Abstract

The crystal structure of ZnZnuA from Escherichia coli reveals two metal binding sites. (i) The primary binding site, His143, is located close the His-rich loop (residues 116–138) and plays a significant role in Zn(II) acquisition. (ii) The secondary binding site involves His224. In this work, we focus on understanding the interactions of two metal ions, Zn(II) and Cu(II), with two regions of ZnuA, which are possible anchoring sites for Zn(II): Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 (primary metal binding site) and Ac-223GHFTVNPEIQPGAQRLHE240-NH2 (secondary metal binding site). The histidine-rich loop (residues 116–138) has a role in the capture of zinc(II), which is then further delivered into other regions of the protein. For both Zn(II) complexes, histidine residues constitute the main anchoring donors. In the longer, His-rich fragment, a tetrahedral complex with four His residues is formed, while in the second ligand, two imidazole nitrogens are involved in zinc(II) binding. In both cases, so-called loop structures are formed. One consists of a 125HxHxExxxExHxH137 motif with seven amino acid residues in the loop between the two central histidines, while the other is formed by a 224HFTVNPEIQPGAQRLH239 motif with 14 amino acid residues in the loop between the two nearest coordinating histidines. The number of available imidazoles also strongly affects the structure of copper(II) complexes; the more histidines in the studied region, the higher the pH in which amide nitrogens will participate in Cu(II) binding.
Short abstract
The interactions of Zn(II) and Cu(II) with two model sequences from Escherichia coli ZnZnuA are studied. Both sequences, which contain the primary binding site, His143, and the His-rich loop, and the region with the secondary binding site, His224, form loop structures when bound to Zn(II).
Introduction
ZnuABC is a Zn(II)-specific uptake system that is responsible for the distribution and excretion of zinc(II) ions. It belongs to the ATP binding cassette (ABC) transporter family,1,2 which consists of three components: (i) the ZnuA periplasmic Zn(II) binding protein, (ii) the ZnuB membrane protein that transports Zn(II) across the cytoplasmic membrane, and (iii) the ZnuC ATPase, which provides energy for ATP hydrolysis for ion transport through the membrane.3
ZnuA is a ZntR-regulated protein that captures Zn(II) ions and delivers them to the ZnuB component. In some bacterial species, ZinT, another zinc(II) binding protein, helps ZnuA to recruit Zn(II) from the periplasmic space.4 Studies of ZnuA homologues confirmed its important role in zinc(II) upake and showed that ZnuA inactivation results in a decrease in virulence in several pathogenic bacteria.3,5−8 Its absence inhibits ZnuB-mediated zinc(II) import.4
The model of a soluble “metallochaperone” or “metallophore”, a peptide or protein that delivers Zn(II) to an adequate membrane transporter, is described quite well for several systems; in some species, both ZnuA and AztC soluble metallochaperones are present.9 Functional similarities exist also in fungal species. The 299 amino acid zincophore Pra1 delivers Zn(II) to the Zrt1 zinc(II) transporter in Candida albicans;10,11 orthologues of pra1 and zrt1 were found in several other fungi, e.g., in Aspergillus fumigatus (aspf2 and zrfC, respectively).12
Several crystal structures of ZnuA from Escherichia coli have been determined (Protein Data Bank (PDB) entries 2PRS, 2PS0, 2PS3, 2PS9, 2OSV, and 2OGW). The crystal structure of ZnZnuA from E. coli reveals two metal binding sites.13−15 The primary metal binding site, His143, is situated in the proximity of the His-rich loop (residues 116–138), located in the N-terminal domain. This long and mobile His-rich loop is a characteristic feature of proteins that belong to the cluster of nine bacterial periplasmic binding proteins (which includes proteins that specifically bind Zn(II) or Mn(II)).16 This loop, together with His143, is proposed to play a significant role in Zn(II) acquisition.13,17 The loop binds zinc(II) with an affinity lower that of the primary site (His143),13,18,19 but it helps in metal acquisition under conditions of severe zinc(II) depletion and regulates Zn(II) import by sensing the high periplasmic levels of this metal.4,19 The structure of this highly mobile site has not yet been fully characterized, but it has been suggested that it may depend on the binding of Zn(II) either in the primary site or to this loop itself.20 This His-rich loop may transfer Zn(II) to the primary metal binding site, or it may facilitate the interaction of ZnuA with ZnuB.21
The second metal binding site in ZnuA is located ∼12 Å from the main metal binding site, and its affinity for Zn(II) is likely lower than that of the first site. The second zinc(II) ion is coordinated by His224, but other ligands have not been identified. Yatsunyk et al. show that the His-rich loop is on the same side of the protein as the second zinc(II) binding site (His224), and any of the amino acid residues present in this loop could be good candidates for additional ligands. Moreover, the structure of ZnZnuA changes with respect to the apoprotein. In the presence of Zn(II), the carbonyl oxygen of His224 interacts with the side chain of Arg237. When the second metal binding site is empty, this interaction is not present; Arg237 is too far from the histidine residue.13 This can suggest some stabilizing effect of the Arg residue on the binding of the second zinc(II) ion to His224.
In this work, we focus on understanding the interactions of Zn(II) and Cu(II) with two regions of ZnuA, which are responsible for metal binding: Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 and Ac-223GHFTVNPEIQPGAQRLHE240-NH2. Although it is well established that ZnuA is a zinc(II) bacterial transporter, there is also some evidence that it may interact with other metal ions [Co(II), Ni(II), Cd(II), Cu(II), and Cu(I)].13 We provide insight into the structural and thermodynamic properties of Cu(II) and Zn(II) complexes with the two chosen Zn(II) binding regions, focusing on the metal binding specificity of these fragments and on the influence of metal binding on their structural properties.
Experimental Section
Materials
Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 and Ac-223GHFTVNPEIQPGAQRLHE240-NH2 peptides (98% pure) were purchased from KareBay and used without further purification. Cu(II) and Zn(II) perchlorate were extrapure products (Sigma-Aldrich). The concentrations of stock solutions of these salts were determined by inductively coupled plasma mass spectrometry. The carbonate-free stock solution of NaOH (0.1 M) was purchased from Sigma-Aldrich and then potentiometrically standardized with potassium hydrogen phthalate as a primary standard. The HClO4 stock solution was prepared by diluting concentrated HCl (Sigma-Aldrich) and then standardized with NaOH. All of the sample solutions were prepared with freshly doubly distilled water. The ionic strength (I) was adjusted to 0.1 M by addition of NaClO4 (Sigma-Aldrich).
Potentiometric Measurements
Potentiometric measurements were performed at a constant temperature of 298 K under an argon atmosphere using a Dosimat 665 Metrohm titrator connected to a Metrohm 691 pH-meter and a Metrohm LL Unitrode glass electrode. Stability constants for proton, Cu(II), and Zn(II) complexes were calculated from titrations (two titrations curves) carried out over the pH range of 2–11 using a total volume of 3 mL. The glass cell was equipped with a magnetic stirring system, a microburet delivery tube, and an inlet–outlet tube for argon. The pH-metric titrations were performed in a water solution of 4 mM HClO4 at a 0.1 M NaClO4 ionic strength. Solutions were titrated with 0.1 M carbonate-free NaOH. Before each measurement, the electrode was calibrated by titration of HClO4 with a strong base under the same experimental conditions as described above. Purities and the exact concentrations of ligand solutions were determined by the Gran method.22 The ligand concentration was 0.5 mM. The metal:ligand ratio was 1:1 for Cu(II) and 1:2 foor Zn(II) complexes [a higher Zn(II):peptide ratio triggered precipitation at pH >5]. HYPERQUAD 2006 and SUPERQUAD programs were used for the stability constant calculations.23 Standard deviations were computed by HYPERQUAD 2006 and refer to random errors only. The constants for hydrolytic Cu(II) and Zn(II) species were used.24,25 The speciation and competition diagrams were computed with the HySS program.26
Mass Spectrometric Measurements
A BrukerQ-FTMS spectrometer (Bruker Daltonik, Bremen, Germany) equipped with an Apollo II electrospray ionization source with an ion funnel was used to record high-resolution mass spectra. The mass spectrometer was operated in positive and negative ion mode with the following parameters: scan range of m/z 150–2000, dry gas of nitrogen, temperature of 170 °C, ion energy of 5 eV, and transfer time of 120 ps. The capillary voltage was optimized to 4500 V to obtain the highest S/N ratio. The small changes in voltage (±500 V) did not significantly affect the optimized spectra. The Cu(II) and Zn(II) complexes (metal:ligand stoichiometries of 1:1 and 1:2, respectively, [ligand]tot = 5 × 10–4 M) were prepared in a 1:1 MeOH/H2O mixture at pH 7. The samples were infused at a flow rate of 3 μL/min. Before each experiment, the instrument was calibrated externally with the Tunemix mixture. Data were processed by application of Compass DataAnalysis 4.0 (Bruker Daltonic).
Spectroscopic Studies
The absorption spectra in the ultraviolet–visible (UV–vis) region were recorded at 298 K on a Cary300 Bio (Varian) spectrophotometer in 1 cm path length quartz cells. Circular dichroism (CD) spectra were recorded on a Jasco J 1500 spectropolarimeter over the range of 180–800 nm using different path lengths (1 and 0.01 cm). The concentrations of solutions used for UV–vis and CD spectroscopic studies were similar to those employed in the potentiometric experiments. CD spectra were recorded using Jasco Spectra Analysis Software. All spectroscopic measurements were recorded in the pH range of 2.5–11.5. The pH was adjusted with appropriate amounts of HClO4 and NaOH solutions. Origin7 was used to process and visualize the obtained spectra.
Nuclear magnetic resonance (NMR) experiments were performed on a Bruker 14.1 T spectrometer at a controlled temperature (±0.1 K) on a Bruker DRX Avance 600 MHz. Suppression of the residual water signal was achieved by excitation sculpting, using a 2 ms long selective square pulse on water.27 Proton resonance assignment was obtained by two-dimensional (2D) TOCSY and NOESY experiments. Spectral processing was performed on a Silicon Graphics O2 workstation using TOPSPIN 3.1 software. The analyzed peptides were dissolved in water with 10% D2O to achieve a final concentration of 0.5–1.0 mM. The pH was adjusted manually by adding HCl or NaOH to reach the desired value. The desired concentration of Zn(II) ions was obtained by using stock solutions of Zn(ClO4)2 in deuterated water.
Results and Discussion
Metal Complexes of the Second Metal Binding Site of ZnuA-Ac-223GHFTVNPEIQPGAQRLHE240-NH2
Metal Binding Stoichiometry of the Cu(II) and Zn(II) Ac-223GHFTVNPEIQPGAQRLHE240-NH2
Electrospray ionization mass spectrometry (ESI-MS) confirmed the purity of the studied ligand and showed the metal binding stoichiometry at pH 7, indicating that only equimolar species with Cu(II) and Zn(II) ions were present. ESI-MS peak assignments were based on the comparison between the precise calculated and experimental m/z values and their isotopic patterns. m/z values of 1047.54 and 698.69 correspond to [L + K]2+ and [L + K]3+ potassium adducts, respectively, of the free ligand (Figure S1). The prevailing signal corresponds to the mononuclear complexes with Cu(II) (e.g., [CuL]2+m/z 1058.53) (Figure S1A) and Zn(II) ions (e.g., [ZnL]3+m/z 706.02) (Figure S1B). The experimental isotopic pattern of the complex is in perfect agreement with the simulated one (Figure S1).
pH-Dependent Coordination Mode of Cu(II) Complexes
Protonation constants of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 ligand are listed in Table S1. The obtained data indicate that the investigated ligand behaves as an H4L acid. The two protonation constants correspond to consecutive protons binding to imidazole nitrogens of two histidine residues: His224 and His239 (pKa = 6.76 and 6.16, respectively). The next two are assigned to the carboxyl groups of Glu residues (pKa = 4.52 and 3.80, respectively). The pKa values obtained from potentiometric titrations are typical for similar systems.28−30
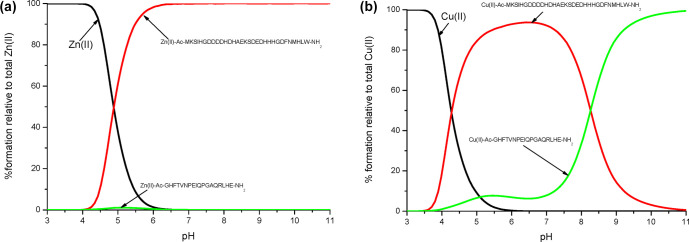
All thermodynamic stability constants and spectroscopic parameters obtained for the Cu(II)-Ac-223GHFTVNPEIQPGAQRLHE240-NH2 system are listed in Table 1. Potentiometric titrations were carried out to evaluate the corresponding stability constants and the pH-dependent species distribution diagram (Table 1 and Figure 1). The combined UV–vis and CD results allowed to conclude the binding mode of Cu(II) and the geometry of these species formed in solution. The first spectroscopically detectable copper(II) complex form observed at acidic pH is CuL, with a maximum at pH 6. It involves two histidine residues in the coordination sphere. The coordination of Cu(II) by two imidazole nitrogens is supported by the CD d–d band in the range of 220–270 nm (Figure S2B and Table 1).31,32 Moreover, the difference in pKa values for the free and bound ligands (6.76 and 5.45, respectively) indicates histidine coordination. Binding of the amide nitrogen for the CuH–1L species, with a maximum concentration at pH 6.8, is provided by the appearance of a d–d band at 538.7 nm, resulting in the [2Nim, 1N–] binding mode (Figure S2B and Table 1). For CuH–2L species (with a maximum at pH 7.5), the number of nitrogen donor atoms remains unchanged (UV–vis d–d band at 601 nm (Figure S2A)), but the replacement of one histidine residue by amide nitrogen takes place, resulting in a [1Nim, 2N–] binding mode. Above pH 9 (CuH–3L species), the blue shift in the UV–vis spectra to 520 nm (Figure S2A and Table 1) and new sets of CD bands (with positive and negative Cotton effect) suggest a [1Nim, 3N–] donor set and a square plane geometry.
Table 1. Potentiometric and Spectroscopic Data for Cu2+-Ac-223GHFTVNPEIQPGAQRLHE240-NH2 Complexes in Aqueous Solution of HClO4 at I = 0.1 M (NaClO4), [L] = 0.0005 M, 298 K, and an M:L Ratio of 1:1 (potentiometric titration, UV–vis, and CD spectroscopy)a.
| UV–vis |
CD |
||||||
|---|---|---|---|---|---|---|---|
| species | log β | pKa | λ (nm) | ε (cm–1 M–1) | λ (nm) | Δε (cm–1 M–1) | proposed donor sets |
| CuHL | 10.83(2) | ||||||
| CuL | 5.38(1) | 5.45 | 258.7 | 0.40 | 2Nim | ||
| CuH–1L | –1.28(2) | 6.66 | 608 | 92.28 | 538.7 | 0.19 | 2Nim, 1N– |
| 254.9 | 2.43 | ||||||
| CuH–2L | –7.69(1) | 6.41 | 601 | 79.67 | 537.5 | 0.16 | 1Nim, 2N– |
| 256.0 | 3.15 | ||||||
| CuH–3L | –16.29(3) | 8.60 | 520 | 89.71 | 647.1 | 0.33 | 1Nim, 3N– |
| 489.2 | –0.56 | ||||||
| 327.3 | 0.36 | ||||||
| 259.6 | 3.88 | ||||||
Values in parentheses are standard deviations on the last significant figure.
Figure 1.
Representative distribution diagram for formation of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 complex with Cu2+ ion at 298 K and I = 0.1 M (NaClO4). [L] = 0.0005 M; M:L molar ratio of 1:1.
Thermodynamic Stability Constants and Zn(II) Binding Sites in Its Complex with Ac-223GHFTVNPEIQPGAQRLHE240-NH2
Studies of zinc(II) complexes were based on the potentiometric (Table 2), mass spectrometric (Figure S1B), and NMR data (Figures 2–5). As mentioned above, the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 ligand forms only equimolar complexes with Zn(II), as shown by experimental and simulated ESI-MS signals (Figure S1B). The titration curves fit best to the formation of the following complexes: ZnHL, ZnL, and ZnH–1L in the pH range of 4–11; the Zn(II) complex formation constants are listed in Table 2. In the first complex detected at low pH, ZnHL, with a maximum concentration at pH 6, most likely, one imidazole nitrogen is coordinated to the zinc(II) ion (a [1Nim] binding mode); however, we cannot exclude an additional coordination via carboxyl groups of glutamic acid residues. In the next species, ZnL, the second histidine residue participates in zinc(II) binding, which is suggested by the decrease of its pKa in the zinc(II) complex in comparison to its pKa in the free ligand (6.76 and 6.24, respectively). Keeping in mind that zinc(II) is normally unable to displace amide protons, the binding modes of Zn(II) species consist of imidazole nitrogens and oxygens from glutamic acid and/or water molecules. The pKa value of 7.79, corresponding to the ZnH–1L species, is most likely associated with the deprotonation of a water molecule.
Table 2. Potentiometric Data and Representative Distribution Diagrams for Zn(II)-Ac-223GHFTVNPEIQPGAQRLHE240-NH2 Complexes in an Aqueous Solution of HClO4 at I = 0.1 M (NaClO4), [L] = 0.0005 M, 298 K, and an M:L Ratio of 1:2a.
Values in parentheses are standard deviations on the last significant figure.
Figure 2.
Superimposition of selected regions of one-dimensional 1H NMR spectra of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 fragment at 1.0 mM in the absence (black) and presence of 0.5 equiv of Zn(II) (blue). The shifts are indicated by arrows.
Figure 5.
Amide region of 1H–1H NOESY 2D NMR spectra of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 fragment at 1.0 mM in the presence of 0.5 equiv Zn(II) at pH 7.2.
To confirm potentiometric analysis of the zinc(II) coordination mode, the zinc(II) binding to the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 ZnuA fragment was investigated by NMR (see Table S2 for the chemical shift assignments at physiological pH). The effect of the metal ion was evaluated by looking at variations of the NMR signals induced by the addition of Zn(II) ions. At pH 5.6, we observed slight chemical shift variations on the two His residues only, as shown by the changes experienced by His Hε signals (Figure 2A). This is consistent with the potentiometric results, which showed that only a small fraction of available Zn(II) is bound to the peptide at this pH. The increase in the pH to the physiological value yielded larger changes on His224 and His239, thus strongly indicating His imidazole coordination to zinc(II) ions (Figure 2B). The larger perturbations of Hε resonances (compared to Hδ) are consistent with metal binding to His Nδ. On the other hand, the results obtained by X-ray structure13 indicate Nε2 rather than Nδ as the zinc(II) donor atom. This behavior might depend on the fact that in aqueous solution the neutral imidazole exists in two different tautomeric forms, having either Nδ or Nε2 available for metal binding. On the other hand, we expect that only one form is present in the crystal structure.
In addition to the effects recorded on His, we also calculated the chemical shift variations for all of the other residues. As shown in Figure 3, zinc(II) binding to this ZnuA fragment causes chemical shift variations, not only on the coordinating histidines but also on Asn228, Glu230, Ile231, and Gln232, suggesting the formation of a turn in the peptide backbone. In fact, the effects recorded on the backbone and side-chain protons are in agreement with conformational changes of the peptide upon metal binding.
Figure 3.
Zn(II)-induced chemical shift variation of the selected proton of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 fragment at 1.0 mM and pH 7.2 in the presence of 0.5 equiv of Zn(II).
Secondary Structure of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 Fragment and Its Cu(II) and Zn(II) Complexes
The secondary structures of the investigated ligand and its copper(II) and zinc(II) complexes were determined by using near-UV CD spectroscopy (Figure S3). The apo ligand in the pH range of 4–11 suggests a nontypical α-helical structure. On the CD spectra, two negative absorption bands at 200 and 230 nm are present (Figure S3A). The second, less intense band at ∼230 nm indicates that the investigated ligand is not completely disordered in aqueous solution. Addition of Cu(II) and Zn(II) ions does not influence the peptide conformation (Figure S3B,C). The CD spectra at pH 7.2 of the apo form and its metal complexes are very similar (Figure S3D).
The structural rearrangement of the ZnuA fragment is also supported by the higher intensity of several NH signals, as indicated in Figure 4. In fact, in flexible and disordered peptides, NH resonances and their correlations are usually weak at physiological pH. The evidence that Zn(II) associations determine an increased intensity of amide protons suggests the stabilization of the specific loop or turn conformation derived from coordination of Zn(II) to the two His residues in the terminal regions (His224 and His239). In addition, although the 1H–1H NOESY experiments did not provide enough connectivities to determine the three-dimensional structure of the complex, NH–NH correlations detected along the -Glu-Ile-Gln- portion suggest the formation of turn structures (Figure 5). In addition, this might explain why two histidine residues, located a considerable distance from each other, can both coordinate the same zinc(II) ion at the same time.
Figure 4.

Superimposition of amide regions of (A) one-dimensional 1H NMR spectra of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 fragment at 1.0 mM in the absence (black) and presence of 0.5 equiv of Zn(II) (blue) at pH 7.2 and (B) 1H–1H TOCSY 2D NMR spectra of the ZnuA fragment at 1.0 mM in the absence (black) and pr esence of 0.5 equiv of Zn(II) (magenta) at pH 7.2.
Metal Complexes of the Primary Metal Binding of ZnuA-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2
Ligand Protonation
Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 consists of 19 possible protonation sites: two lysines, seven histidines, eight aspartic acids, and two glutamic acid residues (Table S3). The first calculated pKa value, 10.62, corresponds to that of the Lys residues. In the studied pH range, the deprotonation of both lysine residues could not be detected. The seven histidine residues are characterized by log β values ranging from 5.95 to 8.17. The obtained protonation constants are in excellent agreement with theoretical expectations based on His protonation equilibria.28,30,33,34 Because of the very large number of acidic residues in the presented sequence, not all protonation constants could be precisely determined. Only four pKa values for acidic residues are collected in Table S3 (pKa range of 4.44–5.15).
Cu(II) Complexes
The stoichiometry of the Cu(II)-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 system is 1:1, as already shown by MS spectra with signals at m/z 953.62, 763.29, and 933.89 for [CuL + 2K]4+, [CuL + 2K]5+, and [CuL]4+ complexes, respectively (Figure S4A). m/z values of 536.66, 625.93, 750.92, and 938.39 correspond to [L + 2K]7+, [L + 2K]6+, [L + 2K]5+, and [L + 2K]4+ ligand species, respectively (Figure S4).
Complex formation constants for the Cu(II)-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 system are listed in Table 3, and the corresponding distribution diagram is plotted on Figure 6. UV–vis and CD spectra are depicted in Figure S5. Cu(II) starts to interact with the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 ligand at pH 3.5, and the first detected complex is [CuH9L]. The first complex species detected by spectroscopic measurements is CuH5L, which dominates at pH 6. Most likely, at this point, all acidic and three histidine residues are deprotonated and CD studies (band at 240 nm) indicate that imidazole nitrogen atoms from histidine residues participate in Cu(II) coordination (Figure S5B). The pKa value for these species (5.77) is significantly decreased in comparison to that of the free ligand (pKa = 6.78), which strongly suggests the involvement of histidine in copper(II) binding. The coordination of the amide nitrogen starts to be seen at pH >6.5 (CuH4L); the d–d bands on the CD spectra at 543, 317, and 241 nm indicate a [2Nim, 1N–] donor set. The lack of changes in the spectra up to pH 9 (CuH3L, CuH2L, CuHL, and CuL species) suggests the same coordination mode (only the dissociation of nonbinding protons from the His residues is present). The three nitrogen atoms in the copper(II) coordination sphere are proven also by UV–vis bands in the range of 590–570 nm (Figure S5A). At pH >9, changes in CD (an intense band for d–d and CT transitions with negative and positive signs of the Cotton effect at 524 and 634 nm, respectively) and UV–vis (displacement of the maximum absorption in the direction of shorter wavelengths: 569 nm → 533 nm) are detected, clearly suggesting a square-planar geometry of the formed complex [2Nim, 2N–].
Table 3. Potentiometric and Spectroscopic Data for Cu2+-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 Complexes in an Aqueous Solution of HClO4 at I = 0.1 M (NaClO4), [L] = 0.0005 M, 298 K, and an M:L Ratio of 1:1 (potentiometric titration, UV–vis, and CD spectroscopy)a.
| UV–vis |
CD |
||||||
|---|---|---|---|---|---|---|---|
| species | log β | pKa | λ (nm) | ε (cm–1 M–1) | λ (nm) | Δε (cm–1 M–1) | proposed donor sets |
| CuH9L | 69.09(3) | ||||||
| CuH8L | 64.36(4) | 4.73 | |||||
| CuH7L | 59.70(4) | 4.66 | |||||
| CuH6L | 54.19(4) | 5.51 | |||||
| CuH5L | 48.42(5) | 5.77 | 553 | 0.12 | 2Nim | ||
| 240 | –4.31 | ||||||
| CuH4L | 42.02(6) | 6.40 | 546 | 0.26 | 2Nim, 1N– | ||
| 317 | –0.12 | ||||||
| 241 | –4.99 | ||||||
| CuH3L | 35.22(6) | 6.80 | 547 | 0.31 | 2Nim, 1N– | ||
| 313 | –0.20 | ||||||
| 241 | –5.56 | ||||||
| CuH2L | 27.94(8) | 7.28 | 594 | 56.62 | 546 | 0.32 | 2Nim, 1N– |
| 318 | –0.26 | ||||||
| 276 | 0.14 | ||||||
| 240 | –6.47 | ||||||
| CuHL | 19.3(1) | 8.64 | 568 | 59.35 | 562 | 0.19 | 2Nim, 1N– |
| 331 | –0.40 | ||||||
| 268 | 0.77 | ||||||
| 240 | –4.74 | ||||||
| CuL | 10.65(9) | 8.65 | 569 | 65.23 | 619 | 0.24 | 2Nim, 1N–/2Nim, 2N– |
| 491 | 0.07 | ||||||
| 334 | –0.61 | ||||||
| 260 | 2.14 | ||||||
| 239 | –2.07 | ||||||
| CuH–1L | 1.4(1) | 9.25 | 533 | 74.14 | 636 | 0.35 | 2Nim, 2N– |
| 524 | –0.26 | ||||||
| 337 | –0.41 | ||||||
| 257 | 3.47 | ||||||
Values in parentheses are standard deviations on the last significant figure.
Figure 6.
Representative distribution diagram for the formation of the complex of Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 with a Cu2+ ion at 298 K and I = 0.1 M (NaClO4). [L] = 0.0005 M; M:L molar ratio of 1:1.
Due to the fact that the investigated ligand contains seven histidine residues in the sequence, all of which constitute potential copper-anchoring sites, it is tempting to perform an additional copper binding analysis. Our suggestions about coordination modes were confirmed by the comparison of the stability constants of Cu(II)-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 complexes with those already reported in the literature,35,36 for macrochelate formation. From the spectroscopic analysis, we did not have evidence that up to four histidine residues can be coordinated to copper(II) to form a macrochelate before the deprotonation of amide nitrogens. We did not observe a UV–vis band at 576 nm (Figure S5A) before the deprotonation of amide nitrogens, which could have indicated a 4Nim coordination mode. Moreover, a comparison with stability constants of macrochelates already reported in the literature for the HuPrP76–114 system indicates that this ligand is more effective in copper(II) binding than the ZnuA first binding site (Ac-MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW-NH2) (Figure S6). The HuPrP76–114 (Ac-PHGGGWGQPHGGGWGQGGGTHSQWNKPSKPKTNMKHMAG-NH2) ligand binds Cu(II) ions with higher affinity, because it forms a very effective macrochelate complex with a set of up to four imidazole nitrogens. This analysis is in good agreement with spectroscopic studies indicating that only two histidine residues are involved in copper binding in the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 system.
Zn(II) Complexes
All of the results presented here show that the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 peptide can form 1:1 complexes with the Zn(II) ion, and no polynuclear or bis complexes have been detected by either potentiometry (Table 4) or mass spectrometry (Figure S4B). The investigated ligand starts to bind Zn(II) ions at pH 4. The first complex is ZnH7L (maximum concentration at pH 5), in which the most likely coordination mode is via one histidine residue [1Nim]. The next species, ZnH5L and ZnH4L, correspond to metal binding by three and four imidazole nitrogen atoms, respectively [the pKa values for the zinc(II) complex (6.17 and 5.89, respectively) are lower than those for free ligand (6.78 and 6.93, respectively)]. Moreover, the decrease in pKa for the Zn(II) complexes (ZnH3L, ZnH2L, and ZnHL) in comparison to the free ligand suggests the occurrence of an exchange between all of the imidazoles, within the 4N coordination. The pKa values of 7.13 and 8.16 (Table 4), corresponding to ZnL and ZnH–1L species, respectively, are associated with the deprotonation of water molecules that fill the coordination sphere of Zn(II).
Table 4. Potentiometric Data and Representative Distribution Diagrams for Zn(II)-Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 Complexes in an Aqueous Solution of HClO4 at I = 0.1 M (NaClO4), [L] = 0.0005 M, 298 K, and an M:L Ratio of 1:2a.
Values in parentheses are standard deviations on the last significant figure.
Additional information about the Zn(II) binding modes was derived by comparing NMR spectra of the ligand in the presence and absence of zinc ions. Metal addition causes selective and severe line broadening and slight chemical shift variation as shown in Figure S7. To identify the peptide regions selectively affected by the Zn(II) ions and to determine the metal donor atoms, we evaluated the effects of Zn(II) in the 2D 1H–1H TOCSY maps.
The large broadening of NMR signals observed for the His aromatic protons strongly indicates the involvement of His in metal binding (Figure 7A). On the other hand, the absence of any or very small effects on Aps and Glu resonances (Figure 7B) excludes the direct coordination of zinc(II) with the carboxylate groups. Because Zn(II) preferentially forms tetrahedral complexes, we expected that only four His residues will be coordinated to the metal. As shown on Figure 7A, all His signals are perturbed upon addition of metal, suggesting the occurrence of an exchange between all of the imidazoles.
Figure 7.

Superimposition of selected regions of 2D 1H–1H NMR TOCSY spectra of the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 fragment at 1.0 mM in the absence (black) and presence of 0.5 equiv of Zn(II) (magenta) at pH 6.0.
Further analysis of the 2D 1H–1H NMR TOCSY spectra shows that amide resonances of Ala128, Glu129, Lys130, Ser131, and Glu133 are influenced, as well (Figure 8). Such behavior is consistent with a structural rearrangement of the region encompassing Glu129 and Glu133 residues. Two His residues (His127 and His135) likely behave as zinc(II)-anchoring sites.
Figure 8.

Superimposition of selected amide regions of 2D 1H–1H NMR TOCSY spectra of the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 fragment at 1.0 mM in the absence (black) and presence of 0.5 equiv of Zn(II) (magenta) at pH 6.0.
Moreover, each of them is one amino acid residue from a glutamic acid (Glu129 and Glu133), which can, in addition, electrostatically stabilize the imidazole binding. Such binding yields to the formation of a stable turn or loop structure composed by seven amino acid residues, 128AEKSDED134
His125 and His137 are successively coordinated to Zn(II), being equally spaced one residue apart from His127 and His135, respectively. This leaves us with a “125HxHxExxxExHxH137” sequence being a crucial site for Zn(II) coordination. Finally, the other three His residues (His119, His136, and His143) participate in the metal coordination sphere to a lesser extent by mutually exchanging with another imidazole.
Near-UV CD Structural Characterization of the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 Peptide and Its Cu(II) and Zn(II) Complexes
The obtained near-UV CD spectra for the Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 system are very similar to the spectra of the second metal binding sequence and its Cu(II) and Zn(II) complexes described in Secondary Structure of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 Fragment and Its Cu(II) and Zn(II) Complexes. Two negative absorption bands at ∼200 and ∼230 nm for the apo ligand suggest an α-helical structure (Figure S8A). Addition of both metal ions to the investigated ligand does not significantly change the set of bands on the CD spectra (Figure S8B–D). This is in good agreement with the conclusions from the NMR spectra; no typical α-helical or β-sheet structure is formed, and a single loop motif cannot be detected by this method.
First versus Second Metal Binding Site
Which of the studied region is more effective in the binding of Zn(II) and Cu(II) ions? To answer this question, it is useful to compare the zinc(II) and copper(II) binding affinities of both ligands on so-called competition plots. The Ac-115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 ligand, involving the histidine-rich loop in the sequence, is a much more effective site in Zn(II) binding (Figure 9A). In the literature,13,17 the primary metal binding sites for Zn(II) ions, involving His143, are also more effective site for zinc ions. The authors conclude that the crystal structure of ZnZnuA reveals two Zn(II) binding sites, and the second site may be filled only after Zn(II) binds to the primary one. From the chemical point of view, this explanation seems to be obvious and confirms our previous observations that with the increase in the number of histidines, metal coordination becomes more effective.,30,33,37 In the case of Cu(II) complexes (Figure 9B), the 115MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW145-NH2 ligand forms more stable complexes at pH 8.5. It is due to the fact that above pH 9, additional amides are involved in the copper(II) coordination sphere, forming a square-planar complex [1Nim, 3N–] in the case of the Ac-223GHFTVNPEIQPGAQRLHE240-NH2 ligand. This finding is also in good agreement with our previous findings that show that the more histidines are present in the peptide sequence, the more likely it is for amides to participate in Cu(II) binding at a higher pH than usual.30,33,34,38
Figure 9.
Competition plots for (A) Zn(II)-Ac-MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW-NH2 and Zn(II)-Ac-GHFTVNPEIQPGAQRLHE-NH2 and (B) Cu(II)-Ac-MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW-NH2 and Cu(II)-Ac-GHFTVNPEIQPGAQRLHE-NH2. Previously calculated stability constants are applied to a theoretical situation in which equimolar amounts of Zn(II), Cu(II), and both ligands are present.
ZnuA versus ZnuD Metal Binding Sites
We compared the metal affinities of specific regions of two bacterial Zn(II) transporters: ZnuA, studied in this work, and the previously studied ZnuD transporter,39 containing a His-rich loop with a function similar to that in ZnuA (Figures S9 and S10). The first ligand in ZnuA (Ac-MKSIHGDDDDHDHAEKSDEDHHHGDFNMHLW-NH2) is a much more effective ligand for zinc(II) than the His-rich loop from ZnuD (Ac-FHDDDNAHAHTHS-NH2) (Figure S9A). The sets of metal binding donors are the same (four histidine residues involved in the coordination sphere) for both ligands, but the specific structural rearrangement of the HxHxExxxExHxH motif and the higher number of histidine residues present in the ZnuA sequence may stabilize the zinc(II) complex. In the case of copper(II) complexes, the situation is reversed: the ZnuD fragment forms more stable complexes than the ZnuA one (Figure S9B). This is due to the fact that at pH 7.5, the ZnuD fragment forms a [3Nim] complex, while only two imidazole nitrogen atoms participate in copper binding in the ZnuA fragment. Moreover, the ZnuD region forms a square-planar complex, including amide residues, at pH values lower than those of the ZnuA fragment. For the second ZnuA fragment (second metal binding site Ac-GHFTVNPEIQPGAQRLHE-NH2), it is obvious that it is the ligand with an affinity for both Zn(II) and Cu(II) that is lower than that of the discussed ZnuD site (Figure S10). This is in good agreement with what can be expected; the fewer histidines participate in metal binding (two imidazoles for the second ZnuA fragment and four for ZnuD), the less stable the complex.
Conclusions
First indentified in E. coli, the ZnuABC system has been shown to be necessary in the capture and transport of zinc(II) ions. The crystal structure of ZnuA from E. coli reveals two metal binding sites: (i) the primary binding site, His143, located close the His-rich loop (residues 116–138), which plays a significant role in Zn(II) acquisition, and (ii) the secondary binding site, encompassing His224. Studies of Zn(II) and Cu(II) complexes with both metal binding sites from ZnuA provide some significant conclusions. (i) The primary metal binding site, which involves the histidine-rich loop, is much more effective site in Zn(II) and Cu(II) binding at physiological pH than the secondary metal binding site. (ii) The structure of both ligands is a nontypical α-helix. (iii) For Zn(II) complexes, histidine residues constitute the main anchoring donors; in the primary metal binding site, a tetrahedral complex with four His residues is present, while for the secondary site, two imidazole nitrogens are involved in zinc(II) binding. In both cases, so-called loop structures are formed, one in the proximity of the primary binding site, a 125HxHxExxxExHxH137 motif with seven amino acid residues in the loop between the two central histidines, and another one that encompasses the second Zn(II) binding site, with a 224HFTVNPEIQPGAQRLH239 motif with 14 amino acid residues in the loop between the two nearest coordinating histidines. (iv) All seven histidine residues present in the His-rich loop are perturbed upon addition of metal, suggesting the occurrence of an exchange among all of the imidazoles. (v) The more histidines in the studied region, the higher the pH at which amide nitrogens will participate in Cu(II) binding.
Acknowledgments
Financial support by the National Science Centre (UMO-2017/26/A/ST5/00363) is gratefully acknowledged. M.R.-Z. is supported by the National Science Centre (UMO-2017/26/A/ST5/00364). This work was also supported by the Italian MIUR, through PRIN (Programmi di Ricerca di Rilevante Interesse Nazionale) Project 2015T778JW_003. The CIRMMP is also gratefully acknowledged for financial support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.inorgchem.9b03298.
Figures S1–S10 and Tables S1–S3 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hantke K. Bacterial zinc transporters and regulators. BioMetals 2001, 14 (3–4), 239–249. 10.1023/A:1012984713391. [DOI] [PubMed] [Google Scholar]
- Hantke K. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 2005, 8 (2), 196–202. 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Patzer S. I.; Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator zur in Escherichia coli. Mol. Microbiol. 1998, 28 (6), 1199–1210. 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Petrarca P.; Ammendola S.; Pasquali P.; Battistoni A. The Zur-Regulated ZinT Protein Is an Auxiliary Component of the High-Affinity ZnuABC Zinc Transporter That Facilitates Metal Recruitment during Severe Zinc Shortage. J. Bacteriol. 2010, 192 (6), 1553–1564. 10.1128/JB.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintilhac A.; Alloing G.; Granadel C.; Claverys J. P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 1997, 25 (4), 727–739. 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- Chen C. Y.; Morse S. A. Identification and characterization of a high-affinity zinc uptake system in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 2001, 202 (1), 67–71. 10.1016/S0378-1097(01)00302-0. [DOI] [PubMed] [Google Scholar]
- Lewis D. A.; Klesney-Tait J.; Lumbley S. R.; Ward C. K.; Latimer J. L.; Ison C. A.; Hansen E. J. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 1999, 67 (10), 5060–5068. 10.1128/IAI.67.10.5060-5068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammendola S.; Pasquali P.; Pistoia C.; Petrucci P.; Petrarca P.; Rotilio G.; Battistoni A. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc Homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 2007, 75 (12), 5867–5876. 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane D. P.; Kumar S.; Yukl E. T. Two ABC Transporters and a Periplasmic Metallochaperone Participate in Zinc Acquisition in Paracoccus denitrificans. Biochemistry 2019, 58 (2), 126–136. 10.1021/acs.biochem.8b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citiulo F.; Jacobsen I. D.; Miramon P.; Schild L.; Brunke S.; Zipfel P.; Brock M.; Hube B.; Wilson D. Candida albicans Scavenges Host Zinc via Pra1 during Endothelial Invasion. PLoS Pathog. 2012, 8 (6), e1002777. 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda D.; Rowinska-Zyrek M. Candida albicans zincophore and zinc transporter interactions with Zn(II) and Ni(II). Dalton Transactions 2018, 47 (8), 2646–2654. 10.1039/C7DT04403H. [DOI] [PubMed] [Google Scholar]
- Wilson D. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics 2015, 7 (6), 979–985. 10.1039/C4MT00331D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunyk L. A.; Easton J. A.; Kim L. R.; Sugarbaker S. A.; Bennett B.; Breece R. M.; Vorontsov I. I.; Tierney D. L.; Crowder M. W.; Rosenzweig A. C. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. JBIC, J. Biol. Inorg. Chem. 2008, 13 (2), 271–288. 10.1007/s00775-007-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Jogl G. Crystal structure of the zinc-binding transport protein ZnuA from Escherichia coli reveals an unexpected variation in metal coordination. J. Mol. Biol. 2007, 368 (5), 1358–1366. 10.1016/j.jmb.2007.02.107. [DOI] [PubMed] [Google Scholar]
- Chandra B. R.; Yogavel M.; Sharma A. Structural analysis of ABC-family periplasmic zinc binding protein provides new insights into mechanism of ligand uptake and release. J. Mol. Biol. 2007, 367 (4), 970–982. 10.1016/j.jmb.2007.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P. A new family of high-affinity ABC manganese and zinc permeases. Res. Microbiol. 2001, 152 (3–4), 231–243. 10.1016/S0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- Banerjee S.; Wei B. X.; Bhattacharyya-Pakrasi M.; Pakrasi H. B.; Smith T. J. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. J. Mol. Biol. 2003, 333 (5), 1061–1069. 10.1016/j.jmb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Desrosiers D. C.; Sun Y. C.; Zaidi A. A.; Eggers C. H.; Cox D. L.; Radolf J. D. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol. Microbiol. 2007, 65 (1), 137–152. 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- Wei B. X.; Randich A. M.; Bhattacharyya-Pakrasi M.; Pakrasi H. B.; Smith T. J. Possible regulatory role for the histidine-rich loop in the zinc transport protein, ZnuA. Biochemistry 2007, 46 (30), 8734–8743. 10.1021/bi700763w. [DOI] [PubMed] [Google Scholar]
- Falconi M.; Oteri F.; Di Palma F.; Pandey S.; Battistoni A.; Desideri A. Structural-dynamical investigation of the ZnuA histidine-rich loop: involvement in zinc management and transport. J. Comput.-Aided Mol. Des. 2011, 25 (2), 181–194. 10.1007/s10822-010-9409-6. [DOI] [PubMed] [Google Scholar]
- Castelli S.; Stella L.; Petrarca P.; Battistoni A.; Desideri A.; Falconi M. Zinc ion coordination as a modulating factor of the ZnuA histidine-rich loop flexibility: A molecular modeling and fluorescence spectroscopy study. Biochem. Biophys. Res. Commun. 2013, 430 (2), 769–773. 10.1016/j.bbrc.2012.11.073. [DOI] [PubMed] [Google Scholar]
- Gran G.; Dahlenborg H.; Laurell S.; Rottenberg M. Determination of the Equivalent Point in Potentiometric Titrations. Acta Chem. Scand. 1950, 4 (4), 559–577. 10.3891/acta.chem.scand.04-0559. [DOI] [Google Scholar]
- Gans P.; Sabatini A.; Vacca A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43 (10), 1739–1753. 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- Vuceta J.; Morgan J. J. HYDROLYSIS OF CU (II). Limnol. Oceanogr. 1977, 22 (4), 742–746. 10.4319/lo.1977.22.4.0742. [DOI] [Google Scholar]
- Pettit L. D.; Gregor J. E.; Kozłowski H.. Complex formation between metal ions and peptides. In Perspectives on Bioinorganic Chemistry; Hay R. W., Dilworth J. R., Nolan K. B., Eds.; JAI Press: London, 1991; Vol. 1, pp 1–41. [Google Scholar]
- Alderighi L.; Gans P.; Ienco A.; Peters D.; Sabatini A.; Vacca A. Hyperquad simulation and speciation (HySS): a utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. 10.1016/S0010-8545(98)00260-4. [DOI] [Google Scholar]
- Hwang T. L.; Shaka A. J. WATER SUPPRESSION THAT WORKS - EXCITATION SCULPTING USING ARBITRARY WAVE-FORMS AND PULSED-FIELD GRADIENTS. J. Magn. Reson., Ser. A 1995, 112 (2), 275–279. 10.1006/jmra.1995.1047. [DOI] [Google Scholar]
- Watly J.; Simonovsky E.; Wieczorek R.; Barbosa N.; Miller Y.; Kozlowski H. Insight into the Coordination and the Binding Sites of Cu2+ by the Histidyl-6-Tag using Experimental and Computational Tools. Inorg. Chem. 2014, 53 (13), 6675–6683. 10.1021/ic500387u. [DOI] [PubMed] [Google Scholar]
- Chiera N. M.; Rowinska-Zyrek M.; Wieczorek R.; Guerrini R.; Witkowska D.; Remelli M.; Kozlowski H. Unexpected impact of the number of glutamine residues on metal complex stability. Metallomics 2013, 5 (3), 214–221. 10.1039/c3mt20166j. [DOI] [PubMed] [Google Scholar]
- Hecel A.; Watly J.; Rowinska-Zyrek M.; Swiatek-Kozlowska J.; Kozlowski H. Histidine tracts in human transcription factors: insight into metal ion coordination ability. JBIC, J. Biol. Inorg. Chem. 2018, 23 (1), 81–90. 10.1007/s00775-017-1512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik-Jankowska T.; Ruta-Dolejsz M.; Wisniewska K.; Lankiewicz L.; Kozlowski H. Copper(II) complexation by human and mouse fragments (11–16) of beta-amyloid peptide. Journal of the Chemical Society-Dalton Transactions 2000, (24), 4511–4519. 10.1039/b006125p. [DOI] [Google Scholar]
- Witkowska D.; Politano R.; Rowinska-Zyrek M.; Guerrini R.; Remelli M.; Kozlowski H. The Coordination of NiII and CuII Ions to the Polyhistidyl Motif of Hpn Protein: Is It as Strong as We Think?. Chem. - Eur. J. 2012, 18 (35), 11088–11099. 10.1002/chem.201200780. [DOI] [PubMed] [Google Scholar]
- Watly J.; Simonovsky E.; Barbosa N.; Spodzieja M.; Wieczorek R.; Rodziewicz-Motowidlo S.; Miller Y.; Kozlowski H. African Viper Poly-His Tag Peptide Fragment Efficiently Binds Metal Ions and Is Folded into an alpha-Helical Structure. Inorg. Chem. 2015, 54 (16), 7692–7702. 10.1021/acs.inorgchem.5b01029. [DOI] [PubMed] [Google Scholar]
- Watly J.; Hecel A.; Rowinska-Zyrek M.; Kozlowski H. Impact of histidine spacing on modified polyhistidine tag - Metal ion interactions. Inorg. Chim. Acta 2018, 472, 119–126. 10.1016/j.ica.2017.06.053. [DOI] [Google Scholar]
- Di Natale G.; Osz K.; Nagy Z.; Sanna D.; Micera G.; Pappalardo G.; Sovago I.; Rizzarell E. Interaction of Copper(II) with the Prion Peptide Fragment HuPrP(76–114) Encompassing Four Histidyl Residues within and outside the Octarepeat Domain. Inorg. Chem. 2009, 48 (9), 4239–4250. 10.1021/ic802190v. [DOI] [PubMed] [Google Scholar]
- Di Natale G.; Osz K.; Kallay C.; Pappalardo G.; Sanna D.; Impellizzeri G.; Sovago I.; Rizzarelli E. Affinity, Speciation, and Molecular Features of Copper(II) Complexes with a Prion Tetraoctarepeat Domain in Aqueous Solution: Insights into Old and New Results. Chem. - Eur. J. 2013, 19 (11), 3751–3761. 10.1002/chem.201202912. [DOI] [PubMed] [Google Scholar]
- Watly J.; Hecel A.; Kolkowska P.; Kozlowski H.; Rowinska-Zyrek M. Poly-Xaa Sequences in Proteins - Biological Role and Interactions with Metal Ions: Chemical and Medical Aspects. Curr. Med. Chem. 2018, 25 (1), 22–48. 10.2174/0929867324666170428104928. [DOI] [PubMed] [Google Scholar]
- Brasili D.; Watly J.; Simonovsky E.; Guerrini R.; Barbosa N. A.; Wieczorek R.; Remelli M.; Kozlowski H.; Miller Y. The unusual metal ion binding ability of histidyl tags and their mutated derivatives. Dalton Transactions 2016, 45 (13), 5629–5639. 10.1039/C5DT04747A. [DOI] [PubMed] [Google Scholar]
- Hecel A.; Rowinska-Zyrek M.; Kozlowski H. Copper(II)-Induced Restructuring of ZnuD, a Zinc(II) Transporter from Neisseria meningitidis. Inorg. Chem. 2019, 58 (9), 5932–5942. 10.1021/acs.inorgchem.9b00265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.