Abstract
Background
Keloids represent chronic fibroproliferative skin disorders in which there is deposition of extracellular components, especially type 1 collagen, fibronectin and elastin, in excessive amounts. NEDD4 is associated with fibrosis found in abnormal wound healing through increased fibroblast proliferation and regulation of type 1 collagen expression. The exact etiology of keloid formation is undefined, but the role of genetic factors was demonstrated.
Objective
To investigate the polymorphism of the NEDD4 gene rs8032158 in a sample of Egyptian patients who have keloids.
Methods
The current case–control study was conducted in 160 unrelated subjects; 100 keloid patients and 60 ages and sex coincided with apparently healthy controls. All subjects underwent a complete history, and weight and length were measured to calculate body mass index (BMI). The Vancouver Scar Scale (VSS) was used to assess keloid severity. An analysis of the polymorphism of the NEDD4 gene rs8032158 T/C was performed using taqman allelic discrimination (real-time PCR).
Results
The rs8032158 CC genotype was observed significantly in keloid patients and increased the risk of keloid development by approximately 2 times (p = 0.003, OR = 1.80). The C allele significantly increased the risk of keloid development by approximately 2 times (P = 0.002, OR = 2.08). The carriers of the CC genotype were significantly associated with severe keloid form and with the highest VSS values.
Conclusion
The polymorphism of the NEDD4 gene rs8032158 could participate in the formation of keloids in the Egyptian population. The NEDD4 rs8032158 CC genotype may have a role in keloid severity.
Keywords: NEDD4, keloid, gene polymorphism
Introduction
The keloid is defined as an aberrant of scar tissue proliferation that occurs at the site of the skin lesion, eg at the site of a surgical incision and trauma, or spontaneously without a history of trauma. The incidence of keloids varies among the different racial groups, and its prevalence has been reported to be nearly 4–6% in the general population, and is more common in women.1 Additionally, the most severe keloids have been observed in the subjects having positive family history, suggesting role of genetic factors in keloid development.2
Clinically, keloids are firm, gummy, shiny and fibrous lesions that fluctuate from pink to skin-colored or red to deep brown. These benign scars are not contagious and are sometimes accompanied by itching. It is mainly the cosmetic aspect of the lesion that causes the referral.3
Histologically, keloids have thick flattened epidermis that was not caused by hyperproliferation, but possibly due to abnormal early terminal differentiation, which affects formation of stratum corneum.4 The superficial dermis was described by lymphocytes and active fibroblasts; the middle dermis had dense extracellular matrix (ECM) with great numbers of fibroblasts, while the deep dermis was poorly cellular and had hyalinized collagen bundles.5
Many studies were conducted to identify the main responsible factors for the occurrence of keloids, however, the mechanism of keloid formation still largely undefined.6 Keloid formation is a complicated and multifactorial process that rests on environmental factors and genetic risk factors.7 Different genetic loci were reported to be associated in keloid predisposition, but the exact genetic deviations involved in the etiology have not yet been known.8
The human gene NEDD4 is situated on chromosome 15q21.3 and includes 30 exons that encode six NEDD4 transcriptional variant (TV) protein variants, all of which have 100% identity from the first part (WW domain) to the end part of the protein, which only differs in the N-terminal region that includes the C2 domain.9 It has been shown that NEDD4 controls the growth of animals by regulating insulin-like growth factor (IGF-1) signaling, the loss of NEDD4 results in reduced signaling of IGF-1 in mice, delayed embryonic development, growth reduction and fetal lethality.10
NEDD4 has been associated with the aetiopathogenesis of fibrosis associated with abnormal wound healing injuries through increased fibroblast proliferation and invasiveness, plus the regulation of the expression of type 1 collagen. Also, the activation of NEDD4 affects p27 protein cellular localization and its stability, suggesting its role in contact inhibition. Additionally, NEDD4 prompts β-catenin aggregation in the cytoplasm and upsurges the transcriptional activity of transforming growth factorβ (TGFβ)-catenin.11
Moreover, the increased NEDD4TV3 in keloid activates signal transducer and activator of transcription factor 3 (STAT3)/nuclear factor-kB (NF-kB) in keloid keratinocytes and fibroblasts resulting in production of many pro-inflammatory cytokines. NEDD4-TV3 also promotes the regulation of insulin-like growth factor receptor signaling (IGF-1R) that was involved in keloid pathogenesis.12
In keloids, few studies investigated the polymorphism of the NEDD4 gene rs8032158 in different populations with controversial results.13–16
The following current study aimed to shed light on the possible association between SNP NEDD4 rs8032158 and the keloid through the evaluation of its gene polymorphism in Egyptian keloid patients, in addition to correlating its estimated results with the available clinical aspects of keloid lesions in those patients.
Materials and Methods
This was a case–control study that was conducted in 100 unrelated keloid patients of both sexes and aged between 15 and 70 years. In addition, 60 healthy unrelated volunteers by age and sex were included who had no family history or keloids (control group). They were selected from the Dermatology Outpatient Clinic, University Hospitals of Menoufia during the period from January 2019 to May 2019. The laboratory part of the study was carried out in the Department of Biochemistry, Faculty of Medicine, University of Menoufia. The study was approved by the Ethical Cometti of Human Right in Research at the University of Menoufia [in accordance with the Declaration of Helsinki in 1975]. Informed written consent (accepted and approved by the Ethical Committee of Human Rights in Research at the University of Menoufia) was obtained from all participants in this study after informing them about the study. Regarding participants under the age of consent, a parent or legal guardian signed the consent forms. Exclusion criteria included any subject who had keloids due to skin disorders, hypertrophic scar syndrome, genetic or chromosomal disorders that included keloids as part of the disorder, eg Rubinstein Taybi syndrome. In addition, pregnant and lactating women were excluded from the study.
The people studied were subjected to a detailed history and thorough general examination. BMI was calculated using weight and height for each individual. BMI is calculated by dividing body mass in kilograms (kg) by the square of body height in meters (m), and is universally expressed in units of kg/m2.17
A dermatological examination was performed to determine the keloid site (for example, face, neck, leg and abdomen). The Vancouver Scar Scale was used to assess keloid severity. In which we evaluate four parameters:18 I–vascularity that is subdivided into normal (0), pink (1), red (2) or purple (3). II-pigmentation; normal (0), hypopigmentation (1) or hyperpigmentation (2). III-Flexibility that was subdivided into normal (0), flexible (1), yield (2), firm (3) or contracture (4). Height IV; (normal), (0–2 mm), (2–5 mm) and more than (5) mm. Every parameter included classified degree in the scale that were added to get a total score range from 0 (descriptive normal skin) to 13 (descriptive the worst imaginable scar). The scars were classified as mild (0–3), moderate (4–7) and severe (8–13).19
Under aseptic condition, 2 mL of venous blood was taken from each subject and then transferred to the EDTA tube for DNA extraction. Genotyping of the NEDD4 gene polymorphism rs 8032158 (T/C):
Extraction of the DNA was done using a Quick-g Mini-pre-DNA genetic DNA purification kit from Zymo Research (USA). Elution and storage of the DNA were then performed at −20 °C for other PCR procedures. The NEDD4 gene was genotyped using an allelic discrimination assay using a real-time PCR technique using an applied TaqMan probe from Biosystems, USA. Thermo Fisher Scientific has provided the Maxima qPCR Master Mix probe, primers and probes. The probe sequence for NEDD4 rs 8032158 was 5ʹTTGGGAAAAAACTAGCTTATAATAACVIC/FAMTATAGGTA GTTTA TAACATTTATTAA-3`. Amount of master mix about 10 microliters were added to 1.25 mL of the genotyping assay of the mixture of primer/probe and amount of water without DNase about 3.75 mL. Also about five microliters of genomic DNA extract were used for each sample and 5 mL of water without DNase for the negative control reaction. The following steps were used: initial denaturation was made for ten minutes at 95 °C, followed by 40 cycles of: denaturation for 15 seconds at 94°C, annealed from the primer for 60 seconds at 50°C, then extended for 2 minutes at 72° C and last extension for 1 minute at 72°C. Analysis of data was done using a real-time 7500 PCR instrument, version 2.0.1, Bio systems applied.
Statistical Analysis
The data were fed to compute them and analyzed by statistical package for the statistical package of social science software (SPSS) version 20 on the computer compatible with the International Business Machines (IBM). Two types of statistics were made; (A): Descriptive statistics: expressed as mean and standard deviation (X ± SD), as well as median and range for quantitative data or number and percentage (No and%) for qualitative data (B): Analytical statistics: Chi test-square (χ2): it was used to study the association between two qualitative variables, Student’s t-test: it was used for the comparison between two groups of normally distributed quantitative variables, Mann–Whitney test (non-parametric test): it was used to the comparison between two groups of quantitative variables not normally distributed, Spearman correlation test: it was used for quantitative variables that were not normally distributed or when one of the variables was qualitative, the P value ≤ 0.05 was statistically significant.
Results
The patients studied were 44 men (44.0%) and 56 women. Their age ranged between 15 and 70 years and the BMI ranged between 20.9 and 39.3 kg/m2. Among them 36 (36.0%) patients were smokers. They were comparable with their controls with respect to their age (P = 0.516), gender (P = 0.602), smoking (P = 0.952) and BMI (P = 0.219). More than half (66, 66.0%) of our patients had a positive family history of keloids. The most frequent site of keloids were the extremities (48, 48%). Half of the cases developed keloids after the operation (50, 50%) (Table 1).
Table 1.
Demographic and Clinical Characteristics of the Studied Subjects
| Demographic Characteristics | Cases (n=100) | Controls (n=60) | Test of Significance | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| Mean ± SD | 35.50±14.01 | 37.70±15.55 | Mann–Whitney=0.65 | 0.516 |
| Median | 32 | 40 | ||
| Range | 15–70 | 10–65 | ||
| Sex [n (%)] | ||||
| Males | 44 (44.0) | 30 (50.0) | χ2=0.54 | 0.602 |
| Females | 56 (56.0) | 30 (50.0) | ||
| Smoking [n (%)] | ||||
| Smokers | 36 (36.0) | 22 (36.7) | χ2=0.01 | 0.952 |
| Non-smokers | 64 (64.0) | 38 (63.3) | ||
| BMI | ||||
| Mean ± SD | 29.91±5.64 | 28.64±4.45 | t=1.24 | 0.219 |
| Median | 29.32 | 29.8 | ||
| Range | 20.9–39.3 | 21.3–38.1 | ||
| Clinical data | ||||
| Family history [No (%)] | ||||
| Present | 66 (66.0) |  |
||
| Absent | 34 (34.0) | |||
| Site of lesion [No (%)] | ||||
| Head and neck | 12 (12.0) |  |
||
| Trunk | 40 (40.0) | |||
| Limbs | 48 (48.0) | |||
| Precipitating factors [No (%)] | ||||
| Spontaneous | 10 (10.0) |  |
||
| Operation | 50 (50.0) | |||
| Burn | 40 (40.0) | |||
| Duration of disease (years) | ||||
| Mean ±SD | 3.88±2.94 |  |
||
| Median | 3 | |||
| Range | 1.25–4 | |||
| VSS | ||||
| Mean ± SD | 7.30±2.91 |  |
||
| Median | 7 | |||
| Range | 2–12 | |||
| VSS classifications No (%) | ||||
| Mild (0–3) | 20 (20.0) |  |
||
| Moderate (4–7) | 32 (32.0) | |||
| Severe (8–11) | 48 (48.0) | |||
Abbreviations: SD, standard deviation; VSS, Vancouver Scar Scale; BMI, body mass index; χ2, Chi-square test; t, Student’s t-test.
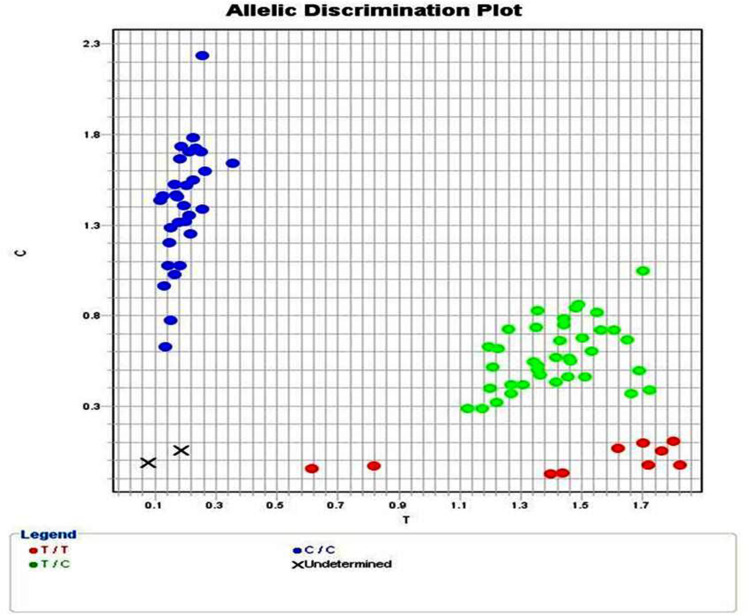
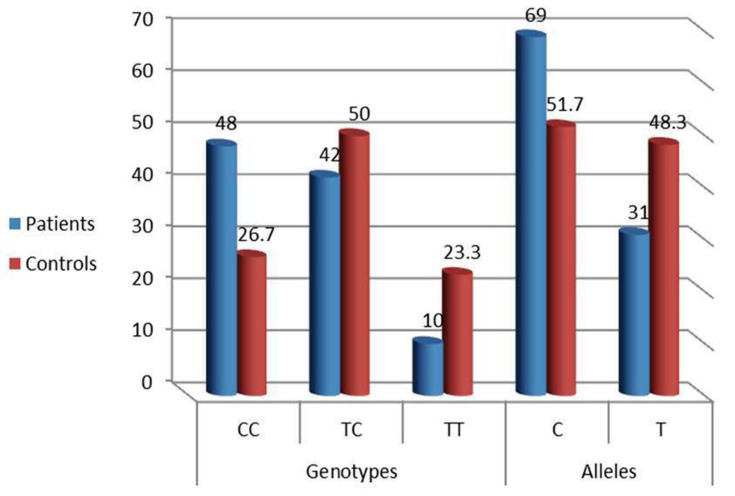
NEDD4 rs8032158 genotypes and allele distribution (Figure 1) were analyzed in all participants. Compared to the wild NEDD4 rs8032158 (TT genotype), the CC genotype was significantly observed in keloid patients [48 (48%) versus 16 (26.2)] and increased the risk of keloid development by approximately 2 times (p = 0.003, OR = 1.80). In addition, C Allele significantly increased the risk of keloid development by approximately 2 times (P = 0.002, OR = 2.08) (Table 2; Figure 2).
Figure 1.
Allelic discrimination plot of (rs8032158) gene polymorphism in both cases and controls [TT genotypes (red dots), TC genotypes (green dots) and CC genotypes (blue dots)].
Table 2.
NEED4 (rs8032158) Genotypes and Allele Frequencies in Keloid Patients and Control Group
| NEDD4 (rs8032158) gene polymorphism | Patients (n=100) No (%) |
Controls (n=60) No (%) |
χ2 | P- value | OR (95% CI) |
|---|---|---|---|---|---|
| Genotypes | |||||
| -CC | 48 (48.0) | 16 (26.7) | 4.62 | 0.010 | NA |
| -TC | 42 (42.0) | 30 (50.0) | |||
| -TT | 10 (10.0) | 14 (23.3) | |||
| Alleles | (n=200) | (n=60) | |||
| -C | 138 (69.0) | 31 (51.7) | 9-61 | 0.002 | 2.08 (1.31-3-32) |
| -T | 62 (31.0) | 29 (48.3) | |||
| -CC vs TC+TT | 7.11 | 0.008 | 2.54 (1.27-5.08) | ||
| -TT vs TC+CC | 5.23 | 0.022 | 0.37 (0.15-0.89) | ||
| -CC vs TC | 4.2 | 0.040 | 2.14 (1.03-4.47) | ||
| -CC vs TT | 8.63 | 0.003 | 1.80 | ||
| -TT vs TC | 2.1 | 0.156 | 0.51 (0.20-0.89) | ||
Note: NEDD4: neural precursor cell expressed developmentally down-regulated protein 4. n: Number. χ2: Chi square test. OR: odds ratio. CI: confidence interval
Figure 2.
Distribution of NEDD 4 rs8032158 genotypes and allele frequencies in keloid patients and control group. [(p=0.003, OR=1.80) and (P=0.002, OR =2.08) respectively].
The relationship between the NEDD4 rs8032158 genotypes and the clinical and personal criteria studied of keloid patients revealed a significant association of the CC genotype with the severe keloid form and with the highest VSS values (P <0.001 for both), it also showed a significant association of CC genotype with smoking (p=0.024) (Table 3).
Table 3.
Expression of NEED4 rs8032158 Genotypes Regarding Demographic and Clinical Data in Keloid Patients.
| Items | NEDD4 rs8032158 Gene Polymorphism in Keloid Patients | Kruskal–Wallis Test | P-value | ||
|---|---|---|---|---|---|
| CC (n=48) Mean± SD | TC (n=42) Mean± SD | TT (n=10) Mean± SD | |||
| Age (years) | 37.71±14.17 | 34.38±14.85 | 29.60±8.23 | 0.80 | 0.455 |
| BMI | 30.48±3.44 | 29.29±3.51 | 29.76±7.43 | 0.50 | 0.608 |
| Duration of disease (years) | 3.15±2.24 | 4.63±3.51 | 4.20±2.94 | 1.76 | 0.239 |
| VSS | 8.96±2.58 | 5.95±2.22 | 5.00±2.83 | 10.81 | <0.001* |
| Sex | n (%) | n (%) | n (%) | χ2 | |
| Males | 18 (37.5) | 20 (47.6) | 4 (40.0) | 0.96 | 0.619 |
| Females | 30 (62.5) | 22 (52.4) | 6 (60.0) | ||
| Smoking | |||||
| Smokers | 16 (33.3) | 18 (42.9) | 8 (80.0) | 7.42 | 0.024* |
| Non-smokers | 32 (66.7) | 24 (57.1) | 2 (20.0) | ||
| Site of lesion | |||||
| Head and neck | 6 (12.5) | 4 (9.5) | 2 (20.0) | 7.79 | 0.100 |
| Trunk | 22 (45.8) | 18 (42.9) | 0 | ||
| Limbs | 20 (41.7) | 20 (47.6) | 8 (80.0) | ||
| Precipitating factors | |||||
| Spontaneous | 6 (12.5) | 4 (9.5) | 0 | 7.90 | 0.095 |
| Operation | 26 (45.2) | 22 (52.4) | 2 (20.0) | ||
| Burn | 16 (33.3) | 16 (38.1) | 8 (80.0) | ||
| Family history | |||||
| Present | 32 (66.7) | 30 (71.4) | 4 (40.0) | 3.57 | 0.167 |
| Absent | 16 (33.3) | 12 (28.6) | 6 (60.0) | ||
| VSS classification | |||||
| Mild | 6 (12.5) | 8 (19.0) | 6 (60.0) | 60.28 | <0.001* |
| Moderate | 2 (4.2) | 28 (66.7) | 2 (20.0) | ||
| Severe | 40 (83.3) | 6 (14.3) | 2 (20.0) | ||
Note: *Significance.
Abbreviations: SD, standard deviation; VSS, Vancouver Scar Scale; BMI, body mass index.
Discussion
Compared to the proliferative scar and other pathological scars, the keloid has a stronger genetic predisposition.20 For identifying the role of NEDD4 in keloid formation, a genome-wide association study (GWAS) has been done and identified an association of a single nucleotide polymorphism (SNP) rs8032158 that is located in the NEDD4 gene on chromosome 15 with keloid development in a Japanese population. The authors investigated 517 patients and 2385 healthy unaffected individuals.7
This study was the first to investigate the polymorphism of the NEDD4 gene rs8032158 T/C in the Egyptian population. We note significant differences between genotypes and allelic distribution of NEDD4 polymorphism rs8032158 T/C between the groups studied. The CC genotype was observed significantly in keloid patients and increased the risk of keloid development by approximately 2 folds. In addition, the C allele significantly increased the risk of keloid development by approximately 2 folds too.
Confirming our result, Yang et al13 investigated 50 Chinese patients with keloids and 52 healthy unaffected subjects. The authors observed a significant link between the T/C NEDD4 polymorphism rs8032158 and the keloid. They revealed a significant increase in the CC genotype in keloid patients, and showed that the rs8032158 T allele was associated with a reduced risk of keloid scarring. Also Fujita et al16 examined 20 skin keloid specimens and found that most of them have the risk C/C genotype in rs8032158.
The NEDD4 gene has been linked to the pathogenesis of fibrosis associated with abnormal wound healing lesions, such as the keloid, through different signaling pathways. In fibroblasts, NEDD4 regulates the expression of collagen and fibronectin. This unusual regulation led to the excessive increase of collagen and fibronectin in the extracellular matrix that could pay to keloid formation and progression.11
In addition, the TV3 increases in keloid and is involved in increase the activation of STAT3/NF-kB in keloid keratinocytes and fibroblasts. Active STAT3/NF-kB is suggested to stimulate inflammation and keloid formation through the production of many proinflammatory cytokines. Alternatively, it is possible that NEDD4-TV3 promotes keloid growth through activation of the mitogenic pathway, because NEDD4-TV3 can act as a regulator of insulin-like growth factor receptor signaling (IGF-1R) that has also been implicated in keloid pathogenesis.12
Also, NEDD4 expresses an ubiquitin E3 ligase enzyme with an HECT domain that participates in the ubiquitin-mediated protein degradation process. It has been reported that the activation of NEDD4 influences the subcellular localization of p27 protein and its stability, suggesting that it plays an important role in contact inhibition. In addition, NEDD4 induces the aggregation of β-catenin in the cytoplasm and increases the transcriptional activity of TGFβ-catenin.11
Therefore, as previously reported,15 we suggest that SNP rs8032158 may have a role in irregular cell proliferation and matrix aggregation observed in keloids.
On the other hand, Zhu et al14 found that the SNP rs8032158 is one of the SNPs that did not have a significant association with keloid formation in their study in 714 keloid patients in the Han China population. The authors found that rs 2271289 located in the intron region of NEDD4 is significantly associated with keloid development. It has been found that rs8032158 has a moderate link imbalance (LD) with rs2271289 according to HapMap3 (the haplotype map of the human genome that describes patterns common human genetics variations). They suggested NEDD4 that could be a common genetic reason for the growth of keloids with multiple populations in terms of Han and Japanese Chinese, although the most important SNPs responsible for keloid formation were not the same among them.
Also, in Chinese Han population, Zhao et al21 analyzed the correlation between 4 NEDD4 SNPs (rs873549, rs2118610, rs1511412, rs2271289) and keloids phenotypes in 309 keloids cases and 1080 control individuals. The authors revealed that rs2271289 is strongly associated development and severity of keloids.
In the current study, there was a significantly higher prevalence of severe degree of keloid in carriers of CC rs8032158 genotypes than TC and TT genotypes. According to our study, Ogawa et al15 reported that the severe keloid group had a significant OR with a dominant inheritance mode of rs8032158 where CT + CC including the risk allele C, compared to TT that does not contain the risk allele C that had significant OR with both controls and mild keloid group.
The relationship between the CC genotype and the C allele and the severe form of keloid could be due to the activation of NF-kB and STAT3 (the inflammation amplifier). This inflammation amplifier activates and induces NF-kB target genes, including chemokines and growth factors22,23 in non-immune cells such as keratinocytes, fibroblasts and endothelial cells.24
The expression of NF-kB target genes such as CCL2 and IL-6 is proinflammatory cytokines that control the inflammatory process in response to trauma or injury. In addition, it was found that CC genotypes cause a higher level of NEDD4 TV3 which, reportedly, acts strongly together with the RIP adapter protein to cause NF-kB activation.16
The role of the NEDD4 TV3 signaling pathway in the aetiopathogenesis of the keloid is evaluated and its potential role as a therapeutic objective is evaluated16. Fujita et al16 studied the immunohistochemical expression of NEDD4 in different samples of normal and human keloid skin using an anti-NEDD4 antibody that identifies all TVs types. They discovered that NEDD4 was equally expressed in keratinocytes and fibroblasts of both keloid and normal skin, but quantitative PCR and electrophoresis showed that NEDD4 TV3 was considerably more profuse in keloid skin in comparison with normal skin tissue. In addition, keloid samples from different anatomical sites had a higher expression of NEDD4 TV3 than in control samples obtained from multiple sites. Therefore, the authors made an anti-NEDD4 antibody defined for amino acids 431–455 of TV3, this antibody recognized NEDD4 TV3. According to the RNA results, there was a large difference between keloid and normal skin during immunohistochemistry using the TV3431-455 antibody and using the pan-NEDD4 antibody. The NEDD4 TV3 signals were also obvious in the central part of the core. In addition, they compared the expression of NEDD4 TV3 in inflammatory lesions and areas of minor inflammation (areas of mature scars) in the same patient. Both keratinocytes and fibroblasts in inflammatory lesions showed a higher expression of NEDD4 TV3 than in areas of mature scars. These results suggest that NEDD4 TV3 is definitely expressed in keratinocytes and keloid fibroblasts, so these data suggest that NEDD4 TV3 could be a potential therapeutic target for easy or excessive healing in keloid patients.16
Conclusions
NEDD4 rs8032158 gene polymorphism could participate in keloid formation in Egyptian population. NEDD4 rs8032158 CC genotype may participate in keloid phenotype and may have a role in development of severe form of keloid. NEDD4 rs8032158 SNP may serve as a biomarker that aids in the prevention and prognosis as well as targeted treatment for keloids.
Funding Statement
No sources of funding were used to conduct this study or prepare this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Halim AS, Emami A, Salahshourifar I, Kannan TP. Keloid scarring: understanding the genetic basis, advances, and prospects. Arch Plast Surg. 2012;39(3):184. doi: 10.5999/aps.2012.39.3.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu W, Zheng X, Yao X, Zhang L. Clinical and epidemiological analysis of keloids in Chinese patients. Arch Dermatol Res. 2015;307(2):109–114. doi: 10.1007/s00403-014-1507-1 [DOI] [PubMed] [Google Scholar]
- 3.O’Sullivan ST, O’Shaughnessy M, O’Connor TP. Aetiology and management of hypertrophic scars and keloids. Ann R Coll Surg Engl. 1996;78(3 pt 1):168–175. [PMC free article] [PubMed] [Google Scholar]
- 4.Limandjaja GC, van den Broek LJ, Waaijman T, et al. Increased epidermal thickness and abnormal epidermal differentiation in keloid scars. Br J Dermatol. 2017;176(1):116–126. doi: 10.1111/bjd.14844 [DOI] [PubMed] [Google Scholar]
- 5.Jiao H, Zhang T, Fan J, Xiao R. The superficial dermis may initiate keloid formation: histological analysis of the keloid dermis at different depths. Front Physiol. 2017;8. doi: 10.3389/fphys.2017.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossiello L, D’Andrea F, Grella R, et al. Differential expression of cyclooxygenases in hypertrophic scar and keloid tissues. Wound Repair Regen. 2009;17(5):750–757. doi: 10.1111/j.1524-475X.2009.00530.x [DOI] [PubMed] [Google Scholar]
- 7.Nakashima M, Chung S, Takahashi A, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42(9):768–771. doi: 10.1038/ng.645 [DOI] [PubMed] [Google Scholar]
- 8.Marneros AG, Norris JE, Watanabe S, Reichenberger E. Clinical genetics of familial keloid. Arch Dermatol. 2001;137(11):1429–1434. [DOI] [PubMed] [Google Scholar]
- 9.Boase NA, Kumar S. NEDD4: the founding member of a family of ubiquitin-protein ligases. Gene. 2015;557(2):113–122. doi: 10.1016/j.gene.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao XR, Lill NL, Boase N, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1(38):ra5. doi: 10.1126/scisignal.1160940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung S, Nakashima M, Zembutsu H, Nakamura Y. Possible involvement of NEDD4 in keloid formation; its critical role in fibroblast proliferation and collagen production. Proc Jpn Acad Ser B. 2011;87(8):563–573. doi: 10.2183/pjab.87.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marneros AG. A role for the E3 ubiquitin ligase NEDD4 in keloid pathogenesis. J Investig Dermatol. 2019;139(2):279–280. doi: 10.1016/j.jid.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Liang Y, Ma X, Su Y, Zhang X. Genetic susceptibility to keloid scarring in Chinese Han population: NEDD4 gene single nucleotide polymorphism. Int J Clin Exp Med. 2017;10(2):4042–4048. [Google Scholar]
- 14.Zhu Z, Wu B, Li P, et al. Association study confirmed susceptibility loci with keloid in the Chinese Han Population. PLoS One. 2013;8(5). [Google Scholar]
- 15.Ogawa R, Watanabe A, Than Naing B, et al. Associations between keloid severity and single-nucleotide polymorphisms: importance of rs8032158 as a biomarker of keloid severity. J Invest Dermatol. 2014;134(7):2041–2043. doi: 10.1038/jid.2014.71 [DOI] [PubMed] [Google Scholar]
- 16.Fujita M, Yamamoto Y, Jiang JJ, et al. NEDD4 is involved in inflammation development during keloid formation. J Investig Dermatol. 2019;139(2):333–341. doi: 10.1016/j.jid.2018.07.044 [DOI] [PubMed] [Google Scholar]
- 17.Kendrick M. Why being “overweight” means you live longer: the way scientists twist the facts. Independent. 2015;15:32. [Google Scholar]
- 18.Fearmonti R, Bond J, Erdmann D, et al. A review of scar scales and scar measuring devices. Eplasty. 2010;10. [PMC free article] [PubMed] [Google Scholar]
- 19.Baryza MJ, Baryza GA. The Vancouver scar scale: an administration tool and its interrater reliability. J Burn Care Rehabil. 1995;16(5):535–538. doi: 10.1097/00004630-199509000-00013 [DOI] [PubMed] [Google Scholar]
- 20.Shih B, Bayat A. Genetics of keloid scarring. Arch Dermatol Res. 2010;302(5):319–339. doi: 10.1007/s00403-009-1014-y [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Liu S, Xie J, et al. NEDD4 single nucleotide polymorphism rs2271289 is associated with keloids in Chinese Han population. Am J Transl Res. 2016;8(2):544–555. [PMC free article] [PubMed] [Google Scholar]
- 22.Atsumi T, Singh R, Sabharwal L, et al. Inflammation amplifier, a new paradigm in cancer biology. Cancer Res. 2014;74(1):8–14. doi: 10.1158/0008-5472.CAN-13-2322 [DOI] [PubMed] [Google Scholar]
- 23.Meng J, Jiang JJ, Atsumi T, et al. Breakpoint cluster region–mediated inflammation is dependent on Casein Kinase II. J Immunol. 2016;197(8):3111–3119. doi: 10.4049/jimmunol.1601082 [DOI] [PubMed] [Google Scholar]
- 24.Ogura H, Arima Y, Kamimura D, Murakami M. The gateway theory: how regional neural activation creates a gateway for immune cells via an inflammation amplifier. Biomed J. 2013;36(6). [DOI] [PubMed] [Google Scholar]