the nine SNPs in our sample set and only two SNPs (WDR12 and LPA markers) were present in our array, and none of them were significantly associated with CV events in our population.
Both SNPs are noncoding intronic variants and their effect on gene function is unknown. We preliminarily explored predictive tools (RegulomeDB [29], HaploReg 4.1 [30] and Match 1.0 Public [31]) to identify functional effects on gene function (gene transcription), but we did not find any significant and consistent results. An in vitro model to test the two variants will be useful to characterize them.
A limitation of our study is indeed the sample size and for this reason we opted for a population-based approach to discover variants that could affect the number of CV events in our sample. Further studies should be focused on increasing the sample size and to use of whole-genome sequencing to discover rare deleterious variants affecting CV risk in dialysis patients. Furthermore, we aim to test these two variants of GLP1R and SCGD using in vitro model to understand their role.
The results of our study reinforce the concept of precision medicine that aims to tailor treatments and prevention to the patient's genetic background in order to identify high-risk patients and to improve the success rate of therapy.
In conclusion, our study identified two SNPs (rs10305445 and rs145292439) in the genes GLP1R and SCGD, respectively, that can contribute to the higher risk of CVD in CKD patients on hemodialysis, allowing better stratification of the CV risk in patients with CKD.
FUNDING
This work was supported by the Sidra Medicine internal research fund (#SDR400002) and from Shire Pharmaceuticals.
AUTHORS’ CONTRIBUTIONS
A.T. and G.V. created the study design and wrote the manuscript. T.A., L.M. and M.S. recruited patients and collected data at San Raffaele Hospital. M.C. and C.B. recruited patients and collected data at San Paolo Hospital. N.P. and F.C. recruited patients and collected data at Cernusco Sul Naviglio Uboldo Hospital. F.P. and A.M. performed blood DNA extraction and contributed to data analysis. S.T. and Le.S. performed the genotyping experiments and gene pathway analysis. M.M. performed bioinformatics and statistical analyses. G.V. contributed to the statistical analysis. A.R. and La.S. contributed to the study design, data discussion and manuscript editing. All authors discussed the results and approved the final submitted version.
CONFLICT OF INTEREST STATEMENT
On behalf of all authors, the corresponding author states that there is no conflict of interest and that the results presented in this article have not been published previously in whole or part, except in abstract format.
Supplementary Material
REFERENCES
- 1. de Jager DJ, Grootendorst DC, Jager KJ. et al. Cardiovascular and noncardiovascular mortality among patients starting dialysis. JAMA 2009; 302: 1782–1789 [DOI] [PubMed] [Google Scholar]
- 2. Ross L, Banerjee D.. Cardiovascular complications of chronic kidney disease. Int J Clin Pract 2013; 67: 4–5 [DOI] [PubMed] [Google Scholar]
- 3. Meeus F, Kourilsky O, Guerin AP. et al. Pathophysiology of cardiovascular disease in hemodialysis patients. Kidney Int 2000; 58(Suppl 7): S140–S147 [DOI] [PubMed] [Google Scholar]
- 4. Bradbury BD, Fissell RB, Albert JM. et al. Predictors of early mortality among incident US hemodialysis patients in the Dialysis Outcomes and Practice Patterns Study (DOPPS). Clin J Am Soc Nephrol 2007; 2: 89–99 [DOI] [PubMed] [Google Scholar]
- 5. Block GA, Klassen PS, Lazarus JM. et al. Mineral metabolism, mortality, and morbidity in maintenance hemodialysis. J Am Soc Nephrol 2004; 15: 2208–22184 [DOI] [PubMed] [Google Scholar]
- 6. Foley RN, Parfrey PS, Harriett JD. et al. Hypocalcemia, morbidity, and mortality in end-stage renal disease. Am J Nephrol 1996; 16: 386–393 [DOI] [PubMed] [Google Scholar]
- 7. Melamed ML, Eustace JA, Plantinga L. et al. Changes in serum calcium, phosphate, and PTH and the risk of death in incident dialysis patients: a longitudinal study. Kidney Int 2006; 70: 351–357 [DOI] [PubMed] [Google Scholar]
- 8. Slinin Y, Foley RN, Collins AJ.. Calcium, phosphorus, parathyroid hormone, and cardiovascular disease in hemodialysis patients: the USRDS waves 1, 3, and 4 study. J Am Soc Nephrol 2005; 16: 1788–1793 [DOI] [PubMed] [Google Scholar]
- 9. Blacher J, Guerin AP, Pannier B. et al. Arterial calcifications, arterial stiffness, and cardiovascular risk in end-stage renal disease. Hypertension 2001; 38: 938–942 [DOI] [PubMed] [Google Scholar]
- 10. Kingsmore SF, Lindquist IE, Mudge J. et al. Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov 2008; 7: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stein EA, Mellis S, Yancopoulos GD. et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med 2012; 366: 1108–1118 [DOI] [PubMed] [Google Scholar]
- 12. Testa A, Leonardis D, Spoto B. et al. A polymorphism in a major antioxidant gene (Kelch-like ECH-associated protein 1) predicts incident cardiovascular events in chronic kidney disease patients: an exploratory study. J Hypertens 2016; 34: 928–934 [DOI] [PubMed] [Google Scholar]
- 13. Rothuizen TC, Ocak G, Verschuren JJ. et al. Candidate gene analysis of mortality in dialysis patients. PLoS One 2015; 10: e0143079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bacgi B, Bagci G, Candan F. et al. The protective effect of MCP-1 -2518 A>G promoter polymorphism in Turkish chronic renal failure patients requiring long-term hemodialysis. Int Urol Nephrol 2015; 47: 551–556 [DOI] [PubMed] [Google Scholar]
- 15. Spoto B, Mattace-Raso F, Sijbrands E. et al. Association of IL-6 and a functional polymorphism in the IL-6 gene with cardiovascular events in patients with CKD. Clin J Am Soc Nephrol 2015; 10: 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shimoyama Y, Mitsuda Y, Tsuruta Y. et al. Polymorphism of Nrf2, an antioxidative gene, is associated with blood pressure and cardiovascular mortality in hemodialysis patients. Int J Med Sci 2014; 11: 726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santoro D, Gagliostro G, Alibrandi A. et al. Vitamin D receptor gene polymorphism and left ventricular hypertrophy in chronic kidney disease. Nutrients 2014; 6: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen YH, Hung SC, Tarng DC.. Length polymorphism in heme oxygenase-1 and cardiovascular events and mortality in hemodialysis patients. Clin J Am Soc Nephrol 2013; 8: 1756–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baragetti I, Norata GD, Sarcina C. et al. 374 T/A RAGE polymorphism is associated with chronic kidney disease progression in subjects affected by nephrocardiovascular disease. PLoS One 2013; 8: e60089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodrigo E, Pich S, Subirana I. et al. A clinical-genetic approach to assessing cardiovascular risk in patients with CKD. Clin Kidney J 2017; 10: 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lluis-Ganella C, Subirana I, Luca G. et al. Assessment of the value of a genetic risk score im improving the estimation of coronary risk. Atheroscl 2012; 222: 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mirza MAI, Larsson A, Lind L. et al. Circulating fibroblast growth factor-23 is associated with vascular dysfunction in the community. Atherosclerosis 2009; 205: 385–390 [DOI] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd-Browm K. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Gen 2007; 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weir BS, Cockerham CC.. Estimating F‐statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- 25. McLaren W, Gil L, Hunt SE. et al. The ensembl variant effect predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mi H, Dong Q, Muruganujan A. et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 2010; 38: D204–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krämer A, Green J, Pollard J Jr. et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014; 30: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boyle AP, Hong EL, Hariharan M. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ward LD, Kellis M.. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matys V, Fricke E, Geffers R. et al. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003; 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hothorn T, Zeileis A.. Partykit: a modular toolkit for recursive partytioning in R. J Machine Learn Res 2015; 16: 3905–3909 [Google Scholar]
- 32. Köttgen A, Pattaro C, Böger CA. et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010; 42: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gudbjartsson DF, Holm H, Indridason OS. et al. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet 2010; 6: e1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gorski M, Tin A, Garnaas M. et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int 2015; 87: 1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin CH, Lee YS, Huang YY. et al. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res 2015; 2015: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Luis DA, Pacheco D, Aller R. et al. Role of the rs6923761 gene variant in glucagone-like peptide 1 receptor gene on cardiovascular risk factors and weight loss after biliopancreatic diversion surgery. Ann Nutr Metab 2014; 65: 259–263 [DOI] [PubMed] [Google Scholar]
- 37. Scott RA, Freitag DF, Li L. et al. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci Transl Med 2016; 8: 341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheen AJ. Pharmacokinetics and clinical use of incretin-based therapies in patients with chronic kidney disease and type 2 diabetes. Clin Pharmacokinet 2015; 54: 1–21 [DOI] [PubMed] [Google Scholar]
- 39. Goldstein JA, Kelly SM, LoPresti PP. et al. SMAD signaling drives heart and muscle dysfunction in a Drosophila model of muscular dystrophy. Hum Mol Gen 2011; 20: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaess BM, Tomaszewski M, Braund PS. et al. Large-scale candidate gene analysis of HDL particle features. PLoS One 2011; 6: e14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.