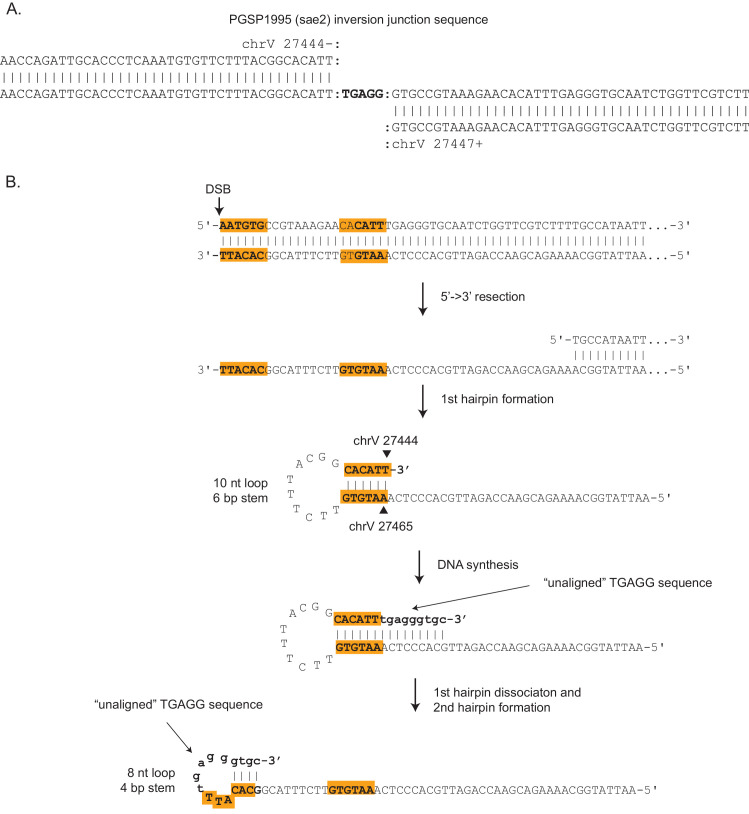
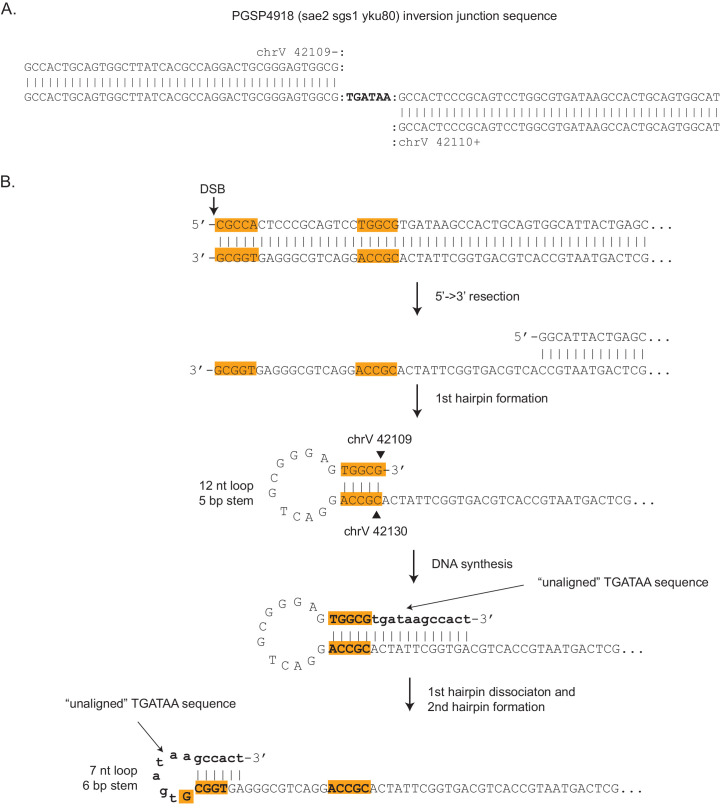
(A) The inversion sequence junction from PGSP1995 (center line), which was present in a GCR from a sae2Δ mutant, shows an unusual feature relative to other inversion junction sequences recovered as it contains a five base TGAGG sequence that does not align to either inverted sequence from chrV. For most inversion junction sequences, the two alignments to chrV overlap, and the overlap indicates the sequences involved in the ssDNA hairpin stem. Thus, this inversion junction sequence must involve a different mechanism of formation. (B) A mechanism of formation of the PGSP1995 inversion junction that is consistent with the junction sequence identified involves two sequential ssDNA hairpins. After resection from a DSB (or other initiating damage), the first hairpin forms by annealing the stem sequences (highlighted in orange) to generate a chrV:27,444_27,465 hairpin with a 10 nt loop and 6 bp stem. DNA synthesis after formation of the first hairpin adds the unaligned TGAGG sequence and bases required for forming the stem of a second hairpin. Dissociation of the first hairpin and formation of a second hairpin (eight nt loop, 4 bp stem) using the newly synthesized bases generates an intermediate that after extension generates an inversion junction sequence identical to the one shown in panel A.