The primary cilium regulates neuronal migration via cyclic AMP production activating protein kinase A at the centrosome.
Abstract
The primary cilium (PC) is a small centrosome-assembled organelle, protruding from the surface of most eukaryotic cells. It plays a key role in cell migration, but the underlying mechanisms are unknown. Here, we show that the PC regulates neuronal migration via cyclic adenosine 3’-5’ monosphosphate (cAMP) production activating centrosomal protein kinase A (PKA). Biosensor live imaging revealed a periodic cAMP hotspot at the centrosome of embryonic, postnatal, and adult migrating neurons. Genetic ablation of the PC, or knockdown of ciliary adenylate cyclase 3, caused hotspot disappearance and migratory defects, with defective centrosome dynamics and altered nucleokinesis. Delocalization of PKA from the centrosome phenocopied the migratory defects. Our results show that the PC and centrosome form a single cAMP signaling unit dynamically regulating migration, further highlighting the centrosome as a signaling hub.
INTRODUCTION
The primary cilium (PC) is a small microtubule-based organelle templated by the centrosome and involved in multiple cellular events, including cell motility (1) and neuronal migration in particular (2–6). How the PC regulates neuronal migration remains largely unknown.
Neuronal migration is essential to the formation of functional neural circuits with defective migration leading to severe brain malformations and involved in psychiatric disorders (7). Most migrating neurons display a cyclic saltatory migration with alternations of somal translocation and pauses. Nucleus and centrosome move forward in a “two-stroke” cycle, with the centrosome moving first within a swelling in the leading process [centrokinesis (CK)] and the nucleus following subsequently [nucleokinesis (NK)] (Fig. 1A). Centrosome dynamics is thus pivotal in the regulation of migration, through microtubular centrosome to nucleus coupling allowing NK (8–12).
Fig. 1. cAMP dynamics in migrating neurons of the postnatal RMS.
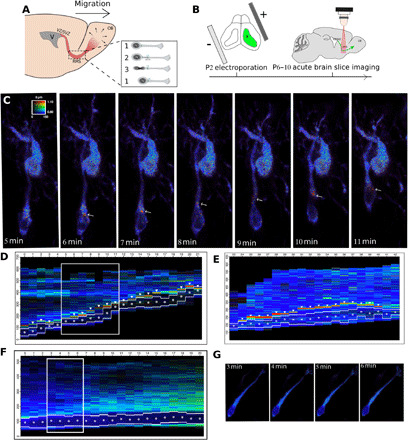
(A) V/SVZ–olfactory bulb (OB) migration pathway and cyclic saltatory migration. 1: pause. 2: leading process extension and centrosome forward movement CK. 3: NK. VZ, ventricular zone; SVZ, sub-ventricular zone. (B) Postnatal experimental procedure. (C) Live two-photon imaging of a representative migrating neuron transfected by Epac-SH187. The white arrows show a dynamic cAMP hotspot present during NK. A nonmigrating transfected cell is present in the field of view. (D) Kymograph of the migrating neuron shown in (C) (time points within the white rectangle) with additional time points showing a second NK (see also movie S1). (E) Kymograph of another migrating neuron transfected by Epac-SH187 (movie S2). All kymographs display the ratiometric measures of the biosensor along the migration direction, with the hotspot indicated by arrowheads. The position of the nucleus is indicated by white lines (borders of the nucleus) and a white dot (center of the nucleus). (F) Kymograph of the nonmigrating neuron transfected by Epac-SH187 shown in (F) (movie S3). (G) Four time points of the cell represented in kymograph. The cell does not migrate but looks healthy.
As the PC is a cyclic adenosine 3′,5′-monophosphate (cAMP)–rich source (13–16), we asked whether it could regulate neuronal migration through cAMP dynamics, in the context of migration along the postnatal rostral migratory stream (RMS) from the ventricular/subventricular zone (V/SVZ) to olfactory bulb (OB) (Fig. 1A).
RESULTS
cAMP dynamics in migrating neurons of the postnatal RMS: A transient centrosomal hotspot during NK
To investigate cAMP dynamics in migrating neurons, we electroporated neonate mice with an intraventricularly injected plasmid to express the Förster resonance energy transfer (FRET)–based cAMP-specific biosensor Epac-SH187 (Fig. 1B) (17). Biosensor-expressing neurons were live imaged in acute sections of the RMS with a two-photon microscope. Ratiometric analysis revealed that a cAMP-rich region (hereafter called hotspot) was present during most NK (Fig. 1, C to E, and movies S1 and S2). Qualitative analysis revealed that 82% of migrating neurons displayed a dynamic hotspot during NK (101 neurons analyzed, 134 NK from 10 mice). This was further confirmed quantitatively by measuring the ratio along the cell profile (see Materials and Methods and figs. S1 and S2). In contrast, the hotspot was predominantly absent during pauses (Fig. 1, F and G, fig. S2, and movie S3). Heatmaps representing the presence of the hotpot in migrating neurons (Fig. 2A) or in nonmigrating neurons (Fig. 2B) show that the frequency of the hotspot is significantly higher in cells exhibiting an NK (Mann-Whitney test (U = 674.75, P < 0.001). This hotspot specifically reflected a cAMP local enrichment, since it was absent from neurons transfected with a version of Epac-SH187 with mutated cAMP-binding site (movie S4). The cAMP hotspot diameter was 0.97 ± 0.10 μm, and the maximum hotspot ratio level was constant during NK, being 52% (±2%) higher than the mean cell ratio (see method in fig. S1). The hotspot usually appeared at the front of the cell body before moving into the leading process, where it remained until after the nucleus performed NK (Fig. 1, C to E; figs. S1 and S2; and movies S1 and S2). As this movement resembled the centrosome movement during migration, we co-electroporated the biosensor and centrin-RFP (red fluorescent protein) to label the centrosome. The hotspot co-localized with the centrosome before and during NK (Fig. 2C). Thanks to biosensor imaging in acute slices (18), we thus discovered a dynamic cAMP hotspot at the centrosome of RMS migrating neurons.
Fig. 2. Occurrence of hotspot and colocalization with centrosome.
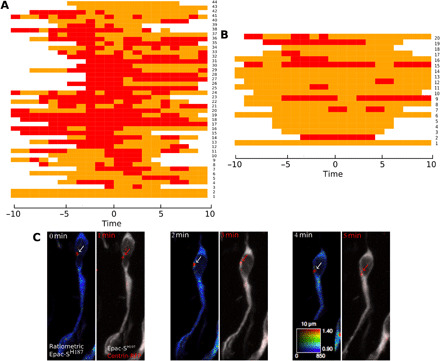
(A) Heatmap of the time course of hotspot presence in multiple migrating neurons. Time 0 is defined as the midpoint of NK. (B) Heatmap of the time course of hotspot presence in multiple nonmigrating neurons. Each row represents a single cell, and each column represents a time point. The presence of the hotspot at a given time point is represented as a red rectangle and its absence as an orange rectangle. White rectangle corresponds to nonrecorded time points. The frequency of the hotspot presence (red) is reduced in nonmigrating neurons as compared to migrating ones. Mann-Whitney test (U = 674.75, P = 0.001). Note that the occasional presence of a hotspot in nonmigrating cells might correspond to an NK that happened just before recording. (C) Live two-photon and confocal imaging of a representative migrating neuron transfected by Epac-SH187 and centrin-RFP. A dynamic cAMP hotspot (white arrows) is present at the centrosome (red arrows).
The PC is necessary for centrosomal hotspot formation through adenylate cyclase 3
The PC is a small rod-shaped organelle linked to the centrosome by its axoneme (Fig. 3A). Embryonic migrating neurons assemble a PC (2, 3, 16), and immunostainings revealed that it is also the case for RMS migrating neurons (Fig. 3B and fig. S3), similar to what was described recently (5). The PC is a cAMP-rich region (13–16), raising the possibility that the cAMP hotspot at the centrosome could be PC dependent. To test this hypothesis, we used two mouse lines in which ablation of the PC can be genetically induced by Cre recombination (Kif3alox/lox and Rpgrip1llox/lox mice). Cre-electroporated mice were compared to green fluorescent protein (GFP)–electroporated mice. Cre electroporation led to an efficient ablation of the PC in both lines (fig. S3). Cotransfection of the biosensor with Cre in floxed mice showed that cilium ablation led to a concomitant disappearance of the hotspot in both lines (Fig. 3C and movies S5 and S6), showing that the PC is necessary for the cAMP hotspot formation.
Fig. 3. PC ablation and AC3 KD lead to disappearance of the cAMP hotspot in the majority of neurons.
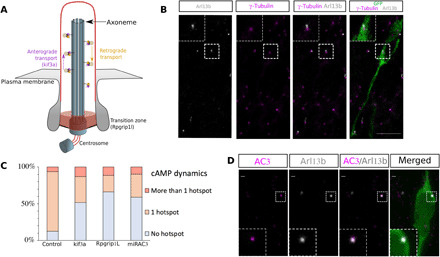
(A) Scheme of the PC. (B) Immunohistochemistry of a GFP-positive RMS neuron (green) in control Kif3alox/lox mice showing a short Arl13b-positive PC (gray) connected to the γ-tubulin–positive centrosome (magenta). The PC length was, on average, 0.64 ± 0.03 μm (N = 3 mice, n = 35 PC). Scale bar: 10 μm. (C) Percentage of neurons with no hotspot, one or more than one hotspot in control or Kif3alox/lox and Rpgrip1lox/lox CRE, or mirAC3-transfected neurons. Control 82% cells with hotspot (N = 10, n = 101) versus CRE-Kif3alox/lox 30% (N = 4, n = 36) and CRE-Rpgrip1llox/lox 20% (N = 7, n = 52) and miRAC3 31% (N = 3, n = 62). (Fisher’s exact test, P < 0.001). (D) Immunocytochemistry of a GFP-positive neuron (green) in control Kif3alox/lox mice showing AC3 subcellular localization (magenta) in the Arl13B-positive PC (gray). Scale bars: 1 μm
The membrane-bound adenylate cyclase 3 (AC3) is the predominant cAMP producer in neuronal PC (19). Immunostainings indeed revealed that AC3 is subcellularly localized in the PC (Fig. 3D). We thus designed a microRNA (miRNA) targeting AC3 mRNA coupled to TdTomato (fig. S4) and co-electroporated it with the biosensor. AC3 knockdown (KD) led to similar disappearance of the hotspot as PC ablation (movie S7 and Fig. 3C), showing that ciliary AC3 produces the hotspot.
Migration defects in the absence of hotspot
To assess whether the hotspot influences migration, we compared the migration of control neurons with the migration of cilium-ablated or AC3 KD neurons, conditions leading to hotspot disappearance. Kif3alox/lox mice were thus transfected with GFP, Cre-GFP, or miRAC3-GFP. Tracking of the GFP cells showed that cilium ablation and AC3 KD both slowed down migration (Fig. 4A and movies S9 to 11) and increased pausing time (Fig. 4B). The frequency of NK was reduced (Fig. 4C), while the speed and distance of NK were unchanged. Moreover, the mutated neurons displayed an altered morphology with their leading process being more often bifurcated than control neurons (fig. S5). To confirm that the migratory phenotype of recombined Kif3alox/lox mice was due to cilium ablation, we performed the same experiment in the PC-ablated Rpgrip1llox/lox mice and observed the same defects (fig. S6).
Fig. 4. Migration defects after PC ablation and AC3 KD.

(A) Speed of neurons electroporated with GFP, CRE-GFP, or miRAC3-GFP in Kif3alox/lox background. GFP: 75.49 ± 3.48 μm/hour; CRE: 55.81 ± 2.74 μm/hour; miRAC3: 58.92 ± 2.35 μm/hour [one-way analysis of variance (ANOVA) (F4, 387 = 7.87, P < 0.001), followed by Tukey test (***P < 0.001)]. ns, not significant. (B) Percentage of pausing time of neurons electroporated with GFP, CRE-GFP, or miRAC3-GFP in Kif3alox/lox background. GFP: 76%; CRE: 87%; miRAC3: 84% (Pearson’s χ2 test = 40.032, P < 0.001). (C) NK frequency of neurons electroporated with GFP, CRE-GFP, or miRAC3-GFP in Kif3alox/lox background. GFP: 3.15 ± 0.21 NK/hour; CRE: 2.06 ± 0.20 NK/hour; miRAC3: 2.27 ± 0.16 NK/hour [one-way Kruskal-Wallis (χ2 = 19.544, P < 0.001, df = 4), followed by Nemenyi (**P < 0.01 and ***P < 0.001)]. The black line represents the median. GFP: N = 3, n = 48; CRE: N = 3, n = 40; miRAC3, N = 3, n = 85.
Defects of centrosome dynamics in the absence of hotspot
Given the centrosomal localization of the hotspot, we wondered whether the hotspot could directly play a role in centrosome dynamics. We cotransfected centrin-RFP with either GFP, Cre-GFP, or miRAC3-GFP in Kif3alox/lox background and cotracked the cell body and centrosome in migrating neurons [movies S12 (GFP) and S13 (CRE)]. The maximum nucleus-centrosome distance reached by the centrosome during CK was reduced in cilium-ablated and AC3 KD neurons, compared to GFP neurons (Fig. 5A). Furthermore, inefficient CK (CK not followed by NK) was increased in cilium-ablated and AC3 KD neurons compared to GFP neurons (Fig. 5B), suggesting that the hotspot is necessary for proper coupling of centrosome and nucleus. Together, our results show that the ciliary-dependent cAMP hotspot regulates migration by a direct action on the centrosome, with consequences on centrosome dynamics and NK.
Fig. 5. Centrosome dynamics defects after PC and AC3 KD.
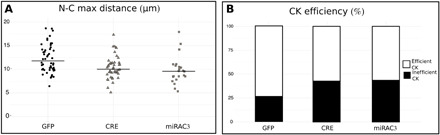
(A) Maximal distance between nucleus and centrosome during CK of neurons electroporated with centrin-RFP and GFP, CRE-GFP, or miRAC3-GFP in Kif3alox/lox background: The maximal distance was reduced in cilium-ablated and AC3 KD neurons [GFP 12.32 ± 0.42 μm versus CRE 10.42 ± 0.35 μm and miRAC3 10.84 ± 0.44 μm; one-way Kruskal-Wallis test (χ2 = 13.54, P = 0.003, df = 3), followed by Nemenyi test (*P < 0.05)]. (B) Percentage of CK efficiency in neurons electroporated with centrin-RFP and GFP, CRE-GFP, or miRAC3-GFP in Kif3alox/lox background: The percentage of inefficient CK was increased in cilium-ablated and AC3 KD neurons. (GFP 26% versus Cre 43% and miRAC3 38%, Pearson’s χ2 test = 10.75, P = 0.01). The black lines represent the median. GFP: N = 3, n = 50; CRE: N = 3, n = 46; miRAC3: N = 3, n = 61.
Centrosomal PKA is the downstream effector of the hotspot
PKA is localized at the centrosome of diverse cell types including neurons (20–22). Immunostaining for the catalytic subunit of PKA (cPKA) showed that it is the case in RMS migrating neurons, where it appears as a diffuse area surrounding the centrosome (Fig. 6A), while it was not detected in the PC (not shown). To test the role of PKA, we electroporated the regulatory subunit of PKA devoid of its centrosomal anchoring domain [dominant-negative PKA (dnPKA)] coupled to GFP, which traps the endogenous cPKA in the cytoplasm (Fig. 6B and fig. S7) (22). Similar to cilium ablation and AC3 KD, dnPKA slowed down migration (Fig. 6C), increased pausing phases (Fig. 6D), and reduced frequency of NK (Fig. 6E and movies S9 and S14).
Fig. 6. PKA is centrosomal, and its delocalization phenocopies cilium ablation.
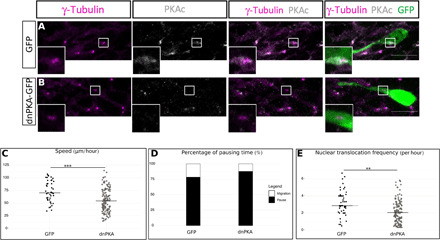
(A) Immunocytochemistry of a GFP-positive neuron showing PKAc subcellular localization (gray) around γ-tubulin–positive centrosome (magenta) (N = 3, n = 43). Scale bar: 10 μm. (B) Immunocytochemistry of a dnPKA-GFP–positive neuron showing a delocalization of PKAc (gray) from the γ-tubulin–labeled centrosome (magenta) (N = 2, n = 30). Scale bar: 10 μm. (C) Speed of neurons electroporated with GFP or dnPKA-GFP in Kif3alox/lox background. GFP: 75.49 ± 3.48 μm/hour; dnPKA: 55.76 ± 1.90 μm/hour [one-way ANOVA (F4,383 = 7.87, P < 0.001), followed by Tukey post hoc test (***P < 0.001)]. (D) Percentage of pausing time of neurons electroporated with GFP or dnPKA-GFP in Kif3alox/lox background. GFP: 76%; dnPKA: 86% (Pearson’s χ2 test = 67.25, P < 0.001). (E) NK frequency of neurons electroporated with GFP or dnPKA-GFP in Kif3alox/lox background. GFP: 3.15 ± 0.21 NK/hour; dnPKA: 2.23 ± 0.12 NK/hour [one-way Kruskal-Wallis (χ2 = 19.57, P < 0.001, df = 4), followed by Nemenyi (***P < 0.001)]. The black line represents the median. GFP: N = 3, n = 48; dnPKA: N = 3, n = 146. **P < 0.01.
Moreover, cotransfection with centrin-RFP revealed that centrosome dynamics was altered: The maximum distance between centrosome and nucleus during CK was reduced as well as the percentage of efficient CK (Fig. 7, A and B). Delocalization of PKA from the centrosome thus phenocopies cilium ablation and AC3 KD, suggesting that centrosomal PKA is the downstream effector of the hotspot. To confirm this result, we cotransfected Kif3alox/lox mice with Cre and dnPKA (fig. S8 and movie S15): The nonadditivity of the phenotypes strongly suggests that PKA indeed acts downstream of the ciliary-produced cAMP to regulate migration at the centrosome.
Fig. 7. Centrosome dynamics defects after PKA delocalization.
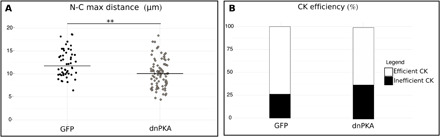
(A) Maximal distance between nucleus and centrosome during CK of neurons electroporated with centrin-RFP and GFP or dnPKA-GFP in Kif3alox/lox background: The maximal distance was decreased in dnPKA neurons [GFP 12.32 ± 0.42 μm versus dnPKA 10.46 ± 0.38 μm; one-way Kruskal-Wallis test (χ2 = 13.54, P = 0.003, df = 3), followed by Nemenyi test (**P < 0.01)]. The black line represents the median. (B) Percentage of CK efficiency in neurons electroporated with centrin-RFP and GFP or dnPKA-GFP in Kif3alox/lox background: The percentage of inefficient CK was increased in dnPKA neurons (GFP 26% versus dnPKA 37%, Pearson’s χ2 test = 6.06, P = 0.027). GFP: N = 3, n = 50; dnPKA: N = 3, n = 86.
The hotspot is also present in adult and embryonic migrating neurons
To test whether this pattern of cAMP dynamics is a general feature of migrating neurons, we assessed its presence in other types of neuronal migration. Radially migrating neurons in the postnatal OB display a transient centrosomal hotspot (Fig. 8A and movie S16) as well as tangentially migrating neurons of the adult RMS (Fig. 8B and movie S17) and radially migrating neurons of the embryonic cortex (Fig. 8C and movie S18). We can thus conclude that the centrosomal cAMP hotspot is a general feature of migrating neurons, independent of age and type of migration.
Fig. 8. The cAMP hotspot is present in different kinds of migrating neurons.
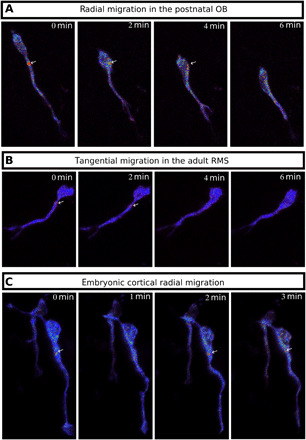
(A) Live two-photon imaging of a radially migrating neuron in the OB transfected by Epac-SH187. The cAMP hotspot is present during NK. (B) Live two-photon imaging of a tangentially migrating neuron in the adult RMS transfected by Epac-SH187. The cAMP hotspot is present during NK. (C) Live two-photon imaging of a radially migrating neuron in the embryonic cortex transfected by Epac-SH187. The cAMP hotspot is present during NK.
DISCUSSION
The importance of the PC for neuronal migration has been inferred from ciliopathies (23) and evidenced in embryonic cortical interneurons (2, 3). The PC is often considered as a signal integrator, with paramount importance for Shh signaling (2, 24). However, the mechanisms by which cilium-mediated signaling is converted into a migratory response are not understood (25). Our data provide such an intracellular mechanism, whereby the centrosome acts as a cAMP/PKA signaling platform downstream of the PC, with AC3-produced cAMP reaching the centrosome, where it locally activates PKA to regulate centrosome dynamics and the subsequent NK. Recently, the PC in postnatal and adult RMS was shown to cyclically emerge from the plasma membrane of migrating neurons before and during NK (5). Notably, this is coincident with the hotspot formation, so that the cyclic emergence of the PC could directly explain the cAMP hotspot cyclicity.
The centrosome as a signaling center is an attractive concept (26). Its importance as a cAMP signaling platform was hypothesized with the discovery of PKA centrosomal localization (20, 21). A study using a centrosome-targeted FRET biosensor in cultured cells reported a low-cAMP microdomain at the centrosome, important for cell cycle progression (27). Regulation of the pattern of division of neural progenitors also involves PKA at the centrosome (22). Here, we report a role of the centrosome as a cAMP signaling platform in a nonmitotic cell, with a functional output on the regulation of migration through local PKA activation. The colocalization of the cAMP hotspot with PKA at the centrosome might ensure that the concentration of cAMP locally exceeds the activation threshold of PKA (28).
This raises the question of the possible targets of PKA at the centrosome. A possible PKA downstream effector could be the Lis1-dynein-nde1-ndeL1 complex, which is localized at the centrosome (10, 29), essential for neuronal migration in diverse types of neurons (11, 30, 31) and involved in nucleus-centrosome coupling (11, 32). PKA phosphorylates Ndel1 and Nde1, which induce the release of Lis1 and dynein from the complex (11, 30), thus allowing their action on microtubule stability (33). Depletion of Lis1 induces a decrease in N-C distances and a decrease in NK frequency (10, 11), which is very similar to the phenotypes of our hotspotless mutants. Lis1 depletion also results in microtubule destabilization (33), potentially triggering a defect of microtubule tension between centrosome and nucleus (34). This suggests that defective Lis1 release consecutive to the absence of the cAMP hotspot might be at the origin of the migratory defects that we observed.
The PC-derived cAMP pathway described here is important for the rhythm of migration but not necessary for migration per se. Ciliary-mutated neurons ultimately reach the OB (not shown) even though at a slower pace. This is consistent with previous works in which ciliary-mutated neurons correctly reach their targets both in mice (35) and in Caenorhabditis elegans (36).
To conclude, we show that the PC and centrosome are functionally linked by cAMP signaling in migrating neurons. We propose that PC and centrosome may be considered as a single cAMP signaling unit, where the PC is a cAMP producer through local AC3, and the centrosome is the effector, through activation of centrosomal PKA by short-range diffusion of ciliary cAMP. This might create a direct link between the PC and the microtubule-organizer function of the centrosome. This cAMP dialog between PC and centrosome might exist in other ciliated cell types to subserve diverse functions in health and disease.
MATERIALS AND METHODS
Mouse lines
Mice were housed in a 12-hour light/dark cycle, in cages containing two females and one male. The postnatal mice were housed in the cages with their parents. Animal care was conducted in accordance with standard ethical guidelines [National Institutes of Health (NIH) publication no. 85-23, revised 1985 and European Committee Guidelines on the Care and Use of Laboratory Animals 86/609/EEC]. The experiments were approved by the local ethics committee (Comité d’Ethique en Expérimentation Animale Charles Darwin C2EA-05 and the French Ministère de l’Education Nationale de l’Enseignement Supérieur et de la Recherche APAFIS#13624-2018021915046521_v5). We strictly performed this approved procedure. The mice used were in a C57BL6-J background. Kif3a conditional knockout (Kif3alox/lox) (37) and Rpgrip1l conditional knockout (Rpgrip1llox/lox) were genotyped according to the original protocols. Kif3a is part of a kinesin motor required for cilium maintenance (38). Rpgrip1l is a protein located at the transition zone of the PC, which mutation leads to ciliopathies in humans (39).
Plasmid constructions
pCaggs-Epac-SH187 was derived from pCDNA3-Epac-SH187 given by K. Jalink (17). Minor modification on the Epac-SH187 sequence was performed to reduce codon redundancy without altering the protein sequence. pCaggs-GFP, pCaggs-CRE-IRES-GFP, and pCaggs-CRE-IRES-Tdtomato were designed in the laboratory. pCIG-PKA RIΔ2-6 wt subunit flagged (call later on dn-PKA) was given by E. Marti (22). pCs2-centrin-RFP was ordered in Addgene (no. 26753). All the plasmids were used at concentrations between 5 to 8 μg/μl (0.01% Fast green) for postnatal electroporation, 2 μg/μl (0.01% Fast green) for in utero electroporation, and 10 μg/μl for adult electroporation.
Biosensor
The Epac-SH187 cAMP biosensor is composed of a part of Epac protein coupled to a donor and an acceptor fluorophore (14). This biosensor displays a high ratio change and excellent photostability for measuring live cAMP concentration in the micromolar range. It switches from a high FRET conformation to a lower FRET conformation upon binding of cAMP. Changes in cAMP concentration were analyzed by ratioing donor (mTurquoise2) and acceptor (td-cpVenus) images and represented by pseudo-colors (see Fig. 1, C and D).
miRNA production
Silencing of AC3 has been performed using BLOCK-iT Pol II miR RNAi Expression Vector kits (Invitrogen) and the RNAi Designer (Invitrogen). The sequences of the single-stranded oligos are AC3.3: top: TGCTGTTAGGATGGAGCACACGGCATGTTTTGGCCACTGACTGACATGCCGTGCTCCATCCTAA; bottom: CCTGTTAGGATGGAGCACGGCATGTCAGTCAGTGGCCAAAACATGCCGTGTGCTCCATCCTAAC. The double-stranded oligos were inserted in a pcDNA 6.2-GW/EmGFP-miR. To produce the pcDNA6.2-GW/Tdtomato-miR, GFP was replaced by tdTomato using Dra1 and Bam HI. The resulting constructions were sequenced before use.
Postnatal electroporation
Postnatal electroporation was performed at postnatal day 2 (P2). The postnatal mice were anesthetized by hypothermia. Pseudo-stereotaxic injection [from lambda medial-lateral (M/L): −1,2; anterior-posterior (A/P): 2; dorsal-ventral (D/V): 2,5-2] using glass micropipette (Drummond Scientific Company, Wiretrol I 50 μl, 5-000-1050) was performed, and 2 μl of plasmid (between 5 and 8 μg/μl) was injected. Animals were subjected to five pulses of 99.9 V during 50 ms separated by 950 ms using the CUY21 SC Electroporator and 10-mm tweezer electrode (CUY650-10, Nepa Gene). The animals were placed on 37°C plates to restore their body temperature before returning with their mother. Animals were considered as fully restored when pups were moving naturally, and their skin color returned to pink.
Acute brain slices
Brain slices from mice aged from P6 to P10 were prepared as previously described (40). Pups were sacrificed by decapitation, and the brain was quickly removed from the skull. Sagittal brain slices (250 μm) were cut with a VT1200S microtome (Leica). Slices were prepared in the ice-cold cutting solution of the following composition: 125 mM NaCl, 0.4 mM CaCl2, 1 mM MgCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, 5 mM sodium pyruvate, 20 mM glucose, and 1 mM kynurenic acid, saturated with 5% CO2 and 95% O2. Slices were incubated in this solution for 30 min at room temperature and then placed in recording solution (identical to the solution used for cutting, except that the Ca2+ concentration was 2 mM and kynurenic acid was absent) for at least 30 min at 32°C before image acquisition.
Time-lapse video microscopy
To analyze cell migration and centrosome dynamics, images were obtained with an inverted SP5D confocal microscope (Leica). Images were taken every 3 min for 2 to 3 hours using a 40×/ 1.25–numerical aperture (N.A.) objective with 1.5 optical zoom.
Biosensor images were acquired with an upright two-photon microscope Leica SP5 MPII with a 25×/ 0.95-N.A. objective, 4× optical zoom, and a GaAsP hybrid detector. The excitation wavelength was set at 850 nm to excite the mTurquoise2 donor. The two emission wavelengths were acquired simultaneously with filters of 479 ± 20 nm and 540 ± 25 nm. Image stacks with 2-μm intervals were taken every minute for 1 hour. The presence of tdTomato, indicative of CRE recombinase or miRNA, was assessed with a confocal head. For simultaneous detection of the centrin-RFP and biosensor, two-photon excitation for the biosensor and confocal head for centrin-RFP were alternated. Stacks were spaced by 1 μm and acquired every 2 min.
The temperature in the microscope chamber was maintained at 32°C for embryos and postnatal imaging or 35°C for P30 imaging, and brain slices were continuously perfused with heated recording solution (see above) saturated with 5% CO2 and 95% O2.
Analyses of neuronal migration
Analyses were performed using ImageJ (NIH Image; National Institutes of Health, Bethesda, MD) software and MTrackJ plugin (41). The nucleus and the centrosome of each cell were tracked manually on each time frame during the whole movie. For cell migration and centrosome movement characteristics, calculation of speed, nuclear translocation frequency, and pausing time were performed using the x,y,t coordinates of the nucleus of each cell.
Cells were excluded from the analysis if they were tracked during less than 30 min or did not perform any nuclear translocation during the whole tracking. A cell was considered as migrating if it performed a distance superior to 6 μm during a 3-min interval.
A CK was defined as a forward movement superior to 2 μm followed by a backward movement superior to 2 μm. Maximal distance between centrosome and nucleus of every CK was considered.
Quantification of biosensor images
Image stacks obtained for donor and acceptor emission were processed with a custom code developed in the IGOR Pro environment (WaveMetrics, Lake Oswego, OR, USA). The maximum intensity was projected vertically to form a two-dimensional image. The fluorescence intensities of donor and acceptor were averaged to build an image indicating biosensor concentration. The fluorescence intensities of donor and acceptor were ratioed for each pixel to report biosensor activation level. For each experiment, ratio values were multiplied by a constant such that baseline ratio was close to 1. These images were combined to produce pseudo-color images, with the ratio used as hue (from blue to red) and the intensity as value, while the saturation was set to the maximum (42).
For each migrating neuron, the ratio was analyzed in one dimension, along its migration direction, and calculated as described in fig S1. A series of anchor points were manually positioned along the length of the cell, and the profile of ratio and intensity was calculated along these line segments, over a total width of four pixels. The intensity profile was annotated manually, marking both sides of the nucleus. The movement of the nucleus during the recording was then used to determine the axis of the migration, by fitting a line to the positions of the center nucleus for all time points. Coordinates in the frame of reference of the image were then converted to a single position along the axis of cell migration. The width of the hotspot was estimated as two times the width parameter of a Gaussian function fitted to the ratio trace.
Immunohistochemistry
P7 to P10 mice were lethally anesthetized using euthasol. Intracardiac perfusion with 4% paraformaldehyde was performed. Brain was postfixed overnight in 4% paraformaldehyde. Three rinses were done with phosphate-buffered saline (PBS) 1× (Gibco, 1400-067). Fifty-micrometer sagittal slices were cut with VT1200S microtome (Leica). Slices were placed 1 hour in a saturation solution (10% fetal bovine serum; 0.5% Triton X-100 in PBS). The primary antibodies used in this study were as follows: GFP (Aves, GFP-1020, 1/1000), PKAc (Cell Signaling Technology, #4782, 1/250), AC3 (Santa Cruz Biotechnology, C-20 sc-588, 1/200), Arl13b (UC Davis/NIH NeuroMab Facility, 75-287, 1/1000), and γ-tubulin (Sigma-Aldrich, T6557, 1/500). The antibodies were diluted in saturation solution. Slices were incubated for 48 to 72 hours at 4°C under agitation with the antibodies. Three rinses were done with PBS 1×. The secondary antibodies used were as follows: anti-chicken immunoglobulin Y (IgY) Alexa Fluor 488 (1/1000; Jackson ImmunoResearch: 703-545-155) against anti-GFP, anti-rabbit IgG Cy5 (1/1000; Jackson ImmunoResearch: 711-175-152) against anti-PKAc, anti-rabbit IgG Cy3 (1/2000; Jackson ImmunoResearch: 711-165-152) against anti-AC3, anti-mouse IgG, Fcγ subclass 1–specific Alexa Fluor 594 (1/2000; Jackson ImmunoResearch: 115-585-205) against anti–γ-tubulin, anti-mouse IgG, and Fcγ subclass 2a–specific Alexa Fluor 647 (1/1000; Jackson ImmunoResearch: 115-605-206) against anti-Arl13b. The antibodies were diluted in saturation solution. Slices were incubated for 1 hour at room temperature under agitation with the secondary antibody solution. Three rinses with PBS 1× were done. Slices were counter-colored with Hoechst and mounted with Mowiol.
To quantify AC3 KD, a PC was considered AC3 negative when it was clearly immunonegative at high magnification (objective: 63×; zoom 3 plus numerical zoom 4). To quantify cPKA delocalization after dnPKA electroporation, the quantification of the centrosome was considered to be devoid of PKA when it was clearly immunonegative (objective: 63×; zoom 3 plus numerical zoom 4).
Statistics
All manipulations and statistical analyses were implemented with R (3.5.1). Normality in the variable distributions was assessed by the Shapiro-Wilk test. Furthermore, the Levene test was performed to probe homogeneity of variances across groups. Variables that failed the Shapiro-Wilk or the Levene test were analyzed with nonparametric statistics using the one-way Kruskal-Wallis analysis of variance (ANOVA) on ranks followed by Nemenyi test post hoc and Mann-Whitney rank sum tests for pairwise multiple comparisons. Variables that passed the normality test were analyzed by means of one-way ANOVA followed by Tukey post hoc test for multiple comparisons or by Student’s t test for comparing two groups. Categorical variables were compared using Pearson’s χ2 test or Fisher’s exact test. All the statistical analyses have been performed on the five groups together (GFP, Cre, miRAC3, dnPKA, and dnPKA/CRE).
A P value of <0.05 was used as a cutoff for statistical significance. Results are presented as the means ± SD or medians, and the given values in the text are the means ± SEM, unless otherwise stated. The statistical tests are described in each figure legend.
In utero electroporation
Timed pregnant embryonic day 15 (E15) C57Bl6 mice were anesthetized with isoflurane. During the experiment, mice were on a 37°C plate. The uterine horns were exposed, and the embryos were injected in the lateral ventricle with a glass micropipette. One microliter of plasmid (2 μg/μl) was injected. The successfully injected animals were then subjected to five pulses of 45 V during 50 ms separated by 950 ms using the CUY21 SC Electroporator and 5-mm tweezer electrode (CUY650-5, Nepa Gene). The uterine horns were then replaced in the belly, and the belly was sewn up. The animals were placed in a cage on 37°C plates to restore their body temperature before returning to their own cage.
Adult electroporation
P21 C57Bl6 animals were anesthetized with a mixture of ketamine and xylazine. Stereotaxic injection (from the bregma M/L: 1; A/P: 0; D/V: 2,1) was performed with a glass micropipette, and 2 μl of the plasmids (10 μg/μl) was injected in the right lateral ventricle. The animals were then removed from the stereotaxic setting and subject to five pulses of 200 V during 50 ms separated by 950 ms using the NEPA21 electroporator with 10-mm tweezer electrode (CUY650-10). The animals were then placed on a 37°C plates to restore their body temperature before returning to their cage.
Acute brain slices of embryonic and adult brains
For imaging of E18 embryos, the mother was sacrificed by cervical dislocation, and the embryos were removed from the horn and placed in an ice-cold cutting solution (same composition as above). The embryos were decapitated, and the brain was quickly removed from the skull. Sagittal brain slices (300 μm) were cut in the same ice-cold solution. Then, the same protocol as above was followed. P30 mice were sacrificed by cervical dislocation. The brain was quickly removed from the skull. Sagittal brain slices (200 μm) were cut with a VT1200S microtome (Leica). Slices were prepared in an ice-cold solution of the following composition: 130 mM potassium gluconate, 15 mM KCl, 2 mM EGTA, 20 mM Hepes, 25 mM glucose, 1 mM CaCl2, and 6 mM MgCl2, supplemented with 0.05 mM D-(-)-2-Amino-5-phosphonopentanoic acid (D-APV) (304 mosmol, pH 7.4 after equilibration) saturated with 5% CO2 and 95% O2 (43). Then, the slices were placed for 3 to 5 min in an ice-cold solution of the following composition: 225 mM d-mannitol, 2.5 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 25 mM glucose, 1 mM CaCl2, and 6 mM MgCl2 (43) saturated with 5% CO2 and 95% O2 before being placed in the recording solution (described above) at 32°C and saturated with 5% CO2 and 95% O2 for at least 30 min.
Supplementary Material
Acknowledgments
We are grateful to K. Jalink for Epac-SH187 plasmid and to E. Marti for dnPKA plasmid. We thank all re-readers including I. Dusart, S. Nédelec, F. Doetsch, and N. Spassky. The experiments were performed in the IBPS imaging facility, and the mice were housed in the IBPS animal facility. We thank C. Fournier-Thibault for lending her electroporator. Funding: this work was supported by the CNRS, INSERM, Fondation Lejeune, Sorbonne University. J.S. was funded by a doctoral fellowship awarded by the IBPS to A.T. and P.V. labs. This work was supported (in part) by the Investissements d’Avenir program managed by the ANR under reference ANR-11-IDEX-0004-02. A.T. and P.V. labs are members of the Bio-Psy Labex. Author contributions: J.S. performed experiments, developed the methodology, analyzed data, and wrote the manuscript; M.C. performed experiments and analyzed data; C.D. and C.F. performed experiments; M.D. performed formal analysis; C.M. participated in the initiation of the work and provided materials; S.S.-M. provided materials; A.T. acquired funding and edited the manuscript; P.V. acquired funding, developed software, supervised the project, and edited the manuscript; and I.C. acquired funding, performed experiments, conceptualized and supervised the project, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eaba3992/DC1
REFERENCES AND NOTES
- 1.Nishimura Y., Kasahara K., Shiromizu T., Watanabe M., Inagaki M., Primary cilia as signaling hubs in health and disease. Adv. Sci. 6, 1801138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoin J.-P., Viou L., Launay P.-S., Luccardini C., Espeso Gil S., Kiyasova V., Irinopoulou T., Alvarez C., Rio J.-P., Boudier T., Lechaire J.-P., Kessaris N., Spassky N., Métin C., Tangentially migrating neurons assemble a primary cilium that promotes their reorientation to the cortical plate. Neuron 76, 1108–1122 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Higginbotham H., Eom T.-Y., Mariani L. E., Bachleda A., Hirt J., Gukassyan V., Cusack C. L., Lai C., Caspary T., Anton E. S., Arl13b in primary cilia regulates the migration and placement of interneurons in the developing cerebral cortex. Dev. Cell 23, 925–938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J., Higginbotham H., Li J., Nichols J., Hirt J., Ghukasyan V., Anton E. S., Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 6, 7857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto M., Sawada M., García-González D., Herranz-Pérez V., Ogino T., Bang Nguyen H., Quynh Thai T., Narita K., Kumamoto N., Ugawa S., Saito Y., Takeda S., Kaneko N., Khodosevich K., Monyer H., Manuel García-Verdugo J., Ohno N., Sawamoto K., Dynamic changes in ultrastructure of the primary cilium in migrating neuroblasts in the postnatal brain. J. Neurosci. 39, 9967–9988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobin J. L., Di Franco M., Eichers E., May-Simera H., Garcia M., Yan J., Quinlan R., Justice M. J., Hennekam R. C., Briscoe J., Tada M., Mayor R., Burns A. J., Lupski J. R., Hammond P., Beales P. L., Inhibition of neural crest migration underlies craniofacial dysmorphology and Hirschsprung’s disease in Bardet–Biedl syndrome. Proc. Natl. Acad. Sci. 105, 6714–6719 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valiente M., Marín O., Neuronal migration mechanisms in development and disease. Curr. Opin. Neurobiol. 20, 68–78 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Bellion A., Baudoin J.-P., Alvarez C., Bornens M., Métin C., Nucleokinesis in tangentially migrating neurons comprises two alternating phases: Forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25, 5691–5699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belvindrah R., Natarajan K., Shabajee P., Bruel-Jungerman E., Bernard J., Goutierre M., Moutkine I., Jaglin X. H., Savariradjane M., Irinopoulou T., Poncer J.-C., Janke C., Francis F., Mutation of the α-tubulin Tuba1a leads to straighter microtubules and perturbs neuronal migration. J. Cell Biol. 216, 2443–2461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T., Serneo F. F., Higgins C., Gambello M. J., Wynshaw-Boris A., Gleeson J. G., Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165, 709–721 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai J.-W., Bremner K. H., Vallee R. B., Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10, 970–979 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Sherpa R. T., Mohieldin A. M., Pala R., Wachten D., Ostrom R. S., Nauli S. M., Sensory primary cilium is a responsive cAMP microdomain in renal epithelia. Sci. Rep. 9, 6523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trivedi N., Ramahi J. S., Karakaya M., Howell D., Kerekes R. A., Solecki D. J., Leading-process actomyosin coordinates organelle positioning and adhesion receptor dynamics in radially migrating cerebellar granule neurons. Neural Develop. 9, 26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore B. S., Stepanchick A. N., Tewson P. H., Hartle C. M., Zhang J., Quinn A. M., Hughes T. E., Mirshahi T., Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc. Natl. Acad. Sci. U.S.A. 113, 13069–13074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J. Y., Falcone J. L., Curci S., Hofer A. M., Direct visualization of cAMP signaling in primary cilia reveals up-regulation of ciliary GPCR activity following Hedgehog activation. Proc. Natl. Acad. Sci. U.S.A. 116, 12066–12071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mick D. U., Rodrigues R. B., Leib R. D., Adams C. M., Chien A. S., Gygi S. P., Nachury M. V., Proteomics of primary cilia by proximity labeling. Dev. Cell 35, 497–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klarenbeek J., Goedhart J., van Batenburg A., Groenewald D., Jalink K., Fourth-generation epac-based FRET sensors for cAMP feature exceptional brightness, photostability and dynamic range: Characterization of dedicated sensors for FLIM, for ratiometry and with high affinity. PLOS ONE 10, e0122513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castro L. R. V., Guiot E., Polito M., Paupardin-Tritsch D., Vincent P., Decoding spatial and temporal features of neuronal cAMP/PKA signaling with FRET biosensors. Biotechnol. J. 9, 192–202 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Bishop G. A., Berbari N. F., Lewis J., Mykytyn K., Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 505, 562–571 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Nigg E. A., Schäfer G., Hilz H., Eppenberger H. M., Cyclic-AMP-dependent protein kinase type II is associated with the Golgi complex and with centrosomes. Cell 41, 1039–1051 (1985). [DOI] [PubMed] [Google Scholar]
- 21.De Camilli P., Moretti M., Donini S. D., Walter U., Lohmann S. M., Heterogeneous distribution of the cAMP receptor protein RII in the nervous system: Evidence for its intracellular accumulation on microtubules, microtubule-organizing centers, and in the area of the Golgi complex. J. Cell Biol. 103, 189–203 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saade M., Gonzalez-Gobartt E., Escalona R., Usieto S., Martí E., Shh-mediated centrosomal recruitment of PKA promotes symmetric proliferative neuroepithelial cell division. Nat. Cell Biol. 19, 493–503 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Valente E. M., Rosti R. O., Gibbs E., Gleeson J. G., Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 10, 27–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bangs F., Anderson K. V., Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veland I. R., Lindbæk L., Christensen S. T., Linking the primary cilium to cell migration in tissue repair and brain development. Bioscience 64, –1115, 1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arquint C., Gabryjonczyk A.-M., Nigg E. A., Centrosomes as signalling centres. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terrin A., Monterisi S., Stangherlin A., Zoccarato A., Koschinski A., Surdo N. C., Mongillo M., Sawa A., Jordanides N. E., Mountford J. C., Zaccolo M., PKA and PDE4D3 anchoring to AKAP9 provides distinct regulation of cAMP signals at the centrosome. J. Cell Biol. 198, 607–621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koschinski A., Zaccolo M., Activation of PKA in cell requires higher concentration of cAMP than in vitro: Implications for compartmentalization of cAMP signalling. Sci. Rep. 7, 14090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., Hirotsune S., A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Bradshaw N. J., Soares D. C., Carlyle B. C., Ogawa F., Davidson-Smith H., Christie S., Mackie S., Thomson P. A., Porteous D. J., Millar J. K., PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J. Neurosci. 31, 9043–9054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu T., Ayala R., Nguyen M.-D., Xie Z., Gleeson J. G., Tsai L.-H., Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Jheng G.-W., Hur S. S., Chang C.-M., Wu C.-C., Cheng J.-S., Lee H.-H., Chung B.-C., Wang Y.-K., Lin K.-H., del Álamo J. C., Chien S., Tsai J.-W., Lis1 dysfunction leads to traction force reduction and cytoskeletal disorganization during cell migration. Biochem. Biophys. Res. Commun. 497, 869–875 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Sapir T., Elbaum M., Reiner O., Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 16, 6977–6984 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertipaglia C., Gonçalves J. C., Vallee R. B., Nuclear migration in mammalian brain development. Semin. Cell Dev. Biol. 82, 57–66 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Shin J., Prescott M., Mair J., Campbell R. E., Roles for primary cilia in gonadotrophin-releasing hormone neurones in the mouse. J. Neuroendocrinol. 26, 18–25 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Li W., Yi P., Zhu Z., Zhang X., Li W., Ou G., Centriole translocation and degeneration during ciliogenesis in Caenorhabditis elegans neurons. EMBO J. 36, 2553–2566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., Goldstein L. S. B., Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Rosenbaum J. L., Witman G. B., Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Arts H. H., Doherty D., van Beersum S. E. C., Parisi M. A., Letteboer S. J. F., Gorden N. T., Peters T. A., Märker T., Voesenek K., Kartono A., Ozyurek H., Farin F. M., Kroes H. Y., Wolfrum U., Brunner H. G., Cremers F. P. M., Glass I. A., Knoers N. V. A. M., Roepman R., Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 39, 882–888 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Yapo C., Nair A. G., Clement L., Castro L. R., Hellgren Kotaleski J., Vincent P., Detection of phasic dopamine by D1 and D2 striatal medium spiny neurons. J. Physiol. 595, 7451–7475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.E. Meijering, O. Dzyubachyk, I. Smal, Methods in Enzymology (Elsevier, 2012), vol. 504, pp. 183–200. [DOI] [PubMed] [Google Scholar]
- 42.Polito M., Vincent P., Guiot E., Biosensor imaging in brain slice preparations. Methods Mol. Biol. 1071, 175–194 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Quiquempoix M., Fayad S. L., Boutourlinsky K., Leresche N., Lambert R. C., Bessaih T., Layer 2/3 pyramidal neurons control the gain of cortical output. Cell Rep. 24, 2799–2807.e4 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eaba3992/DC1


