Abstract
Berries of genus Vaccinium are rich in flavonoids and proanthocyanidins (PAs). We studied the PA composition and biosynthesis in bilberry (Vaccinium myrtillus L.) tissues and during fruit development. Soluble PAs, analyzed by UHPLC–MS/MS, were most abundant in stem and rhizome with the mean PA polymerization level varying between 4 and 6 in all tissues. Both A- and B-type PAs were present in all tissues. Procyanidin subunits were more common than prodelphinidin subunits in PAs. During fruit ripening, the amount of procyanidin subunits decreased while prodelphinidin subunits and F3′5′H expression increased, indicating a shift in biosynthesis toward the delphinidin branch of the flavonoid pathway. Epicatechin was the most abundant flavan-3-ol in all tissues. Expression of ANR and three isolated LAR genes, analyzed by qRT-PCR, showed connection to accumulation of PAs and flavan-3-ols biosynthesized from different flavonoid branches. Insoluble PAs accumulated during berry development, suggesting that PAs are not recycled after biosynthesis.
Keywords: flavan-3-ols, flavonoid biosynthesis, fruit development, gene expression, proanthocyanidins, Vaccinium myrtillus
Introduction
Genus Vaccinium contains economically important cultivated berry species, including blueberries (e.g. V. corymbosum and V. angustifolium), American cranberry (V. macrocarpon), and wild berry species, such as bilberry (V. myrtillus) and lingonberry (V. vitis-idaea).1 The berries of Vaccinium plants are among the best dietary sources of anthocyanins but they also contain high levels of other health-beneficial flavonoids, namely flavonols and proanthocyanidins (PAs).2
PAs, also known as condensed tannins, are oligomers (2–10 subunits) and polymers (>10 subunits) of flavan-3-ols. PAs are linked to many bioactivities, such as anti-inflammatory and antioxidant properties, and have also been associated with the reduced risks of cardiovascular disease, cancer, and type 2 diabetes.3 PAs show a high degree of structural diversity deriving from the myriad of flavan-3-ol monomers, types of linkage between the monomers (A- and B-type linkages and multiple possible linkage positions), and the degree of polymerization (DP). PA oligomers and polymers consist of one terminal flavan-3-ol unit linked with one or more extension flavan-3-ol units by B-type (most commonly 4ß → 8 or 4ß → 6) and/or A-type (typically with an additional 2ß → O → 7) bonds. Although PAs containing A-type linkages are more rare, both A- and B-type PAs have earlier been detected in Vaccinium berries.4,5
PAs are widely present in various fruits and berries as well as seeds, flowers, and leaves. Like all flavonoids, the biosynthesis of PAs requires products from both the shikimate pathway and the acetate/malonate pathway.6 PA subunits are biosynthesized via the flavonoid pathway sharing the same upstream pathway with anthocyanins (Figure S1). Two specific enzymes in the flavonoid pathway catalyze the formation of two groups of PA monomers, 2,3-trans-flavan-3-ols (catechins) and 2,3-cis-flavan-3-ols (epicatechins).7 Leucoanthocyanidin reductase (LAR) converts leucoanthocyanidins to 2,3-trans-flavan-3-ols (most commonly catechin and gallocatechin) and anthocyanidin reductase (ANR) converts anthocyanidins to 2,3-cis-flavan-3-ols (epicatechin and epigallocatechin).8LAR and ANR genes have been characterized from many plant species but with variable copy numbers. It has been shown that LAR can have a role in the regulation of the relative proportions of terminal and extension units of PAs and therefore affecting the degree of PA oligomerization.8 Furthermore, recent results on overexpression of Camellia sinensis ANR and LAR in tobacco indicated that flavan-3-ol carbocations are also involved in PA biosynthesis.9 The substrates for LAR and ANR are derived from both cyanidin (flavonoid 3′-hydroxylase, F3′H) and delphinidin (flavonoid 3′5′-hydroxylase, F3′5′H) branches of the flavonoid pathway but the specificities toward procyanidin (PC) and prodelphinidin (PD) substrates vary between enzymes/isoforms.10,11 Catechin and epicatechin are produced from the cyanidin branch while gallocatechins and epigallocatechins are produced from the delphinidin branch (Figure S1). The current evidence from several studies suggests that the biosynthesis of PA monomers takes place in the cytosol followed by transportation to vacuoles, similar to other flavonoids.12 Different models have been proposed for the polymerization/oligomerization of PAs in the vacuole but it is still not clear whether the process is enzymatic or nonenzymatic.7,8 It is also unclear whether PAs can further be metabolized after their biosynthesis.
Previous studies have generally shown higher levels of soluble PAs during the early stages of fruit development with a decreasing trend toward maturity in many fruits, including Vaccinium species.13−15 As astringent compounds, PAs are suggested to protect unripe fruits against early feeding and there is a need to abolish astringency from ripe fruits to make them attractive for seed dispersers.16 Because PAs have an effect on taste, mouth feel, and astringency, their biosynthesis has been studied in many fruits and berries. However, detailed studies on the PA composition and biosynthesis in bilberry are still lacking.
In the present study, we have analyzed the PA composition and biosynthesis in bilberry tissues and during bilberry fruit development. The composition of soluble PAs in five different developmental stages of bilberry fruits in addition to leaf, stem, and rhizome was analyzed with ultra-high-performance liquid chromatography (UHPLC)–MS/MS methods targeting PC and PD subunits separately. Additionally, a spectrophotometric acid butanol assay and UHPLC–HRMS/MS were used to further investigate cell wall-bound insoluble PAs and A-type PAs, respectively, to provide comprehensive PA profiles in bilberry. The biosynthesis of PAs was further investigated by identifying specific PA biosynthetic genes from bilberry and analyzing their expression in the same tissues.
Materials and Methods
Plant Material
The tissue samples were collected from wild V. myrtillus L. plants growing in natural forest stand in Oulu (65° 01′ N, 25° 28′ E), Finland. Five developmental stages of bilberry fruit (Figure 1) were collected from June to August 2011, while leaf, stem, and rhizome samples were collected in mid-July 2011, as described earlier.17 The fruit developmental stages were S1, flower collected June 7 (anthesis); S2, small unripe green fruit (15 days after anthesis); S3, large unripe green fruit just before coloring began (28 days after anthesis); S4, ripening purple fruit (34 days after anthesis); and S5, fully ripe blue fruit (55 days after anthesis).17 Immediately after collection, all samples were frozen in liquid nitrogen and stored at −80 °C until they were used for RNA extraction and chemical analyses.
Figure 1.
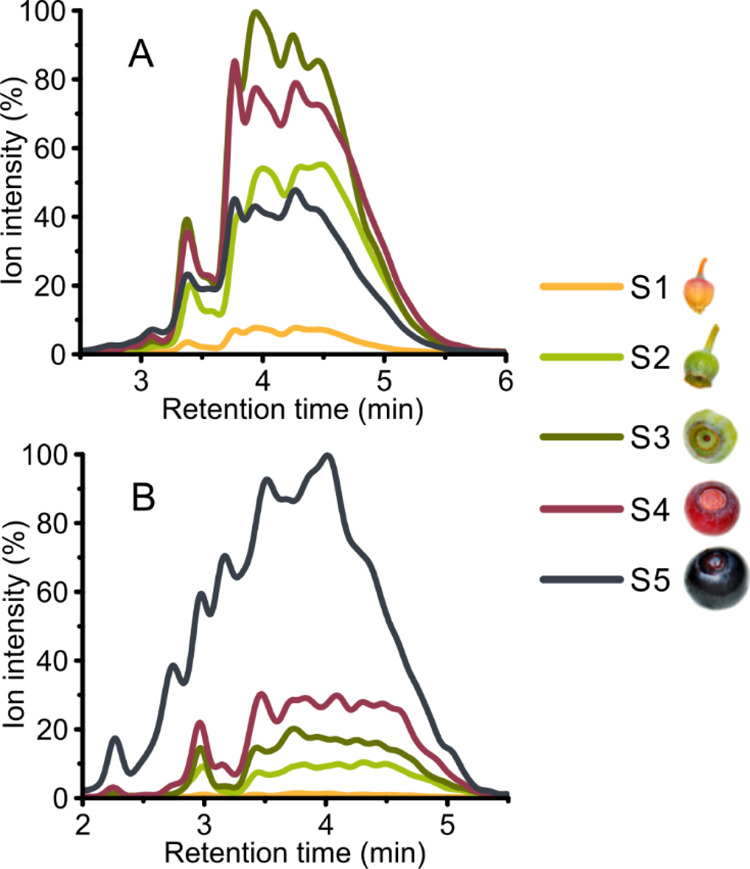
Two-dimensional PA fingerprints of PC units (A) and PD units (B) present in oligomeric and polymeric PAs at different fruit developmental stages determined by UHPLC-DAD–ESI-3Q-MS. (A) Sum fingerprint of PC-type terminal and extension units revealed by PC-specific SRM methods (m/z 287 → 125 and m/z 289 → 245) and (B) sum fingerprint of PD-type terminal and extension units revealed by PD-specific SRM methods (m/z 303 → 125 and m/z 305 → 125). S1, flower; S2, small unripe green fruit; S3, large unripe green fruit; S4, ripening purple fruit; and S5, fully ripe blue fruit.
For chemical analyses, the tissues were further freeze-dried using a vacuum freeze-drier (Edwards High Vacuum International, West Sussex, England). The tissues (11–46 berries from each developmental stage, 32 leaves, 50 cm of rhizome, and 70 cm of stem) were ground using a ball mill. Ground plant materials from the same tissue were pooled together prior to extraction.
Chemicals and Reagents
Water used for UHPLC–MS and acid butanol analyses was purified using a Millipore Synergy UV (Merck KGaA, Darmstadt, Germany) water purification system. ACS-grade acetone was obtained from VWR International S.A.S. (Fontenay-sous-Bois, France) and LC–MS-grade acetonitrile and formic acid from Fluka Analytical (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). Catechin, epicatechin, gallocatechin, and epigallocatechin were purchased from Extrasynthese (Genay, France).
UHPLC-DAD–ESI-3Q-MS
The samples were prepared for the analyses as described by Suvanto et al.,18 with the exception that the lyophilized extracts were dissolved in 1 mL of water, filtered using a syringe filter (0.2 μm, PTFE, 13 mm, VWR International GmbH, Darmstadt, Germany), and diluted 1:4 (v/v) with water. The last two developmental stages of the fruit, S4 and S5, were further diluted 1:4 (v/v) with water.
Quantitative analyses for soluble PAs and flavan-3-ols and data processing were performed with methods developed by Engström et al.19 and further described by Malisch et al.20 and James et al.21 A Waters Acquity UPLC system (Waters Corporation, Milford, MA, USA) consisting of a binary solvent manager, a sample manager, a column oven, and a photodiode array detector (190–500 nm), equipped with an Acquity UPLC BEH Phenyl column (1.7 μm particle size, 100 × 2.1 mm i.d., Waters Corporation), was used. The system was coupled with a Xevo TQ triple quadrupole mass spectrometer (Waters Corporation).
Aqueous (0.1%) formic acid (A) and acetonitrile (B) were used as eluents, the injection volume was 5 μL, and the gradient was as follows: 0.0–0.5 min, 0.1% B in A; 0.5–5.0 min, 0.1–30.0% B in A (linear gradient); 5.0–6.0 min, 30.0–45.0% B in A (linear gradient); and column wash and stabilization. The flow rate was kept at a constant 0.5 mL min–1. For MS, the negative full scan mode was used with ions detected between m/z 150–2000 in addition to the MS/MS transitions described by Engström et al.19 The temperature of the ESI source was 150 °C, desolvation temperature was 650 °C, and capillary voltage was −3.4 kV. N2 was used both as a desolvation and cone gas (1000 L h–1 and 100 L h–1, respectively). Argon was used as a collision gas. Quantitative data processing was done using the TargetLynx software (Version 4.1, Waters Corporation), and the calibration curves were created using procyanidin- and prodelphinidin-rich fractions (59–434 and 159–1112 μg mL–1, respectively) prepared by Sephadex LH-20 column purification. The mean degree of polymerization (mDP) of the PAs was calculated based on the ratios of observed PA terminal and extension units and calibrated using the same PA fractions. Flavan-3-ols were quantified using pure compounds (catechin 40–24,800 ng mL–1, epicatechin 42–26,000 ng mL–1, gallocatechin 42–26,300 ng mL–1, and epigallocatechin 40–24,800 ng mL–1).
UHPLC-DAD–ESI-Orbitrap-MS
To qualitatively and semiquantitatively assess the PAs, UHPLC-DAD–ESI-Orbitrap-MS was used. The extracts used for the analyses were the same as for UHPLC-DAD–ESI-3Q-MS.
The system consisted of an identical Acquity UPLC system as described for UHPLC-DAD–ESI-3Q-MS, coupled with a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). All chromatographic conditions were as described for UHPLC-DAD–ESI-3Q-MS. For MS, a heated ESI source (H-ESI II, Thermo Fisher Scientific Inc.) was operated in the negative ion mode with the following parameters: spray voltage −3.0 kV, sheath gas (N2) flow rate 60 (arbitrary units), aux gas (N2) flow rate 20 (arbitrary units), sweep gas flow rate 0 (arbitrary units), and capillary temperature 380 °C. For a full scan, a resolution of 35,000 was used in the Orbitrap detector, the automatic gain used was 3 × 106, and the mass range was set at m/z 150–2000. To verify the identifications, full scan coupled with data-dependent MS/MS (Top N) analyses were run with a loop count and Top N value of 5, a resolution of 17,500, stepped normalized collision energies of 30, 50, and 80 eV, and an automatic gain of 1 × 105. For the semiquantitative analysis of dimeric and trimeric PAs, extracted ion chromatograms of the theoretical m/z values (±0.10) of singly charged ions were integrated. Pierce ESI Negative Ion Calibration Solution (Thermo Fischer Scientific Inc.) was used for the calibration of the detector and the data were processed with the Thermo Xcalibur Qual Browser software (Version 3.0.63, Thermo Fisher Scientific Inc.).
Acid Butanol Assay
Cell wall-bound insoluble PAs were quantified using an acetone-enhanced butanol–HCl–iron assay.22 After extraction of plant tissue (3–20 mg) with acetone/water (4:1, v/v) using the same method as for UHPLC–MS samples, the extraction residue was washed once using acetone and twice using CH2Cl2/MeOH (1:1, v/v). Each washing step included mixing after solvent addition by vortexing (30 min), centrifugation (10 min, 7800g), and removal of the supernatant. Water was then added to the residue, and the remaining organic solvents were first evaporated using nitrogen and then under vacuum. Finally, the residues were frozen, lyophilized, and stored in −20 °C until analyzed.
For measurements, 1 mL of the freshly prepared modified acetone–butanol–HCl–iron reagent as described by Grabber et al.22 was added to the samples. The samples were mixed for 5 min and incubated in 70 °C for 3 h. A 200 μL aliquot of the sample was measured at 550 nm using a microplate spectrophotometer (Thermo Multiskan Ascent, Thermo Fischer Scientific Inc.). Quantification was performed against PA-enriched standard fractions19 that were prepared and measured identically to the samples.
Isolation of RNA and cDNA Preparation
Total RNA was isolated from bilberry tissues as described previously.23 The cDNA was synthesized from total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. The cDNA was purified from contaminating genomic DNA using the method described by Jaakola et al.24
Isolation and Sequence Analysis of Bilberry LAR Genes
The amplification of the LAR sequences was achieved from bilberry fruit cDNA with gene-specific primers that were designed based on the sequences found in V. macrocarpon sequence databases.25 The polymerase chain reactions (PCR) were performed using Phusion DNA polymerase (Thermo Fisher Scientific Inc.). The amplified PCR products were gel-purified with an E.Z.N.A Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA) and ligated into a pJET cloning vector using a CloneJET PCR Cloning Kit (Thermo Fisher Scientific Inc.). Sequencing reactions were performed using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and the sequencing was done at the UiT, the Arctic University of Norway sequencing facility on a 3130xl Genetic Analyzer (Applied Biosystems). The full-length nucleotide sequences of VmLAR1a, VmLAR1b, and VmLAR2 were deposited to GenBank under accession numbers MT013357, MT013358, and MT013359, respectively.
For alignment and phylogenetic analysis, amino acid sequences of previously characterized LAR proteins of other dicot species were obtained from GenBank and aligned with V. myrtillus LAR sequences using Clustal Omega program. A phylogenetic tree was constructed using the neighbor-joining method with the MEGA software (Version 6.06). The reliability of the tree was evaluated by a bootstrap analysis with 1000 replicates.
Relative Quantification of Gene Expression
Real-time quantitative reverse-transcription PCR (qRT-PCR) analyses were performed with a LightCycler 480 instrument and software (Roche Applied Sciences, Indianapolis, IN, USA). The transcript abundances of bilberry PA biosynthesis specific genes VmANR (GenBank Accession no. FJ666338) and three genes encoding LAR (VmLAR1a, VmLAR1b, and VmLAR2) as well as genes important for the branch point of flavonoid biosynthesis, VmF3′H and VmF3′5′H, were detected using a LightCycler SYBR Green I Master qPCR kit (Roche). The qRT-PCR conditions were an initial incubation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s. The gene-specific primer sequences used for the qRT-PCR analysis are shown in Table S1. Glyceraldehyde-3-phosphate dehydrogenase (VmGAPDH; GenBank Accession no. AY123769) was used as a reference gene for the relative quantification of PCR products. The results were calculated with LightCycler 480 software (Roche), using the calibrator-normalized PCR efficiency-corrected method (Technical note no. LC 13/2001, Roche). The amplification of only one product in qRT-PCR was confirmed by a melting curve analysis and sequencing. The analyses were performed with three to five replicates.
Statistical Analysis
SigmaPlot 14.0 (Systat Software Inc., San Jose, CA, USA) was used for the statistical analyses of the gene expression results. Significant differences between the tissues were calculated by one-way analysis of variance with pairwise post-hoc comparisons by Tukey’s test. Statistical significance was defined at p < 0.05. Correlation analysis was performed using R version 4.0.0 in RStudio version 1.2.5033 with the package corrplot.
Results
Soluble PAs in Bilberry Tissues
In order to acquire profiles of soluble PAs in bilberry tissues, including fruits at different developmental stages, methods utilizing UHPLC–MS/MS were used.19 These methods reveal the concentrations of PC and PD subunits in PAs, along with information on the mDP of the PAs. The selected reaction monitoring (SRM) PA methods utilize the electrospray ionization chamber of the mass spectrometer to cleave monomer-linking bonds of the PAs using three different optimized cone voltages, resulting in quinone methide cleavage depolymerization products, which are further fragmented in the collision cell and filtered using the final quadrupole to eliminate false positives. The concentrations of soluble PAs are presented both per dry weight and per unit (i.e. a single fruit or a leaf) where applicable. The latter is included to demonstrate the accumulation of PAs per berry during development and to therefore enable conclusions on the fate of PAs in a single fruit throughout its development.
Our results indicate PAs as the main group of polyphenols in many bilberry tissues (Figure S2). Generally, our study showed a high total PA concentration in the stem and rhizome while lower levels were detected in the leaf and fruit samples (Table 1). PC subunits were the highest in the PAs of the stem, while the rhizome PAs had the highest PD subunit content. Leaves contained markedly lower concentration of both PC and PD subunits. Overall, higher content of PC subunits was observed compared to PD subunits in all bilberry tissues.
Table 1. Content of Soluble PAs and mDP Level in Bilberry Tissues Determined by UHPLC-DAD–ESI-3Q-MS.
| S1a | S2 | S3 | S4 | S5 | leaf | stem | rhizome | |
|---|---|---|---|---|---|---|---|---|
| mg/g DW | ||||||||
| PC subunits | 8.64 | 35.03 | 27.01 | 14.70 | 6.27 | 16.91 | 75.72 | 63.50 |
| PD subunits | 0.16 | 0.73 | 0.61 | 0.69 | 1.63 | 1.16 | 1.56 | 2.97 |
| total PAs | 8.80 | 35.76 | 27.62 | 15.39 | 7.90 | 18.08 | 77.29 | 66.47 |
| μg/unitb | ||||||||
| PC subunits | 49.42 | 459.97 | 727.90 | 627.47 | 346.60 | 99.44 | ||
| PD subunits | 0.90 | 9.62 | 16.44 | 29.42 | 90.22 | 6.85 | ||
| total PAs | 50.31 | 469.59 | 744.34 | 656.89 | 436.82 | 106.29 | ||
| calculated values | ||||||||
| PC/PD ratio | 55.14 | 47.83 | 44.29 | 21.33 | 3.84 | 14.52 | 48.40 | 21.40 |
| mDP | 4.5 | 6.4 | 5.8 | 5.2 | 6.6 | 4.0 | 5.6 | 4.5 |
Fruit developmental stages: S1, flower; S2, small unripe green fruit; S3, large unripe green fruit; S4, ripening purple fruit; and S5, fully ripe blue fruit.
μg per individual fruit or leaf.
In fruits, variation in PA content during development was detected with the highest PA content found in early/unripe developmental stages (S2 and S3) of the fruit (Table 1). The accumulation of PC and PD subunits in PAs followed variable patterns during fruit development. The concentration of PCs peaked in unripe berries, while the level of PDs elevated during fruit development being the highest in ripe berries (Table 1, Figure 1). Figure 1 shows a detailed overlook of the change in the profiles of the PC and PD subunits of PAs during the fruit development as measured by the SRM methods.
The PC/PD ratio decreased throughout the fruit development, dropping from 55.14 in S1 stage to 3.84 in S5 stage (Table 1). Notably, a three-fold increase from S4 to S5 fruit was detected in PD subunit concentration when quantified per fruit.
Mean Degrees of Polymerization (mDPs) of the PAs
The mDPs of the soluble PAs was fairly low and constant between bilberry tissues with the lowest measured in leaves (Table 1). During the fruit development, just a slight increase was detected in the ripe berry. However, when terminal and extension PC and PD subunits were examined separately, clear differences could be found in the PA polymerization degree. Although the proportion of PC terminal units to PC extension units stays mostly the same during fruit development, suggesting a quantitative rather than a qualitative change in the PC subunit proportions (Figure S3), there is a rapid increase in PD extension units from S4 to S5 fruits while the PD terminal units stay relatively similar both quantitatively and qualitatively. This suggests that the slight increase in mDP in ripe berries can be attributed to the increase in PD extension units in the PAs.
Flavan-3-ols
The concentrations of flavan-3-ols (which are the immediate biosynthetic precursors to PAs) indicated epicatechin as the most abundant flavan-3-ol in all bilberry tissues (Table 2). During the fruit development, especially at fruit ripening, the amount of epicatechins and epigallocatechins decreased, with the effect being more pronounced in epicatechins. Conversely, catechins and gallocatechins accumulated to ripe fruits when quantified per fruit. Similar to the PC/PD ratio, the ratio of (epi)catechins to (epi)gallocatechins dropped during the development of the fruit (from 17.08 in S1 to 3.16 in S5). The total cis/trans ratio was most heavily on the cis side in the rhizome, followed by leaves and S2 and S3 berries (Table 2). The lowest cis/trans ratios were found in the fully ripe S5 berries and stem. Overall, the cis/trans ratio remained fairly constant during the fruit development, with a sharp increase in the ratio of trans flavan-3-ols in the ripe fruit. The same effect was observed for both catechin/epicatechin and gallocatechin/epigallocatechin ratio.
Table 2. Content of Flavan-3-ols in Bilberry Tissues Determined by UHPLC-DAD–ESI-3Q-MS.
| S1a | S2 | S3 | S4 | S5 | leaf | stem | rhizome | |
|---|---|---|---|---|---|---|---|---|
| mg/g DW | ||||||||
| catechin | 0.04 | 0.10 | 0.04 | 0.02 | 0.03 | 0.02 | 0.88 | 0.12 |
| epicatechin | 2.55 | 6.98 | 4.39 | 1.59 | 0.75 | 2.63 | 19.50 | 13.10 |
| gallocatechin | 0.05 | 0.08 | 0.05 | 0.05 | 0.09 | 0.06 | 0.54 | 0.08 |
| epigallocatechin | 0.10 | 0.48 | 0.26 | 0.21 | 0.16 | 0.46 | 1.69 | 5.74 |
| total flavan-3-ols | 2.74 | 7.64 | 4.74 | 1.88 | 1.03 | 3.16 | 22.61 | 19.04 |
| μg/unitb | ||||||||
| catechin | 0.25 | 1.27 | 1.05 | 0.93 | 1.67 | 0.10 | ||
| epicatechin | 14.59 | 91.74 | 118.44 | 67.96 | 41.44 | 15.45 | ||
| gallocatechin | 0.30 | 1.03 | 1.31 | 2.34 | 4.74 | 0.33 | ||
| epigallocatechin | 0.57 | 6.33 | 7.02 | 9.17 | 8.90 | 2.69 | ||
| total flavan-3-ols | 15.71 | 100.37 | 127.83 | 80.40 | 56.75 | 18.57 | ||
| calculated values | ||||||||
| (epi)catechin/(epi)gallocatechin ratio | 17.08 | 12.63 | 14.34 | 5.99 | 3.16 | 5.16 | 9.12 | 2.27 |
| cis/trans flavan-3-ol ratio | 27.86 | 42.56 | 53.03 | 23.57 | 7.85 | 42.79 | 14.85 | 94.31 |
Fruit developmental stages: S1, flower; S2, small unripe green fruit; S3, large unripe green fruit; S4, ripening purple fruit; and S5, fully ripe blue fruit.
μg per individual fruit or leaf.
A-Type Linkages in Bilberry PAs
Our semiquantitative results obtained by UHPLC-DAD–ESI-Orbitrap-MS analyses showed that PAs with at least one A-type linkage were detected in all bilberry tissues (Table S2). The highest proportion of PAs containing at least one A-type linkage was detected in the rhizome. Furthermore, dimeric and trimeric A-type PCs were more abundant than dimeric and trimeric B-type PCs in the rhizome. The UHPLC-DAD–ESI-Orbitrap-MS method was selected for detecting PAs with A-type linkages because the UHPLC-DAD–ESI-3Q-MS methods used in this study for soluble PA quantification do not fully detect all monomeric PA subunits with A-type linkages. The used UHPLC–MS/MS methods are optimized to cleave B-type bonds in PAs, resulting in some A-type bonds possibly staying intact. The mDPs of the PAs are possibly underestimated slightly because of the same reason. However, as each A-type-linkage-containing PA most often contained only one A-type linkage, the overall inaccuracies in the results are minor.
Insoluble PAs in Bilberry
Because soluble PAs do not give full overview of PA accumulation in tissues, insoluble PAs were also determined. The cell wall-bound insoluble PAs were measured using microplate spectrophotometry to complete the quantification of total PAs in the tissues. The highest amounts of soluble PAs were found in the rhizome followed by the stem with lower levels present in leaves and fruits (Table 3). Their concentrations showed an increased accumulation during bilberry fruit development especially toward fully ripe fruit when quantified per fruit (Table 3).
Table 3. Content of Insoluble PAs in Bilberry Tissues Determined by the Acid Butanol Assay.
| S1a | S2 | S3 | S4 | S5 | leaf | stem | rhizome | ||
|---|---|---|---|---|---|---|---|---|---|
| insoluble PAs | mg/g DW | 42.18 | 38.89 | 35.22 | 26.99 | 42.91 | 28.92 | 57.19 | 79.90 |
| μg/unitb | 241.45 | 510.83 | 949.40 | 1151.90 | 2373.03 | 170.11 |
Fruit developmental stages: S1, flower; S2, small unripe green fruit; S3, large unripe green fruit; S4, ripening purple fruit; and S5, fully ripe blue fruit.
μg per individual fruit or leaf.
Identification of Bilberry PA Biosynthetic Genes
In order to connect PA accumulation to biosynthesis, genes specifically attending to PA biosynthesis were identified from bilberry. A search from Vaccinium sequence databases for homologues of LAR genes led to the identification of three sequences. They were isolated in full-length from V. myrtillus and named VmLAR1a, VmLAR1b, and VmLAR2. At the amino acid level, VmLAR1a and VmLAR1b showed 74% sequence identity to each other, while VmLAR2 shared a sequence identity of 61 and 65% with VmLAR1a and VmLAR1b, respectively. Multiple sequence alignment analysis showed that the predicted amino acid sequences of the isolated bilberry LARs had high homology with other members of dicot LAR proteins (Figure S4). All the bilberry LAR sequences displayed the RFLP, ICCN, and THD motifs as well as NADP-binding site GXXGXXG, which all are reported to be conserved among LAR proteins,26,27 verifying the three bilberry sequences as LAR proteins. Phylogenetic analysis demonstrated that LAR1 and LAR2 proteins grouped as their own clusters including V. myrtillus LARs with the exception of sequences of Rosaceae species, which all grouped together (Figure S4). Clustering of Rosaceae PA biosynthetic genes together has been reported previously by Henry-Kirk et al.28 Our search from Vaccinium databases25,30 identified only one ANR gene sequence that has already been reported previously for bilberry.29
Expression Patterns of PA Biosynthetic Genes
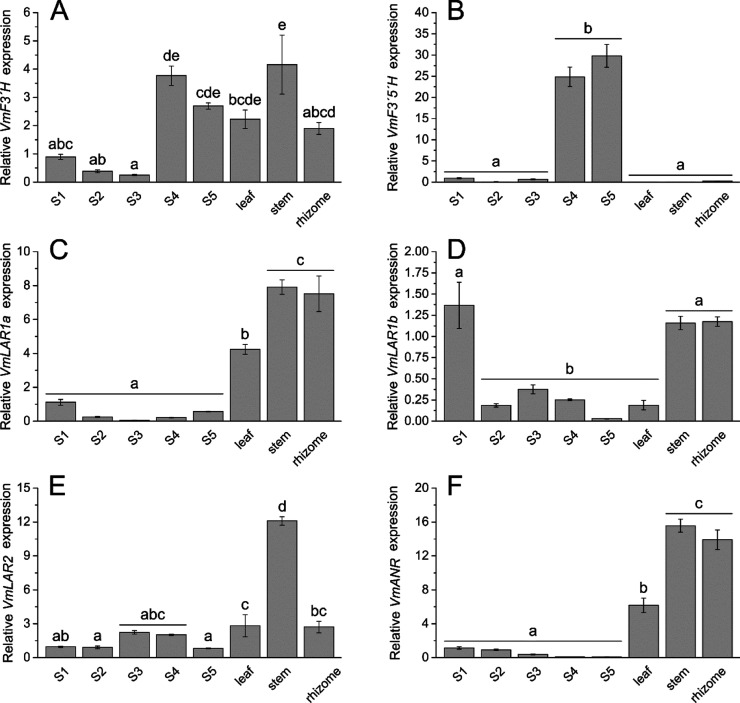
The expression of PA biosynthetic genes in the bilberry fruit at five different developmental stages as well as in the leaf, stem, and rhizome was analyzed by qRT-PCR (Figure 2). For most genes, the expression was relatively high in the stem and rhizome. Instead, VmF3′5′H had an exceptionally high expression in the late developmental stages of the berry (Figure 2B). The three LAR genes showed variable expression patterns during bilberry fruit development. The expression of VmLAR1a and VmLAR1b was high in flowers. However, while VmLAR1a expression first decreases toward the large unripe stage (S3) and then increases toward fruit ripening, the expression of VmLAR1b increases toward the large unripe stage and then decreases at fruit ripening (Figure 2C,D). VmLAR2 expression during fruit development also decreased toward fruit ripening showing somewhat an opposite trend compared to VmLAR1a (Figure 2E). The expression of VmANR was high in the stem and rhizome but also in leaves and the expression in the fruit showed a decreasing trend throughout the development (Figure 2F).
Figure 2.
Transcript abundance of flavonoid biosynthetic genes VmF3′H (A), VmF3′5′H (B), VmLAR1a (C), VmLAR1b (D), VmLAR2 (E), and VmANR (F) determined by qRT-PCR in bilberry tissues. Fruit developmental stages: S1, flower; S2, small unripe green fruit; S3, large unripe green fruit; S4, ripening purple fruit; and S5, fully ripe blue fruit. Values represent means ± SE of three to five biological replicates. Values with no shared letters show statistically significant difference (one-way analysis of variance, pairwise post-hoc comparisons with Tukey’s test, p < 0.05).
Discussion
The total quantity of soluble PAs was the highest in the bilberry rhizome and stem but was also high in the unripe fruit, likely to protect the maturation of the seeds by inhibiting early feeding.16 Overall, PC subunits were prevalent in all bilberry tissues indicating higher biosynthetic activity of the cyanidin branch of the flavonoid pathway. The PD concentration, however, peaked in the ripe fruit indicating that the delphinidin branch of the flavonoid pathway is increasingly providing substrates for PA biosynthesis toward fruit ripening. This was also confirmed by the noticeably elevated expression of VmF3′5′H in S4 and S5 fruits. The result is consistent with earlier studies on Vaccinium berries showing activation of the delphinidin branch and the expression of F3′5′H during the ripening stage.13,15 Thus, our results may indicate that PD-based PAs can have another role in bilberry fruits besides protection from early feeding. It should be noted that PD-rich PA mixtures and their monomeric constituents are on average associated with higher antioxidant and anthelmintic activities compared to their PC counterparts,31−33 owing to the additional hydroxyl group in the B-ring and the resulting pyrogallol structure. Thus, increased amounts of PDs in ripe bilberries increase their nutritional value. Even though the bilberry leaf had lower absolute quantity of PD subunits compared to the stem and rhizome (when quantified against dry weight of the tissue), a smaller PC/PD ratio was observed in the leaves.
The mDPs of the PAs remained relatively constant in all bilberry tissues and during fruit development, suggesting that in bilberry the mDP is not a significant contributing factor in deterring early feeding of fruits. According to present knowledge, no all-encompassing generalizations between the polymerization degree of PAs and their bioactivities can be made. For example, PAs with a higher degree of polymerization are typically linked with higher antimicrobial activity, while their ability to reduce inflammation or act as antioxidants is not as clear.34 However, our results demonstrate that the mDP value does not describe the true DP distribution of PAs in tissue. Salminen35 has already earlier remarked that the same mDP value can be achieved from PA profiles, which are completely unlike one another as it does not give information of the true size distribution of the PAs in a sample. Our results show that while the single numeric mDP value was fairly similar in all tissue samples, the MS/MS methods revealed that the true distribution of PC and PD terminal and extensions units varied markedly between the tissues. We found a rapid increase in PD extension units from S4 to S5 fruits while the PD terminal units stay relatively similar both quantitatively and qualitatively (Figure S3). This suggests that the slight increase in mDP at ripe berries can be attributed to the increase in PD extension units in the PAs. Therefore, also the quick increase in PD-rich PAs especially in quantity, but also in the degree of polymerization, in the ripe fruit supports our data of higher PD subunit amount toward fruit ripening. Furthermore, the fingerprints of the PC and PD terminal and extension units also showed that while the profiles of the PC terminal and extension units were similar throughout fruit development and in the stem and rhizome, the PC subunits in the leaf had larger dispersity. Similar results were observed in the PD subunits as the stem and rhizome had very similar profiles, while the leaf showed larger dispersity. Thus, it is important to consider the distribution of both PC and PD terminal and extension units to get a true overview of the PAs in any plant tissue.
The quantitative results of flavan-3-ols (catechin, epicatechin, gallocatechin, and epigallocatechin) per fruit correlated moderately with their corresponding soluble PAs (PCs and PDs) during berry maturation (Figure S5A). Levels of epicatechins and catechins, which were the prevalent flavan-3-ols in all bilberry tissues, correlated with the concentrations of PC subunits, dropping during the final maturation stage. The same trend has earlier been observed in, for example, strawberry fruits (Fragaria × ananassa Duch.),36 grapes (Vitis vinifera),37 and grape seeds.38 Correspondingly, the levels of gallocatechin seem to correlate those of PD subunits, reaching their maximum levels in the fully ripe berry. As for other plant tissues, especially PC subunits and epicatechin shared trends in their quantities (Figure S5B).
Our study identified A-type PAs in all bilberry tissues. Our results are in agreement with previous studies, as A-type PAs have been reported to appear prevalently in bilberry, lingonberry, bog bilberry (V. uliginosum), and cranberry (V. oxycoccos), whereas common B-type PAs are prominent in blueberry.4
Our results on insoluble PA accumulation to ripe berries strongly suggest that the PAs are not further modified or degraded after they are biosynthesized but are merely transferred to the cell wall constantly during fruit development and maturation. It can be assumed that in the cell wall, PAs become insoluble and their function as a deterrent for feeding is diminished once the fruits are ripe and ready for seed dispersal. Therefore, the content of both soluble and insoluble PAs should always be considered to avoid drawing false conclusions on the presence or fate of PAs in plant tissues. It should be noted, however, that due to the insoluble PAs having been measured using a different method than the soluble PAs and the possible underestimation of PAs due to A-type linkages as described earlier, the ratio of soluble-to-insoluble PAs should be considered as an estimation.
Earlier, variable copy number of both LAR and ANR genes has been reported in different plant species. For example, in Arabidopsis and Medicago truncatula, only one ANR gene exists10 while two ANR genes have been identified in the apple genome.40 The presence of only one ANR gene in the bilberry genome indicates that the enzyme utilizes both PC and PD precursors for epicatechin biosynthesis. In M. truncatula, a single LAR gene exists, while in Arabidopsis, functional LARs are totally lacking and PAs consist exclusively of epicatechin units limited to seed coat.8 However, many plants produce both epicatechin- and catechin-based PAs with one or more copies of LAR genes in their genomes. For example, two LAR genes exist in grapevine and apple genomes11 while three LAR genes, similarly to bilberry, were recently reported in C. sinensis.39 Based on our phylogenetic analysis, bilberry LARs seem to be homologous for C. sinensis LARs.
The ANR and all three LAR genes showed high expression in the stem and rhizome, which is in accordance with the highest PA levels found in these tissues in our study. In light of the PA content during bilberry fruit development, the results may suggest that from the three identified LAR genes, VmLAR1a has substrate specificity toward leucodelphinidins, while VmLAR1b and VmLAR2 prefer leucocyanidin as a substrate. The profile of VmANR expression showed correlation with the detected epicatechin and epigallocatechin levels (Figure S5), demonstrating a decreasing trend toward the mature fruit. Overall, even though VmANR had relatively high expression in the nonberry tissues, all tissue samples showed a heavy and fairly similar bias toward 2,3-cis-flavan-3-ols as opposed to their trans counterparts, indicating high ANR enzyme activity (Figure 2 and Table 3). The role of LARs in PA biosynthesis is not totally understood and according to the recent knowledge, in addition to biosynthesis of 2,3-trans-flavan-3-ols (catechin and gallocatechin), LAR genes have been indicated to have a role in regulation of PA polymerization.8
To conclude, PAs are currently considered as end products of the flavonoid pathway and our results are in agreement with this presumption. The PA levels seemingly decrease at the onset of ripening when only soluble PAs are taken into account. However, their transport to the cell wall should be considered, as a notable portion of PAs appears as cell wall-bound insolubles. Overall, our results demonstrate the necessity of using valid and comprehensive methodology to avoid overlooking a part of the PAs and possibly drawing false conclusions from incomplete results. Furthermore, the detailed PA fingerprints achieved by our MS/MS methods are extremely useful in accurately predicting bioactivities, as many of them are affected by the PC/PD ratio and the distribution of PAs of different degrees of polymerization, among other factors.34,35,41−43
Acknowledgments
The authors thank Saku Valkamaa for conducting the laboratory work for the acid butanol assay and Annika Lehtimäki for her work with the PA-enriched standard fractions used for the same analyses.
Glossary
Abbreviations Used
- 3Q-MS
triple quadrupole mass spectrometry
- ANR
anthocyanidin reductase
- DP
degree of polymerization
- ESI
electrospray ionization
- F3′H
flavonoid 3′-hydroxylase
- F3′5′H
flavonoid 3′5′-hydroxylase
- (HR)MS/MS
(high resolution) tandem mass spectrometry
- LAR
leucoanthocyanidin reductase
- mDP
mean degree of polymerization
- PA
proanthocyanidin
- PC
procyanidin
- PD
prodelphinidin
- qRT-PCR
real-time quantitative reverse transcription PCR
- SRM
selected reaction monitoring
- U(H)PLC
ultra(-high)-performance liquid chromatography
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.0c02158.
PA biosynthetic pathway; UV chromatograms (λ = 280 nm) of the bilberry tissue samples from the UHPLC-DAD–ESI-3Q-MS analyses; fingerprints of PCs and PDs in bilberry tissues determined from the UHPLC-DAD–ESI-3Q-MS analyses, showing terminal and extension units separately; comparison of the deduced amino acid sequences of LAR genes from bilberry with other species; correlation matrices of measured concentrations and expressions among berry and all tissue samples; gene-specific primers used for quantitative real-time PCR analysis; and heatmap of the semiquantitative relative concentrations of dimeric and trimeric PAs with A- and B-type bonds (PDF)
This work was financially supported by the Niemi Foundation and Osk. Huttunen Foundation to K.K.
The authors declare no competing financial interest.
Supplementary Material
References
- Karppinen K.; Zoratti L.; Nguyenquynh N.; Häggman H.; Jaakola L. On the developmental and environmental regulation of secondary metabolism in Vaccinium spp. berries. Front. Plant Sci. 2016, 7, 655. 10.3389/fpls.2016.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttä-Riihinen K. R.; Kamal-Eldin A.; Mattila P. H.; González-Paramás A. M.; Törrönen A. R. Distribution and contents of phenolic compounds in eighteen Scandinavian berry species. J. Agric. Food Chem. 2004, 52, 4477–4486. 10.1021/jf049595y. [DOI] [PubMed] [Google Scholar]
- de la Iglesia R.; Milagro F. I.; Campión J.; Boqué N.; Martínez J. A. Healthy properties of proanthocyanidins. BioFactors 2010, 36, 159–168. 10.1002/biof.79. [DOI] [PubMed] [Google Scholar]
- Määttä-Riihinen K. R.; Kähkönen M. P.; Törrönen A. R.; Heinonen I. M. Catechins and procyanidins in berries of Vaccinium species and their antioxidant activity. J. Agric. Food Chem. 2005, 53, 8485–8491. 10.1021/jf050408l. [DOI] [PubMed] [Google Scholar]
- Grace M. H.; Esposito D.; Dunlap K. L.; Lila M. A. Comparative analysis of phenolic content and profile, antioxidant capacity, and anti-inflammatory bioactivity in wild Alaskan and commercial Vaccinium berries. J. Agric. Food Chem. 2014, 62, 4007–4017. 10.1021/jf403810y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen J.-P.; Karonen M. Chemical ecology of tannins and other phenolics: we need a change in approach. Funct. Ecol. 2011, 25, 325–338. 10.1111/j.1365-2435.2010.01826.x. [DOI] [Google Scholar]
- Tian L.; Pang Y.; Dixon R. A. Biosynthesis and genetic engineering of proanthocyanidins and (iso)flavonoids. Phytochem. Rev. 2008, 7, 445–465. 10.1007/s11101-007-9076-y. [DOI] [Google Scholar]
- Liu C.; Wang X.; Shulaev V.; Dixon R. A. A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants 2016, 2, 16182. 10.1038/nplants.2016.182. [DOI] [PubMed] [Google Scholar]
- Wang P.; Liu Y.; Zhang L.; Wang W.; Hou H.; Zhao Y.; Jiang X.; Yu J.; Tan H.; Wang Y.; et al. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 2020, 101, 18–36. 10.1111/tpj.14515. [DOI] [PubMed] [Google Scholar]
- Xie D.-Y.; Sharma S. B.; Dixon R. A. Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch. Biochem. Biophys. 2004, 422, 91–102. 10.1016/j.abb.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Pfeiffer J.; Kühnel C.; Brandt J.; Duy D.; Punyasiri P. A. N.; Forkmann G.; Fischer T. C. Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus x domestica Borkh.) and other crops. Plant Physiol. Biochem. 2006, 44, 323–334. 10.1016/j.plaphy.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Zhao J. Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. 10.1016/j.tplants.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Jaakola L.; Määttä K.; Pirttilä A. M.; Törrönen R.; Kärenlampi S.; Hohtola A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. 10.1104/pp.006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya I. O.; Vorsa N. Flavonoid composition over fruit development and maturation in American cranberry, Vaccinium macrocarpon Ait. Plant Sci. 2004, 167, 1043–1054. 10.1016/j.plantsci.2004.06.001. [DOI] [Google Scholar]
- Primetta A. K.; Karppinen K.; Riihinen K. R.; Jaakola L. Metabolic and molecular analyses of white mutant Vaccinium berries show down-regulation of MYBPA1-type R2R3 MYB regulatory factor. Planta 2015, 242, 631–643. 10.1007/s00425-015-2363-8. [DOI] [PubMed] [Google Scholar]
- Harborne J.Phytochemistry of fruits and vegetables: an ecological overview. In Phytochemistry of Fruits and Vegetables; Tomas-Barberan F. A., Ed.; Oxford University Press: New York, 1997; pp 353–367. [Google Scholar]
- Karppinen K.; Hirvelä E.; Nevala T.; Sipari N.; Suokas M.; Jaakola L. Changes in the abscisic acid levels and related gene expression during fruit development and ripening in bilberry (Vaccinium myrtillus L.). Phytochemistry 2013, 95, 127–134. 10.1016/j.phytochem.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Suvanto J.; Nohynek L.; Seppänen-Laakso T.; Rischer H.; Salminen J.-P.; Puupponen-Pimiä R. Variability in the production of tannins and other polyphenols in cell cultures of 12 Nordic plant species. Planta 2017, 246, 227–241. 10.1007/s00425-017-2686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström M. T.; Pälijärvi M.; Fryganas C.; Grabber J. H.; Mueller-Harvey I.; Salminen J.-P. Rapid qualitative and quantitative analyses of proanthocyanidin oligomers and polymers by UPLC-MS/MS. J. Agric. Food Chem. 2014, 62, 3390–3399. 10.1021/jf500745y. [DOI] [PubMed] [Google Scholar]
- Malisch C. S.; Lüscher A.; Baert N.; Engström M. T.; Studer B.; Fryganas C.; Suter D.; Mueller-Harvey I.; Salminen J.-P. Large variability of proanthocyanidin content and composition in sainfoin (Onobrychis viciifolia). J. Agric. Food Chem. 2015, 63, 10234–10242. 10.1021/acs.jafc.5b04946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. M.; Ma D.; Mellway R.; Gesell A.; Yoshida K.; Walker V.; Tran L.; Stewart D.; Reichelt M.; Suvanto J.; et al. Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol. 2017, 174, 154–171. 10.1104/pp.16.01962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabber J. H.; Zeller W. E.; Mueller-Harvey I. Acetone enhances the direct analysis of procyanidin- and prodelphinidin-based condensed tannins in Lotus species by the butanol–HCl–iron assay. J. Agric. Food Chem. 2013, 61, 2669–2678. 10.1021/jf304158m. [DOI] [PubMed] [Google Scholar]
- Jaakola L.; Pirttilä A. M.; Halonen M.; Hohtola A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 2001, 19, 201–204. 10.1385/mb:19:2:201. [DOI] [PubMed] [Google Scholar]
- Jaakola L.; Pirttilä A. M.; Vuosku J.; Hohtola A. Method based on electrophoresis and gel extraction for obtaining genomic DNA-free cDNA without DNase treatment. Biotechniques 2004, 37, 744–748. 10.2144/04375bm06. [DOI] [PubMed] [Google Scholar]
- Polashock J.; Zelzion E.; Fajardo D.; Zalapa J.; Georgi L.; Bhattacharya D.; Vorsa N. The American cranberry: first insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 2014, 14, 165. 10.1186/1471-2229-14-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J.; Downey M. O.; Harvey J. S.; Ashton A. R.; Tanner G. J.; Robinson S. P. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L.; Vimolmangkang S.; Wei G.; Zhou H.; Korban S. S.; Han Y. Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 2015, 6, 243. 10.3389/fpls.2015.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry-Kirk R. A.; McGhie T. K.; Andre C. M.; Hellens R. P.; Allan A. C. Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. J. Exp. Bot. 2012, 63, 5437–5450. 10.1093/jxb/ers193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskimäki J. J.; Hokkanen J.; Jaakola L.; Suorsa M.; Tolonen A.; Mattila S.; Pirttilä A. M.; Hohtola A. Flavonoid biosynthesis and degradation play a role in early defence responses of bilberry (Vaccinium myrtillus) against biotic stress. Eur. J. Plant Pathol. 2009, 125, 629–640. 10.1007/s10658-009-9511-6. [DOI] [Google Scholar]
- Nguyen N.; Suokas M.; Karppinen K.; Vuosku J.; Jaakola L.; Häggman H. Recognition of candidate transcription factors related to bilberry fruit ripening by de novo transcriptome and qRT-PCR analyses. Sci. Rep. 2018, 8, 9943. 10.1038/s41598-018-28158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. R.; Fryganas C.; Ramsay A.; Mueller-Harvey I.; Thamsborg S. M. Direct anthelmintic effects of condensed tannins from diverse plant sources against Ascaris suum. PLoS One 2014, 9, e97053 10.1371/journal.pone.0097053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice-Evans C.; Miller N.; Paganga G. Antioxidant properties of phenolic compounds. Trends Plant Sci. 1997, 2, 152–159. 10.1016/s1360-1385(97)01018-2. [DOI] [Google Scholar]
- Quideau S.; Deffieux D.; Douat-Casassus C.; Pouységu L. Plant polyphenols: chemical properties, biological activities, and synthesis. Angew. Chem., Int. Ed. 2011, 50, 586–621. 10.1002/anie.201000044. [DOI] [PubMed] [Google Scholar]
- Neilson A. P.; O’Keefe S. F.; Bolling B. W. High-molecular-weight proanthocyanidins in foods: overcoming analytical challenges in pursuit of novel dietary bioactive components. Annu. Rev. Food Sci. Technol. 2016, 7, 43–64. 10.1146/annurev-food-022814-015604. [DOI] [PubMed] [Google Scholar]
- Salminen J.-P. Two-dimensional tannin fingerprints by liquid chromatography tandem mass spectrometry offer a new dimension to plant tannin analyses and help to visualize the tannin diversity in plants. J. Agric. Food Chem. 2018, 66, 9162–9171. 10.1021/acs.jafc.8b02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaby K.; Mazur S.; Nes A.; Skrede G. Phenolic compounds in strawberry (Fragaria x ananassa Duch.) fruits: composition in 27 cultivars and changes during ripening. Food Chem. 2012, 132, 86–97. 10.1016/j.foodchem.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Lee C. Y.; Jaworski A. Major phenolic compounds in ripening white grapes. Am. J. Enol. Vitic. 1989, 40, 43–46. [Google Scholar]
- Kennedy J. A.; Matthews M. A.; Waterhouse A. L. Changes in grape seed polyphenols during fruit ripening. Phytochemistry 2000, 55, 77–85. 10.1016/s0031-9422(00)00196-5. [DOI] [PubMed] [Google Scholar]
- Wang P.; Zhang L.; Jiang X.; Dai X.; Xu L.; Li T.; Xing D.; Li Y.; Li M.; Gao L.; et al. Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 2018, 247, 139–154. 10.1007/s00425-017-2771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Vimolmangkang S.; Soria-Guerra R. E.; Korban S. S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. 10.1093/jxb/err415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatew B.; Stringano E.; Mueller-Harvey I.; Hendriks W. H.; Carbonero C. H.; Smith L. M. J.; Pellikaan W. F. Impact of variation in structure of condensed tannins from sainfoin (Onobrychis viciifolia) on in vitro ruminal methane production and fermentation characteristics. J. Anim. Physiol. Anim. Nutr. 2016, 100, 348–360. 10.1111/jpn.12336. [DOI] [PubMed] [Google Scholar]
- Huang X. D.; Liang J. B.; Tan H. Y.; Yahya R.; Long R.; Ho Y. W. Protein-binding affinity of Leucaena condensed tannins of differing molecular weights. J. Agric. Food Chem. 2011, 59, 10677–10682. 10.1021/jf201925g. [DOI] [PubMed] [Google Scholar]
- Zeller W. E.; Sullivan M. L.; Mueller-Harvey I.; Grabber J. H.; Ramsay A.; Drake C.; Brown R. H. Protein precipitation behavior of condensed tannins from Lotus pedunculatus and Trifolium repens with different mean degrees of polymerization. J. Agric. Food Chem. 2015, 63, 1160–1168. 10.1021/jf504715p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.