Abstract
A series of five rationally designed decapeptides [DEHGTAVMLK (DP1), THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH)] have been studied for their interactions with Cu(II) and Mn(II) ions. The peptides, constructed including the most prevalent amino acid content found in the cell-free extract of Deinococcus radiodurans (DR), play a fundamental role in the antioxidant mechanism related to its exceptional radioresistance. Mn(II) ions, in complex with these peptides, are found to be an essential ingredient for the DR protection kit. In this work, a detailed characterization of Cu(II) systems was included, because Cu(II)–peptide complexes have also shown remarkable antioxidant properties. All peptides studied contain in their sequence coordinating residues that can bind effectively Mn(II) or Cu(II) ions with high affinity, such as Asp, Glu, and His. Using potentiometric techniques, NMR, EPR, UV–vis, and CD spectroscopies, ESI-MS spectrometry, and molecular model calculations, we explored the binding properties and coordination modes of all peptides toward the two metal ions, were able to make a metal affinity comparison for each metal system, and built a structural molecular model for the most stable Cu(II) and Mn(II) complexes in agreement with experimental evidence.
Short abstract
Five rationally designed decapeptides reconstituted from the cell-free extract of Deinococcus radiodurans have been precisely analyzed in terms of their coordination properties toward Mn(II) and Cu(II). The results provide new insight to enhance our understanding of the impact of metal complexes in the protection of the bacterium from various damaging agents such as ionizing radiation, ultraviolet radiation, and oxidative stress and novel information useful for exploiting this extraordinary ability in future biotechnological applications.
Introduction
The mechanism of Deinococcus radiodurans (DR) radioprotection is assumed to be based on a concomitant involvement of key elements, including a number of regulators that can repair DNA efficiently, a rapid cellular cleansing, a high antioxidant capacity of its enzymatic systems [superoxide dismutases (SOD) and catalases], and a well-controlled Mn/Fe metal homeostasis, together with non-enzymatic systems involving complexes of Mn(II) with small molecules such as carotenoids, nucleosides, peptides, and orthophosphate groups that provide cellular protection against harmful reactive oxygen species (ROS).1−5 In particular, catalytic Mn complexes could neutralize the O2•– radical by mimicking the SOD enzyme, thus shielding proteins against oxidative damage and preserving their activity.1,6−8 Despite the fact that Mn complexes, taken individually, have a lower catalytic activity compared with antioxidant enzymatic systems, when considered as a whole they could outperform antioxidant enzymes in protecting proteins against ROS damage.9−11 Cell-free extracts of DR showed the majority of cellular manganese was bound in small Mn(II) complexes with peptides strongly implicated in shielding the proteome from oxidation. In particular, these complexes showed remarkable antioxidant properties selectively protecting cytosolic proteins from ROS induced by ionizing radiation (IR), thereby preserving the functionality of cytoplasmic enzymes, including those needed to repair DNA and, consequently, indispensable for DR survival.2 The hydrolyzed DR ultrafiltrates were found to be enriched in peptides containing aspartic acid (Asp, D), glutamic acid (Glu, E), histidine (His, H), glycine (Gly, G), alanine (Ala, A), and methionine (Met, M), but not proline (Pro, P).2,12 DP1 (DEHGTAVMLK) is a synthetic reconstituted decapeptide rationally designed by Daly et al. considering the most abundant amino acids in the hydrolyzed DR ultrafiltrate.2 The DP1–Mn(II) system has been reported to protect the structure and function of surface proteins exposed to massive ionizing radiation doses and to preserve the proteome integrity also under desiccation conditions, ultraviolet radiation, and other forms of oxidative stress. Moreover, the antioxidant properties of the DP1–Mn(II) complex were evaluated in vivo in mice subjected to radiation injury, revealing their extraordinary potency without presenting any toxic or immunogenic effect.13 Recently, this kind of complex has also been successfully applied in the development of viral vaccines based on an IR-inactivation approach.14−16
The aim of this paper is focused on the molecular characterization of the complexes formed by Mn(II) and five synthetic decapeptides [DEHGTAVMLK (DP1) and four derivatives thereof, THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH)], whose sequences reflect the natural amino acid abundance found in DR cell-free extracts but differ in the order of residues.
We were interested in finding evidence of formation of complexes between Mn(II) and DR peptides, which according to previous studies is one of the key elements in the radioprotection mechanism,2,12−14 and in identifying the sequences with the highest affinity for the metal ion. The study was also extended to the determination of interactions of the peptides with Cu(II), another bivalent metal ion, whose complexes are much more spectroscopically informative compared to Mn(II), allowing us to gain insight and a better understanding of the coordination abilities of these selected sequences. Furthermore, several Cu(II)–peptide complexes, in particular those containing histidine residues, demonstrated antioxidant properties.17−21 For example, the tripeptide Gly-His-Lys, a naturally occurring component of the plasma and liver cell growth factor, in complex with Cu(II) demonstrated to possess a SOD-mimetic activity, and a simple modification of the peptide sequence, to His-Gly-Lys increased its activity ≤223-fold.22,23 Likewise, the complex of Cu(II) with the first four amino acids of the N-terminus of human albumin Asp-Ala-His-Lys exhibited SOD-like activity by significantly inhibiting superoxide formation.21 The occurrence and the position of histidine residues along the peptide chain are crucial for the metal chelation ability, affinity, and stability of these species,24,25 together with the direct interaction of the other coordinating residues and a possible shielding or stabilizing effect exerted by amino acid side chains that can stabilize or protect the complexes against water molecules.26 In this study, the coordinating residues Asp, Glu, and His in the order -DEH- were scrutinized for Mn(II) and Cu(II) coordination ability depending on their position along the sequence. In particular, in DP1, the active metal binding site DEH motif is located in the first portion of the peptide, in Term-DEH, the motif is at the end, and in Mid-DEH, it is in the middle. In the Bis-DEH peptide, the -DEH- sequence is inserted twice, in the N- and C-terminal parts, separated by four glycine residues, whereas in DP2, a random scrambled version of DP1 was chosen to evaluate the ability of His, placed far apart with respect to the other two coordinating residues, Asp and Glu, to compete for metal ions. The coordination properties of the five peptides toward Cu(II) and Mn(II) were tested by using potentiometry, NMR, EPR, UV–vis, and CD spectroscopies, and ESI-MS spectrometry. Molecular modeling techniques were further used to elaborate three-dimensional models of the metal–peptide complexes on the basis of the experimental evidence.
Experimental Section
Materials
The five decapeptides [DEHGTAVMLK (DP1), THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH)] were synthesized by Elim Biopharmaceuticals (Hayward, CA).
All of the chemicals used in this work were purchased from Sigma-Aldrich or POCH and used without any further purification. Cu(ClO4)2 and Mn(ClO4)2 stock solutions were standardized by ICP-AES along with the complexometric titrations with standardized Na2H2EDTA and murexide. The HClO4 solution was prepared by dilution of concentrated HClO4 and then standardized with NaOH. A carbonate-free NaOH (0.1 M) solution was standardized by titration with potassium hydrogen phthalate (POCH). The ionic strength (I) was adjusted to 0.1 M by addition of NaClO4 (Sigma-Aldrich).
Potentiometric Measurements
The potentiometric titrations were performed at a constant temperature of 298 K in an argon atmosphere as the inert gas, using the automatic titrator system Titrando 905 (Methrom), connected to a combined glass electrode (Mettler Toledo InLab Semi-Micro). The electrode was calibrated daily in terms of hydrogen ion concentration by using a HCl solution. The pH-metric titrations were performed in a water solution of 4 mM HClO4 at a 0.1 M NaClO4 ionic strength. Starting volumes of ∼1 mM ligand solutions were 3 mL, and the exact concentration of the ligand was determined using Gran’s method.27 Metal:ligand molar ratios were 1:1.1. The potentiometric data, collected over a pH range of 2.0–11.0, were refined with the HYPERQUAD 200628 and SUPERQUAD29 programs, which use nonlinear least-squares methods. The species distribution diagrams were generated using a computer HYSS program.30
Ultraviolet–Visible (UV–vis) and Circular Dichroism (CD) Spectroscopies
The absorption spectra in the UV–vis region were recorded at 298 K on a Cary300 Bio (Varian) spectrophotometer in 1 cm path length quartz cells. CD spectra were recorded on a Jasco J 715 spectropolarimeter over the range of 220–800 nm using different path lengths (1 and 0.1 cm). Sample solutions for UV–vis and CD spectroscopic studies had compositions similar to those employed in the potentiometric experiments. All measurements were recorded in the pH range of 2.5–11.5. The pH was adjusted with appropriate amounts of HCl and NaOH solutions. Absorptivities (ε, M–1 cm–1) were calculated at the pH value of the maximum concentration of the considered species, as indicated by the potentiometric distribution diagrams. All of the used solutions in this study were deaerated. CD spectra were analyzed using Jasco Spectra Analysis Software (version 1.53.04). Data were processed using Origin 7.0.
Electron Paramagnetic Resonance (EPR) Spectroscopy
EPR spectra were recorded using a Bruker ELEXSYS E500 CW-EPR spectrometer equipped with an NMR teslameter (ER 036TM) and a frequency counter (E 41 FC) at X-band frequency, at 77 K and room temperature. The peptide concentration was 1 mM, and the metal:ligand molar ratio 1:1.1. The solution for EPR experiments was prepared using ethylene glycol (5%) as a cryoprotectant. EPR parameters were obtained by using the Bruker WinEPR SimFonia program and Doublet new (EPR OF S = 1/2) program by A. Ozarowski (National High Field Magnetic Laboratory, University of Florida, Gainesville, FL).
ESI-MS Spectrometry
Electrospray ionization mass spectrometry (ESI-MS) data were recorded on a Bruker Q-FTMS spectrometer. The following instrumental parameters were used: scan range, m/z 200–1500; dry gas, nitrogen; temperature, 170 °C; capillary voltage, 4500 V; ion energy, 5 eV. The capillary voltage was optimized to the highest signal:noise ratio. Spectra were recorded in the positive and negative ion modes. Samples (with 1:1.1 metal:ligand stoichiometry and 10–5 M ligand) were prepared by using a MeOH/H2O mixture (50:50, v/v) as the solvent, at pH 7.4 and 9.0. The instrument was calibrated using the Tunemix mixture (BrukerDaltonik) in the quadratic regression mode. The overall charge of the analyzed ions was calculated on the basis of the distance between isotopic peaks. Data were processed by application of the Compass Data Analysis 4.0 (Bruker Daltonik) program.
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR measurements were performed on a Bruker Ascend 400 MHz spectrometer equipped with a 5 mm automated tuning and matching broad band probe (BBFO) with z-gradients, as previously described.31,32
NMR experiments were carried out using a 2 mM concentration of the ligands in a 90:10 (v/v) H2O/D2O mixture at 298 K in 5 mm NMR tubes. Two-dimensional (2D) 1H–13C heteronuclear correlation spectra (HSQC) were recorded using a phase-sensitive sequence employing Echo-Antiecho-TPPI gradient selection with a heteronuclear coupling constant JXH of 145 Hz and shaped pulses for all 180° pulses on the f2 channel with decoupling during acquisition. Sensitivity improvement and gradients in back-inept were also used. Relaxation delays of 2 s and 90° pulses of ∼10 μs were employed in all of the experiments. Solvent suppression for one-dimensional (1D) 1H and 2D 1H–1H TOCSY experiments was carried out by using excitation sculpting with gradients. The spin-lock mixing time of TOCSY experiments was obtained with MLEV17. 1H–1H TOCSY spectra were recorded using mixing times of 60 ms. A combination of 1D and 2D TOCSY, HSQC experiments was used to assign the signals of both free and metal-bound ligands at different pH values. All NMR data were processed with TopSpin (Bruker Instruments) software and analyzed by Sparky 3.11 and MestRe Nova 6.0.2 (Mestrelab Research S.L.) programs.
Molecular Model Calculations
Molecular mechanics geometry optimization was performed within HyperChem 8.0.3 (Hypercube, Inc., Gainesville, FL) as previously described for similar systems.25,33 The optimization procedure of the structural features of 4N Cu(II)-{D1 Nterm, E2 N–amide, H3 Nim, N–amide} (for DP1) and 3N Cu(II)-{T1 Nterm, H2 Nim, N–amide} (for DP2) systems has been modeled on the basis of the geometry of the X-ray crystal structures of aqua-((aspartic acid)-alanyl-histidinyl-lysinyl)-copper(II) chloride hexahydrate (The Cambridge Crystallographic Data Centre, CSD entry OZEBOL) that were used as the template,34 whereas for Mn(II), the 3NO {T1 Nterm, D5 O–carb, E6 O–carb, H7 Nim, G10 O–term-carb, nH2O} (for Mid-DEH) coordination mode has been based on the structure of MNTR mutant E11K complexed with Mn2+ in octahedral geometry (Protein Data Bank entry 4HX4).35 Spatial positions and distances between the metal ion and its donor (N and/or O) atoms and dihedral angles were constrained to the values found for the selected crystal structures. Additional constraints based on experimental evidence were also imposed in the calculations. In particular, in Cu(II)-albumin-like systems, cross-correlation ROEs and consequently H–H spatial proximity were evaluated using diamagnetic Ni(II) to probe paramagnetic Cu(II).26,36 The geometry of the rest of the peptides was optimized without any constraints allowing all of the atoms, bonds, and dihedral angles to change simultaneously to reach the lowest overall energy. Models were generated by using Chimera.37
Results and Discussion
Cu(II) and Mn(II) complexes with DEHGTAVMLK (DP1), THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH) have been investigated by means of a combination of potentiometric, spectroscopic (NMR, UV–vis, EPR, and CD), and spectrometric (ESI-MS) techniques. Preliminary NMR and EPR results of the Mn(II) coordination ability of DP1 and DP2 peptides, at physiological pH, have already been reported.31 In this paper, the study has been performed and completed over a wide pH range, including also the coordination properties of Cu(II) ions.
The stoichiometry and the stability constants of the observed species together with the pH-dependent deprotonation steps for both free and metal-bound peptides have been characterized by potentiometric titrations. Spectroscopic data have supported and confirmed the potentiometric results and allowed us, moreover, to identify the binding donor atoms involved in the coordination process.
Because of the paramagnetic character of Cu(II) and Mn(II), the NMR studies have been performed with the progressive addition of substoichiometric amounts of metal ions to the peptide solutions to avoid large broadening of the signals. This method allows us to localize the metal binding sites along the amino acid sequence depending on the selective relaxation effect experienced by the nuclei that are closer to the paramagnetic metal center.32,38
In the pH range investigated, a total of five protonation steps have been determined by potentiometric titrations for all of the peptides, except Bis-DEH (for which seven protonations were observed), as reported in Table 1; the representative distribution diagrams as a function of pH are given in Figure S1.
Table 1. Protonation Constants (log β) and pK Values of the Peptides at 298 K and a 0.1 M NaClO4 Ionic Strengtha.
| peptide | species | log β | log K | |
|---|---|---|---|---|
| DP1 | [HL]− | 9.62(3) | Lys | |
| [H2L] | 17.47(5) | 7.85 | Nterm | |
| [H3L]+ | 23.94(5) | 6.47 | His | |
| [H4L]2+ | 28.14(7) | 4.20 | Glu | |
| [H5L]3+ | 31.20(6) | 3.06 | Asp | |
| DP2 | [HL]− | 9.86(1) | Lys | |
| [H2L] | 16.87(2) | 7.01 | Nterm | |
| [H3L]+ | 22.78(2) | 5.91 | His | |
| [H4L]2+ | 27.48(2) | 4.70 | Glu | |
| [H5L]3+ | 31.37(2) | 3.89 | Asp | |
| Term-DEH | [HL]− | 10.82(1) | Lys | |
| [H2L] | 18.94(2) | 8.16 | Nterm | |
| [H3L]+ | 25.96(2) | 7.02 | His | |
| [H4L]2+ | 30.27(2) | 4.31 | Glu | |
| [H5L]3+ | 33.73(2) | 3.46 | Asp | |
| Mid-DEH | [HL]− | 10.52(1) | Lys | |
| [H2L] | 17.85(2) | 7.33 | Nterm | |
| [H3L]+ | 24.14(2) | 6.29 | His | |
| [H4L]2+ | 28.20(2) | 4.06 | Glu | |
| [H5L]3+ | 31.75(2) | 3.55 | Asp | |
| Bis-DEH | [HL]3– | 8.41(1) | 8.41 | Nterm |
| [H2L]2– | 15.82(1) | 7.41 | His | |
| [H3L]− | 22.49(1) | 6.67 | His | |
| [H4L] | 27.18(2) | 4.69 | Glu | |
| [H5L]+ | 31.23(2) | 4.05 | Glu | |
| [H6L]2+ | 34.59(2) | 3.36 | Asp | |
| [H7L]3+ | 37.26(2) | 2.67 | Asp |
Standard deviations (3σ values) are given in parentheses.
For all of the peptides, the lowest pK values refer to the carboxylic group of Asp (pK values of 2.67–3.89) and Glu (pK values of 4.05–4.70) residues. According to the amino acid sequence, double pK values are found for Asp (pK values of 2.67 and 3.36) and Glu (pK values of 4.05 and 4.69) residues in Bis-DEH. Since the deprotonation steps overlap, the first and second protonation constants have not been assigned to the specific aspartic or glutamic acid residues. Nevertheless, comparing the pK values of the Bis-DEH peptide to the corresponding values of the DP1 and Term-DEH analogues, with which it shares the first and the last three residues (DEH), respectively, we can assign the deprotonations in the following order: Asp-1, Asp-8, Glu-2, and Glu-9.
For all the five sequences analyzed, the next deprotonation step involves an imidazole nitrogen atom on the His (pK values of 5.91–7.41) residue. Also in this case, the comparison of pK values for the two His residues of Bis-DEH (pK values 6.67 and 7.41) with those of the aforementioned DP1 and Term-DEH peptides suggests that the order of deprotonation could most probably be His-3 and His-10.
Their deprotonation constants, slightly higher with respect to those of the other two peptides, can be attributed to the different charge of Bis-DEH. In fact, in this case, the two consecutive His protons are released from the positive (LH4+) and neutral (LH3) species, while for DP1 and Term-DEH, the corresponding species is doubly positively charged (LH42+).
The next deprotonation involves the terminal amino group (Nterm) (pK values of 7.33–8.41), which is less basic than the ε-amino group of the Lys (pK values of 9.62–10.82) residue due to the former’s proximity to the electron-withdrawing carbonyl group of the first peptidic bond. The deprotonation of the ε-amino group of Lys (not present in Bis-DEH) occurs, in fact, at higher pH, in agreement with what has been reported in the literature for peptides containing this amino acid.32,39,40
A complete 1H and 13C resonance assignment for all the five peptides has been made by a combination of 1D 1H and 2D 1H–1H TOCSY, 1H–1H ROESY, and heteronuclear 1H–13C HSQC experiments (Table S1). None of the peptides adopt a folded structure, because only short-range (aa–aa+1) and not medium- nor long-range H–H cross-correlation ROE signals were detected. Nevertheless, the range of the parts per million chemical shift related to peptide backbone N–H protons is, in all cases, larger than that assigned to a completely unfolded peptide, suggesting a partial structural arrangement of the peptides in solution.41
Copper Complexes
Electrospray ionization mass spectrometry (ESI-MS) showed the metal binding stoichiometry. All of the peptides form Cu(II) mononuclear complexes with different degrees of protonation. The complete set of complex formation constants is reported in Table 2. For all Cu(II)–peptide complexes studied here, with a free Nterm group and a His residue on the peptide chain, one can find that both Nterm and Nim atoms may play the anchoring site role in the binding of Cu(II) ions, together or separately. It should be mentioned that the formation of the coordination isomers in equilibrium in solution cannot be excluded.42,43
Table 2. Protonation Constants (log β) and pK Values of the Cu(II)–Peptide Systems at 298 K and a 0.1 M NaClO4 Ionic Strengtha.
| peptide | species | log β | log K | |
|---|---|---|---|---|
| DP1 | [CuH2L]2+ | 22.83(6) | ||
| [CuHL]+ | 18.66(4) | 4.17 | Glu | |
| [CuL] | 14.19(2) | 4.47 | N–amide | |
| [CuLH–1]− | 9.11(2) | 5.08 | N–amide | |
| [CuLH–2]2– | 0.11(4) | 9.00 | Lys | |
| [CuLH–3]3– | –10.05(4) | 10.16 | Owater | |
| DP2 | [CuHL]+ | 20.81(2) | ||
| [CuL] | 15.90(4) | 4.91 | Glu | |
| [CuLH–1]− | 8.17(4) | 7.73 | N−amide | |
| [CuLH–2]2– | –1.26(4) | 9.43 | N−amide | |
| [CuLH–3]3– | –11.13(5) | 9.87 | Lys | |
| Term-DEH | [CuH2L]2+ | 24.17(3) | ||
| [CuHL]+ | 18.83(2) | 5.34 | Nterm | |
| [CuL] | 12.46(2) | 6.37 | N−amide | |
| [CuLH–1]− | 3.62(4) | 8.84 | N−amide | |
| [CuLH–2]2– | –5.22(3) | 8.84 | N−amide | |
| [CuLH–3]3– | –16.27(4) | 11.05 | Lys | |
| Mid-DEH | [CuH2L]2+ | 22.16(3) | ||
| [CuHL]+ | 17.57(1) | 4.59 | Nterm | |
| [CuL] | 11.86(1) | 5.71 | N−amide | |
| [CuLH–1]− | 3.09(2) | 8.77 | N−amide | |
| [CuLH–2]2– | –6.38(2) | 9.47 | N−amide | |
| [CuLH–3]3– | –17.58(2) | 11.20 | Lys | |
| 1:1 Bis-DEH | [CuH2L]2+ | 22.53(1) | ||
| [CuL] | 12.87(1) | 9.66 | Nterm, 2 × N−amide | |
| [CuLH–1]− | 7.28(2) | 5.59 | ||
| [CuLH–2]2– | –0.45(4) | 7.73 | His | |
| 2:1 Bis-DEH | [CuH2L] | 22.53(1) | ||
| [CuL]2– | 12.87(1) | 9.66 | Nterm, 2 × N−amide | |
| [CuLH–1]3– | 7.28(2) | 5.59 | ||
| [Cu2LH–2]2– | 4.34(9) | |||
| [Cu2LH–3]3– | –2.83(5) | 7.17 | N−amide | |
| [Cu2LH–4]4– | –11.14(7) | 8.31 | N−amide | |
| [Cu2LH–5]5– | –22.35(7) | 11.21 | Owater |
Standard deviations (3σ values) are given in parentheses.
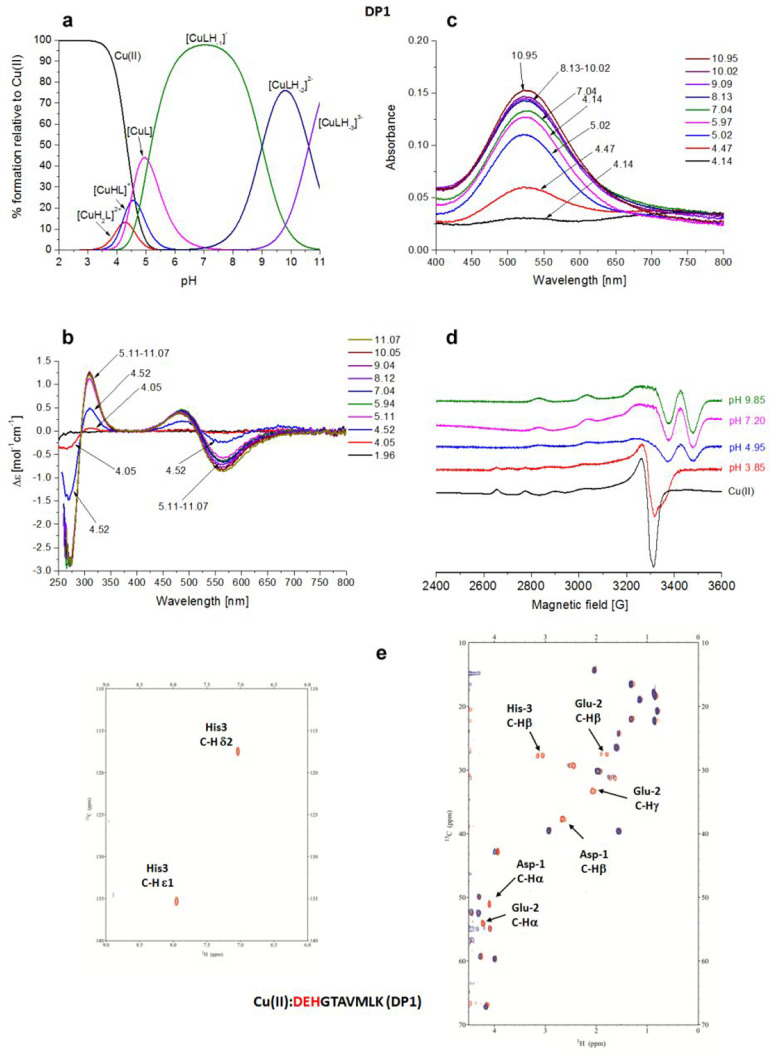
Cu(II)–DEHGTAVMLK (DP1) System
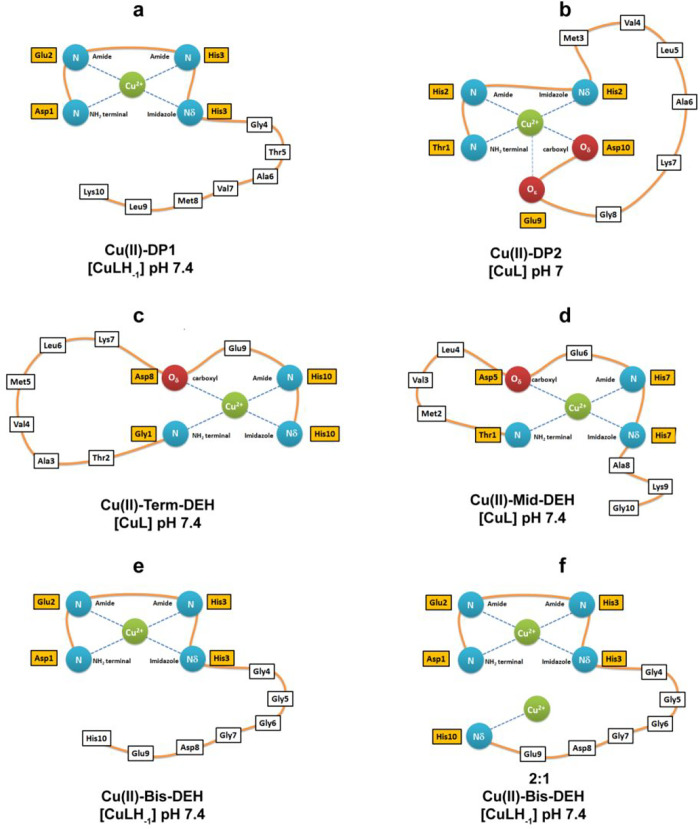
The distribution diagram in Figure 1a shows that [CuH2L]2+ is the first Cu(II) complex species formed by DP1, with the involvement of Nim of His-3 and Nterm on the first Asp-1 residue. The formation of [CuHL]+ occurs by the deprotonation of glutamic acid, which is a nonbinding residue (the pK value of 4.17 is almost the same as in the free ligand). The spectroscopic characteristic of these complexes is not accessible due to the low abundance of these species (12% and 25%, respectively). The next predominant forms are [CuL] and [CuLH–1]− in which the deprotonation process and consequent coordination involve two successive amide nitrogens (pK values of 4.47 and 5.08, respectively) relative to His-3 and Glu-2 residues. The latter represents the main species from pH 5 to 9. The 4N complex {Nterm, Nim, 2N–amide}, with the simultaneous formation of three fused chelate rings and the saturation of the coordination plane, is the most stable one formed among the Cu(II)–peptide complexes (log K* = −14.83).24 The next species, under basic conditions, [CuLH–2]2–, is almost certainly formed by the deprotonation of Lys-10 (pK of 9.0 in comparison to pK of 9.62 for the free ligand), whereas the last pK corresponds to the deprotonation of a water molecule, which is likely interacting with the complex.
Figure 1.
Cu(II)–DEHGTAVMLK (DP1) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 1:1.1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 1:1.1 Cu(II):L molar ratio, I = 0.1 M NaClO4). (e) Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free DP1 (red) and the Cu(II)–DP1 system at a 0.02:1 molar ratio (blue), and pH 4.3. Disappearing peaks are labeled.
All of the spectroscopic methods strongly support the formation of only one type of 4N complex. The bands in the CD spectra (Figure 1b) clearly indicate the formation of an albumin-like binding mode {Nterm, Nim, 2N–amide}, starting at pH ∼4.5, with a typical negative Δε value around 560 nm and a positive Δε value of 485 nm for the copper d–d electronic transitions44−46 (Table S2). In addition, a charge transfer band at 310 nm can be ascribed to the coordination of deprotonated amido nitrogens to Cu(II), 2N–amide → Cu(II) LMCT (ligand-to-metal charge transfer), while an additional negative band present at 272 nm is due to an Nterm → Cu(II) LMCT.47−49 UV–vis spectra (Figure 1c) confirm the data obtained from CD and potentiometric titrations as indicated by the typical single band centered at 525 nm, whose λmax value is close to the computed value of 530 nm as determined by the empirical formula related to the coordination of 4N atoms {Nterm, Nim, 2N–amide} to Cu(II).44 This band, due to formation of the 4N complex, is clearly present from pH 4.47 to the end of UV–vis measurements. The parameters obtained from EPR spectra of the Cu(II)–DP1 system (A|| = 208.7 G, and g|| = 2.18) provide evidence for a 4N coordination sphere around the Cu(II) ion (Figure 1d and Table S2). The DP1–Cu(II) system has been scrutinized also via NMR techniques to determine the specific anchoring site along the sequence. The titration of DP1 with an increasing amount of Cu(II), at metal:peptide molar ratios from 0:1 to 0.02:1, has been monitored at pH 7 where the main species CuLH–1 is present (Figure S1). The results, i.e., disappearance of selected signals in 1D 1H and 2D 1H–13C HSQC and 1H–1H TOCSY spectra, confirmed the involvement of the first three residues, Asp-1, Glu-2, and His-3, in metal ion coordination, whereas the resonances of all of the other residues were unaffected. The more intense decrease of the intensity of histidine signal Hε1, with respect to Hδ2, indicates that Cu(II) is bound to Nδ1, in agreement with the formation of a highly thermodynamically stable six-membered {Nim, N–} chelate ring, instead of larger and less stable ones that could result with the involvement of Nε1. This coordination mode was confirmed from pH 4.3 (Figure 1e) until a basic pH was reached.
In the ESI-MS spectra, the most intense signals come from the free ligand variously protonated (m/z 548.75 and 1098.48, corresponding to [L-2H+]2– and [L-3H+]−, respectively) and its Cu(II) complexes (m/z 579.21 and 1159.39, corresponding to [CuL-4H+]2– and [CuL-3H+]−, respectively), in a 1:1 Cu(II):L molar ratio (Figure S3).
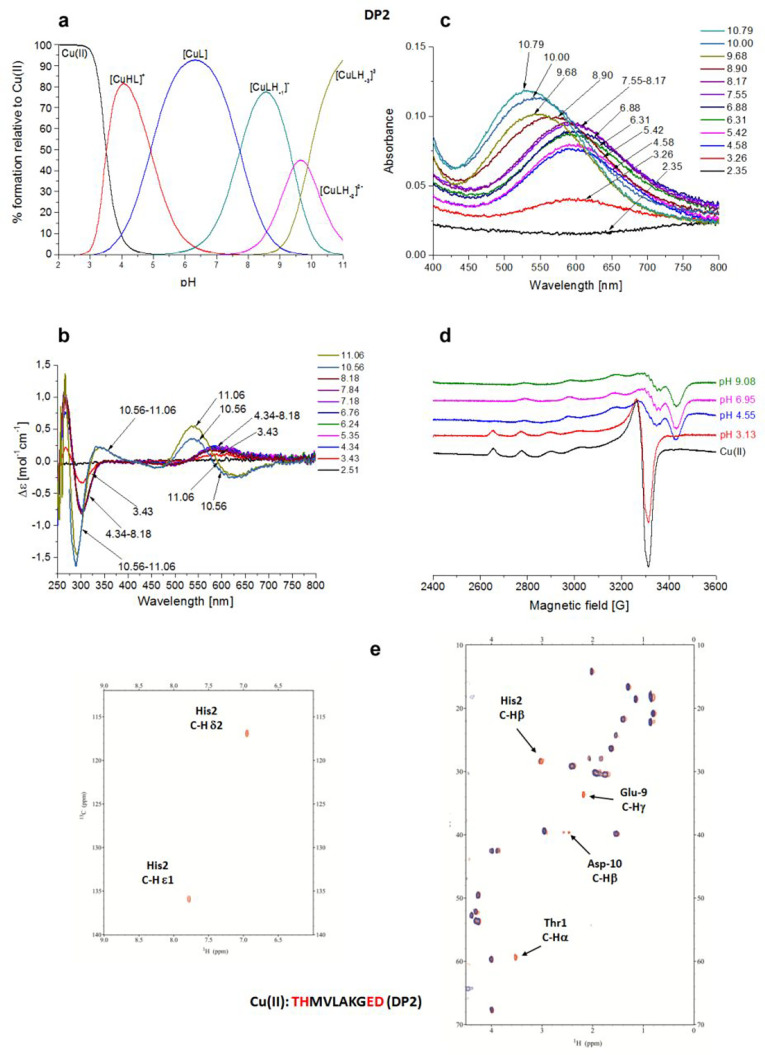
Cu(II)–THMVLAKGED (DP2) System
The DP2 peptide is a scrambled version of DP1, with the coordinating amino acids Glu-9 and Asp-10 located far from His-2. Potentiometric titrations (Table 2 and Figure 2a) show that the first detected species is [CuHL]+, which dominates the solution from pH 3 to 5.5, reaching almost 90% of the total Cu(II) bound at pH 4.2.
Figure 2.
Cu(II)–THMVLAKGED (DP2) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 1:1.1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 1:1.1 Cu(II):L molar ratio, I = 0.1 M NaClO4). (e) Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free DP2 (red) and the Cu(II)–DP2 system at a 0.02:1 molar ratio (blue) and pH 7. Disappearing peaks are labeled.
In this species, three nitrogens, {Nterm, Nim, N–amide}, are involved in Cu(II) coordination, and this is in agreement with the finding that when a His residue is in the second position with respect to the free Nterm in the peptide chain, the simultaneous involvement of the indicated nitrogen donors in Cu(II) ion binding is observed.43 In the next species, the deprotonation of Glu results in the formation of [CuL], which is the major species at physiological pH, existing from pH 3.2 to 10. The very high stability of this complex (log K* = −6.88) results from the formation of a pair of fused five- and six-membered chelate rings.24 With a pK of 7.73, one can observe the deprotonation of N−amide, the [CuLH–1]− is still of the 3N type, but the coordination mode changes to {Nterm, 2N–amide}, which is in agreement with CD spectra, where starting from pH 8.18 a blue shift of the N–amide → Cu(II) MLCT band from 300 to 290 nm is observed, indicating the involvement of the second N–amide donor, instead of Nim. The deprotonation of the third amide group results in the formation of a 4N [CuLH–2]2– species. The last species, [CuLH–3]3–, is related to Lys residue deprotonation.
UV–vis and CD spectra (Figure 2b,c and Table S2), in agreement with potentiometric data, showed the formation of the 3N complex from pH ∼3.2, indicated by the band at 595 nm matching exactly with the estimated value for a 3N {Nterm, Nim, N–amide} donor set.44 Up to pH 8.2, the absorbance of this complex is only growing. The formation of a 4N {Nterm, 3N–amide} complex is observed above pH 9.7 (UV–vis band at 530 nm).50 The slight difference from the estimated λmax for this species (515 nm) can be due to the presence of an axial binding group. At pH >10, the CD spectra, in accordance with the other techniques employed in the analysis, exhibit an oppositely signed curve for the d–d transition with the negative band centered at 620 nm and the positive one at 537 nm. The EPR experiments (Figure 2d and Table S2) resulted in the spectra characterized by parameters corresponding to both 3N and 4N complexes (A|| = 190.0 G, and g|| = 2.25). The formation of a 3N complex at physiological pH is confirmed by NMR titration experiments (Figure S2). At pH 7, the species under investigation should be [CuL] in which the coordination sphere of Cu(II) involves the set {T1 Nterm, H2 Nim, H2 N–amide}. In agreement with this hypothesis, a selective disappearance of the signals is confined to only these two residues (Figure 2e).
In particular for Thr-1, the selective disappearance in the HSQC spectrum of the cross-correlation peak corresponding to Cα–Hα (attached to Nterm) and not Cβ–Hβ (same behavior for Hα–Hα and Hβ–Hβ in the TOCSY spectrum) is undoubtedly a clear indication of the paramagnetic Cu(II) ion bound to Nterm. Other proofs about of the formation of 3N species comes from the disappearance of the aromatic signals for Hδ2 and Hε1 together with the Hβ protons of the His-2 residue, confirming the binding of its Nim. Moreover, from NMR analysis, a secondary interaction of the Cu(II) ion with the carboxylate moiety of Glu-9 and Asp-10 is detected, from pH 4.25 to basic values, as evidenced by the selective disappearance from the HSQC spectrum of their side chain C–H correlation peaks (γ and δ, respectively). With an increase in pH to 10, the change of the Cu(II) coordination sphere to {Nterm, 3N–amide} can also be detected from NMR spectra. The change is assisted by a severe broadening and disappearance of the signals on the nuclei located in the plane of the complex, i.e., Thr-1, His-2, Met-3, and Val-4 residues, that are directly affected by the paramagnetic Cu(II) ion bound to their deprotonated amide nitrogens (Figure S4). The histidine aromatic protons Hδ2 and Hε1 are also still absent, because the formation of the square planar complex forces the imidazole ring to stay on the coordination plane.
In the ESI-MS spectra, the most intense signals are due to the variously protonated free ligand (m/z 1098.49, corresponding to [L-3H+]−) and its Cu(II) complex (m/z 579.21 and 1159.39, corresponding to [CuL-4H+]2– and [CuL-3H+]−, respectively), in a 1:1 Cu(II):L molar ratio (Figure S5).
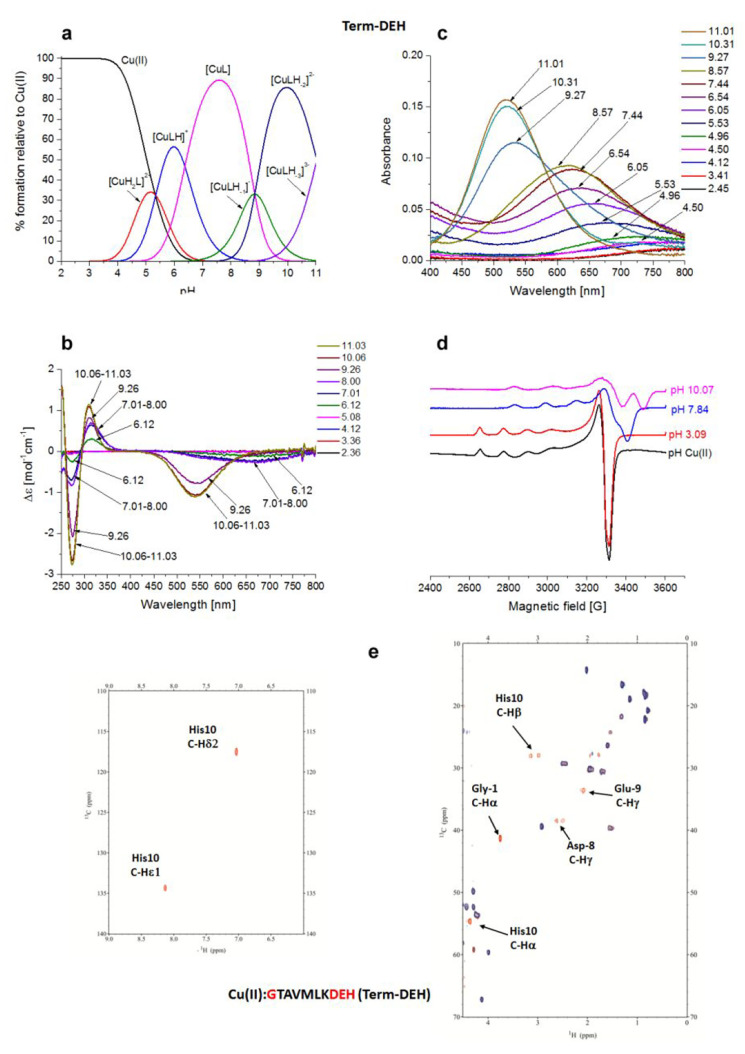
Cu(II)–GTAVMLKDEH (Term-DEH) System
Term-DEH differs from DP1 in the location of the amino acid portion -DEH-, which is now present at the C-terminal part of the sequence (relative to the N-terminus in DP1).
The first species identified by potentiometric titrations (Figure 3a), starting form pH 3.5, is [CuH2L]2+ in which His-10 anchored the metal binding site with its imidazole nitrogen.
Figure 3.
Cu(II)–GTAVMLKDEH (Term-DEH) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 1:1.1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 1:1.1 Cu(II):L molar ratio, I = 0.1 M NaClO4). (e) Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free Term-DEH (red) and the Cu(II)–Term-DEH complex at a 0.02:1 molar ratio (blue) and pH 7.4. Disappearing peaks are labeled.
For 1N complexes, the anchoring of Cu(II) at the C-terminal His residue is preferred over N-terminal binding.51 The EPR parameters (A|| = 140.0 G, and g|| = 2.32) are in good agreement with 1N coordination. However, the λ value of 710 nm (Figure 3d and Table S2) for the maximum absorption of this species (pH ∼5) seems to be more in agreement with {Nim, COO–} coordination (estimated λ = 731 nm), whereas the expected value for a Cu(II) complex with only a {1Nim} configuration is 760 nm (see below).44
The blue shift observed in the UV–vis spectra above pH 4.96, as well as the increase in the A|| parameter (160.0 G), together with the decrease in the g|| value (2.29) suggests the formation of a complex with an additional nitrogen atom bound. The negative band present at 270 nm is in the range of an Nterm → Cu(II) LMCT (260–280 nm).52,53
It corresponds to the formation of the next species, [CuHL]+, where the Nterm belonging to the Gly-1 residue takes part in the metal coordination with the formation of a macrochelate system involving the Cu(II) ion, Nim of His-10 and Nterm {Nterm, Nim,}. At physiological pH, the main species is [CuL], which starts appearing from pH 5, with its predominant presence between pH 6.5 and 9. Here the pK value suggests the deprotonation of an amide hydrogen (pK of 6.37) with a 3N {Nterm, Nim, N–amide} coordination mode. Changes in the EPR parameters (A|| = 169.0 G, and g|| = 2.23) together with the presence of a band with maximum absorption at 625 nm in the UV–vis spectra are evidence for three nitrogen atoms in the Cu(II) coordination sphere. Via comparison of the λ = 625 nm found for this species in UV–vis spectra to the estimated value (λ = 595 nm), one can suppose the participation of a COO– group in complex formation. The [CuL] complex shows a LMCT band at 320 nm due to the presence of {Nim, N–amide} in the metal ion coordination sphere.
With an increase in pH, two more amide deprotonations occur with the formation of a 3N [CuLH–1]− species with the probable coordination mode {Nim, 2N–amide} or {Nterm, 2N–amide}, and the most intense 4N [CuLH–2]2– species, for which the proposed binding mode is {Nterm, 3N–amide} or {Nim, 3N–amide}.43 The [CuLH–1]− complex is still 3N, but the coordination mode changes to {Nterm/Nim, 2N–amide}; this is in agreement with the CD spectra, where starting from pH 8.18 we observed a blue shift of the N–amide → Cu(II) LMCT band from 320 to 310 nm. Most likely, the equilibrium between two forms is reached: the first, in which the Nterm and the adjacent amides are bound to Cu(II), and the second, with the Nim and the amides involved in the binding. The EPR parameters (A|| = 204.0 G, and g|| = 2.185), as well as the estimated value οf λ = 521 nm, matching perfectly with that found at 520 nm for the absorbance of the 4N species, strongly support the proposed coordination modes. The last species, [CuLH–3]3–, comes from the deprotonation of a noncoordinating Lys residue. The NMR titration of Term-DEH, by increasing the substoichiometric Cu(II):ligand molar ratio, was performed at physiological pH 7.4, where the maximum level of formation of the main species [CuL] occurs. The 1D 1H spectra are shown in Figure S2, while a snapshot of the comparison of 2D 1H–13C HSQC NMR spectra of the free and Cu(II)-bound peptide is given in Figure 3e.
NMR studies clearly indicate the involvement, at this pH, of His-10, precisely through its Nim and N–amide, and Gly-1 N-terminus, as evidenced by the selective disappearances of aromatic (imidazolic Hε1 and Hδ2) and aliphatic signals (Hα and Hβ) of His-10 and the aliphatic resonances (Hα) of Gly-1. Moreover, a decrease in the intensity of the signals of Asp-8 and Glu-9 was also clearly detected. When considering a more acidic pH of 5.12, the NMR spectra indicated that Gly-1 is not taking part in the coordination of Cu(II) because its signals are unaffected by the paramagnetic metal. Only Nim of His-10 is playing a main role in the coordination sphere of the metal within the [CuH2L]2+ species, being in agreement with spectroscopic parameters. Also, at this pH, the signals of Asp-8 are experiencing paramagnetic relaxation enhancement. The acquisition performed at pH 6.30, to collect more details for the main species [CuLH–1]−, also revealed the involvement of Gly-1 through its Nterm, in addition to Nim and O–carb, as in the case of the [CuL] species already discussed. Under more basic conditions, pH 9, the system is compatible with a {Nterm, Nim, 2N–amide, O–carb} coordination sphere around Cu(II), involving Gly-1 (Nfigterm), His-10 (Nim, N–amide), and Glu-9 (N–amide) in a plane, and probably Asp-8 (O–carb) in apical coordination. The increase in pH to 10 in NMR spectra is in agreement with a switch of coordination to the {Nterm, 3N–amide} type.43
Via ESI-MS spectrometry, the prevailing signals correspond to the differently protonated free ligand (m/z 548.75 and 1098.48, corresponding to [L-2H+]2– and [L-3H+]−, respectively) and Cu(II) complexes (m/z 579.21 and 1159.39, corresponding to [CuL-4H+]2– and [CuL-3H+]−, respectively) in a 1:1 Cu(II):L molar ratio (Figure S6).
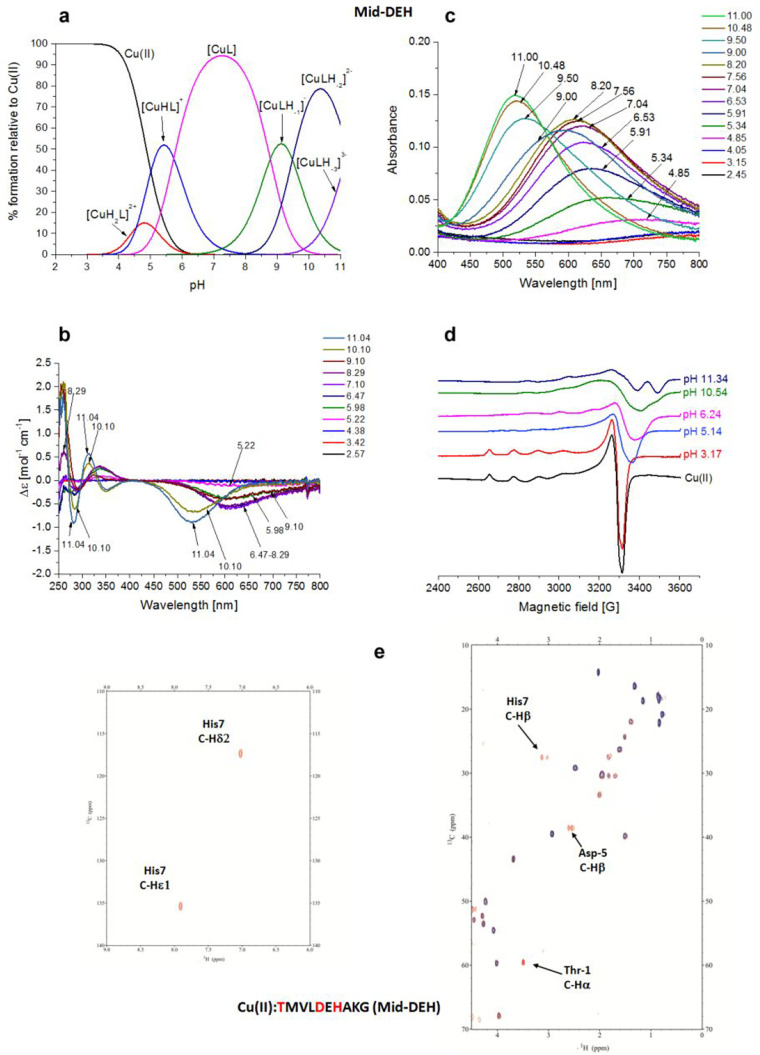
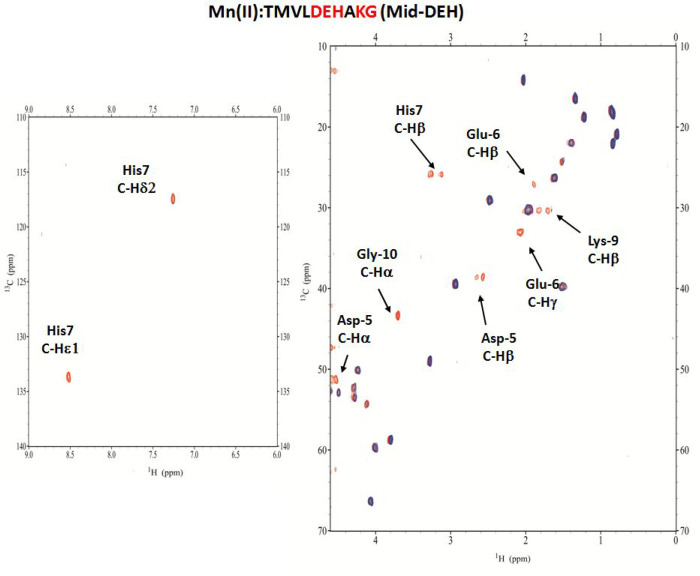
Cu(II)–TMVLDEHAKG (Mid-DEH) System
The Mid-DEH peptide contains the -D5E6H7- residues in the middle of the primary sequence. As for the Term-DEH peptide, for Mid-DEH the first species identified by potentiometric titrations (Figure 4a) is [CuH2L]2+, which starts to appear from pH 3.5, with His-7 playing the key role in anchoring the metal ion through its imidazole nitrogen.
Figure 4.
Cu(II)–TMVLDEHAKG (Mid-DEH) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 1:1.1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 1:1.1 Cu(II):L molar ratio, I = 0.1 M NaClO4). (e) Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free Mid-DEH (red) and the Cu(II)–Mid-DEH system at a 0.02:1 molar ratio (blue) and pH 7. Disappearing peaks and those with major broadening are labeled.
The presence of a band with a maximum absorption at 715 nm in the UV–vis spectra suggests a single nitrogen atom in the metal ion coordination sphere. This finding is additionally supported by the EPR parameters (A|| = 151.9 G, and g|| = 2.323), as well as the presence of characteristic CD bands at 235 and 340 nm [Nim → Cu(II) MLCT].54 The UV–vis band with a λmax at 715 nm, like in the case of the Cu(II)–Term-DEH system, supports the participation of also oxygen donor(s) from carboxylate group(s) in metal binding. In the next species, [CuHL]+, the engagement of Nterm on Thr-1 occurs, with the formation of macrochelate loops with the {Nterm, Nim} coordination mode.51 The spectroscopic values (Table S2), similar to those of Term-DEH, indicated the involvement of a COO– group in complex formation. The NMR spectra recorded at pH 4.9 and 5.5 give, in fact, evidence of the disappearance of signals related to His-7, Asp-5, and Gly-10. The involvement of Nterm of Thr-1 in the binding is noticeable in NMR spectra recorder at pH 7.2. Here the major species identified is [CuL], and as in the previous similar Cu(II)–Term-DEH system, it should be a 3NO form with the {Nterm, Nim, N–amide, O–carb} coordination mode (Figure S2 and Figure 4e).
The involvement of N–amide in the binding of Cu(II) ions is confirmed by the appearance of a new CT transition in CD spectra at 285 nm [N–amide → Cu(II)].55 The EPR parameters suggest the formation of a 3N species (A|| = 169 G, and g|| = 2.23).
In particular, NMR studies identified the nitrogen donors as Nterm of Thr-1, Nim and N–amide of His-7, and Ocarb from a carboxylate moiety of Asp-5. The slight decrease in the intensity of the signals related to Glu-6 was also found. The following species are [CuLH–1]− and [CuLH–2]2– that are derived the deprotonation of two more nitrogen amides, giving a 3N and 4N species with the coordination modes {Nim, 2N–amide} and {Nim, 3N–amide}, respectively, with the latter dominating the basic region up to pH 9.5. The blue shift of the d–d transition energy to 520 nm as well as the EPR parameters (A|| = 204 G, and g|| = 2.185) is in agreement with a 4N coordination mode above pH 9. The HSQC spectra recorded at pH 9 showed the reappearance of the correlation signals related to α nuclei of Thr-1 and β nuclei of Asp-5, whereas the disappearance of the α of Asp-5 and α, β, δ, and ε1 of His-7 are mostly in agreement with the formation of a square planar complex around Cu(II) and the involvement of the -D5E6H7- motif in a {Nim, 3N–amide} coordination mode.43 The last species identified is [CuLH–3]3– in which the deprotonation of a nonbinding Lys (pK = 11.20) residue occurs, as suggested from the spectroscopic parameters that remained unchanged with respect to the previous species. ESI-MS spectrometry shows a peak corresponding to a differently protonated free ligand (m/z 548.75 and 1098.48, corresponding to [L-2H+]2– and [L-3H+]−, respectively) and 1:1 Cu(II) complexes (m/z 579.21 and 1159.39, corresponding to [CuL-4H+]2– and [CuL-3H+]−, respectively) (Figure S7).
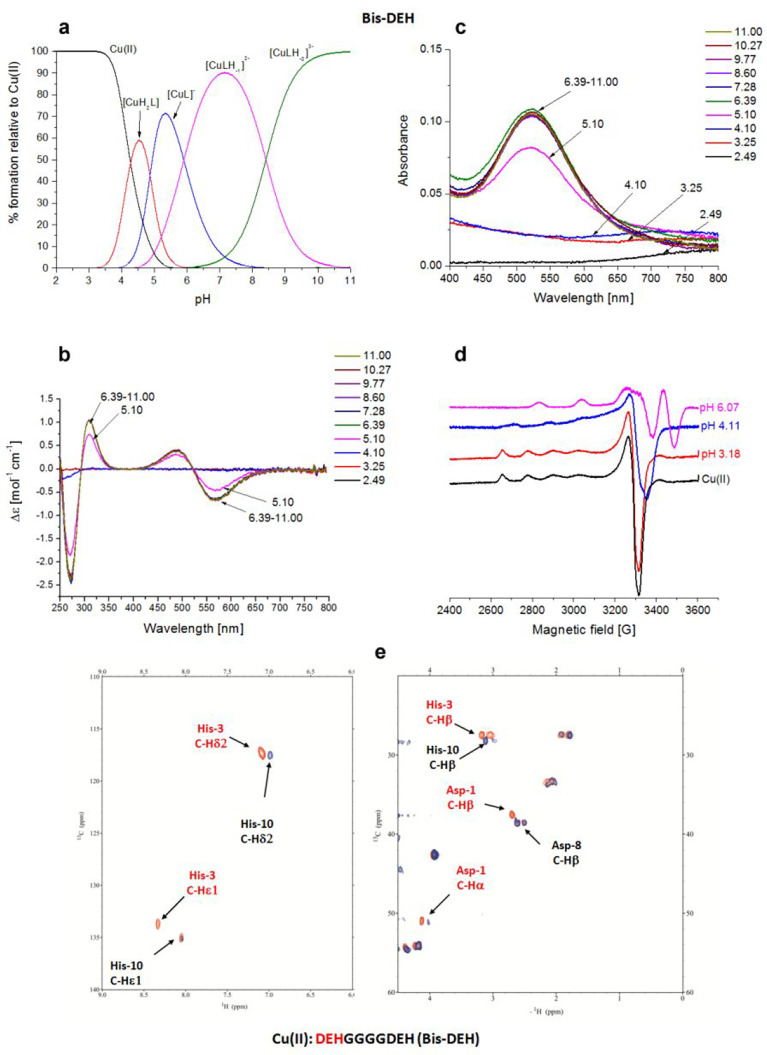
Cu(II)–DEHGGGGDEH (Bis-DEH) System
The Bis-DEH peptide contains a double repetition of the DEH amino acid pattern, spaced out by four glycine residues. The typical ATCUN (amino-terminal Cu and Ni binding) sequence NH2-X1Y2H3(DEH) in the N-terminal position should have a higher affinity compared to that of the same sequence in the C-terminus. The potentiometric results (Figure 5a) indicated that [CuH2L]2+ is the first species that appears at pH ∼3.5 and can be assigned as an {Nim} species. The UV–vis absorption band (λ = 728 nm) suggests a coordination mode also involving a carboxylate oxygen {NIm, COO–} (731 nm) (Table S2).
Figure 5.
Cu(II)–DEHGGGGDEH (Bis-DEH) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 1:1.1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 1:1.1 Cu(II):L molar ratio, I = 0.1 M NaClO4). (e) Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free Bis-DEH (red) and the Cu(II)–Bis-DEH complex at a 0.2:1 molar ratio (blue) and pH 8.9. Disappearing peaks are those marked in red.
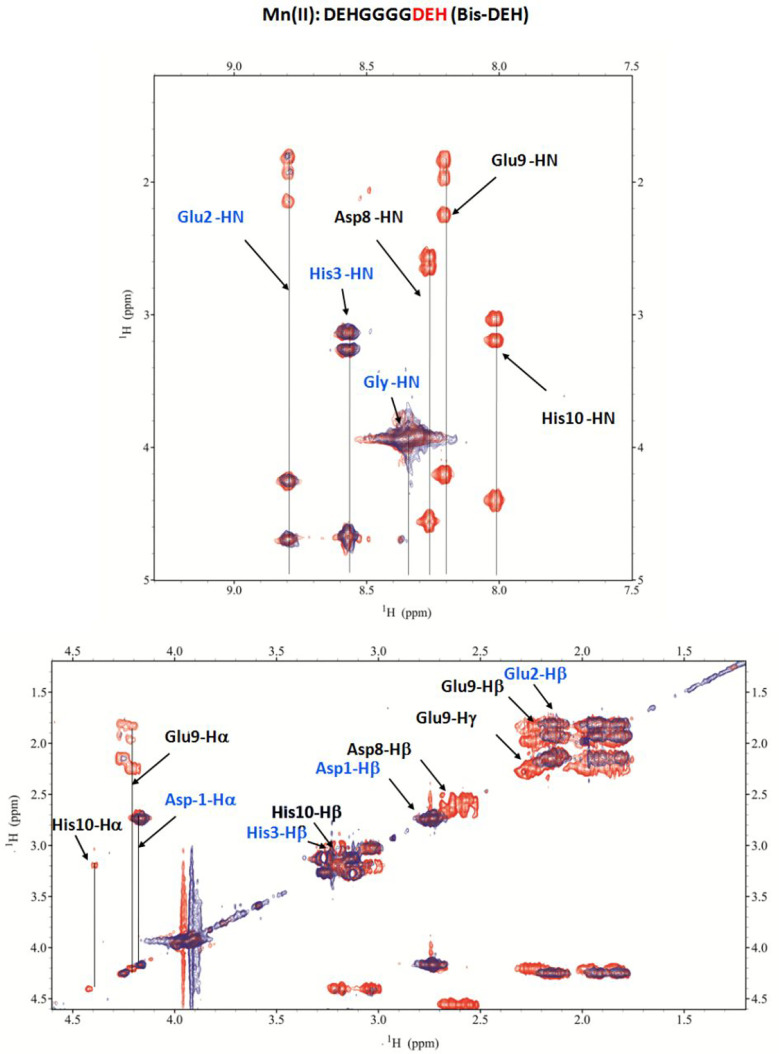
By increasing the pH, the [CuL] species appears starting from pH 4,, but it is almost immediately followed by [CuLH–1]− emerging from pH 4.5, and dominating the solution over the pH range of 6–8.5, reaching its maximum concentration around physiological pH. Spectroscopic evidence indicates the formation of a stable 4N albumin-like complex (log K* = −14.31) involving the first three residues with a chromophore around Cu(II) of the type {D1 Nterm, E2 N–amide, H3 Nim, N–amide}. As it was for the Cu(II)–DP1 system, a CT band at 310 nm can be ascribed to the coordination of deprotonated amido nitrogens to Cu(II), 2N–amide → Cu(II) LMCT, and an additional negative band present at 270 nm is due to an Nterm → Cu(II) LMCT. NMR experiments performed at acidic pH, with the a 0.2:1 Cu(II):Bis-DEH molar ratio, resulted in a general broadening and disappearance of almost all signals, except for glycines, so the attribution of the donor atom nuclei and the type of coordination mode cannot be ambiguous. The NMR titrations performed at pH 7.2 with substoichiometric addition of Cu(II) (Figure S2) showed the decrease in the intensity of the signal of both His-3 and His-10, but the subsequent NMR acquisition of the Cu(II)–Bis-DEH system at pH 8 and 8.9 is in perfect agreement with the attribution of a square planar albumin-like system for Cu(II) species. In fact, the resonances affected by the paramagnetic metal are exactly those of Nterm Asp-1 (α), Glu-2 (α), and His-3 (α, β, δ1, and ε1), and not those of the identical sequence of Asp-8, Glu-9, and His-10 at the C-terminus (Figure 5e).
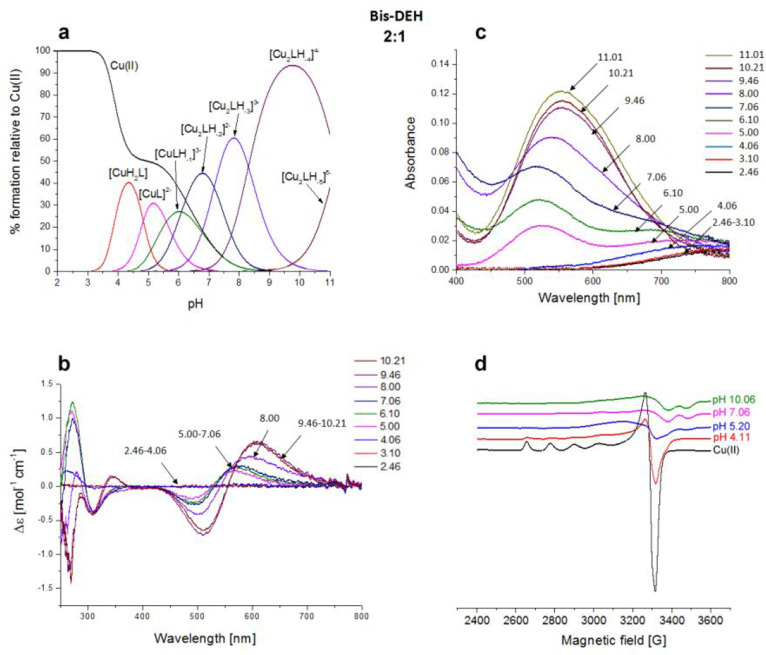
Moreover, the concomitant disappearance of the side chain signals related to Asp-1 (β) and Glu-2 (β and γ) can be attributed to the fact that they can be blocked above and below the coordination plane experiencing a metal relaxation effect.25,26 The next species found through potentiometric titrations is [CuLH–2]2– in which the albumin-like coordination mode of Cu(II) is maintained because no changes appear in the UV–vis, CD, and EPR spectra. At the same time, in NMR a shift of the aromatic signals of noncoordinating His-10 is detected up to pH 7, being in agreement with the deprotonation of its imidazole nitrogen. Via ESI-MS spectrometry at a 1:1 Cu(II):Bis-DEH molar ratio, the prevailing signals correspond to the equimolar Cu(II) complex (Figure S8), whereas at a 2:1 molar ratio, together with the presence of 1:1 species, the 2:2 complexes were also observed (Figure S9). The presence of a second DEH motif in this peptide provides the possibility of binding another metal ion. The spectroscopic studies performed in a 2:1 molar ratio, in comparison to 1:1, for the Cu(II)–Bis-DEH complex showed differences in the CD and UV–vis spectra above pH 8, indicating the presence of a new metal binding site. The potentiometric titrations of the 2:1 Cu(II):Bis-DEH molar ratio system were analyzed using the stability constants of mononuclear complexes obtained for the 1:1 molar ratio system as constant values. As a result, a satisfying fit of the potentiometric curves was obtained using a model containing both mono- and dinuclear complexes. As shown in Figure 6a, the mononuclear species dominate in solution in the low-pH range, up to pH 6, while at higher pH values, dinuclear complexes are formed. The UV–vis spectroscopic parameters, with a λmax at 525 nm (over the pH range 5–7), clearly correspond to a 4N {Nterm, Nim, 2N–amide} coordination of dominating [CuLH–1]3– species (Table S2 and Figure 6c). The same indication comes from CD spectra, with maxima at 500 and 560 nm, in comparison to a 1:1 Cu(II):Bis-DEH molar ratio (Table S2 and Figure 6b). The binding of the second Cu(II) ion leads to the formation of [Cu2LH–2]2–, with the involvement of His-10 and the coordination mode of 1N {Nim}, as reflected by a weak d–d band centered at 690 nm. With an increase in pH, the red shift in the d–d band region for the UV–vis and CD spectra is observed. It clearly indicates the deprotonation and involvement of N–amide atoms in Cu(II) binding, changing the coordination from {Nterm, Nim, 2N–amide}{Nim} to {Nterm, Nim, 2N–amide}{Nim, N–amide} in [Cu2LH–3]3–, and {Nterm, Nim, 2N–amide}{Nim, 2N–amide} in [Cu2LH–4]4–. The d–d band centered at 555 nm is most probably the result of averaging of 4N and 3N coordination modes of Cu(II) ions and is in agreement with our hypothesis. In the EPR spectra with 2 equiv of Cu(II) (Figure 6d), a partial vanishing of the spectra suggests the formation of dinuclear species above pH 5.
Figure 6.
Cu(II)–DEHGGGGDEH (Bis-DEH) system. (a) Species distribution of Cu(II) complexes, at 298 K and a 0.1 M NaClO4 ionic strength. [Cu(II)]tot = 1 mM; 2:1 Cu(II):L molar ratio. (b) CD spectra as a function of pH. (c) UV–vis spectra as a function of pH. (d) EPR spectra as a function of pH (conditions for CD, UV–vis, and EPR spectroscopy: [L] = 1 mM, 2:1 Cu(II):L molar ratio, I = 0.1 M NaClO4).
Manganese Complexes
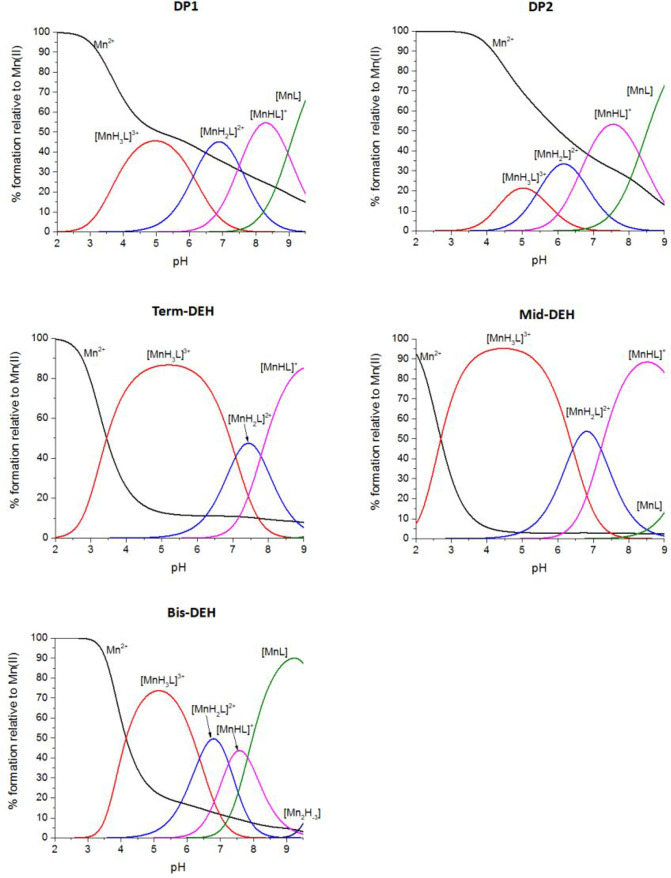
The complex formation constants for Mn(II) with all the five peptides are listed in Table 3, and the corresponding distribution diagrams are shown in Figure 7. Table S3 reports the coordination mode for the Mn(II)–peptide species. EPR spectra obtained over the whole range of pH are shown in Figure S10.
Table 3. Protonation Constants (log β) and pK Values of the Mn(II)–Peptide Systems at 298 K and a 0.1 M NaClO4 Ionic Strengtha.
| peptide | species | log β | log K | |
|---|---|---|---|---|
| DP1 | [MnH3L]3+ | 27.53(4) | ||
| [MnH2L]2+ | 21.42(4) | 6.11 | His | |
| [MnHL]+ | 13.92(5) | 7.5 | Nterm | |
| [MnL] | 4.96(5) | 8.96 | Lys | |
| DP2 | [MnH3L]3+ | 25.93(5) | ||
| [MnH2L]2+ | 20.53(4) | 5.40 | His | |
| [MnHL]+ | 13.94(4) | 6.59 | Nterm | |
| [MnL] | 5.65(4) | 8.29 | Lys | |
| Term-DEH | [MnH3L]3+ | 30.24(8) | ||
| [MnH2L]2+ | 23.16(8) | 7.08 | His | |
| [MnHL]+ | 15.37(8) | 7.79 | Nterm | |
| Mid-DEH | [MnH3L]3+ | 29.61(8) | ||
| [MnH2L]2+ | 23.19(8) | 6.42 | His | |
| [MnHL]+ | 15.98(8) | 7.21 | Nterm | |
| [MnL] | 6.18(9) | 9.8 | Lys | |
| Bis-DEH | [MnH3L]+ | 26.61(3) | ||
| [MnH2L] | 20.24(3) | 6.37 | His | |
| [MnHL]− | 12.99(3) | 7.25 | His | |
| [MnL]2– | 5.17(2) | 7.82 | Nterm |
Standard deviations (3σ values) are given in parentheses.
Figure 7.
Species distribution of Mn(II) complexes with DEHGTAVMLK (DP1), THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH), at 298 K and a 0.1 M NaClO4 ionic strength. [Mn(II)]tot = 0.5 mM; 1:1.1 Mn(II):L ratio.
Mn(II)–DEHGTAVMLK (DP1) System
The distribution diagram in Figure 7 shows that [MnH3L]3+ is the first Mn(II) complex species formed, in which Asp-1 and Glu-2 are the residues coordinating the metal ion already from pH <3, and with the maximum concentration [<50% of the Mn(II) in solution] at pH 5. The deprotonation of His-3Nim, in the next species, [MnH2L]2+, occurs with a pK of 6.11 (Table 3), which is 0.39 log unit lower in comparison to that of the free ligand (Table 1). The deprotonation of Nterm (pK of 7.5) leads to the formation of the [MnHL]+ complex, followed by the last species [MnL] deriving from the deprotonation of the ε-amino group of the Lys side chain. NMR acquisitions of the Mn(II)–DP1 system performed at pH 5 are consistent with the coordination of Mn(II) {3O–carb} with the carboxylate groups of Asp-1, Glu-2, and most likely with the terminal COO– of Lys-10, because its Hα proton experiences a severe intensity loss. At this pH, the interaction of His-3 is just starting, but the more intense decrease of the imidazolic signal Hδ2, with respect to Hε1, indicates that Mn(II) prefers the binding with Nε2 instead of Nδ1 that occurred for the Cu(II) ion. NMR titrations of DP1 with an increasing amount of Mn(II) at pH 7.2 showed the metal ion bound to the carboxylate oxygen of Asp-1 and Glu-2 moieties and to Nim of His-3 {Nim, 2O–carb} in agreement with the potentiometric data.31 The new NMR acquisitions of the Mn(II)–DP1 system at more basic pH values are consistent with the same coordination environment. The involvement of Nterm related to Asp-1 should also be considered for the {Nterm, Nim, 2O–carb} species. The octahedral coordination sphere, suggested by EPR spectroscopy,31 can be completed by water molecules. Analysis performed with ESI-MS techniques revealed the presence of signals deriving from the complexes with 1:1, and also 1:2, stoichiometry.31
Mn(II)–THMVLAKGED (DP2) System
As with DP1, also with the DP2 peptide, in the first species [MnH3L]3+, the two acidic Glu-9 and Asp-10 residues are already deprotonated and coordinated to the Mn(II) ion (Figure 7). The deprotonation of His-2, with the formation of a [MnH2L]2+ species, occurs with a pK of 5.4 (Table 3), and that of Nterm, with a pK of 6.59, is related to the next [MnHL]+ species. The last species, with a pK of 8.29, derived from the deprotonation of Nε of Lys, as in the previous case. Mn(II) titrations of DP2, followed by 1D and 2D NMR spectroscopy, were performed at pH 7.2 and showed that the metal anchoring side is Asp-10.31 In fact, a larger decay progression detected for β protons of this residue (compared to the trend of γ protons of Glu-9 and the δ2 proton of His-2) is a clear indication that Asp-10 is playing a central role in complex formation, through the metal ion binding with the oxygen donor of the side chain, and likely with the terminal carboxylate group. Glu-9 is also clearly involved in the coordination. A deeper NMR analysis of the Mn(II)–DP2 system showed, in addition, that His-2, whose aromatic resonances are affected by selective paramagnetic relaxation, is playing a role in the coordination with Mn(II) anchored to Glu-9 and Asp-10 residues in the C-terminal part of the sequence. Presumably, this is due to the presence of chemical exchanges between metal–peptide conformations in an intermediate regime with respect to the NMR time scale.
NMR acquisitions performed at more acidic pH, in the range of dominance of [MnH3L]3+ species, are consistent with the metal anchoring side located at the C-terminal portion of the sequence in a {3O–carb} coordination mode, whereas at basic pH (pH 9.5), the coordination of Mn(II) became analogous to the one found at pH 7.2, because only the deprotonation of a noncoordinating Lys was detected. EPR spectroscopy, performed at various pH values, showed a line pattern related with an octahedral coordination sphere around Mn(II), and ESI-MS spectrometric data showed peaks associated with mononuclear Mn(II) complexes.31
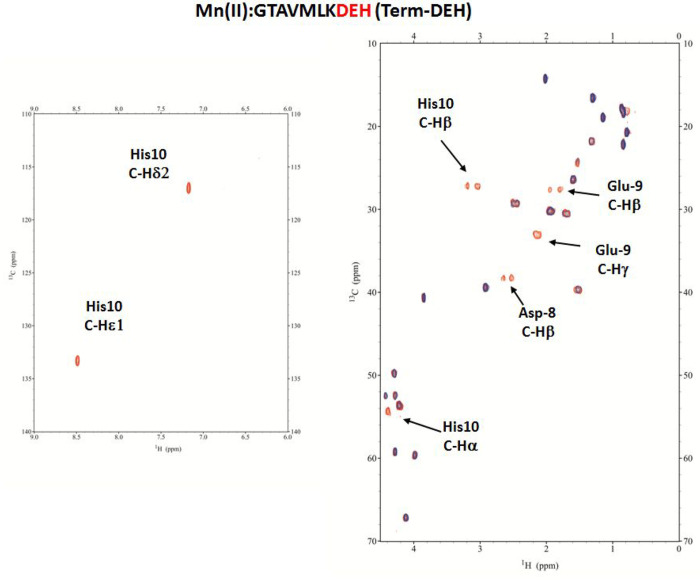
Mn(II)–GTAVMLKDEH (Term-DEH) System
The species with [MnH3L]3+ stoichiometry, present from pH 2 to 8.5, and reaching its maximum concentration around pH 5, is the first species identified by potentiometric titrations. The Mn(II) ion is coordinated to Term-DEH through Asp-8, Glu-9, and His-10 residues, as confirmed by NMR spectroscopy. The additions of a substoichiometric amount of paramagnetic Mn(II) to the peptide resulted in the spectra given in Figure S11 and Figure 8, in which the selective relaxation of the signals of Asp, Glu, and His highlighted the specific residues involved in the metal coordination.
Figure 8.
Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free Term-DEH (red) and the Mn(II)–Term-DEH system at a 0.1:1 molar ratio (blue) and pH 5. Disappearing peaks are labeled.
In particular, the severe broadening of Hβ compared to Hα of Asp-8, Hγ compared to Hβ of Glu-9, and the disappearance of the signals belonging to His-10, in particular of Hα and only one of the two Hβ, are consistent with a {3O–carb} sphere, with the metal ion being bound through the oxygen donors of carboxylate moieties of Asp-8, Glu-9, and terminal COO– of His-10. This coordination blocked the side chain on His-10 in a position that differentiates the two β protons, bringing one closer to the metal ion, experiencing its paramagnetic effect. The deprotonation of His in the next species, [MnH2L]2+, allows also Nim to participate in metal binding {Nim, 2O–carb}.
The NMR acquisitions at pH 7.4 for the Mn(II)–Term-DEH system confirm such a coordination sphere around Mn(II). With an increase in the pH until the [MnHL]+ species prevails, the potentiometric data indicated the deprotonation and coordination of Nterm. This is confirmed by NMR spectra recorded at pH 9.2, where a clear disappearance of the Hα signal related to Gly-1 can only be due to the binding of Mn(II) also to the deprotonated Nterm of this residue, {Nterm, Nim, 2O–carb}. ESI-MS measurements are consistent with mononuclear (1:1) and dinuclear (2:1) Mn–L species (Figure S12). The EPR spectra of hexa-aqua Mn(II) and Mn(II)–Term-DEH (Figure S10) systems are similar, with a six-line 55Mn (I = 5/2; 100% abundance) pattern centered at g around 2, indicating that in Mn(II)–peptide species there is no significant deviation from a spherical symmetry. The decrease in the intensity of the paramagnetic Mn(II) signal with an increase in pH, together with the increase in intensity of the forbidden transitions relative to the allowed transitions, can be related to a decrease in the order of the ligand field around Mn(II) with a change in the symmetry of the complex when the metal moves from a totally hydrated environment to various new donors.56,57
Mn(II)–TMVLDEHAKG (Mid-DEH) System
Potentiometric data (Table 3) indicated that the first species, [MnH3L]3+, starts appearing at pH <2, with its maximum concentration around pH 4.5. Mn(II) is bound here to Asp, Glu, and also His, as confirmed by the NMR titrations (Figure S11) performed at pH 5. Moreover, the 2D 1H–13C HSQC spectra, recorded at the same pH (Figure 9), showed, additionally, a clear involvement of the terminal carboxylate oxygen of Gly-10. In fact, its Hα signal disappeared, together with some other signals of the adjacent Lys-9 residue, as a consequence of its proximity to the paramagnetic metal ion. The coordination mode for [MnH3L]3+ is {Nim, 3O–carb}.
Figure 9.
Aromatic and aliphatic region of the 1H–13C HSQC spectrum of free Mid-DEH (red) and the Mn(II)–Mid-DEH system at a 0.1:1 molar ratio (blue) and pH 5. Disappearing peaks are labeled.
The next two species, [MnH2L]2+ and [MnHL]+, dominate at pH >7, and here, the deprotonation involved Nim of His-7 and Nterm of Thr-1. Accordingly, NMR spectra acquired at pH 7.2 (Figure S13) showed an intensity loss related to Asp-5, Glu-6, His-7, and Gly-10 signals, but also to Thr-1, that is participating to the metal binding through the Nterm donor atom. The Mn(II) ion is bound to Mid-DEH with the formation of a macrochelate system {Nterm, Nim, 3O–carb} that forces the peptide to fold around the metal ion. Due to this new conformational arrangement, some nuclei of other residues, as Leu-4, Ala-8, and Lys-9, placed in a space close to the metal ion, experienced its paramagnetic effect and lose their NMR signals. The last species identified by potentiometry is [MnL], which resulted from the deprotonation of the amino group of the Lys-9 side chain, in agreement with NMR experiments performed at high pH (revealing the shift related to the Hγ proton as a consequence of the Nε deprotonation). ESI-MS spectra indicated the presence of mono- and dinuclear species, i.e., [MnL]2+ and [Mn2L-2H+]2+ (Figure S14). EPR spectra (Figure S10) showed for the Mid-DEH peptide, similarly to the previous system, an octahedral coordination mode around Mn(II).
Mn(II)–DEHGGGGDEH (Bis-DEH) System
Because Mn(II) ions have no tendency to form stable square planar complexes involving the terminal nitrogen and the deprotonated amide atoms, as in the case of Cu(II), the ATCUN sequence (NH2-D1E2H3) should not have a higher affinity for Mn(II), in comparison to the identical sequence in the C-terminal portion (D8E9H10-COO–). The NMR study showed that in the first species identified by the potentiometric technique, [MnH3L]3+, the complex formation arises from the involvement of the last three residues, i.e., through the oxygen donors of carboxylate moieties located in the side chain of Asp-8, Glu-9, and the COO– terminal group attached to His-10 (Figure S11 and Figure 10) {3O–carb}. The Nim of His-10 at this pH does not participate in metal ion binding.
Figure 10.
Aromatic and aliphatic region of the 1H–1H TOCSY spectrum of free Bis-DEH (red) and the Mn(II)–Bis-DEH complex at a 0.05:1 molar ratio (blue) and pH 5. Disappearing peaks are labeled in black color, and unaffected peaks are colored blue.
The following species is [MnH2L]2+, which begins to appear at pH 4, and with its maximum concentration at pH 6.8. The pK value of 6.37 suggests that a histidine residue is being deprotonated. In this species, Nim of His-10 is also participating in metal binding {Nim, 3O–carb}, as in the case of the species related to Term-DEH found, however, at a higher pH of 7.4. His-3 loses its proton in the following [MnHL]+ species with a pK of 7.25, while in the [MnL] species, which is the main one at basic pH, there is the deprotonation of the Nterm group, with a pK of 7.82.
The NMR acquisitions performed in the pH range of 6–8 showed a dramatic increase in the relaxation effect for the signals related to both DEH sequences, i.e., in the N- and C-terminal parts of the peptide (Figure S15). This behavior is compatible with the simultaneous involvement of both DEH fragments in the coordination of one metal ion, with the formation of a macrochelate complex of MnL stoichiometry {2Nim, 4O–carb}, or of two different simultaneous complexes between Mn(II) and each DEH sequence, with the formation of a binuclear complex with Mn2L stoichiometry {Nterm, Nim, 2O–carb} for N-terminal species and {Nim, 3O–carb} for C-terminal species. Even though the potentiometric data reveal the formation of dinuclear species only at pH >9, ESI-MS results support both of these hypotheses already from pH 7.4 (Figure S16).
Final Remarks
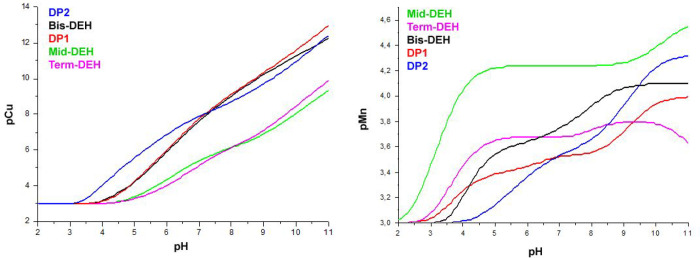
To compare the global binding affinity of each peptide with respect to Cu(II) or Mn(II) ions, under given experimental conditions, log K* and pM values were calculated.33,58
The former can be used to compare the stability constants of complexes with different stoichiometry and degree of protonation. In the case of 1:1 species, the log K* value refers to the following equilibrium:
where n is the maximum degree of protonation of the ligand and j is the degree of protonation of the complex; charges are omitted for the sake of simplicity.
pM is the negative logarithm of the concentration of the free metal ion, in the presence of the ligand, under normalized experimental conditions:
where in our case Cu(II):L or Mn(II):L ratios are 1:1, and with the concentrations used for potentiometric analysis (Figure 11).
Figure 11.
pCu and pMn values of peptides as a function of pH.
pM can be used to compare the relative strength of the complexes, independently of their number and their stoichiometry. As pM depends on pH and on the concentration of both the metal and the ligand, to compare the results to other systems, the calculations should be performed under the same conditions. Generally, the calculation of the pM value for metal chelation therapy (MCT) uses the values found at a physiological pH of 7.4, and metal and ligand concentrations of 10–6 and 10–5 mol/L, respectively.
Table 4 collects the calculated parameters for Cu(II) and Mn(II) systems at the concentrations used for potentiometric analysis, and for MCT relevant conditions.
Table 4. Log K* and pM Values for the Studied Systems at pH 7.4.
| log K* |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| species | DP1 | species | DP2 | species | Term-DEH | species | Mid-DEH | species | Bis-DEH | |
| 1N | CuH2L | –1.11 | CuH2L | –1.79 | CuH2L | –1.98 | CuH2L | 0.04 | ||
| 2N | CuHL | –7.13 | CuHL | –6.57 | ||||||
| 3N | CuL | –9.75 | CuL | –6.88 | CuL | –13.5 | CuL | –12.28 | CuL | –9.62 |
| 4N | CuLH–1 | –14.83 | CuLH–3 | –23.02 | CuLH–1 | –22.34 | CuLH–2 | –21.05 | CuLH–1 | –15.21 |
| DP1 | DP2 | Term-DEH | Mid-DEH | Bis-DEH | |
|---|---|---|---|---|---|
| pCu (pH 7.4)a ([L] = 1.1 × 10–3, [Cu(II)] = 10–3) | 8.34 | 8.23 | 5.58 | 5.74 | 8.23 |
| pCu (pH 7.4)b ([L] = 10–5, [Cu(II)] = 10–6) | 14.65 | 14.41 | 9.11 | 9.44 | 14.43 |
| pMn (pH 7.4)a ([L] = 0.55 × 10–3, [Mn(II)] = 0.5 × 10–3) | 3.54 | 3.58 | 3.69 | 4.24 | 3.81 |
| pMn (7.4)b ([L] = 10–5, [Mn(II)] = 10–6) | 6.05 | 6.04 | 6.08 | 6.53 | 6.13 |
Concentrations used for potentiometric analysis.
MCT physiologically relevant conditions.
Moreover, we compared the binding ability of the five peptides toward the same metal ion by plotting the competition diagrams (Figures S17 and S18). These plots can be calculated by means of the speciation models obtained by potentiometry and assuming that no mixed-ligand species is formed and, in this case, refer to hypothetical solutions in which a couple of peptides are present at the same time and compete in the binding with the metal ion.
Competition plots and pCu give the evidence that the weakest ligands are Term-DEH and Mid-DEH. The stability constants log K* for their major species [CuL], at physiological pH, are −13.5 and −12.28, respectively. The interesting feature of these species is the formation of large macrochelate loops, with Cu(II) bound to histidine and a distant N-terminal amino group donor (Figure 12c,d).
Figure 12.
Scheme of the Cu(II) coordination sphere for all peptides at physiological pH.
DP1 and Bis-DEH containing the N-terminal sequence D1E2H3 (albumin-like sequence, ATCUN motif also known as NTS) exhibit particular affinity for Cu(II) resulting from the formation of three fused chelate rings and a flat 4N coordination sphere around the metal ion (Figure 12a and Figure S19a). However, the strongest ligand from acidic to nearly neutral pH (7.2) is DP2, which binds Cu(II) more efficiently than the albumin-like sequence in DP1 or Bis-DEH, because it is able to form, already from the starting complex, a 3N species {T1 Nterm, H2 Nim, H2 N–amide} and/or a coordination isomer 3NO species {T1 Nterm, H2 Nim, H2 N–amide and E9, D10 O–carb}, as it was better evidenced by NMR measurements (Figure 12a,b and Figure S19b,c). The log K* of −6.88 for this species is ∼3 orders of magnitude higher than for the corresponding [CuL] species in Cu(II)–DP1 or −Bis-DEH systems (Table 4).
Instead, from pH 7.2, the thermodynamically stable albumin-like 4N coordination mode starts to be more efficient with the formation of [CuLH–1] species for both DP1 and Bis-DEH (log K* values of −14.83 and −15.21, respectively) (Figure 12a,e). The change in coordination mode from 3N {Nterm, Nim, N−amide} to 4N {Nterm, 3N−amide} for Cu(II)-DP2 is energetically disadvantaged (log K* = −23.02). Upon examination of the two strongest Cu(II) binder peptides, DP1 and Bis-DEH, for the first Cu(II) 1N species, identified at pH <5, Bis-DEH is slightly, ∼10-fold, more attractive toward the metal ion (log K* of 0.04 compared to −1.11 for DP1). The reason for this difference can be the presence of the second DEH-COO– motif in the C-terminal region, rich in oxygen donors from three carboxylate groups, and another histidine residue. In most peptides, histidine plays a role as the Cu(II) anchoring side at acidic pH; thus, the presence of two histidines would result in an internal metal competition also if only one, i.e., that in the ATCUN motif, is a stronger chelator. In any case, the contributions of both DEH motifs seem to be cumulative, and Bis-DEH turned out to be the best chelator at acidic pH. This hypothesis would justify the results obtained from NMR titration of Cu(II) to Bis-DEH performed at acidic pH, where a severe broadening of both DEH fragments was evidenced.
When the pH is increased from 5 to 9, in the range of existence of [CuLH–1] species, the binding of Cu(II) to DP1 is preferred (log K* = −14.83) over binding to Bis-DEH (log K* = −15.21).
When the pH is increased above 9, the difference in affinity with DP1 > Bis-DEH becomes more remarkable due to the presence of a Lys residue in DP1 (absent in Bis-DEH), which can influence the stability of the complex formed at basic pH.26,59 The values of the equilibrium constants for actual complex formation in terms of log K* are useful to compare the stability of similar 4N species with analogue ligands when coordination follows deprotonation. Considering the log K* of −14.44 reported in the literature for the ATCUN motif NH2-DAHK-Am from the N-terminal sequence of human serum albumin, we can observe a similar value (log K* = −14.83) for DP1 NH2-DEHGTAVMLK. The slight differences in Cu(II) affinities for these two motifs can result from differences in acidities of the peptide nitrogens of Glu with respect to Ala.60 The comparison of the pCu value of 14.65 for DP1 with a list of those 250 ligands normally used or projected in chelation therapy shows the strength of this complex; it is lower with respect to those of only a few chelators, such as o-Trensox (pCu = 22.9), Alaternin (pCu = 16.5), L-DOPA and some derivatives (pCu = 15.1–15.2), and a new chelation agent recently proposed by our group, abbreviated as SC (pCu = 16.5).61,62 The different charge in these complexes should be taken into consideration to exploit their redistribution in lipophilic (neutral complexes) or hydrophilic (charged complexes) environments. The peptide stability, moreover, is a critical aspect for drug development and therapeutic applications. The use of d-isomers instead of l-isomers will offer the opportunity to design stable therapeutic d-peptides with the ability to circulate longer inside the organism and may render them orally available and less immunogenic. Our previous results demonstrated an equal coordination ability of d-DP1 and d-DP2 compared to their l-forms and their stability over time.31 In addition, the enzyme protection ability of amino acids in their d- and l-forms was found to be the same against inactivation by irradiation.12
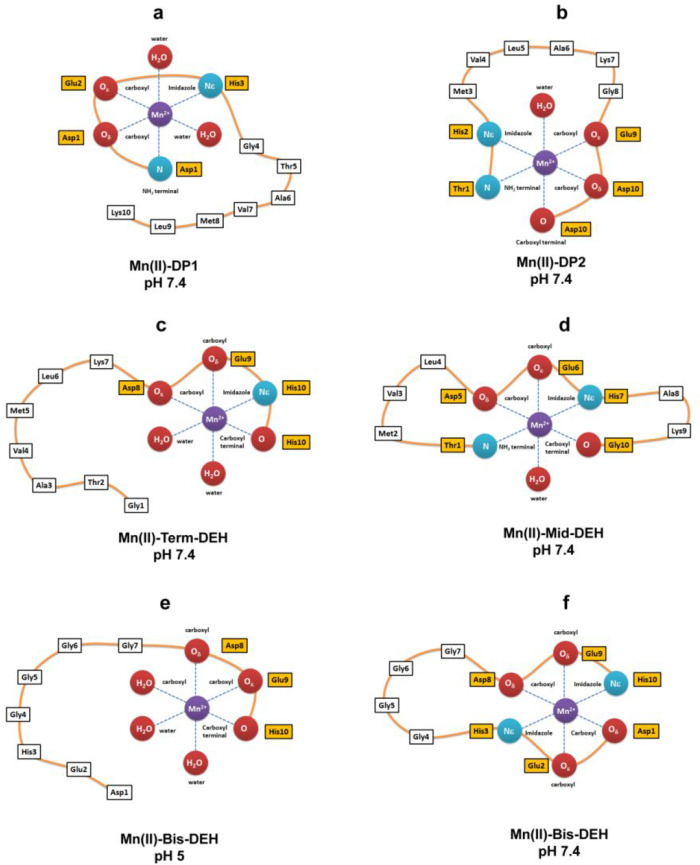
The scheme of the Mn(II) coordination sphere for all of the peptides at physiological pH is reported in Figure 13.
Figure 13.
Scheme of the Mn(II) coordination sphere for all peptides at physiological pH.
The competition plots related to the Mn(II)–peptide system are shown in Figure S18, and the pMn for the whole pH range is reported in Figure 11. Among all of the peptides, Mid-DEH is the most effective Mn(II) binder over the entire pH range. The -DEH- motif in the central position results in better metal chelation ability (Figure 13d and Figure S19d). The C-terminal DEH motif in Term-DEH favors Mn(II) binding at acidic pH, with the metal ion bound by three carboxylate groups and Nim, whereas only at basic pH, with the deprotonation of Nterm, does the N-terminal DEH motif in DP1 start to be competitive in metal binding with respect to Term-DEH. The two DEH motifs in Bis-DEH showed an almost additive contribution, especially up to pH 6.5. Indeed, from this pH on, the pM of Bis-DEH is greater than those related to Term-DEH and DP1. The affinity of Mn(II) for DP1 and DP2 peptides is opposite with respect to what was found for Cu(II). DP1 proved to be a better chelator from acidic to physiological pH, whereas at more basic pH, DP2 proved to be a more effective Mn(II) binder.
A pMn of 6.53 calculated under CMT physiological conditions for the strongest Mn(II)–Mid-DEH complex is higher than values of 90% of the ligands normally used, or proposed, in metal chelation therapy,61,63 and also higher than those of the Mn(II)–peptide complexes, derived from Ypk9, a vacuolar Mn(II)-transporting ATPase protein encoded from a Parkinson’s disease gene whose role is in manganese efflux and detoxification (pMn = 6.01).33
Conclusion
D. radiodurans is a source of several biotechnological potential applications: environmental bioremediation of contaminated sites and nuclear wastes, human bioprotection (prophylactic measure before radiation exposure or for preventing radiation toxicity after nuclear accidents or military activity), biomedical applications (radiotherapy and nuclear medicine), and the formulation of personal care products (antiaging cosmetics and sunscreens).64 In particular, the remarkable capacity of synthetic DR antioxidant complexes made of selected peptides and Mn(II) ions to scavenge IR-induced ROS was demonstrated in several studies in vitro and in vivo.2,12,13 Moreover, in a study performed by Gupta et al., the Mn(II)–DP1 complexes tested in mice proved to be nontoxic and able to efficiently prevent radiation damage.13
These results open new avenues for clinical applications of safe Mn(II)–peptide complexes as a promising countermeasure for acute radiation syndrome for humans, whereas other applications such as the development of inactivated vaccines have already been pursued.14,15
For these purposes, a detailed characterization of the metal−peptide interaction is needed.
In this paper, five rationally designed decapeptides derived from the cell-free extract of D. radiodurans, namely, DEHGTAVMLK (DP1), THMVLAKGED (DP2), GTAVMLKDEH (Term-DEH), TMVLDEHAKG (Mid-DEH), and DEHGGGGDEH (Bis-DEH), have been minutely analyzed for their coordination properties toward Cu(II) and Mn(II) ions via complementary techniques such as potentiometry, NMR, EPR, UV–vis, and CD spectroscopies, and ESI-MS spectrometry. The results may provide new insight for our understanding of the complex radioprotection mechanism in DR and novel information useful for exploiting its extraordinary ability in future biotechnological applications.
Acknowledgments
M.O., K.P., and E.G.-K. acknowledge the Polish National Science Centre (NCN, UMO 2015/19/B/ST5/00413) for financial support. M.P. acknowledges Thomas J. Lamkin (Air Force Research Laboratory, Wright Patterson Air Force Base, OH 45433) for the initial supply of peptides. The results were obtained within the frame of COST NECTAR Action CA18202.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.inorgchem.9b03737.
Figures S1–S19 and Tables S1–S3 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Slade D.; Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol. Biol. Rev. 2011, 75 (1), 133–91. 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J.; Gaidamakova E. K.; Matrosova V. Y.; Kiang J. G.; Fukumoto R.; Lee D. Y.; Wehr N. B.; Viteri G. A.; Berlett B. S.; Levine R. L. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 2010, 5 (9), e12570 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peana M.; Chasapis C. T.; Simula G.; Medici S.; Zoroddu M. A. A Model for Manganese interaction with Deinococcus radiodurans proteome network involved in ROS response and defense. J. Trace Elem. Med. Biol. 2018, 50, 465–473. 10.1016/j.jtemb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Santos S. P.; Yang Y.; Rosa M. T. G.; Rodrigues M. A. A.; De La Tour C. B.; Sommer S.; Teixeira M.; Carrondo M. A.; Cloetens P.; Abreu I. A.; Romao C. V. The interplay between Mn and Fe in Deinococcus radiodurans triggers cellular protection during paraquat-induced oxidative stress. Sci. Rep. 2019, 9 (1), 17217. 10.1038/s41598-019-53140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. Z.; Wang W. Z.; He J. Y.; Ma Y.; Xiao F. Z.; He S. Y. Antioxidative system of Deinococcus radiodurans. Res. Microbiol. 2020, 171, 45. 10.1016/j.resmic.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Daly M. J. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 2009, 7 (3), 237–45. 10.1038/nrmicro2073. [DOI] [PubMed] [Google Scholar]
- Culotta V. C.; Daly M. J. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid. Redox Signaling 2013, 19 (9), 933–44. 10.1089/ars.2012.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald F. S.; Fridovich I. The scavenging of superoxide radical by manganous complexes: in vitro. Arch. Biochem. Biophys. 1982, 214 (2), 452–63. 10.1016/0003-9861(82)90049-2. [DOI] [PubMed] [Google Scholar]
- Krisko A.; Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (32), 14373–7. 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnese K.; Gralla E. B.; Valentine J. S.; Cabelli D. E. Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (18), 6892–7. 10.1073/pnas.1203051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana H.; Hazari P. P.; Mishra A. K. Radioprotective efficacy of GSH based peptidomimetic complex of manganese against radiation induced damage: DT(GS)2Mn(II). Free Radical Biol. Med. 2019, 145, 161–174. 10.1016/j.freeradbiomed.2019.09.023. [DOI] [PubMed] [Google Scholar]
- Berlett B. S.; Levine R. L. Designing antioxidant peptides. Redox Rep. 2014, 19 (2), 80–6. 10.1179/1351000213Y.0000000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Gayen M.; Smith J. T.; Gaidamakova E. K.; Matrosova V. Y.; Grichenko O.; Knollmann-Ritschel B.; Daly M. J.; Kiang J. G.; Maheshwari R. K. MDP: A Deinococcus Mn2+-Decapeptide Complex Protects Mice from Ionizing Radiation. PLoS One 2016, 11 (8), e0160575 10.1371/journal.pone.0160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayen M.; Gupta P.; Morazzani E. M.; Gaidamakova E. K.; Knollmann-Ritschel B.; Daly M. J.; Glass P. J.; Maheshwari R. K. Deinococcus Mn(2+)-peptide complex: A novel approach to alphavirus vaccine development. Vaccine 2017, 35 (29), 3672–3681. 10.1016/j.vaccine.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Tobin G. J.; Tobin J. K.; Gaidamakova E. K.; Wiggins T. J.; Bushnell R. V.; Lee W. M.; Matrosova V. Y.; Dollery S. J.; Meeks H. N.; Kouiavskaia D.; Chumakov K.; Daly M. J. A novel gamma radiation-inactivated sabin-based polio vaccine. PLoS One 2020, 15 (1), e0228006 10.1371/journal.pone.0228006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidamakova E. K.; Myles I. A.; McDaniel D. P.; Fowler C. J.; Valdez P. A.; Naik S.; Gayen M.; Gupta P.; Sharma A.; Glass P. J.; Maheshwari R. K.; Datta S. K.; Daly M. J. Preserving immunogenicity of lethally irradiated viral and bacterial vaccine epitopes using a radio- protective Mn2+-Peptide complex from Deinococcus. Cell Host Microbe 2012, 12 (1), 117–124. 10.1016/j.chom.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso A.; Carvalho T.; Rousselot-Pailley P.; Correia Dos Santos M. M.; Delgado R.; Iranzo O. Effect of the Peptidic Scaffold in Copper(II) Coordination and the Redox Properties of Short Histidine-Containing Peptides. Chem. - Eur. J. 2015, 21 (37), 13100–11. 10.1002/chem.201501715. [DOI] [PubMed] [Google Scholar]
- Bonomo R. P.; Impellizzeri G.; Pappalardo G.; Purrello R.; Rizzarelli E.; Tabbì G. Co-ordinating properties of cyclopeptides. Thermodynamic and spectroscopic study on the formation of copper (II) complexes with cyclo (Gly-His) 4 and cyclo (Gly-His-Gly) 2 and their superoxide dismutase-like activity. J. Chem. Soc., Dalton Trans. 1998, (22), 3851–3858. 10.1039/a804481c. [DOI] [Google Scholar]
- Timari S.; Cerea R.; Varnagy K. Characterization of CuZnSOD model complexes from a redox point of view: redox properties of copper(II) complexes of imidazole containing ligands. J. Inorg. Biochem. 2011, 105 (8), 1009–17. 10.1016/j.jinorgbio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Bellia F.; La Mendola D.; Maccarrone G.; Mineo P.; Vitalini D.; Scamporrino E.; Sortino S.; Vecchio G.; Rizzarelli E. Copper(II) complexes with β-cyclodextrin-homocarnosine conjugates and their antioxidant activity. Inorg. Chim. Acta 2007, 360 (3), 945–954. 10.1016/j.ica.2006.07.028. [DOI] [Google Scholar]
- Bar-Or D.; Rael L. T.; Lau E. P.; Rao N. K.; Thomas G. W.; Winkler J. V.; Yukl R. L.; Kingston R. G.; Curtis C. G. An analog of the human albumin N-terminus (Asp-Ala-His-Lys) prevents formation of copper-induced reactive oxygen species. Biochem. Biophys. Res. Commun. 2001, 284 (3), 856–62. 10.1006/bbrc.2001.5042. [DOI] [PubMed] [Google Scholar]
- Pickart L.; Vasquez-Soltero J. M.; Margolina A. GHK-Cu may prevent oxidative stress in skin by regulating copper and modifying expression of numerous antioxidant genes. Cosmetics 2015, 2 (3), 236–247. 10.3390/cosmetics2030236. [DOI] [Google Scholar]
- Tegoni M.; Valensin D.; Toso L.; Remelli M. Copper chelators: chemical properties and bio-medical applications. Curr. Med. Chem. 2014, 21 (33), 3785–818. 10.2174/0929867321666140601161939. [DOI] [PubMed] [Google Scholar]
- Kozłowski H.; Bal W.; Dyba M.; Kowalik-Jankowska T. Specific structure-stability relations in metallopeptides. Coord. Chem. Rev. 1999, 184 (1), 319–346. 10.1016/S0010-8545(98)00261-6. [DOI] [Google Scholar]
- Zoroddu M. A.; Medici S.; Peana M. Copper and nickel binding in multi-histidinic peptide fragments. J. Inorg. Biochem. 2009, 103 (9), 1214–20. 10.1016/j.jinorgbio.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Medici S.; Peana M.; Nurchi V.; Zoroddu M. The involvement of amino acid side chains in shielding the nickel coordination site: an NMR study. Molecules 2013, 18 (10), 12396–12414. 10.3390/molecules181012396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gran G.; Dahlenborg H.; Laurell S.; Rottenberg M. Determination of the equivalent point in potentiometric titrations. Acta Chem. Scand. 1950, 4 (4), 559–577. 10.3891/acta.chem.scand.04-0559. [DOI] [Google Scholar]
- Gans P.; Sabatini A.; Vacca A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43 (10), 1739–1753. 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- Gans P.; Sabatini A.; Vacca A. Superquad - An improved general program for computation of formation-constants from potentiometric data. J. Chem. Soc., Dalton Trans. 1985, (6), 1195–1200. 10.1039/dt9850001195. [DOI] [Google Scholar]
- Alderighi L.; Gans P.; Ienco A.; Peters D.; Sabatini A.; Vacca A. Hyperquad simulation and speciation (HySS): a utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. 10.1016/S0010-8545(98)00260-4. [DOI] [Google Scholar]
- Peana M.; Medici S.; Pangburn H. A.; Lamkin T. J.; Ostrowska M.; Gumienna-Kontecka E.; Zoroddu M. A. Manganese binding to antioxidant peptides involved in extreme radiation resistance in Deinococcus radiodurans. J. Inorg. Biochem. 2016, 164, 49–58. 10.1016/j.jinorgbio.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Peana M. F.; Medici S.; Ledda A.; Nurchi V. M.; Zoroddu M. A. Interaction of Cu(II) and Ni(II) with Ypk9 protein fragment via NMR studies. Sci. World J. 2014, 2014, 656201. 10.1155/2014/656201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remelli M.; Peana M.; Medici S.; Delogu L. G.; Zoroddu M. A. Interaction of divalent cations with peptide fragments from Parkinson’s disease genes. Dalton Trans 2013, 42 (17), 5964–74. 10.1039/C2DT32222F. [DOI] [PubMed] [Google Scholar]
- Hureau C.; Eury H.; Guillot R.; Bijani C.; Sayen S.; Solari P.-L.; Guillon E.; Faller P.; Dorlet P. X-ray and Solution Structures of CuIIGHK and CuIIDAHK Complexes: Influence on Their Redox Properties. Chem. - Eur. J. 2011, 17 (36), 10151–10160. 10.1002/chem.201100751. [DOI] [PubMed] [Google Scholar]
- McGuire A. M.; Cuthbert B. J.; Ma Z.; Grauer-Gray K. D.; Brunjes Brophy M.; Spear K. A.; Soonsanga S.; Kliegman J. I.; Griner S. L.; Helmann J. D.; Glasfeld A. Roles of the A and C sites in the manganese-specific activation of MntR. Biochemistry 2013, 52 (4), 701–13. 10.1021/bi301550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. E.; Klewpatinond M.; Abdelraheim S. R.; Brown D. R.; Viles J. H. Probing Copper2+ Binding to the Prion Protein Using Diamagnetic Nickel2+ and 1H NMR: The Unstructured N terminus Facilitates the Coordination of Six Copper2+ Ions at Physiological Concentrations. J. Mol. Biol. 2005, 346 (5), 1393–1407. 10.1016/j.jmb.2004.12.043. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25 (13), 1605–12. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Peana M.; Medici S.; Nurchi V. M.; Crisponi G.; Lachowicz J. I.; Zoroddu M. A. Manganese and cobalt binding in a multi-histidinic fragment. Dalton Trans 2013, 42 (46), 16293–301. 10.1039/c3dt51091c. [DOI] [PubMed] [Google Scholar]
- Peana M.; Zdyb K.; Medici S.; Pelucelli A.; Simula G.; Gumienna-Kontecka E.; Zoroddu M. A. Ni(II) interaction with a peptide model of the human TLR4 ectodomain. J. Trace Elem. Med. Biol. 2017, 44, 151–160. 10.1016/j.jtemb.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Remelli M.; Peana M.; Medici S.; Ostrowska M.; Gumienna-Kontecka E.; Zoroddu M. A. Manganism and Parkinson’s disease: Mn (II) and Zn (II) interaction with a 30-amino acid fragment. Dalton Transactions 2016, 45 (12), 5151–5161. 10.1039/C6DT00184J. [DOI] [PubMed] [Google Scholar]
- Magri A.; Munzone A.; Peana M.; Medici S.; Zoroddu M. A.; Hansson O.; Satriano C.; Rizzarelli E.; La Mendola D. Coordination Environment of Cu(II) Ions Bound to N-Terminal Peptide Fragments of Angiogenin Protein. Int. J. Mol. Sci. 2016, 17 (8), 1240. 10.3390/ijms17081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnagy K.; Szabó J.; Sóvágó I.; Malandrinos G.; Hadjiliadis N.; Sanna D.; Micera G. Equilibrium and structural studies on copper(II) complexes of tetra-, penta- and hexa-peptides containing histidyl residues at the C-termini. J. Chem. Soc., Dalton Trans. 2000, (4), 467–472. 10.1039/a907342f. [DOI] [Google Scholar]
- Kozłowski H.; Kowalik-Jankowska T.; Jeżowska-Bojczuk M. Chemical and biological aspects of Cu2+ interactions with peptides and aminoglycosides. Coord. Chem. Rev. 2005, 249 (21), 2323–2334. 10.1016/j.ccr.2005.04.027. [DOI] [Google Scholar]
- Sigel H.; Martin R. B. Coordinating properties of the amide bond. Stability and structure of metal ion complexes of peptides and related ligands. Chem. Rev. 1982, 82 (4), 385–426. 10.1021/cr00050a003. [DOI] [Google Scholar]
- Tsangaris J. M.; Martin R. B. Visible circular dichroism of copper(II) complexes of amino acids and peptides. J. Am. Chem. Soc. 1970, 92 (14), 4255–60. 10.1021/ja00717a020. [DOI] [PubMed] [Google Scholar]
- Stanyon H. F.; Cong X.; Chen Y.; Shahidullah N.; Rossetti G.; Dreyer J.; Papamokos G.; Carloni P.; Viles J. H. Developing predictive rules for coordination geometry from visible circular dichroism of copper(II) and nickel(II) ions in histidine and amide main-chain complexes. FEBS J. 2014, 281 (17), 3945–54. 10.1111/febs.12934. [DOI] [PubMed] [Google Scholar]
- Ösz K.; Bóka B.; Várnagy K.; Sóvágó I.; Kurtán T.; Antus S. The application of circular dichroism spectroscopy for the determination of metal ion speciation and coordination modes of peptide complexes. Polyhedron 2002, 21 (21), 2149–2159. 10.1016/S0277-5387(02)01151-8. [DOI] [Google Scholar]
- Mendola D. L.; Magri A.; Hansson O.; Bonomo R. P.; Rizzarelli E. Copper(II) complexes with peptide fragments encompassing the sequence 122–130 of human doppel protein. J. Inorg. Biochem. 2009, 103 (5), 758–65. 10.1016/j.jinorgbio.2009.01.017. [DOI] [PubMed] [Google Scholar]
- La Mendola D.; Magri A.; Santoro A. M.; Nicoletti V. G.; Rizzarelli E. Copper(II) interaction with peptide fragments of histidine-proline-rich glycoprotein: Speciation, stability and binding details. J. Inorg. Biochem. 2012, 111, 59–69. 10.1016/j.jinorgbio.2012.02.027. [DOI] [PubMed] [Google Scholar]
- Sóvágó I.; Sanna D.; Dessí A.; Várnagy K.; Micera G. EPR and potentiometric reinvestigation of copper(II) complexation with simple oligopeptides and related compounds. J. Inorg. Biochem. 1996, 63 (2), 99–117. 10.1016/0162-0134(95)00185-9. [DOI] [Google Scholar]
- Pettit L. D.; Pyburn S.; Bal W.; Kozlowski H.; Bataille M. A study of the comparative donor properties to Cu of the terminal amino and imidazole nitrogens in peptides. J. Chem. Soc., Dalton Trans. 1990, (12), 3565–3570. 10.1039/dt9900003565. [DOI] [Google Scholar]
- Pappalardo G.; Impellizzeri G.; Bonomo R. P.; Campagna T.; Grasso G.; Saita M. G. Copper(ii) and nickel(ii) binding modes in a histidine-containing model dodecapeptide. New J. Chem. 2002, 26 (5), 593–600. 10.1039/b110655d. [DOI] [Google Scholar]
- Daniele P. G.; Prenesti E.; Ostacoli G. Ultraviolet-circular dichroism spectra for structural analysis of copper(II) complexes with aliphatic and aromatic ligands in aqueous solution. J. Chem. Soc., Dalton Trans. 1996, (15), 3269–3275. 10.1039/DT9960003269. [DOI] [Google Scholar]
- Kowalik-Jankowska T.; Ruta-Dolejsz M.; Wiśniewska K.; Łankiewicz L.; Kozłowski H. Copper(II) complexation by human and mouse fragments (11–16) of β-amyloid peptide. J. Chem. Soc., Dalton Trans. 2000, (24), 4511–4519. 10.1039/b006125p. [DOI] [Google Scholar]
- Kowalik-Jankowska T.; Kadej A.; Kuczer M.; Czarniewska E. Copper(II) complexes of the Neb-colloostatin analogues containing histidine residue structure stability biological activity. Polyhedron 2017, 134, 365–375. 10.1016/j.poly.2017.06.023. [DOI] [Google Scholar]
- Stich T. A.; Lahiri S.; Yeagle G.; Dicus M.; Brynda M.; Gunn A.; Aznar C.; Derose V. J.; Britt R. D. Multifrequency Pulsed EPR Studies of Biologically Relevant Manganese(II) Complexes. Appl. Magn. Reson. 2007, 31 (1–2), 321–341. 10.1007/BF03166263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon D. M.; Brophy M. B.; Bowman S. E.; Stich T. A.; Drennan C. L.; Britt R. D.; Nolan E. M. Manganese binding properties of human calprotectin under conditions of high and low calcium: X-ray crystallographic and advanced electron paramagnetic resonance spectroscopic analysis. J. Am. Chem. Soc. 2015, 137 (8), 3004–16. 10.1021/ja512204s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski H.; Luczkowski M.; Remelli M. Prion proteins and copper ions. Biological and chemical controversies. Dalton Trans 2010, 39 (28), 6371–85. 10.1039/c001267j. [DOI] [PubMed] [Google Scholar]
- Folcarelli S.; Battistoni A.; Carri M. T.; Polticelli F.; Falconi M.; Nicolini L.; Stella L.; Rosato N.; Rotilio G.; Desideri A. Effect of Lys-->Arg mutation on the thermal stability of Cu,Zn superoxide dismutase: influence on the monomer-dimer equilibrium. Protein Eng., Des. Sel. 1996, 9 (4), 323–5. 10.1093/protein/9.4.323. [DOI] [PubMed] [Google Scholar]
- Zablotna E.; Kret A.; Jaskiewicz A.; Olma A.; Leplawy M. T.; Rolka K. Introduction of alpha-hydroxymethyamino acid residues in substrate specificity P1 position of trypsin inhibitor SFTI-1 from sunflower seeds retains its activity. Biochem. Biophys. Res. Commun. 2006, 340 (3), 823–8. 10.1016/j.bbrc.2005.12.074. [DOI] [PubMed] [Google Scholar]
- Tosato M.; Di Marco V. Metal Chelation Therapy and Parkinson’s Disease: A Critical Review on the Thermodynamics of Complex Formation between Relevant Metal Ions and Promising or Established Drugs. Biomolecules 2019, 9 (7), 269. 10.3390/biom9070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurchi V. M.; de Guadalupe Jaraquemada-Pelaez M.; Crisponi G.; Lachowicz J. I.; Cappai R.; Gano L.; Santos M. A.; Melchior A.; Tolazzi M.; Peana M.; Medici S.; Zoroddu M. A. A new tripodal kojic acid derivative for iron sequestration: Synthesis, protonation, complex formation studies with Fe(3+), Al(3+), Cu(2+) and Zn(2+), and in vivo bioassays. J. Inorg. Biochem. 2019, 193, 152–165. 10.1016/j.jinorgbio.2019.01.012. [DOI] [PubMed] [Google Scholar]
- Lachowicz J. I.; Nurchi V. M.; Crisponi G.; Cappai I.; Cappai R.; Busato M.; Melchior A.; Tolazzi M.; Peana M.; Garribba E.; Zoroddu M. A.; Coni P.; Pichiri G.; Aaseth J. para-Aminosalicylic acid in the treatment of manganese toxicity. Complexation of Mn2+ with 4-amino-2-hydroxybenzoic acid and its N-acetylated metabolite. New J. Chem. 2018, 42 (10), 8035–8049. 10.1039/C7NJ04648K. [DOI] [Google Scholar]
- Jin M.; Xiao A.; Zhu L.; Zhang Z.; Huang H.; Jiang L. The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express 2019, 9 (1), 138. 10.1186/s13568-019-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.