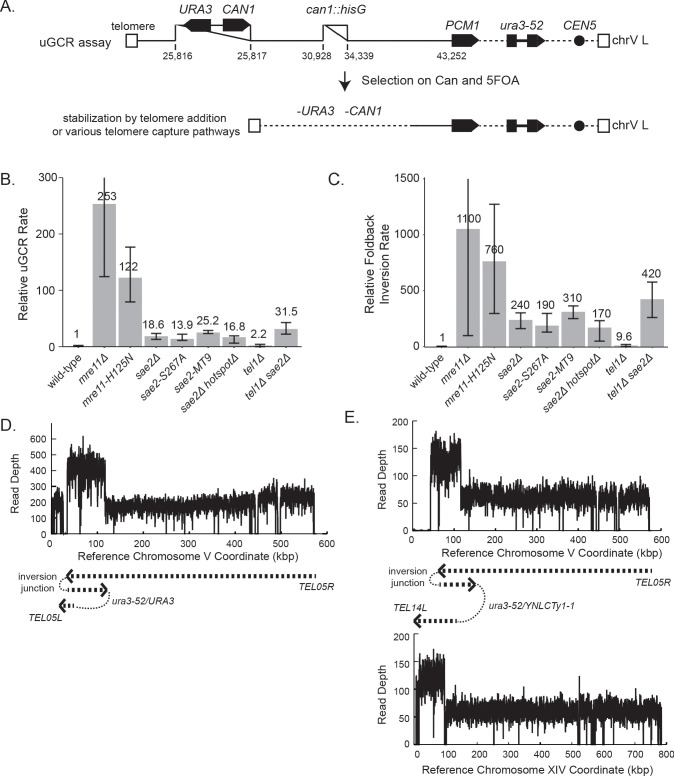
Figure 1. GCRs recovered in uGCR strains with sae2 defects are primarily hairpin-mediated foldback inversions.
(A) Diagram of the left arm of chromosome V (chrV L) containing the uGCR assay. The normal CAN1 locus is deleted by substitution with a hisG fragment and a cassette containing the CAN1 and URA3 genes has been inserted into the YEL068C gene. Coordinates for the modified chrV L are reported as the reference genome coordinates for unmodified regions or reported as positions relative to the centromeric coordinate of the insertion site for inserted elements, for example chrV:34,339–110 is a position in the inserted hisG fragment 110 bases telomeric to the insertion site at chrV 34,339. Simultaneous selection against CAN1 and URA3 by canavanine (Can) and 5-fluoroorotic acid (5FOA) selects for GCRs that ultimately lose both CAN1 and URA3 and are stabilized by addition or capture of a telomere. (B) The relative uGCR rate for strains with mre11 or sae2 defects are displayed with error bars corresponding to the 95% confidence intervals. GCR rates are reported in Supplementary file 1. (C) The relative foldback inversion GCR rate for strains with mre11 or sae2 defects are displayed as in panel B. (D) Example read depth plot determined by WGS for chrV from a foldback inversion GCR resolved by the ura3-53/URA3 homology-mediated rearrangement. Thick-hashed arrows underneath the plot indicate the connectivity between the portions of the GCR that map to the different regions of the reference chromosome. (E) Example read depth plot for chrV and chrXIV from a foldback inversion GCR resolved by a ura3-52/YNLCTy1-1 homology-mediated rearrangement displayed as in panel D, showing the duplication of the region of chrXIV between TEL14L and YNLCTy1-1.