Abstract
To advance the scientific understanding of bacteria-driven mercury (Hg) transformation processes in natural environments, thermodynamics and kinetics of divalent mercury Hg(II) chemical speciation need to be understood. Based on Hg LIII-edge extended X-ray absorption fine structure (EXAFS) spectroscopic information, combined with competitive ligand exchange (CLE) experiments, we determined Hg(II) structures and thermodynamic constants for Hg(II) complexes formed with thiol functional groups in bacterial cell membranes of two extensively studied Hg(II) methylating bacteria: Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132. The Hg EXAFS data suggest that 5% of the total number of membranethiol functionalities (Mem-RStot = 380 ± 50 μmol g–1 C) are situated closely enough to be involved in a 2-coordinated Hg(Mem-RS)2 structure in Geobacter. The remaining 95% of Mem-RSH is involved in mixed-ligation Hg(II)-complexes, combining either with low molecular mass (LMM) thiols like Cys, Hg(Cys)(Mem-RS), or with neighboring O/N membrane functionalities, Hg(Mem-RSRO). We report log K values for the formation of the structures Hg(Mem-RS)2, Hg(Cys)(Mem-RS), and Hg(Mem-RSRO) to be 39.1 ± 0.2, 38.1 ± 0.1, and 25.6 ± 0.1, respectively, for Geobacter and 39.2 ± 0.2, 38.2 ± 0.1, and 25.7 ± 0.1, respectively, for ND132. Combined with results obtained from previous studies using the same methodology to determine chemical speciation of Hg(II) in the presence of natural organic matter (NOM; Suwannee River DOM) and 15 LMM thiols, an internally consistent thermodynamic data set is created, which we recommend to be used in studies of Hg transformation processes in bacterium–NOM–LMM thiol systems.
Introduction
Methylmercury (MeHg), a highly toxic pollutant that poses a serious risk to humans and ecosystems, is primarily produced by microbial methylation of inorganic divalent mercury, Hg(II).1−3 In addition to the activity of the methylating organisms, MeHg formation is controlled by the bioavailability and biouptake of Hg(II) species,4 where reactions with reduced sulfur (including inorganic sulfide and organic thiol functional groups) are of utmost importance.1,5
Thiol groups associated with the outer or inner cell membranes (Mem-RSH) have been proposed to play an important role in the uptake of Hg(II) by bacteria capable of methylation and reduction.6−9 Membrane associated thiols have been suggested as being a part of the critical step for Hg(II) uptake.10,11 In contrast, adsorption of Hg(II) onto the outer cell membrane has also been proposed to inhibit the uptake by bacteria by lowering the aqueous Hg(II) concentration, as demonstrated for Hg(II) methylation6,7 and reduction.8,9 Thiol functional groups of the Hg(II) transporters MerC and MerT, associated with the inner cell membrane, are known to form well-defined chemical structures with Hg(II).12 Current models for microbial uptake of metals, such as the free-ion activity model, biotic ligand model, and surface complexation models13−18 build on the assumption that the free Hg(II) ions or Hg(II) complexes in solution are in chemical equilibrium with Hg(II) bonded to the outer cell membrane functional groups.
In spite of the indicated importance of the Mem-RSH functional group and its complexation with Hg(II), only a few studies have determined chemical structures formed between Hg(II) and Mem-RSH functional groups in the outer membrane.9,19 There is so far no reported data on the thermodynamic stability of these structures, with reports limited to the distribution (partitioning coefficient) of Hg(II) between the aqueous phase and the cell surface.20−22 This lack of information is partly explained by the challenge to quantitate Mem-RSH functionalities. One principal type of methodology builds on the use of fluorescent probes forming covalent, nonreversible bonds with thiols, followed by either fluorescence spectroscopy23,24 or potentiometry25 measurements to quantify the thiols. The accuracy of this method is dependent on matrix effects affecting the fluorescence and its quenching.24 Another possibility is to use Hg LIII-edge extended X-ray absorption fine structure (EXAFS) spectroscopy, which is element specific and essentially matrix independent. The concentration of thiol groups is determined by a titration method, where the concentration of Mem-RSH can be calculated from the coordination number and bond-lengths of Hg–O/NR and Hg–SR structures formed at stepwise additions of Hg(II).19 The method has been previously used to determine the concentration of RSH groups associated with natural organic matter (NOM) functional groups, using either MeHg or Hg as the probe.26−28
Here, we use a competitive ligand exchange (CLE) method where cysteine (Cys) is added as a competitor with Mem-RSH for Hg(II), to establish thermodynamic constants for complexes formed between Hg(II) and Mem-RSH functional groups of Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132, two Gram-negative bacteria frequently used in assay studies of Hg(II) transformation reactions. The concentration of the Hg(Cys)2 complex in equilibrium with Hg(II) complexes formed with Mem-RSH is determined by liquid chromatography inductively coupled plasma mass spectrometry (LC-ICPMS). The method has previously been used to determine stability constants (log K) for the bonding of Hg(II) to 15 different LMM-RSH, as well as with NOM associated thiols (NOM-RSH).28,29 Mercury LIII-edge EXAFS data, complemented with sulfur K-edge X-ray absorption near-edge structure (XANES) data, were used to characterize the cell membrane sulfur groups to determine the concentration of Mem-RSH groups, and to provide direct spectroscopic evidence for structures of Hg(II) complexes formed with Mem-RSH functionalities. Combined with data from previous studies,28,29 the overarching goal is to provide an internally consistent set of log K values for well-defined Hg(II) complexes and structures formed with LMM-RSH, NOM-RSH, and bacterial Mem-RSH functionalities. We recommend applying this thermodynamically consistent data set to studies of microbial driven Hg(II) transformation processes in the presence of LMM thiols and NOM to improve the molecular scale understanding of the processes of Hg(II) bacterial uptake, reduction/oxidation, and methylation in a relatively complex environment.
Materials and Methods
Bacterial Cultures and Membrane Isolation
Geobacter sulfurreducens PCA30 and Desulfovibrio desulfuricans ND132 were harvested at late exponential phase (OD660 ∼0.5) as described in previous studies.31,32 The membrane isolation procedure was modified from standard protocols33 to expose all accessible thiol groups on both inner and outer membranes.33,34 Extracellular metabolites were separated from the membranes in the assay buffer solutions by filtration using 0.2 μm filters.35
Details about bacterial cultures, membranes, and extracellular metabolite isolation are given in the Supporting Information.
Membrane and Cell Sample Preparation and Characterization
The total organic carbon (TOC) content of the whole cells and isolated membranes were determined by an elemental analyzer-isotope ratio mass spectrometer (Flash 2000, Thermo Fisher Scientific). The sulfide concentration in the assay media for both Geobacter and ND132 was determined by the methylene blue method (LOD = 0.3 μM).36 Sulfur K-edge XANES data were collected on freeze-dried samples of whole cells, membranes, and extracellular metabolites. Mercury LIII-edge EXAFS analysis was conducted on freeze-dried and frozen membrane samples.
In a first set of samples, aliquots of Hg(NO3)2 stock solutions were added to the membranes of Geobacter and ND132 to get a final Hg(II) concentration of 32 and 610–1220 μmol g–1 C, respectively. After 2 h of reaction, the membrane suspensions were frozen at −80 °C, freeze-dried, and pressed into 5 mm diameter pellets for later use. In a second set of samples, isolated Geobacter membranes were added an Hg(II) content of 8–834 μmol g–1 C. After 2 h of reaction, the samples were ultracentrifuged (at 160 000g) and the pellets were rinsed with deoxygenated Milli-Q water three times before being frozen at −80 °C in 5 mm diameter Teflon holders. Both sets of samples were stored at −80 °C until X-ray absorption spectroscopy analyses.
Competitive Ligand Exchange Experiments
The isolated membranes were suspended in deoxygenated NaClO4 (10 mM) solutions in 15 mL polypropylene tubes (Sarstedt) to make a final membrane concentration of 1–19 mg C L–1 for Geobacter and 1–27 mg C L–1 for ND132. Aliquots of Hg(NO3)2 stock solution were added to yield a final concentration of 0.5 μM allowed to react for 24 h with the cell membrane sample. Although the reaction of Hg(II) with Mem-RSH is expected to be kinetically controlled, 24 h should with marginal be sufficient to allow equilibrium to be reached according to previous studies of Hg(II) with thiols of NOM, cells and membranes.19,21,28,37 Finally, aliquots of Cys stock solution were added to obtain a final concentration of 2.0 μM. This solution was then sampled for total Hg(II) (Hgtot) and Hg(Cys)2 analysis at the time of 1, 24, 48 and 72 h. Parallel experiments with 0.5 μM Hg(II) and 2.0 μM Cys prepared in deoxygenated Milli-Q water in the absence of membrane were used as controls. All experiments were duplicated. The reaction vessels were protected from light by aluminum foil and maintained at 25 ± 1 °C with a thermostat in the N2 filled glovebox. To avoid introducing potential interference, no pH buffer was added and pH was maintained at ∼4.0.
Total Hg and Hg(Cys)2 Analyses
The Hgtot concentration was determined by combustion atomic absorption spectrometry (CAAS) using a direct mercury analyzer (DMA-80, Milestone, RSD = 6%). The concentration of Hg(Cys)2 was determined by LC-ICPMS (RSD = 10%).29
X-ray Absorption Spectroscopy Analyses
We applied sulfur K-edge XANES and Hg LIII-edge EXAFS to characterize the sulfur chemistry at the bacterial cells and membranes as well as to quantify the membrane-associated thiol groups. Details about these two methods are given in the Supporting Information.
Thermodynamic Calculations
Thermodynamic calculations were conducted in the software R38 using the PHREEQC package39,40 and following the protocol used in a previous study of the complexation Hg(II) with NOM-RSH functional groups.28 In Table S1, reactions and constants expected to dominate the Hg(II) chemical speciation in bacterium–NOM–LMM thiol systems are listed. Experimental uncertainties in the reported log K values were propagated using the first-order Taylor series method calculated by the R package errors.41
Results and Discussion
Sulfur Speciation of Bacteria Characterized by Sulfur K-Edge XANES
Total organic carbon (TOC) content accounted for 51 and 41% of the membrane mass of Geobacter and ND132, respectively, which is in agreement with a previous study.42 Sulfur K-edge XANES spectra (Figure S1 and Table S2) of whole cell and membrane samples demonstrated a high dominance of Org-SRED, representing the sum of reduced organic S functionalities organic disulfide (RSSR), monosulfide (RSR), and thiol (RSH) that cannot be separated by this methodology. Notably, our membrane samples are expected to include both outer and inner membrane structures. A similarly shaped XANES spectrum, highly dominated by Org-SRED, was previously reported for the outer membrane of the bacterium Shewanella oneidensis.19 In contrast, oxidized sulfur such as sulfonate and sulfate were the dominant species in the extracellular solution (Figure S1). Given the reported excretion of LMM thiols from Geobacter(35) and ND13243 grown under similar conditions as in this study, it is suggested that some Org-SRED in the extracellular solution was represented by LMM thiols. It should be noted that a small amount of inorganic sulfide was detected by S XANES in the whole cell samples. Not surprisingly, the sulfide contribution to total S in the sulfate-reducing bacterium ND132 sample was almost twice as high as in that the iron-reducing bacterium Geobacter. Notably, sulfide was not detected in the membrane samples, in the extracellular solution, or in the bacteria assay media of any of the two bacteria, using the methylene blue method. This result is in agreement with previous research showing that sulfide was undetectable under incubation in low sulfate concentration media.31,32,35
Chemical Structures of Hg(II) and Quantitation of Mem-RSH Functionalities on Bacteria Membranes Using Mercury LIII-Edge EXAFS
As expected from previous studies of NOM28,44 and of bacteria membranes,8,45,46 we observed a significant loss of Hgtot (∼10–64%) added to the bacterial membrane samples for EXAFS experiments (Table 1). These losses are explained mainly by Hg(II) reduction to Hg(0) caused by reductive functional groups in the membrane structures. Details about losses and processes causing them in different types of experiments are discussed further in connection to Figures S2 and S3 in the Supporting Information. The final determined Hg(II) concentration, remaining in the samples after the losses, was used when interpreting EXAFS data. Because the Hg LIII-edge EXAFS results (Table 1 and Figure S4) for membranes of Geobacter did not reveal any clear differences between freeze-dried and frozen samples, data for freeze-dried and frozen samples were combined in the analyses of our results.
Table 1. First Coordination Shell Model Fits to Hg LIII-Edge EXAFS R-Space Data for the Isolated Membranes of Geobacter and ND132 with Different Concentrations of Hg(II)a.
| first S shell |
first O shell |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hgadded | Hgtot | CN | R (Å) | σ2 (Å2) | CN | R (Å) | σ2 (Å2) | ΔE0 (eV) | k-range (Å–1) | |
| Geobacter | 8b | 4 | 2.00 | 2.34 | 0.003 | 7.4 | 2.7–13.5d | |||
| 32c | 28 | 1.43 | 2.37 | 0.003 | 0.48 | 2.08 | 0.003 | 10.1 | 2.7–9.8d | |
| 83b | 55 | 1.13 | 2.36 | 0.003 | 0.87 | 2.08 | 0.003 | 9.7 | 2.7–13.5d | |
| 324c | 215 | 0.73 | 2.39 | 0.003 | 0.64 | 2.04 | 0.003 | 7.3 | 2.7–12.0e | |
| 834b | 406 | 0.61 | 2.39 | 0.003 | 0.70 | 2.03 | 0.003 | 8.9 | 2.7–12.1d | |
| ND132 | 610c | 397 | 0.59 | 2.40 | 0.006 | 0.78 | 2.06 | 0.003 | 9.8 | 2.7–13.1e |
| 1220c | 436 | 0.58 | 2.39 | 0.004 | 0.83 | 2.05 | 0.003 | 8.6 | 2.7–12.6e | |
Hgadded denotes the theoretical added Hg concentration and Hgtot denotes the final determined total Hg concentration (unit, μmol g–1 C) in samples subjected to EXAFS measurements. CN denotes coordination number, R denotes bond distance, ΔE0 denotes edge energy shift, and σ2 denotes the Debye–Waller factor which is constrained to 0.003–0.01 Å2; the amplitude reduction factor, S02, is fixed to 0.9 for all samples.
Frozen samples.
Freeze-dried samples.
Seven knots used in spline fit of k3 weighted data.
Eight knots used in spline fit of k3 weighted data.
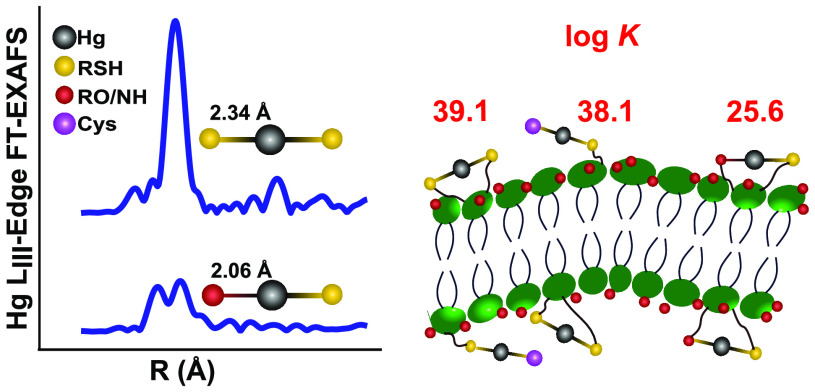
The average Hg–S bond distance formed at the outer and inner membranes, represented by the peak at ∼1.9 Å in the Fourier transformed (FT) spectra (Table 1 and Figure S4), varied from 2.34 to 2.39 Å in the five characterized samples. This distance range is in line with previous EXAFS studies of Hg(II) added to the cell envelope (supposedly the outer cell membrane) of Geobacter with a reported Hg–S bond distance of 2.32–2.38 Å.19 At the lowest addition of Hg(II), corresponding to 4 μmol g–1 C, the reported distance of 2.34 Å is well in agreement with the previously determined 2-coordinated Hg(NOM-RS)2 structure in organic soils and in streamwater DOC.27,28 This distance is also in agreement with the Hg–S distance in the range 2.32–2.35 Å reported for the Hg(LMM-RS)2 structure formed between Hg(II) and the LMM thiols Cys, Pen, and GSH.47−49 Combined with the information illustrated by the FT peak at ∼4 Å, representing the sum of three- and four-legged multiple scattering paths at the double Hg–S distance (Figure S4), EXAFS data provide evidence for a 2-coordinated, linear structure (close to 180° angle) with the formula Hg(Mem-RS)2 at this relatively low Hg(II) concentration. At higher additions of Hg(II), a first-coordination shell RO/N contribution (O and N ligands cannot with certainty be separated by EXAFS data) was prevalent with the Hg–O/N bond distance varying between 2.03 and 2.08 Å, in agreement with previous Hg LIII-edge EXAFS studies of Geobacter (2.08 Å)19 as well as with studies of NOM samples (2.04–2.09 Å).27 Due to increasing Hg–O/N contribution with increasing Hg(II) addition, the first-shell peak was getting broader and shifted to shorter distances until a well-resolved peak dominated by the Hg–O/N distance appeared at ∼1.6 Å (corresponding to a bond length of 2.03 Å after correction for phase shift) for samples with Hg(II) concentrations of 215 and 406 μmol g–1 C (Table 1 and Figure S4). The Hg–S bond length was slightly longer (2.36–2.39 Å) at the higher additions of Hg(II). One possible explanation for the longer Hg–S distance may be some contribution from 3-coordinated structures, Hg(Mem-RS)3, known to have an average bond length varying in the range 2.40–2.51 Å for Hg(II) complexes with LMM-RS,48,49 but it could likewise be due to the involvement of other types of thiols having slightly longer Hg–S distances for 2-coordinated structures. Distortion of the structure due to steric hindrance effects could also affect bond lengths.
If 3-coordinated structures indeed would be of importance, they are expected to increase with decreasing Hg(II)/RSH ratios, which was not the case. The bond length (2.34 Å) and the multiple scattering peak clearly point at a high dominance of the linear Hg(Mem-RS)2 structure at the lowest Hg(II) addition. Further, although less informative than bond lengths (due to a larger error of ± 15% and a covariation with the Debye–Waller factor), CN would be expected to increase with Hg(II) addition if the Hg(Mem-RS)3 made up a significant contribution, which was not the case (Table 1). In addition, the occurrence of first shell Hg–O/N bond lengths of 2.03–2.08 Å (Table 1) is suggesting 2-coordinated structures. Some of these bonds may involve mixed structures of Hg–S and Hg–O/N bonds in the first coordination shell. In summary, our EXAFS data points at a high dominance of 2-coordinated Hg(II) at the membranes of Geobacter with a shift from two Mem-RS to a mixture of Mem-RS and Mem-RO/N functionalities with increasing Hg(II) addition. Our results agree with the Hg EXAFS study of Geobacter cell envelopes conducted by Mishra et al.19 In their study the Hg–S distance decreased from 2.38 Å at < 5% Hg(II) saturation of Mem-RSH groups (our calculation based on data reported in the study) to a minimum of 2.32 Å when all thiol groups were more than saturated by Hg(II) and RO/N groups were dominant (at 2.06 Å). Similar to our interpretation, the authors argued that 3-coordinated Hg(Mem-RS)3 structures would only make a minor contribution (if any) in the cell envelope of Geobacter.19 In a recent study,46 Hg Lα1 fluorescence HR XANES data were collected on the outer membrane of Geobacter added Hg(II) corresponding to less than 0.1% Mem-RSH (our calculation based on data in the report). A model developed from 2-, 3-, and 4-coordinated Hg(II) model compound structures was used to interpret their data. The authors suggested that added Hg(II) was combining with membrane associated thiols in a mixture of 2- and 3-coordinated structures at pH 6.8, but the average coordination could not be specified more in detail than to an “average coordination number of 2–3”. Because the method used is only semiquantitative, it is difficult to compare this result with the well-constrained Hg–S bond lengths of 2.34 ± 0.01 Å as determined by EXAFS in our study (at 5% Hg(II) saturation of Mem-RS) and 2.38 ± 0.01 Å (at < 5% saturation of Mem-RSH) in the study of Mishra et al. at pH 7.0.
For ND132, our Hg EXAFS data were only collected at relatively high Hg(II) concentrations at which the Mem-RSH functional groups were fully saturated and bonding to RO/N groups made significant contributions (Table 1 and Figure S4). The Hg–S bond distance determined in the ND132 membrane samples (2.39 and 2.40 Å, respectively, for the two samples) was in fair agreement with the distance of 2.35–2.43 Å reported for Hg(II) bonded to ND132 cell spheroplasts,9 which notably are devoid of the outer membrane and cannot be directly compared with our findings. Because we did not study the Hg speciation at this bacterium at sufficiently low Hg(II) additions, we have no information about possible 3-coordinated structures at low Hg(II) saturation of Mem-RSH groups.
EXAFS determined CN values of Hg–S and Hg–O/N have previously been used to calculate the concentration of NOM-associated thiol groups based on an approach where all thiols groups are assumed to be involved in a 2-coordinated Hg(NOM-RS)2 structure before Hg(II) is involved in the bonding with RO/N groups, Hg(NOM-RO)2.26−28 Using this model, we were not able to simulate the Mem-RSH data on Geobacter as determined by Hg EXAFS (Figure S5a). It is expected that the bacteria membranes are more rigid than NOM structures and that the washing procedure is removing dissolved biomolecules and smaller fragments. Therefore, we propose a refined model to fit the Hg EXAFS data for the membranes (see the details in the Supporting Information). As a theoretical basis for our model, we suggest that because of steric effects only a limited number of Mem-RSH groups (designated Mem-RIISH) are located sufficiently close to each other to form a linear Hg(Mem-RIIS)2 structure. The large majority of the membranethiols (designated Mem-RISH) are instead expected to be involved in a 2-coordinated complex, Hg(Mem-RISRO), composed of a mixture of Mem-RISH and neighboring O/N functional groups.
With this model, the concentration of Mem-RIIStot ([Mem-RIISH] + [Mem-RIIS–]) was calculated to be 20 ± 5 μmol g–1 C (± SD) (corresponding to 2.1 × 10–12 μmol cell–1) and the concentration of Mem-RIStot ([Mem-RISH] + [Mem-RIS–]) was calculated to be 360 ± 50 μmol g–1 C (corresponding to 3.6 × 10–11 μmol cell–1) for Geobacter membranes. Thus, the total concentration of thiol groups at the membranes, Mem-RStot ([Mem-RIIStot] + [Mem-RIStot]), was estimated to be 380 ± 50 μmol g–1 C (corresponding to 3.8 × 10–11 μmol cell–1). As shown in Figure S5b, the model showed a reasonable fit to the experimental Hg EXAFS data.
Using the same model, the thiol concentration of membranes of ND132 was estimated to be 19 ± 5, 330 ± 40, and 350 ± 40 μmol g–1 C (corresponding to 1.5 × 10–10, 2.7 × 10–9 and 2.8 × 10–9 μmol cell–1) for Mem-RIIStot, Mem-RIStot, and Mem-RStot, respectively. Given the limited Hg EXAFS data for this bacterium, our estimate rests on the assumption that the concentration of Mem-RIIStot made up 5% of Mem-RStot also for ND132 (as determined for Geobacter membranes). If the total S accounts for ∼1% of bacterial cell mass (dry weight),42 Mem-RStot will account for 55% of total S and for 62% of Org-SRED (as determined by S K-edge XANES) in membranes of Geobacter; whereas for ND132 Mem-RStot accounts for 48% of total S and 72% of Org-SRED.
Thiol functional group concentrations in membranes of Geobacter and ND132 determined here and in previous studies are listed in Table S3. Mishra et al.19 used two different methods to determine the Geobacter cell envelope concentration of thiols, assumed to represent the outer membrane concentration. They reported a concentration of 1000 ± 300 μmol g–1 dry cells, detected as fluorescence from qBBr used as the thiol probe, which would correspond to 2000 μmol g–1 C if we assume a TOC mass concentration of 50%. They also reported a thiol concentration of 67.8 ± 22.8 μmol g–1 wet weight using a potentiometric titration method, with and without blocking of thiol groups with qBBr.19 This thiol concentration would correspond to 550 μmol g–1 C assuming 50% TOC content and a 4.2:1 wet/dry mass ratio (adopted from their study). Our estimate of the Mem-RSH concentration of 380 ± 50 μmol g–1 C thus corresponds fairly well with the potentiometric titration data (550 μmol g–1 C), while our estimate is lower than the estimate using qBBr as fluorescence probe. In another study using the qBBr fluorescence method, Thomas et al.46 obtained a Geobacter cell envelope concentration of 55.5 ± 1.3 μmol g–1 wet cells, corresponding to 466 μmol g–1 C (assuming 50% TOC content and 4.2:1 wet/dry mass ratio19). It should be pointed out that while our Mem-RSH estimate is expected to include both inner and outer membranes, Mishra et al. and Thomas et al. determined the cell surface thiols, which likely involves mostly the outer membranethiols.19,46
Notably, our results, as well as the results of Mishra et al. and Thomas et al. deviate largely from the report by Rao et al.50 on the membranethiols of Geobacter. Reported per cell, our determined concentration of Mem-RSH of 3.8 × 10–11 μmol cell–1 is 3 orders of magnitude higher than the thiol concentration of 3.4 × 10–14μmol cell–1 reported by Rao et al. We think the difference is due to methodology, where the fluorescence-labeling method using ThioGlo-1 as the probe for thiols seems to underestimate the thiol functionalities as compared to using EXAFS determinations and qBBr as a probe.
For ND132, our estimated Mem-RStot concentration of 2.8 × 10–9 μmol cell–1 can only be compared with one other study in which TFP-4 was used as thiolate fluorescence probe. Wang et al.9 reported a thiol concentration in the isolated cell membranes of 2.8 × 10–12 μmol cell–1 and in the spheroplast of 1.6 × 10–11 μmol cell–1 (recalculated based on Avogadro’s constant). Thus, their reported membranethiol concentrations are 2–3 orders lower than ours. In addition to differences in methodology, it should also be noted that differences in culture media, growth phase and growth conditions may also lead to differences in membranethiol concentrations.51,52
Competitive Ligand Exchange Experiments and Thermodynamic Calculations
Thermodynamic constants (log K) for the formation of Hg(II) complexes with Mem-RSH were calculated from equilibrium concentrations of Hgtot and Hg(Cys)2 in the CLE experiments and by the chemical reactions in Table S1. The concentration of Hgtot and Hg(Cys)2 was determined by CAAS and LC-ICPMS, respectively, and the sum of the concentrations of Hg(Mem-RIIS)2, Hg(Cys)(Mem-RIS) and Hg(Mem-RISRO) was calculated as the difference between Hgtot and Hg(Cys)2. The concentration of Hg(Cys)2 did not change significantly with the selected reaction times of 1, 24, 48 and 72 h (p > 0.05, ANOVA, Tukey-HSD) in our CLE-experiments (Figure S6). Thus, we conclude that chemical equilibrium between Hg(Cys)2 and Hg(II) complexed by membrane functional groups was attained already after 1 h. This result can be compared with a previous study, in which Cys was added to a solution with Hg(NOM-RS)2 complexes. Full equilibrium was demonstrated to be achieved already within minutes.28 Thus, we could safely use all our data determined at the reaction times of 1 to 72 h in thermodynamic calculations.
The major chemical reactions considered in our model, and their thermodynamic constants (log K’s), are listed in Table S1. The pKa value of Mem-RSH of Geobacter was set to 9.5 ± 0.2.19 The proposed dominant membrane associated species were Hg(Mem-RIIS)2 (reaction 9, Table S1), Hg(Cys)(Mem-RIS) (reaction 10, Table S1), and Hg(Mem-RISRO) (reaction 11, Table S1), based on Hg LIII-edge EXAFS experiments (Table 1 and Figure S4). Because of difficulties to separate chemical structures in the second coordination shell (e.g., Hg–C bond distances), there is no EXAFS data on the formation of a mixed complex, Hg(Cys)(Mem-RIS) in the CLE experiments. However, based on previously reported experimental support for the mixed Hg(Cys)(NOM-RS) complex (reaction 8, Table S1),28 and mixed Hg(II)-LMM thiol complexes,29,53−55 as well as on calculated linear free-energy relationships (LFER) for mixed ligand complex formation,56 the Hg(Cys)(Mem-RIS) complex is likely to form in the Hg(II)–Cys–bacterium membrane system. In studies of the Hg(II)–Cys–NOM system using CLE experiments, Song et al.28 showed that the log K values for the formation of Hg(NOM-RS)2 and Hg(Cys)(NOM-RS) were positively correlated, which is expected according to linear free energy relationships (LFER).57 In fact, the two log K values were very similar when the differences in the pKa values of Cys and NOM-RSH were taken into account.28 Therefore, in order to include the Hg(Cys)(Mem-RIS) species in our model, the log K difference between Hg(Mem-RIIS)2 and Hg(Cys)(Mem-RIS) was set to the same value as the difference determined between Hg(NOM-RS)2 and Hg(Cys)(NOM-RS),28 as calculated by eq 1:
| 1 |
where −0.5 is the difference in pKa between Mem-RSH and NOM-RSH (Table S1). Similarly, the difference in log K between Hg(Mem-RIIS)2 and Hg(Mem-RISRO) was calculated by eq 2:
| 2 |
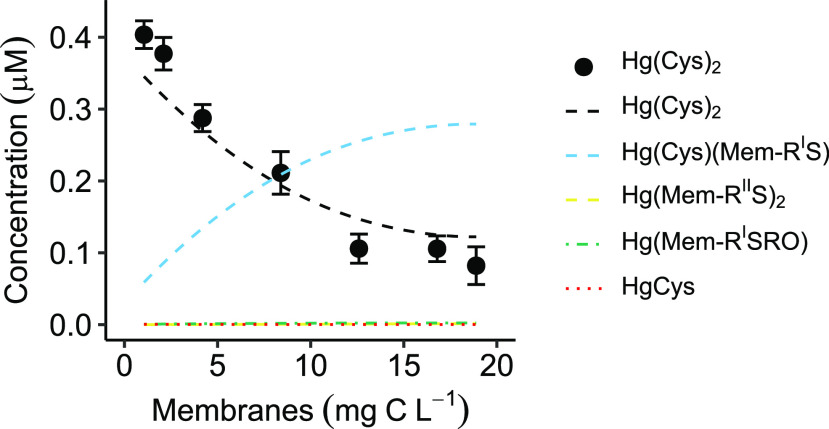
The log K values of Hg(Mem-RIIS)2 (reaction 9, Table S1), Hg(Cys)(Mem-RIS) (reaction 10, Table S1), and Hg(Mem-RISRO) (reaction 11, Table S1) were then calculated to be 39.1 ± 0.2, 38.1 ± 0.1, and 25.6 ± 0.1 (± SD), respectively. The model was optimized by the software R package PHREEQC and a minimum merit-of-fit, Σ(model – experiment)2/Σexperiment2, was obtained at 2.5%, with experiment denoting the measured concentration of Hg(Cys)2 and model denoting the modeled Hg(Cys)2 concentration (Figure 1). Note that the log K values represent an average of possibly several different types of thiol groups associated with the bacterium inner and outer membrane.
Figure 1.
Experimentally determined Hg(Cys)2 concentration (± SD, n = 4) as a function of Geobacter membrane concentration in the competitive ligand exchange experiment. pH ∼4.0 and I = 10 mM NaClO4. Different concentrations of membranes (1–19 mg C L–1 corresponding to 0.4–7.1 μM Mem-RStot) were pre-equilibrated in 0.5 μM Hg(NO3)2 for 24 h followed by the addition of Cys to reach a final concentration of 2 μM. The concentration of Hg(Cys)2 was measured at equilibrium. Dashed lines demonstrate the modeled concentrations, optimized by R software using R package PHREEQC to minimize the merit-of-fit, Σ(model – experiment)2/Σexperiment2, at 2.5%.
Our determined log K value of the mixed Hg(Cys)(Mem-RIS) complex can be compared with the LFER calculated by Dyrssen and Wedborg.56 They proposed a statistical relationship between the log K values of mixed ligation complex HgAB (Hg2+ + A– + B– ⇌ HgAB) and complexes HgA2 and HgB2: log KHgAB = log2 + 0.5(logKHgA2 + logKHgB2). Using this relationship, the log K of Hg(Cys)(Mem-RIS) is calculated to be 38.6 (based on the log K of 37.5 for Hg(Cys)2 and 39.1 for Hg(Mem-RIIS)2), which is reasonably close to our result of 38.1.
For the membranes of ND132, the log K’s for Hg(Mem-RIIS)2, Hg(Cys)(Mem-RIS), and Hg(Mem-RISRO) were calculated to be 39.2 ± 0.2, 38.2 ± 0.1, and 25.7 ± 0.1, respectively, at a merit-of-fit of 5% (Figure S7). Similar to the case of Geobacter, the pKa value for ND132 Mem-RSH was set to 9.5 ± 0.2.9,19,25 It should be pointed out that the Hgtot concentration in the ND132 membrane CLE experiment was unavailable and it was therefore set to the average concentration obtained in the Geobacter membrane CLE experiment (0.4 μM), after accounting for Hg(II) losses. Therefore, the uncertainly (SD) of the log K values may be larger than 0.2 for ND132. Yet, our results indicate that the log K values for the formation of Hg(II) complexes with membrane thiol functional groups are quite similar for the two different bacteria. As shown by calculations in the Supporting Information, the log K of Hg–thiol complexes formed with Cys, NOM, and membranes only differ by a maximum 0.5 log units at pH values when thiols are protonated.
An Internally Consistent Chemical Speciation Model for Bacterium–LMM Thiol–NOM Systems and its Environmental Implications
Cellular uptake is a membrane passing process and a critical step for bacterial Hg(II) methylation. Although the mechanism of the molecular processes of Hg(II) bacterial uptake and methylation remain unclear, it has been demonstrated in laboratory31,32,35 and mesocosm studies58−60 that the chemical speciation of Hg(II) in soils, sediments, and waters plays an important role in MeHg formation and subsequent uptake in organisms. Several field studies have reported significant, positive relationships between porewater concentrations of Hg(II) and rates of MeHg formation in marine systems,61−63 and tundra lakes.64 Identification of the Hg(II) species responsible for these relationships has proven to be more difficult.65,66 A major difficulty is that several Hg(II) species are expected to be bioavailable at different rates of uptake, which needs to be known, or at least estimated from experimental systems, if field data are to be simulated. Thermodynamic models have further been biased by including uncertain stability constants and nonconfirmed chemical species.5 Finally, because most models have been constructed from a range of thermodynamic experiments conducted with variable methodology, very few thermodynamic models are internally consistent.
In this study, we provide membrane thiol concentrations of Geobacter and ND132, and we have identified their surface complexes formed with Hg(II), i.e., Hg(Mem-RIIS)2, Hg(Cys)(Mem-RIS), and Hg(Mem-RISRO), and their corresponding thermodynamic constants. By combining these parameters with constants previously reported for complexes formed between Hg(II) and 15 naturally occurring LMM thiols,29 as well as with thiol groups associated with Suwannee River NOM,28 we can calculate the chemical speciation of Hg(II) in contact with bacteria under nonsulfidic environmental conditions. Because the experimentally determined parameters combined in such a model are all derived by the same CLE methodology, the data set represents an internally consistent thermodynamic model that can be used to simulate the chemical speciation of Hg(II) in reasonably complex systems relevant for nonsulfidic conditions in the environment (Table S1). Because Cys is used as a competitive ligand in all the CLE experiments, constants determined for the reaction between Hg(II) and functional groups associated with bacterial membranes, LMM thiols, and NOM all rely on a log K of 37.5 set for reaction 4 (Table S1).29 The strength of this approach is that, although it can be argued that there are other reported values on the constant for reaction 4 in the literature, the model is internally consistent and will thus accurately describe the distribution of Hg(II) among membranes, LMM thiols, and NOM.
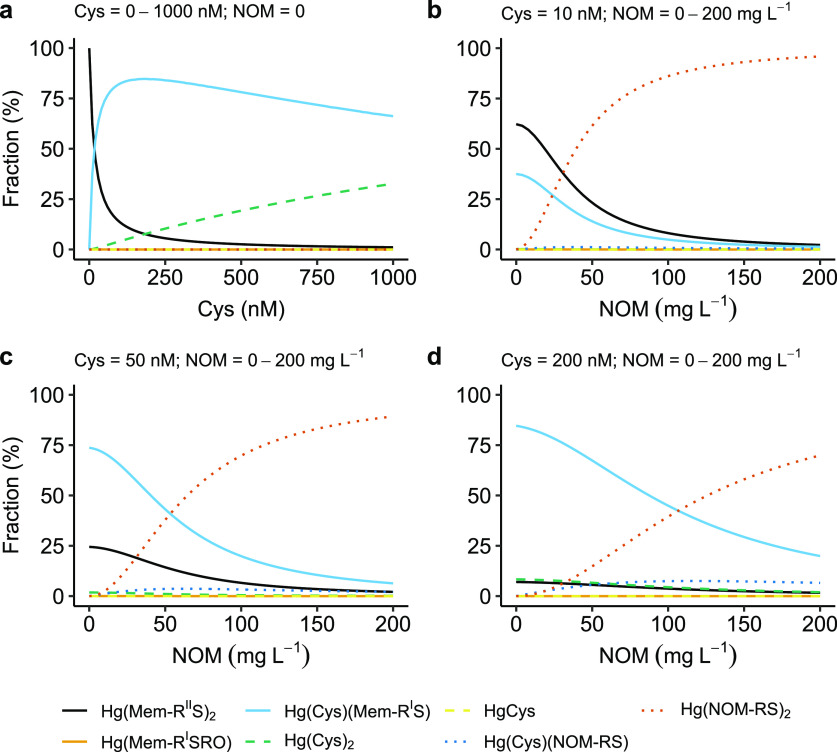
In Figure 2, the chemical speciation of Hg(II) in a system with the bacterium Geobacter is illustrated as a function of varying concentrations of NOM(aq) and the LMM thiol Cys. The amino acid Cys is known to be produced by Geobacter, reaching concentrations on the order of 100 nM in incubation experiments.35 The Hg(II) concentration was fixed at 1 ng L–1, which is a common level in freshwaters, and pH and the ionic strength were fixed at 7.0 and 10 mM, respectively. The cell abundance of Geobacter was set to 2 × 108 cells mL–1, a number typically selected for conventional bacterial incubation experiments. Because the outer and inner membranes were not separated in our experiments, the outer membrane thiols were for simplicity assumed to represent 50% of Mem-RStot (1.9 × 10–11 μmol cell–1). This means we assume the densities of thiols are equal (μmol g–1) at both the inner and outer membranes and that they contribute equally to our reported density per cell.
Figure 2.
Hg(II) speciation as a function of Cys and Suwannee River natural organic matter (NOM) concentration under environmentally relevant condition: Hg(II) = 1 ng L–1, Cys = 0–1000 nM, cell abundance = 2 × 108 cells mL–1, NOM = 0–200 mg L–1 (corresponding to 0–1.5 μM NOM-RSH based on the NOM-RSH concentration of 7.5 μmol g–1 NOM28), pH = 7, and ionic strength = 10 mM.
Figure 2a illustrates that in absence of NOM, Hg(II) is complexed by the cell membrane mainly in the structure Hg(Mem-RIIS)2 when the Cys concentration is less than ∼20 nM. When the Cys concentration exceeds ∼20 nM, the Hg(Cys)(Mem-RIS) structure will gradually take over and dominate even up to 1 μM Cys. Figure 2b–d further demonstrates the effect of NOM on the speciation of Hg(II) in this system. When the concentration of both Cys and NOM are relatively low (e.g., 10 nM Cys and 20 mg L–1 NOM corresponding to 150 nM NOM-RSH according to Song et al.28), all three complexes, Hg(Mem-RIIS)2, Hg(Cys)(Mem-RIS), and Hg(NOM-RS)2, make significant contributions. With increased NOM concentration, the Hg(NOM-RS)2 complex will gradually take over as the dominant species. The NOM concentrations at which Hg(Cys)(Mem-RIS) and Hg(NOM-RS)2 concentrations are approximately equal are 25, 50, and 100 mg L–1 at 10, 50, and 200 nM Cys, respectively. All these combinations are expected to cover relevant conditions in soils and sediments, depending on microbial activity and redox potential.
The model demonstrates a very significant association of Hg(II) with cell membranes, even at high concentrations of NOM and LMM thiols in aqueous phase covering the 10 nM to 1 μM range, conditions expected to be encountered in soils and sediments. Notably, under the circumstances in soils and waters when LMM concentrations exceed 10 nM,67 Hg(II) is expected to be dominated by complexes formed as a mixture of Mem-RSIH and various types of LMM thiols at bacterium membrane surfaces. Although Cys, NOM-RSH, and Mem-RSH have similar affinities for Hg(II) at neutral pH (when thiols are kept protonated), the 2-coordinated Hg(Mem-RIIS)2 structure only dominates at very low LMM thiol concentrations. This is because Mem-RIIStot only contributes to 5% of Mem-RStot. It should be noted that a potential complex composed of a mixture of Hg(II) with NOM-RSH and Mem-RSH ligation is not included in this model. The rationale for excluding this species is that such a complex may be constrained by steric hindrance effects.
Overall, our results point at the quantitative importance of a mixed ligation between LMM thiols and Mem-RSH functionalities under environmentally relevant conditions. The question that remains to be answered is what role such a complex may have for the internalization of Hg(II) and its methylation in Geobacter. The bonding of Hg(II) to membrane functional groups was not included in the study of Adediran et al.35 on the same bacteria. They demonstrated that the thermodynamically weaker complexes involving O and Cl ligands showed larger methylation rate constants than 2-coordinate complexes involving only LMM thiols but noted that the weaker complexes are less likely to be quantitatively important in natural environments under thermodynamic equilibrium conditions. Also our modeling shows that complexes involving O and Cl bonds in the first coordination shell are too weak to be quantitatively important at a Hg(II) concentration of 1 ng L–1. Under such environmentally relevant conditions, the uptake and methylation rate of complexes formed with thiols associated with LMM molecules, NOM, and bacterial membranes will regulate MeHg formation.
We recommend the use of this internally consistent thermodynamic model in future experimental studies aiming at resolving the mechanisms of Hg(II) uptake and transformation in bacterium–NOM–LMM thiol systems, in the presence of Geobacter and ND132. In order to be able to specifically address the question whether cell membrane associated thiol functionalities inhibit, stimulate, or are indifferent to Hg-species cellular uptake, a further differentiation of thiol groups associated with specific locations at membranes is required. Notably, our chemical modeling (Figure 2) does not apply to sulfidic conditions, where aqueous Hg–S complexes and solid HgS(s) phases need to be included. To get an internally consistent data set covering also sulfidic conditions, the thermodynamics of, e.g., Hg(Cys)2 and aqueous Hg–S complex formation need to be related in the same experiment.
Acknowledgments
The authors acknowledge Dr. Yuwei Wang at Department of Environmental Sciences, Rutgers University, for assistance on the cell abundance estimation; Dr. Chenyan Ma at Beijing Synchrotron Radiation Facility (Beamline 4B7A) for assistance with the sulfur K-edge XANES spectroscopy measurements; and Sofia Diaz-Moreno and Ann-Kathrin Geiger at Diamond Light Source (Beamline I20-scanning, project SP9157) for assistance at the DLS beamline and sample handling. This work was funded by the Swedish Research Council (VR) project Sino-Swedish Mercury Management Framework—SMaReF (2013-6978), the VR project (621-2014-5370), and Carl Trygger Foundation CTS 17:423 to U.S. and by the Kempe Foundations (JCK-1501, SMK-2745, SMK-1243).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.0c01751.
Description of bacterial growth media, membrane isolation, S K-edge XANES and Hg LIII-edge EXAFS experiments, Mem-RSH concentration calculations, experimental losses of Hg(II), and thermodynamic calculations of reactions with thiols in protonated form; list of selected chemical reactions and thermodynamic constants, S K-edge XANES result, and reported Mem-RSH concentrations in previous studies; S K-edge XANES and Hg LIII-edge EXAFS spectra and model fits, Mem-RSH calculation model, loss of Hg(II), CLE experimental results, and Hg(II)–Mem-RSH thermodynamic stabilities of ND132 (PDF)
Author Present Address
∥ G.A.A.: Department of Soil and Environment, Swedish University of Agricultural Science, SE-750 07 Uppsala, Sweden.
The authors declare no competing financial interest.
Supplementary Material
References
- Hsu-Kim H.; Kucharzyk K. H.; Zhang T.; Deshusses M. A. Mechanisms Regulating Mercury Bioavailability for Methylating Microorganisms in the Aquatic Environment: A Critical Review. Environ. Sci. Technol. 2013, 47, 2441–2456. 10.1021/es304370g. [DOI] [PubMed] [Google Scholar]
- Parks J. M.; Johs A.; Podar M.; Bridou R.; Hurt R. A.; Smith S. D.; Tomanicek S. J.; Qian Y.; Brown S. D.; Brandt C. C.; et al. The Genetic Basis for Bacterial Mercury Methylation. Science 2013, 339, 1332–1335. 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- Podar M.; Gilmour C. C.; Brandt C. C.; Soren A.; Brown S. D.; Crable B. R.; Palumbo A. V.; Somenahally A. C.; Elias D. A. Global Prevalence and Distribution of Genes and Microorganisms Involved in Mercury Methylation. Sci. Adv. 2015, 1, e1500675 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnell O.; Watras C. J. Microbial Mercury Methylation in Aquatic Environments: A Critical Review of Published Field and Laboratory Studies. Environ. Sci. Technol. 2019, 53, 4–19. 10.1021/acs.est.8b02709. [DOI] [PubMed] [Google Scholar]
- Skyllberg U. Competition among Thiols and Inorganic Sulfides and Polysulfides for Hg and MeHg in Wetland Soils and Sediments under Suboxic Conditions: Illumination of Controversies and Implications for MeHg Net Production. J. Geophys. Res. 2008, 113, 1–14. 10.1029/2008JG000745. [DOI] [Google Scholar]
- Graham A. M.; Bullock A. L.; Maizel A. C.; Elias D. A.; Gilmour C. C. Detailed Assessment of the Kinetics of Hg-Cell Association, Hg Methylation, and Methylmercury Degradation in Several Desulfovibrio Species. Appl. Environ. Microbiol. 2012, 78, 7337–7346. 10.1128/AEM.01792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. R.; Lu X.; Zhao L.; An J.; He J. Z.; Pierce E. M.; Johs A.; Gu B. Effects of Cellular Sorption on Mercury Bioavailability and Methylmercury Production by Desulfovibrio Desulfuricans ND132. Environ. Sci. Technol. 2016, 50, 13335–13341. 10.1021/acs.est.6b04041. [DOI] [PubMed] [Google Scholar]
- Hu H.; Lin H.; Zheng W.; Rao B.; Feng X.; Liang L.; Elias D. A.; Gu B. Mercury Reduction and Cell-Surface Adsorption by Geobacter Sulfurreducens PCA. Environ. Sci. Technol. 2013, 47, 10922–10930. 10.1021/es400527m. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Schaefer J. K.; Mishra B.; Yee N. Intracellular Hg(0) Oxidation in Desulfovibrio Desulfuricans ND132. Environ. Sci. Technol. 2016, 50, 11049–11056. 10.1021/acs.est.6b03299. [DOI] [PubMed] [Google Scholar]
- Lin H.; Hurt R. A.; Johs A.; Parks J. M.; Morrell-Falvey J. L.; Liang L.; Elias D. A.; Gu B. Unexpected Effects of Gene Deletion on Interactions of Mercury with the Methylation-Deficient Mutant ΔhgcAB. Environ. Sci. Technol. Lett. 2014, 1, 271–276. 10.1021/ez500107r. [DOI] [Google Scholar]
- Thomas S. A.; Tong T.; Gaillard J.-F. Hg(II) Bacterial Biouptake: The Role of Anthropogenic and Biogenic Ligands Present in Solution and Spectroscopic Evidence of Ligand Exchange Reactions at the Cell Surface. Metallomics 2014, 6, 2213–2222. 10.1039/C4MT00172A. [DOI] [PubMed] [Google Scholar]
- Barkay T.; Miller S. M.; Summers A. O. Bacterial Mercury Resistance from Atoms to Ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Slaveykova V. I.; Wilkinson K. J. Predicting the Bioavailability of Metals and Metal Complexes: Critical Review of the Biotic Ligand Model. Environ. Chem. 2005, 2, 9–24. 10.1071/EN04076. [DOI] [Google Scholar]
- Aristilde L.; Xu Y.; Morel F. M. M. Weak Organic Ligands Enhance Zinc Uptake in Marine Phytoplankton. Environ. Sci. Technol. 2012, 46, 5438–5445. 10.1021/es300335u. [DOI] [PubMed] [Google Scholar]
- Flynn S. L.; Szymanowski J. E.; Fein J. B. Modeling Bacterial Metal Toxicity Using a Surface Complexation Approach. Chem. Geol. 2014, 374, 110–116. 10.1016/j.chemgeo.2014.03.010. [DOI] [Google Scholar]
- Zhao C.-M.; Wilkinson K. J. Biotic Ligand Model Does Not Predict the Bioavailability of Rare Earth Elements in the Presence of Organic Ligands. Environ. Sci. Technol. 2015, 49, 2207–2214. 10.1021/es505443s. [DOI] [PubMed] [Google Scholar]
- Zhao C.-M.; Campbell P. G. C.; Wilkinson K. J. When Are Metal Complexes Bioavailable?. Environ. Chem. 2016, 13, 425–433. 10.1071/EN15205. [DOI] [Google Scholar]
- Fein J. B. Advanced Biotic Ligand Models: Using Surface Complexation Modeling to Quantify Metal Bioavailability to Bacteria in Geologic Systems. Chem. Geol. 2017, 464, 127–136. 10.1016/j.chemgeo.2016.10.001. [DOI] [Google Scholar]
- Mishra B.; Shoenfelt E.; Yu Q.; Yee N.; Fein J. B.; Myneni S. C. B. Stoichiometry of Mercury-Thiol Complexes on Bacterial Cell Envelopes. Chem. Geol. 2017, 464, 137–146. 10.1016/j.chemgeo.2017.02.015. [DOI] [Google Scholar]
- Daughney C. J.; Siciliano S. D.; Rencz A. N.; Lean D.; Fortin D. Hg(II) Adsorption by Bacteria: A Surface Complexation Model and Its Application to Shallow Acidic Lakes and Wetlands in Kejimkujik National Park, Nova Scotia, Canada. Environ. Sci. Technol. 2002, 36, 1546–1553. 10.1021/es010713x. [DOI] [PubMed] [Google Scholar]
- Dunham-Cheatham S.; Farrell B.; Mishra B.; Myneni S.; Fein J. B. The Effect of Chloride on the Adsorption of Hg onto Three Bacterial Species. Chem. Geol. 2014, 373, 106–114. 10.1016/j.chemgeo.2014.02.030. [DOI] [Google Scholar]
- Johnson C. R.; Fein J. B. The Effect of Metal Loading on Bacterial Hg Adsorption. Chem. Geol. 2018, 498, 106–114. 10.1016/j.chemgeo.2018.09.017. [DOI] [Google Scholar]
- Yi L.; Li H.; Sun L.; Liu L.; Zhang C.; Xi Z. A Highly Sensitive Fluorescence Probe for Fast Thiol-Quantification Assay of Glutathione Reductase. Angew. Chem., Int. Ed. 2009, 48, 4034–4037. 10.1002/anie.200805693. [DOI] [PubMed] [Google Scholar]
- Joe-Wong C.; Shoenfelt E.; Hauser E. J.; Crompton N.; Myneni S. C. B. Estimation of Reactive Thiol Concentrations in Dissolved Organic Matter and Bacterial Cell Membranes in Aquatic Systems. Environ. Sci. Technol. 2012, 46, 9854–9861. 10.1021/es301381n. [DOI] [PubMed] [Google Scholar]
- Yu Q.; Szymanowski J.; Myneni S. C. B.; Fein J. B. Characterization of Sulfhydryl Sites within Bacterial Cell Envelopes Using Selective Site-Blocking and Potentiometric Titrations. Chem. Geol. 2014, 373, 50–58. 10.1016/j.chemgeo.2014.02.027. [DOI] [Google Scholar]
- Qian J.; Skyllberg U.; Frech W.; Bleam W. F.; Bloom P. R.; Petit P. E. Bonding of Methyl Mercury to Reduced Sulfur Groups in Soil and Stream Organic Matter as Determined by X-Ray Absorption Spectroscopy and Binding Affinity Studies. Geochim. Cosmochim. Acta 2002, 66, 3873–3885. 10.1016/S0016-7037(02)00974-2. [DOI] [Google Scholar]
- Skyllberg U.; Bloom P. R.; Qian J.; Lin C.-M.; Bleam W. F. Complexation of Mercury(II) in Soil Organic Matter: EXAFS Evidence for Linear Two-Coordination with Reduced Sulfur Groups. Environ. Sci. Technol. 2006, 40, 4174–4180. 10.1021/es0600577. [DOI] [PubMed] [Google Scholar]
- Song Y.; Jiang T.; Liem-Nguyen V.; Sparrman T.; Björn E.; Skyllberg U. Thermodynamics of Hg(II) Bonding to Thiol Groups in Suwannee River Natural Organic Matter Resolved by Competitive Ligand Exchange, Hg LIII-Edge EXAFS and 1H NMR Spectroscopy. Environ. Sci. Technol. 2018, 52, 8292–8301. 10.1021/acs.est.8b00919. [DOI] [PubMed] [Google Scholar]
- Liem-Nguyen V.; Skyllberg U.; Nam K.; Björn E. Thermodynamic Stability of Mercury(II) Complexes Formed with Environmentally Relevant Low-Molecular-Mass Thiols Studied by Competing Ligand Exchange and Density Functional Theory. Environ. Chem. 2017, 14, 243–253. 10.1071/EN17062. [DOI] [Google Scholar]
- Caccavo F.; Lonergan D. J.; Lovley D. R.; Davis M.; Stolz J. F.; McInerney M. J. Geobacter Sulfurreducens Sp. Nov., a Hydrogen-and Acetate-Oxidizing Dissimilatory Metal-Reducing Microorganism. Appl. Environ. Microbiol. 1994, 60, 3752–3759. 10.1128/AEM.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J. K.; Morel F. M. High Methylation Rates of Mercury Bound to Cysteine by Geobacter Sulfurreducens. Nat. Geosci. 2009, 2, 123–126. 10.1038/ngeo412. [DOI] [Google Scholar]
- Schaefer J. K.; Rocks S. S.; Zheng W.; Liang L.; Gu B.; Morel F. M. M. Active Transport, Substrate Specificity, and Methylation of Hg(II) in Anaerobic Bacteria. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 8714–8719. 10.1073/pnas.1105781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrini S. M.; Haigh R.; Freestone P. P. Fractionation by Ultracentrifugation of Gram Negative Cytoplasmic and Membrane Proteins. Bio-Protocol 2014, 4, e1287 10.21769/BioProtoc.1287. [DOI] [Google Scholar]
- Gorby Y. A.; Beveridge T. J.; Blakemore R. P. Characterization of the Bacterial Magnetosome Membrane. J. Bacteriol. 1988, 170, 834–841. 10.1128/JB.170.2.834-841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adediran G. A.; Liem-Nguyen V.; Song Y.; Schaefer J. K.; Skyllberg U.; Björn E. Microbial Biosynthesis of Thiol Compounds: Implications for Speciation, Cellular Uptake, and Methylation of Hg(II). Environ. Sci. Technol. 2019, 53, 8187–8196. 10.1021/acs.est.9b01502. [DOI] [PubMed] [Google Scholar]
- Cline J. D. SPECTROPHOTOMETRIC DETERMINATION OF HYDROGEN SULFIDE IN NATURAL WATERS1. Limnol. Oceanogr. 1969, 14, 454–458. 10.4319/lo.1969.14.3.0454. [DOI] [Google Scholar]
- Dunham-Cheatham S.; Mishra B.; Myneni S.; Fein J. B. The Effect of Natural Organic Matter on the Adsorption of Mercury to Bacterial Cells. Geochim. Cosmochim. Acta 2015, 150, 1–10. 10.1016/j.gca.2014.11.018. [DOI] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; 2020.
- Parkhurst D. L.; Appelo C. A. J.. Description of Input and Examples for PHREEQC Version 3–A Computer Program for Speciation, Batch-Reaction, One-Dimensional Transport, and Inverse Geochemical Calculations; Techniques and Methods; U.S. Geological Survey, 2013; Book 6. [Google Scholar]
- Charlton S. R.; Parkhurst D. L. Modules Based on the Geochemical Model PHREEQC for Use in Scripting and Programming Languages. Comput. Geosci. 2011, 37, 1653–1663. 10.1016/j.cageo.2011.02.005. [DOI] [Google Scholar]
- Ucar I.; Pebesma E.; Azcorra A. Measurement Errors in R. R J. 2019, 10, 549–557. 10.32614/RJ-2018-075. [DOI] [Google Scholar]
- Barton L. L.; Goulhen F.; Bruschi M.; Woodards N. A.; Plunkett R. M.; Rietmeijer F. J. M. The Bacterial Metallome: Composition and Stability with Specific Reference to the Anaerobic Bacterium Desulfovibrio Desulfuricans. BioMetals 2007, 20, 291–302. 10.1007/s10534-006-9059-2. [DOI] [PubMed] [Google Scholar]
- Liem-Nguyen V.; Huynh K.; Gallampois C.; Björn E. Determination of Picomolar Concentrations of Thiol Compounds in Natural Waters and Biological Samples by Tandem Mass Spectrometry with Online Preconcentration and Isotope-Labeling Derivatization. Anal. Chim. Acta 2019, 1067, 71–78. 10.1016/j.aca.2019.03.035. [DOI] [PubMed] [Google Scholar]
- Jiang T.; Skyllberg U.; Wei S.; Wang D.; Lu S.; Jiang Z.; Flanagan D. C. Modeling of the Structure-Specific Kinetics of Abiotic, Dark Reduction of Hg(II) Complexed by O/N and S Functional Groups in Humic Acids While Accounting for Time-Dependent Structural Rearrangement. Geochim. Cosmochim. Acta 2015, 154, 151–167. 10.1016/j.gca.2015.01.011. [DOI] [Google Scholar]
- Lin H.; Morrell-Falvey J. L.; Rao B.; Liang L.; Gu B. Coupled Mercury-Cell Sorption, Reduction, and Oxidation on Methylmercury Production by Geobacter Sulfurreducens PCA. Environ. Sci. Technol. 2014, 48, 11969–11976. 10.1021/es502537a. [DOI] [PubMed] [Google Scholar]
- Thomas S. A.; Mishra B.; Myneni S. C. Cellular Mercury Coordination Environment, and Not Cell Surface Ligands, Influence Bacterial Methylmercury Production. Environ. Sci. Technol. 2020, 54, 3960–3968. 10.1021/acs.est.9b05915. [DOI] [PubMed] [Google Scholar]
- Jalilehvand F.; Leung B. O.; Izadifard M.; Damian E. Mercury(II) Cysteine Complexes in Alkaline Aqueous Solution. Inorg. Chem. 2006, 45, 66–73. 10.1021/ic0508932. [DOI] [PubMed] [Google Scholar]
- Leung B. O.; Jalilehvand F.; Mah V. Mercury(II) Penicillamine Complex Formation in Alkaline Aqueous Solution. Dalton Trans 2007, 4666–74. 10.1039/b711436b. [DOI] [PubMed] [Google Scholar]
- Mah V.; Jalilehvand F. Glutathione Complex Formation with Mercury(II) in Aqueous Solution at Physiological pH. Chem. Res. Toxicol. 2010, 23, 1815–23. 10.1021/tx100260e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao B.; Simpson C.; Lin H.; Liang L.; Gu B. Determination of Thiol Functional Groups on Bacteria and Natural Organic Matter in Environmental Systems. Talanta 2014, 119, 240–247. 10.1016/j.talanta.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yu Q.; Fein J. B. Controls on Bacterial Cell Envelope Sulfhydryl Site Concentrations: The Effect of Glucose Concentration during Growth. Environ. Sci. Technol. 2017, 51, 7395–7402. 10.1021/acs.est.7b01047. [DOI] [PubMed] [Google Scholar]
- Fein J. B.; Yu Q.; Nam J.; Yee N. Bacterial Cell Envelope and Extracellular Sulfhydryl Binding Sites: Their Roles in Metal Binding and Bioavailability. Chem. Geol. 2019, 521, 28–38. 10.1016/j.chemgeo.2019.04.026. [DOI] [Google Scholar]
- Hilton B.; Man M.; Hsi E.; Bryant R. NMR Studies of Mercurial-Halogen Equilibria. J. Inorg. Nucl. Chem. 1975, 37, 1073–1077. 10.1016/0022-1902(75)80707-X. [DOI] [Google Scholar]
- Pei K. L.; Sooriyaarachchi M.; Sherrell D. A.; George G. N.; Gailer J. Probing the Coordination Behavior of Hg 2+, CH 3 Hg+, and Cd 2+ towards Mixtures of Two Biological Thiols by HPLC-ICP-AES. J. Inorg. Biochem. 2011, 105, 375–381. 10.1016/j.jinorgbio.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Pyreu D. F.; Ryzhakov A. M.; Kozlovskii E. V.; Gruzdev M. S.; Kumeev R. S. Mixed-Ligand Complex Formation of Mercury (II) Ethylenediaminetetraacetate with Cysteine and Methionine in Aqueous Solution. Inorg. Chim. Acta 2011, 371, 53–58. 10.1016/j.ica.2011.03.013. [DOI] [Google Scholar]
- Dyrssen D.; Wedborg M. The Sulphur-Mercury(II) System in Natural Waters. Water, Air, Soil Pollut. 1991, 56, 507–519. 10.1007/BF00342295. [DOI] [Google Scholar]
- Wells P. R. Linear Free Energy Relationships. Chem. Rev. 1963, 63, 171–219. 10.1021/cr60222a005. [DOI] [Google Scholar]
- Jonsson S.; Skyllberg U.; Nilsson M. B.; Westlund P.-O.; Shchukarev A.; Lundberg E.; Björn E. Mercury Methylation Rates for Geochemically Relevant Hg(II) Species in Sediments. Environ. Sci. Technol. 2012, 46, 11653–11659. 10.1021/es3015327. [DOI] [PubMed] [Google Scholar]
- Jonsson S.; Skyllberg U.; Nilsson M. B.; Lundberg E.; Andersson A.; Björn E. Differentiated Availability of Geochemical Mercury Pools Controls Methylmercury Levels in Estuarine Sediment and Biota. Nat. Commun. 2014, 5, 4624. 10.1038/ncomms5624. [DOI] [PubMed] [Google Scholar]
- Zhu W.; Song Y.; Adediran G. A.; Jiang T.; Reis A. T.; Pereira E.; Skyllberg U.; Björn E. Mercury Transformations in Resuspended Contaminated Sediment Controlled by Redox Conditions, Chemical Speciation and Sources of Organic Matter. Geochim. Cosmochim. Acta 2018, 220, 158–179. 10.1016/j.gca.2017.09.045. [DOI] [Google Scholar]
- Hammerschmidt C. R.; Fitzgerald W. F. Geochemical Controls on the Production and Distribution of Methylmercury in Near-Shore Marine Sediments. Environ. Sci. Technol. 2004, 38, 1487–1495. 10.1021/es034528q. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt C. R.; Fitzgerald W. F. Methylmercury Cycling in Sediments on the Continental Shelf of Southern New England. Geochim. Cosmochim. Acta 2006, 70, 918–930. 10.1016/j.gca.2005.10.020. [DOI] [Google Scholar]
- Hollweg T. A.; Gilmour C. C.; Mason R. P. Mercury and Methylmercury Cycling in Sediments of the Mid-Atlantic Continental Shelf and Slope. Limnol. Oceanogr. 2010, 55, 2703–2722. 10.4319/lo.2010.55.6.2703. [DOI] [Google Scholar]
- Hammerschmidt C. R.; Fitzgerald W. F.; Lamborg C. H.; Balcom P. H.; Tseng C.-M. Biogeochemical Cycling of Methylmercury in Lakes and Tundra Watersheds of Arctic Alaska. Environ. Sci. Technol. 2006, 40, 1204–1211. 10.1021/es051322b. [DOI] [PubMed] [Google Scholar]
- Benoit J. M.; Gilmour C. C.; Mason R. P.; Heyes A. Sulfide Controls on Mercury Speciation and Bioavailability to Methylating Bacteria in Sediment Pore Waters. Environ. Sci. Technol. 1999, 33, 951–957. 10.1021/es9808200. [DOI] [Google Scholar]
- Drott A.; Lambertsson L.; Björn E.; Skyllberg U. Importance of Dissolved Neutral Mercury Sulfides for Methyl Mercury Production in Contaminated Sediments. Environ. Sci. Technol. 2007, 41, 2270–2276. 10.1021/es061724z. [DOI] [PubMed] [Google Scholar]
- Liem-Nguyen V.; Skyllberg U.; Björn E. Thermodynamic Modeling of the Solubility and Chemical Speciation of Mercury and Methylmercury Driven by Organic Thiols and Micromolar Sulfide Concentrations in Boreal Wetland Soils. Environ. Sci. Technol. 2017, 51, 3678–3686. 10.1021/acs.est.6b04622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.