Abstract
Two different drug micro-carriers consisting of doxorubicin-dextran (DOX-D)- and camptothecin-modified carboxymethyl cellulose (CPT-CMC)-loaded nucleic acid-stabilized microcapsules, MC-1 and MC-2, or two different nanocarriers consisting of nucleic-acid-locked doxorubicin (DOX)- and camptothecin (CPT)-loaded metal–organic framework nanoparticles, NMOF-1 and NMOF-2, are coupled to auxiliary constitutional dynamic networks, CDNs, for the triggered release of the drugs. CDN “S” composed of four constituents AA′’, AB′, BA′, and BB′, and two hairpin structures, H1 and H2, leads to the CDN “S”-guided unlocking of the MC-1/MC-2 carriers and the release of DOX-D and CPT-CMC or of the NMOF-1 and NMOF-2 carriers that release DOX and CPT, respectively. The unlocking processes are activated by the cleavage of H1 and H2 by BB′ and BA′, respectively, to yield fragmented strands that unlock the gating units of the microcapsules/NMOFs carriers. In the presence of miRNA-155 or miRNA-124, dictated orthogonal reconfiguration of CDN “S” into CDN “X” or “Y” proceeds. The miRNA-155 stimulates the reconfiguration of CDN “S” to CDN “X”, where AA′ and BB′ are upregulated, and AB′ and BA′ are downregulated, leading to the enhanced release of DOX-D or DOX from the microcapsule/NMOFs carriers, and to the concomitant inhibition of the release of CPT-CMC or CPT from the respective carriers. Similarly, the miRNA-124-triggered reconfiguration of CDN “S” to CDN “Y” results in the BA′-guided cleavage of H2 and the preferred release of CPT-CMC or CPT from the respective carriers. The miRNA-triggered CDN-driven unlocking of the carriers stimulates the amplified and selective release of the drugs from the microcapsules/NMOFs carriers.
Keywords: microcapsule, metal−organic framework, nanoparticle, nanomedicine, miRNA-155, miRNA-124
Chemical processes in cells are often driven by complex constitutional dynamic networks (CDNs) that are triggered by environmental stimuli.1−4 The dynamic networks represent equilibrated systems of constituents that can be upregulated or downregulated by external triggers and, thus, the control over the contents of the constituents might lead to emerging functionalities. For example, a [2 × 2] CDN consists of a dynamically equilibrated system composed of four constituents AA′, AB′, BA′, and BB′. The stabilization of one of the constituents by an auxiliary trigger T, e.g., AA′-T, is accompanied by the reconfiguration of the CDN into a new equilibrated CDN where AA′-T is upregulated on the expense of the constituents BA′ and AB′ (that share components with AA′). The upregulation of AA′ and downregulation of constituents BA′ and AB′ are concomitantly accompanied by the upregulation of the constituent BB′ (that does not share components with AA′). Different molecular and macromolecular5 CDN systems revealing dynamic switchable adaptive properties were reported, and different auxiliary triggers such as pH,6 light,7 Lewis bases, and ions8 were used to reconfigure the dynamically equilibrated systems. Nonetheless, these systems suffered from the lack of versatile paradigm of system design, scalability, operation in bioinspired aqueous environments, and, particularly, limited translation for practical applications.
The base-sequences in nucleic acids encode important structural and functional information into the biopolymer. This sequence-specific information embedded in nucleic acids resulted in the development of nucleic-acid-based catalysts, DNAzymes9−11 or nucleoapzymes,12 and sequence-specific oligonucleotide for the binding of low-molecular-weight or macromolecular ligands, aptamers.13,14 These features of nucleic acids provide a rich “tool-box” for the development of the field of DNA nanotechnology15−17 and, particularly, the design of reconfigurable DNA nanostructures (switches),18,19 operating DNA machines,20,21 and the assembly of materials revealing switchable properties,22,23e.g., hydrogels.24−27 By the coupling of stimuli-responsive DNA switches to nanoparticles, e.g., SiO2 or metal–organic framework nanoparticles,28−31 and the design of DNA-based stimuli-responsive microcapsules,32−34 “smart” carriers for the controlled release of drugs were developed.
In addition, the structural and functional information embedded in oligonucleotides was recently used to develop nucleic acid-based CDNs.35 The assembly of DNA-based CDNs of variable complexities [2 × 2; 3 × 2; 3 × 3] was reported, and their reconfigurations by fuel/antifuel strands, K+-ions-stabilized G-quadruplexes/crown ether,36 and light37 were demonstrated. The adaptive and reversible control over the contents of the signal-triggered reconfigured CDNs was demonstrated, and the hierarchical control of the composition of CDNs triggered by external stimuli was highlighted.36 In addition, feedback-driven CDNs, intersubstitution of CDNs,38 and control over emerging catalytic functions of the CDNs were reported.39 The applications of CDNs are, however, scarce. Recently, we reported on the CDN-guided aggregation of Au NPs or CdSe/ZnS quantum dots, resulting in switchable, plasmonic, catalytic (e.g., oxidation of dopamine), or chemiluminescence resonance energy transfer (CRET) functions.40 Here, we wish to report on the use of CDNs for the programmed release of two anticancer drugs from micro-/nanocarriers consisting of stimuli-responsive drug-loaded microcapsules and metal–organic framework nanoparticles. We use two different microRNAs (miRNAs) as triggers to unlock the carriers and to release the drugs. Specifically, we use the breast cancer miRNA-155 biomarker and the papillary thyroid carcinoma miRNA-124 biomarker to unlock the respective carriers and to selectively release either doxorubicin (DOX) or camptothecin (CPT) from the carriers. The major conclusion of the study is that while the parent CDN, in the absence of the miRNAs, releases at a moderate yield both drugs, the miRNAs-triggered reconfiguration of the parent CDN yields CDNs that selectively release the doxorubicin (by miRNA-155) or camptothecin (by miRNA-124). The results might have important implications for the development of personalized chemotherapeutic medicine, controlled by specific overexpressed miRNAs.
Results and Discussion
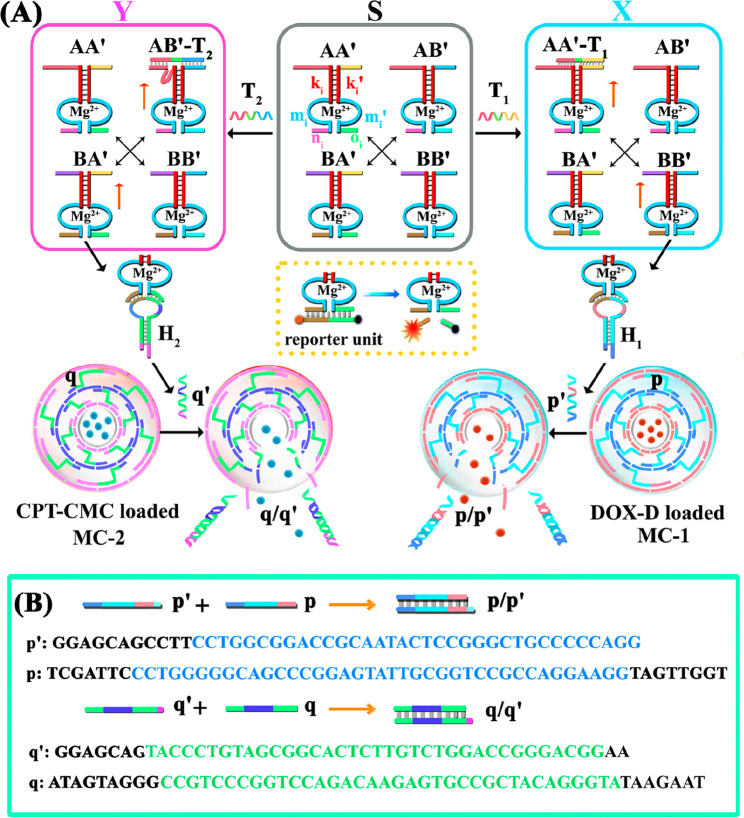
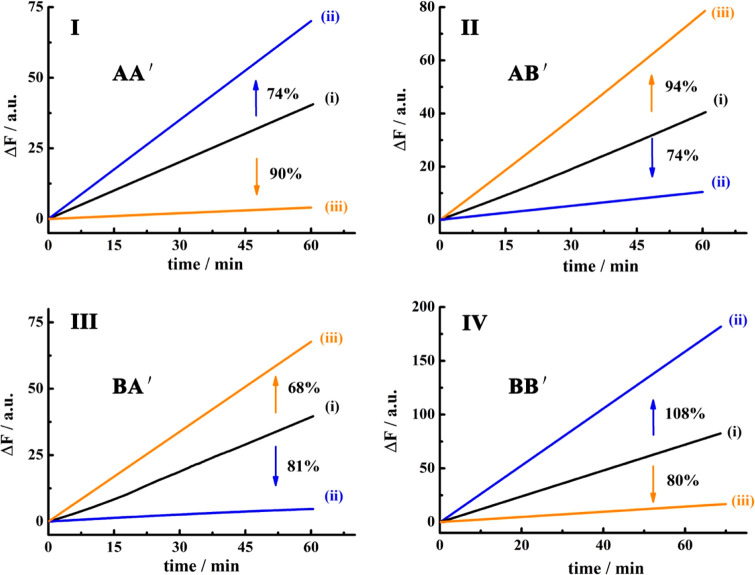
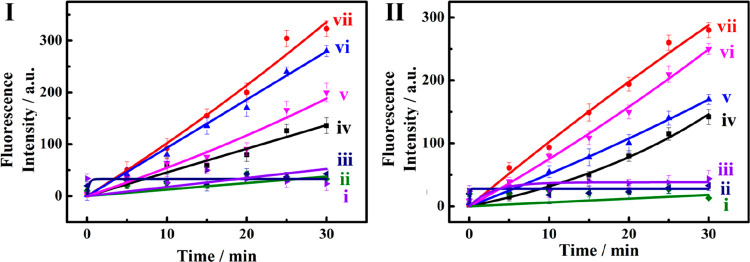
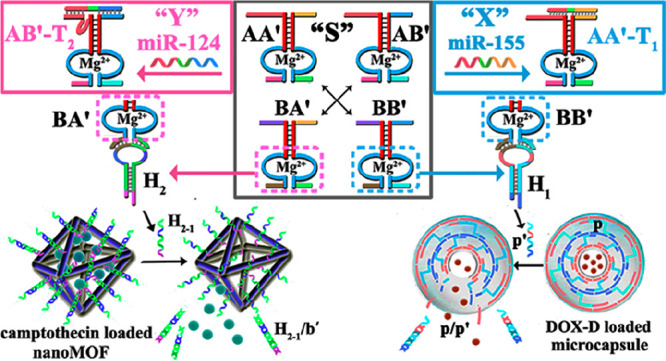
Figure 1A outlines schematically the configuration of a constitutional dynamic network, CDN “S”, that undergoes guided reconfiguration in the presence of miRNA-155 (being a biomarker for breast cancer) or miRNA-124 (that is a biomarker for papillary thyroid carcinoma) to yield CDN “X” or CDN “Y” that stimulates the controlled opening of two different microcapsules that release doxorubicin dextran, DOX-D, or camptothecin-CMC, CPT-CMC. The CDN “S” consists of four constituents AA′, AB′, BA′, and BB′. Each of the constituents is composed of a duplex domain, ki/ki′, that is conjugated to a quasiloop structure, mi/mi′, terminated by single-strand tethers, ni/oi. Subjecting CDN “S” to miRNA-155 results in the hybridization of the biomarker to the constituent AA′. The stabilization of AA′ leads to its upregulation and to the reconfiguration of CDN “S” into “X”, where the constituent BB′ is, concomitantly, upregulated, and the constituents AB′ and BA′ are downregulated. Similarly, subjecting CDN “S” to miRNA-124 results in the hybridization of the biomarker to the constituent AB′ and leading to the reconfiguration of CDN “S” into CDN “Y”, where the constituent AB′ is upregulated together with BA′, while the constituents AA′ and BB′ are downregulated. The quasiloop domains mi/mi′ and the conjugated arms ni/oi represent Mg2+-ion-dependent DNAzyme sequences, and their catalytic activities toward the cleavage of the respective fluorophore-quencher-modified substrates provide four different reporter units, that allow the calculation of the quantitative contents of the constituents in the different CDNs upon using appropriate calibration curves. Figure 2 shows the rates of cleavage of the respective substrates by the four Mg2+-ion-dependent DNAzymes associated with the constituents of CDN “S”, curves (i), after subjecting CDN “S” to miRNA-155 trigger (T1) and its reconfiguration into CDN “X”, curves (ii), and upon treatment of CDN “S” with the miRNA-124 trigger (T2) and the reconfiguration of CDN “S” into CDN “Y”, curves (iii). Using the appropriate calibration curves (Figure S1), the contents (concentrations) of the constituents in the different CDNs were evaluated (Table S1). Evidently, the subjection of CDN “S” to T1 is accompanied by the upregulation of AA′ and BB′ by 74% and 108%, respectively. Also, the concentrations of AB′ and BA′ in CDN “X” are significantly lower compared to the concentrations of AB′ and BA′ in the parent CDN “S”. In addition, the T2-guided reconfiguration of CDN “S” to CDN “Y” is accompanied by the upregulation of AB′ and BA′ by 94% and 68%, whereas the constituents of AA′ and BB′ are downregulated by 90% and 80%, respectively, indicating that the concentration of these constituents in the equilibrated mixture of CDN “Y” is very low. The contents of the constituents in CDNs “S”, “X”, and “Y” were, also, evaluated by quantitative electrophoretic experiments; see Figure S2 and the accompanying discussion. The contents of the constituents in the different CDNs derived from the electrophoretic experiments are provided (in brackets) in Table S1. Very good agreement exists between the concentration values of the constituents in the CDNs evaluated by the activities of the DNAzyme reporter units and the electrophoretically derived concentrations.
Figure 1.
Scheme for the miRNA-stimulated constitutional dynamic networks (CDNs)-guided release of loads from DNA-based microcapsules. (A) The T1 (miRNA-155)-triggered reconfiguration of CDN “S” to CDN “X” leads to the guided unlocking of the DOX-D-loaded microcapsules (MC-1). The T2 (miRNA-124)-triggered reconfiguration of CDN “S” to CDN “Y” leads to the guided unlocking of the CPT-CMC-loaded microcapsules (MC-2). (B) Unlocking principle of p-bridged DOX-D-loaded MC-1 and the q-bridged CPT-CMC-loaded MC-2 through the hybridization of the p/p′ and q/q′. The sequences shown in the scheme are listed from 5′ to 3′.
Figure 2.
Time-dependent fluorescence changes generated by the DNAzyme reporter units associated with the different constituents in (i) CDN “S”, (ii) CDN “X”, (iii) CDN “Y”. Panel I, constituent AA′; panel II, constituent AB′; panel III, constituent BA′; panel IV, constituent BB′. The percentages of upregulated/downregulated contents of the constituents to miRNA driven transitions of CDN “S” to CDN “X” and CDN “S” to CDN “Y” are indicated in the panels.
The concept of CDN-guided release of loads from the drug-loaded microcapsules is depicted in Figure 1. Two different microcapsules were prepared: microcapsules 1 (MC-1) were loaded with doxorubicin-dextran, DOX-D, whereas microcapsules 2 (MC-2) were loaded with camptothecin-CMC, CPT-CMC. The microcapsules consist of shells composed of layered nucleic acids cross-linked by nucleic acid bridges p for capsules MC-1 and q for MC-2. The CDN systems are subjected to two hairpin structures, H1 and H2. The ribonucleobase-functionalized hairpin H1 acts as substrate of constituent BB′, and its cleavage leads to a fragment p′ that is complementary to the interlayer bridges of the shell associated with MC-1. As a result, the BB′ cleavage of H1 leads to p′ that displaces the bridges p in the form of p′/p. The displacement of the MC-1 bridging unit degrades the MC-1 microcapsules, leading to the release of DOX-D. That is, the release of DOX-D by CDN “S” is inefficient, due to the low content of AA′ and BB′. The T1 (miRNA-155)-guided reconfiguration of CDN “S” into CDN “X”, where the upregulation of AA′ is accompanied by the upregulation of BB′, is anticipated to enhance the CDN “X”-driven release of DOX-D from the MC-1 microcapsules. Similarly, the hairpin H2 includes a ribonucleobase-functionalized loop that is cleaved by the Mg2+-ion-dependent DNAzyme associated with the BA′ constituent present in the different CDNs. The cleavage of H2 yields the fragment q′ that displaces the bridging units q that stabilize the interlayers of the shells associated with the MC-2 microcapsules. The displacement of the bridging units in the form of q′/q separates the microcapsules and results in the release of the camptothecin drug linked to carboxymethyl cellulose, CPT-CMC. The miRNA-124 T2-triggered reconfiguration of CDN “S” into CDN “Y” upregulates the constituent AB′, and concomitantly upregulates the constituent BA′. In turn, the constituents AA′ and BB′ are downregulated. Accordingly, the cleavage of H2 by the constituent BA′ is enhanced in the presence of CDN “Y”, resulting in an improved release of CPT-CMC from the microcapsules MC-2 as compared to the release of the load stimulated by BA′, present in CDN “S”.
The preparation of the nucleic acid-based drug-loaded microcapsules followed the method previously reported by our laboratory. (The schematic preparation of microcapsules is shown in Figure S3).32−34 CaCO3 microparticles were loaded with DOX-D or CPT-CMC, and the particles were functionalized with a primary positively charged poly(allylamine hydrochloride) layer. Subsequently, using a layer by layer deposition process, the duplexes (1)/(2) or (4)/(5) were deposited on the primer layer, where (2) and (5) (p and q) provide the bridging units between the shell layers. Subsequently, the (1)/(2) or (4)/(5) functionalized layers were reacted with (2)/(3) or (5)/(6) to yield the second layer. Using this process, shells consisting of a controlled number of layers were prepared. In the present study, we constructed 6 layers to stabilize the respective shells. (Note that the bridges (2)/p and (5)/q include the sequences to be displaced by the cleaved hairpin H1 or H2 (p′ and q′), respectively). The resulting DNA-coated CaCO3 microparticles were etched with EDTA to remove the CaCO3 cores and yield the, respective drug-loaded microcapsules. The SEM images and confocal microscopy fluorescence images of the microcapsules are shown in Figure S4. The loading of DOX on the dextran polymer was evaluated spectroscopically to be 60 DOX units per dextran chain (see the Supporting Information, Figure S5). Also, the loading of camptothecin units on the CMC polymer was evaluated to be 45 camptothecin units per polymer chain. According to the concentration of the microcapsules by flow cytometry and the appropriate calibration curves of DOX-D and CPT-CMC (Figure S6), we calculated that the average loadings of DOX-D and CPT-CMC in the capsules are about 2.2 × 10–14 mol and 0.8 × 10–14 mol, respectively. The concentration of the microcapsules was estimated by flow cytometry experiments to be ca. 2000 capsules per 1 μL solution. We then examined the miRNA-stimulated release of the DOX-D and CPT-CMC loads from the respective microcapsules (for the primary characterization of the release of two different fluorescent dyes, see Figure S7, panel I, in the Supporting Information and the accompanying discussion).
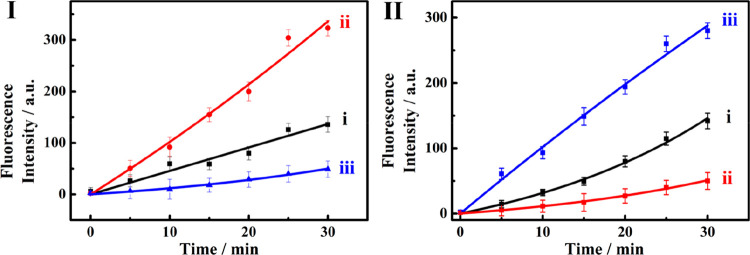
Figure 3I shows the time-dependent release of DOX-D from the MC-1 microcapsules upon interaction with CDN “S” (curve i), and upon triggering CDN “S” with trigger T1 (miRNA-155) (curve ii) or T2 (miRNA-124) (curve iii). The T1-guided reconfiguration of CDN “S” into CDN “X” leads to a significantly ca. 3-fold enhanced release of DOX-D from the MC-1 microcapsules (curve ii). In addition, interacting CDN “S” with trigger T2 depletes the release of DOX-D from MC-1. These results are consistent with the fact that the constituents AA′ and BB′ are upregulated upon the treatment of CDN “S” with T1, resulting in the enhanced cleavage of H1 and the release of DOX-D. However, subjecting CDN “S” to trigger T2 leads to CDN “Y” where AA′ and BB′ are downregulated, and thus, the release of DOX-D under these conditions is almost depleted. These results demonstrate the miRNA-155-triggered selection of the unlocking of the microcapsules MC-1. Also, Figure 3II shows the activity of the different CDNs toward the unlocking of microcapsules MC-2 and the release of CPT-CMC. Figure 3II, curve i, shows the time-dependent release of CPT-CMC upon interacting CDN “S” with H2. Subjecting CDN “S” to the trigger T2 (miRNA-124) leads to the formation of equilibrated CDN “Y”, where the constituent AB′ is upregulated and, concomitantly, constituent BA′ is upregulated. Figure 3II, curve iii, shows the time-dependent release of CPT-CMC in the presence of CDN “Y”. The release of CPT-CMC is ca. 3-fold enhanced as compared to the release of the load by CDN “S”, consistent with the upregulation of BA′, that participates in the cleavage of H2 that leads to the unlocking of the MC-2 microcapsules (cf. Figure 1). Figure 3II, curve ii, depicts the time-dependent release of CPT-CMC upon the treatment of CDN “S” with the trigger T1 (miRNA-155), in the presence of H2. The release of CPT-CMC is almost depleted under these conditions, implying that the T1-guided reconfiguration of CDN “S” to CDN “X” depletes the constituents BA′ and AB′, leading to inhibited release of the CPT-CMC load. The results reveal an important feature of the CDNs systems. CDN “S” is nonselective and stimulates the release of DOX-D and CPT-CMC from the two kinds of microcapsules at moderate rates. The miRNA-triggered transitions of CDN “S” into CDN “X” or CDN “Y” not only enhance the release of DOX-D or CPT-CMC from the respective microcapsules but also introduce selectivity reflected by the depletion of the release of CPT-CMC upon the T1-enhanced release of DOX-D, and the depletion of the release of DOX-D upon the T2-induced enhanced release of CPT-CMC. Such network-guided functions, where auxiliary triggers dictate selective dynamically controlled chemical transformations, frequently operate in natural systems. It should be noted that the dots shown in curves i–iii in Figure 3, panels I and II, represent the experimental time-dependent fluorescence changes upon the CDNs-stimulated release of the drugs. The solid lines correspond to a fit of the experimental results to a first-order drug release profile (r = 0.9685 for MC-1 and r = 0.9901 for MC-2).
Figure 3.
(I) Time-dependent release of the DOX-D from the MC-1 microcapsule driven by (i) CDN “S”, (ii) CDN “X”, (iii) CDN “Y”. (II) Time-dependent release of the CPT-CMC from the MC-2 microcapsule driven by (i) CDN “S”, (ii) CDN “X”, (iii) CDN “Y”. Error bars derived from N = 3 experiments.
We further note that CDNs shown in Figure 1 respond selectively to one of two alternative triggers (miRNA-155; miRNA-124) to unlock the respective microcapsules. This selectivity originates from the dictated stabilization of either AA′ of CDN “X” by miRNA-155 (T1), or the stabilization of AB′, associated with CDN “Y”, by miRNA-124 (T2). This selectivity is demonstrated by subjecting CDN “S” to two foreign miRNAs: miRNA-145; miRNA-16, Figure S8. These two miRNAs have no effect on the unlocking of the different drug-loaded microcapsules. Furthermore, we note that the all-DNA microcapsules shown in Figure 1 reveal structural stabilities against deoxyribonuclease I (DNase I) digestion on the time-scale of the drug release processes (Figure S9). We find that the microcapsules subjected to 0.5 U/mL DNase I exhibit an intact structure for 60 min, and almost no degradation can be detected in the electrophoretic gel. This stability of the DNA microcapsule coatings against DNase I may be attributed to the rigidified cross-linked structures that provide a barrier for the DNase activity.
The parent composition of the constituents in CDN “S” was designed to include an initial equimolar concentration of the constituents (each 0.5 μM). Under these conditions, the miRNA-155-triggered reconfiguration of CDN “S” into CDN “X” led to a ca. 2-fold increase in the release of DOX-D from MC-1, as compared to CDN “S”. Similarly, the miRNA-124-triggered reconfiguration of CDN “S” to CDN “Y” led to a 2-fold enhanced release of CPT-CMC from the MC-2, as compared to the release from CDN “S”. We note, however, that the degree of enhancement of the release of the respective drugs can be optimized by altering the equilibrated composition of the constituents in the parent CDN “S”. For example, by the modified engineering of one of the components comprising CDN “S”, to yield CDN “P”, the initial concentrations of the equilibrated constituents in CDN “S” were altered to include AA′, 0.70 μM; AB′, 0.24 μM; BA′, 0.26 μM; BB′, 0.73 μM (see the Supporting Information, Table S2). Under these conditions, the miRNA-124-triggered reconfiguration of the parent CDN “P” to CDN “Q” led to 4-fold inhibition of the release of DOX-D from the MC-1, while only a 2-fold inhibition of the release of DOX-D by CDN “Y” as compared to CDN “S” was originally achieved. Similarly, a 4-fold enhancement was observed in the release of CPT-CMC from MC-2, as compared to the release profiles of the drugs from the capsules by CDN “P”, while only a 2-fold enhancement in releasing CPT-CMC was originally observed upon the miRNA-guided reconfiguration of CDN “S” to CDN “Y”. (For the experimental results and associated discussion see Figures S10–S15). These results imply that by the appropriate optimization of the structures of the constituents in the CDNs, the control over the selectivity and efficiency of the released drugs from the respective microcapsules can be tuned. The miRNAs-triggered CDNs-guided release of the drugs from the respective microcapsules was, so far, examined in pure buffer solutions, and thus, the effects of biological fluids on the reconfiguration of the CDNs and the release rates would be interesting. Figure S16 depicts the effect of the reconfiguration of CDN “S” into CDN “X” or CDN “Y”, in the presence of miRNA-155 or miRNA-124, respectively, on the release profiles of the two drugs in 10% plasma solutions. We find that the release profiles of the drugs under these conditions are very similar to those observed in pure buffers.
It would be of interest to validate, as a proof-of-concept, the functionality of the miRNA-responsive doxorubicin on specific cancer cells and compare the cytotoxicity of the drug-loaded microcapsules toward normal epithelial cells or foreign cancer cells. Toward this goal, we subjected the MDA-MB-231 breast cancer cells (that include overexpressed miRNA-155), HpeG2 liver cancer cells, and normal MCF-10A breast cells to the DOX-D-loaded miRNA-155-responsive microcapsules and evaluated the permeation and cytotoxicity of the microcapsules toward the different cells. We find that the microcapsules permeate effectively into the cancer cells while their permeation into the epithelial cell is inefficient. For example, Figure S17 shows the uptake of the DOX-D-loaded miRNA-155-responsive microcapsules into the MDA-MB-231 breast cancer cells, the MCF-10A normal breast cells, and the HpeG2 liver cancer cells. Evidently, the permeation of the drug-loaded microcapsules into the two cancer cells is ca. 3-fold more efficient as compared to the normal cells. Accordingly, the three kinds of cells were subjected to CDN “S”, the hairpin H1 or H2, and the MC-1 (capsules loaded with DOX-D), for a time interval of 2 days, and the cytotoxicity of the MC-1 microcapsules toward the cells and control systems was examined (Figure 4). The cytotoxicity of the MC-1 toward the three kinds of cells treated with the MC-1 and the hairpin H1 (but in the absence of CDN “S”) is very low (entry II) in Figure 4. That is, despite the effective permeation of MC-1 into the cancer cell, a very low cytotoxicity is observed. These results are consistent with the fact that the CDN “S” is missing in the microcapsules, and thus, the cleavage of H1 and the unlocking of the microcapsules and release of the DOX-D are prohibited. The minor cytotoxicity may be attributed to minute leakage of DOX-D from the capsules. Similarly, entry III presents the important control experiment where all three kinds of cells are subjected to MC-1, the CDN “S”, and hairpin H2. Only a minute cytotoxic effect on all three kinds of cells is observed. That is, the overexpressed miRNA-155 cannot trigger, in the presence of H2, the enhanced release of DOX-D required to impose a significant cytotoxic effect on all three kinds of cells. Figure 4, entry IV, shows the cytotoxicity stimulated by the MC-1, H1, and cotransfected CDN “S”. Only, minute cell death of the normal MCF-10A is observed (ca. 5%), and a moderate cell death of the HpeG2 liver cancer cells is observed (15%) owing to the effective cell uptake of cancer cells and the controlled release of drugs from CDN “S”, while effective cell death of the MDA-MB-231 breast cancer cells (ca. 30%) is detected. These results and, particularly, the selectivity toward the MDA-MB-231 cancer cells originate from the overexpressed miRNA-155 that unlocks the MC-1 and releases the drug in the specific cancer cells. That is, entry III in Figure 4 emphasizes that CDN “S” cannot induce cytotoxicity toward any of the cells since MC-1 are loaded only with H2 that cannot unlock MC-1. In turn, entry IV in Figure 4 presents the cytotoxicity of the CDN “S” and the DOX-D-loaded MC-1 upon permeation into the three types of cells. While no cytotoxic effect is observed toward the normal MCF-10A breast cells (consistent with the low permeation of MC-1 into the cells), distinct differences in the cytotoxicity toward HpeG2, liver cancer cells, ca. 15% motility, and significantly higher cytotoxicity toward the MDA-MB-231 breast cancer cells, 30% motility, are observed. The higher cytotoxicity toward the MDA-MB-231 cells is attributed to the miRNA-155 overexpressed in these cells that transform CDN “S” to CDN “X”, where the cleavage of H1 and the drug release from MC-1 are enhanced. The low cytotoxicity toward the HpeG2 liver cancer cells, relatively low-expressed miRNA-155, is explained by the low-yield cleavage of H1 by CDN “S”. These results demonstrate the significance of the miRNA-guided release of the drug in the specific cancer cell. While the cellular cytotoxicities of the CDN “S” and the MC-1/MC-2 are at preliminary steps, the rapid incorporation of the CDN and MC-1/MC-2 in cancer cells suggests that the translation of these structures into the in vivo system could be accomplished by invasive injection of the component in the form of suspensions or in the presence of appropriate carriers. Nonetheless, such experiments will require future efforts. It should be noted that the CDN-driven cytotoxicity of the system is moderate. Increasing the doses of MC-1 and H1, as well as increasing the incubation time intervals between the CDN/H1/MC-1 and the cancer cells, could enhance the cytotoxic effect.
Figure 4.
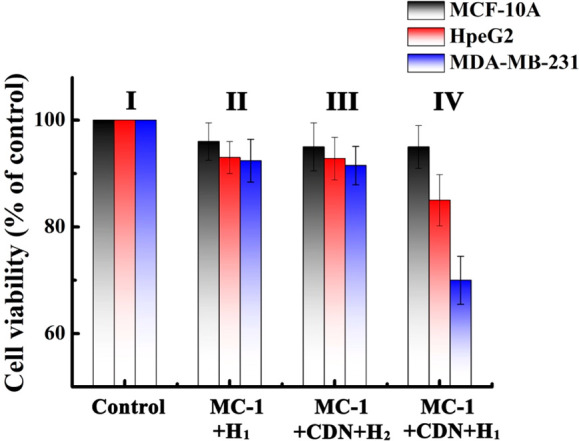
Cytotoxicity of the CDN “S”/H1/MC-1 toward MCF-10A, HpeG2, and MDA-MB-231 cells and appropriate control systems. Panel I: no addition of the components to the cells. Panel II: MC-1 and H1 were incorporated in all kinds of kinds of cells (No CDN added). Panel III: MC-1, the CDN “S”, and H2 were incorporated into the cell (no H1). Panel IV: MC-1, CDN “S”, and H1 were incorporated into all three kinds of cells. The cell viability was monitored after a time interval of 2 days at the respective conditions. Error bars derived from N = 3 experiments.
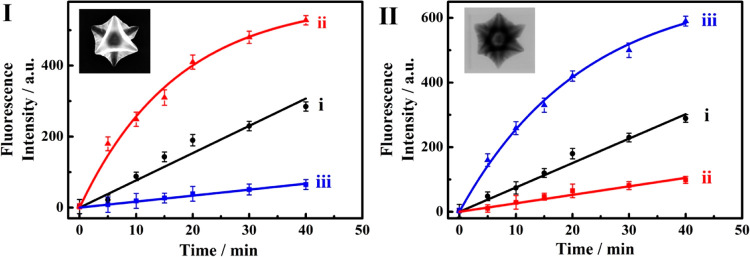
In addition, the miRNAs-triggered CDNs-guided release of the two drugs from a different carrier system, namely, metal–organic framework nanoparticles, was explored. Our laboratory has developed a versatile method to synthesize porous nucleic acid-functionalized UiO-68 metal–organic framework nanoparticles, NMOFs, loaded with drugs and locked by means of different stimuli-responsive gates. Different triggers29,31 such as pH,30 ATP,41 or VEGF were used to unlock the NMOFs and release the drugs. In the present study, we synthesized two kinds of drug-loaded gated NMOFs that substitute the microcapsules MC-1 and MC-2 discussed earlier. One kind of NMOFs carrier included the loading of the nucleic acid, (7)-functionalized UiO-68 with doxorubicin, DOX, and the locking of the NMOFs by the hybridization with (7′), NMOF-1. The second kind of NMOF included the loading of the nucleic acid, (8)-functionalized UiO-68 NMOFs, with camptothecin, CPT, and the locking of the NMOFs through the formation of duplex gates with (8′), NMOF-2. For the detailed synthesis and characterization of the two kinds of NMOFs drug carriers, see the Supporting Information, Figures S18 and S19 and the accompanying discussion. The DOX-loaded NMOF-1 and CPT-loaded NMOF-2 were then subjected to the miRNA-triggered reconfiguration of CDN “S” into CDN “X”, in the presence of miRNA-155, or to the reconfiguration of CDN “S” into CDN “Y”, in the presence of miRNA-124, to induce the miRNA-guided release of drugs from the respective DOX-loaded NMOFs-1 and CPT-loaded NMOF-2. For a detailed discussion and description of the miRNA-triggered CDN-guided release of the drugs from NMOFs-1 and NMOFs-2, see Figure S20. Figure 5I depicts the time-dependent release of the DOX by the different CDNs: The BB′-driven release of DOX by the CDN “S” is shown in curve i. The miRNA-155 reconfiguration of CDN “S” to CDN “X” results in the enhanced release of DOX from the NMOFs-1 (curve ii). A 2-fold increase in the release rate is observed, and we estimate that after a time interval of 40 min ca. 22.8 nmol mg–1 of the loaded drugs were released from the NMOFs by CDN “S”, and ca. 42.9 nmol mg–1 of the drugs were released by CDN “X”. This is consistent with the stabilization of the constituent AA′ by miRNA-155 and the concomitant upregulation of BB′. The increase in the contents of the constituent BB′ results in the enhanced cleavage of hairpin H1 yielding the fragmented strand L1, (9) that unlocks the DOX-loaded NMOFs and stimulates the release of DOX. Subjecting CDN “S” to miRNA-124 results in the very inefficient release of DOX from the DOX-loaded NMOFs (Figure 5, panel I, curve iii). We estimate that only ca. 5.2 nmol mg–1 of the DOX were released after a time interval of 40 min from the NMOFs by CDN “Y”. That is, subjecting CDN “S” to the two miRNAs regulates the release of DOX from the NMOFs. While treatment of the CDN “S” with miRNA-155 enhances the release of DOX as compared to the parent CDN “S”, the miRNA-124-triggered reconfiguration of CDN “S” to CDN “Y” depletes the release of DOX from the NMOFs as compared to the moderate release of DOX stimulated by CDN “S”. This result is consistent with the fact that treatment of CDN “S” with the trigger miRNA-124 (T2) reconfigures CDN “S” into CDN “Y” where the constituent BB′ is downregulated, resulting in the inefficient release of DOX. Similarly, Figure 5, panel II depicts the release of CPT from the miRNA-124-responsive NMOFs-2 in the presence of the different CDNs. The time-dependent release of CPT stimulated by the constituent BA′ associated with CDN “S” is shown in curve i. Using the respective calibration curve (Figure S5), we estimate that after a time interval of 40 min ca. 9.7 nmol mg–1 of the CPT were released by CDN “S”. The T2-triggered reconfiguration of CDN “S” into CDN “Y” (T2 = miRNA-124) results in the time-dependent release of CPT from the NMOFs shown in Figure 5II, curve iii. An almost 3-fold enhanced release of CPT is observed, and after a time interval of 40 min, ca. 28.3 nmol mg–1 CPT were released from the NMOFs. The enhanced release of CPT by CDN “Y” is consistent with the T2-triggered stabilization of the constituent AB′-T2 in CDN “Y” and the accompanying upregulation of BA′. The upregulated BA′ enhances the cleavage of H2 that results in the unlocking of the NMOFs by L2, (10), and the release of CPT. Figure 5II, curve ii, shows the time-dependent release of CPT triggered by CDN “X”, generated by the treatment of CDN “S” with miRNA-155. An inefficient release of CPT is observed, and a ca. 4-fold decrease in the release efficiency of CPT by CDN “X” as compared to the release by CDN “S” is observed. After a time interval of 40 min ca. 2.6 nmol mg–1 of the CPT were released by CDN “X”, as compared to the release of 9.7 nmol mg–1 of CPT by the parent CDN “S”. This result is consistent with the fact that the reconfiguration of CDN “S” to CDN “X” (by miRNA-155) overexpressed AA′ and BB′, while it downregulated AB′ and BA′. The downregulation of BA′ retards the release of CPT from the respective carriers. While in the absence of the miRNAs triggers (CDN “S”), the two drugs were released from the two carriers at a moderate rate, triggering of the CDN “S” with either miRNA-155 (to yield CDN “X”) or miRNA-124 (to yield CDN “Y”) forward the enhanced CDN-guided release of DOX or CPT, respectively. Concomitantly to the miRNA-CDNs-driven enhanced release of the specific drugs, we observe, however, that the miRNA-driven release of the drug from one of the carriers depresses and almost completely depletes the release of the other drug present in the carrier mixture. Such miRNA-triggered CDN-induced selective release of a drug might be of significance for personalized therapeutic medicine. It should be noted that the dots shown in curves i–iii in Figure 5, panels I and II, represent the experimental time-dependent fluorescence changes upon the CDNs-stimulated release of the drugs. The solid lines correspond to a fit of the experimental results to a first-order drug release profile (r = 0.9876 for miRNA-155-responsive NMOFs and r = 0.9904 for miRNA-124-responsive NMOFs).
Figure 5.
(I) Time-dependent release of the DOX from miRNA-155-responsive NMOF driven by (i) CDN “S”, (ii) CDN “X”, (iii) CDN “Y”. (II) Time-dependent release of the camptothecin from the miRNA-124-responsive NMOF driven by (i) CDN “S”, (ii) CDN “X”, (iii) CDN “Y”. The insets in panels I and II correspond to the SEM and TEM images of the DOX-and CPT-loaded NMOFs, respectively. Error bars derived from N = 3 experiments.
Conclusions
The study has introduced the application of miRNA-responsive constitutional dynamic networks (CDNs) as functional units that control and dictate the guided release of one or two anticancer drugs (DOX-D/DOX or CPT-CMC/CPT). While in the absence of miRNA, inefficient release of the two drugs is observed, the miRNAs-triggered reconfiguration of CDN “S” into CDN “X” or CDN “Y” leads to the enhanced release of either DOX-D/DOX or CPT-CMC/CPT. Most importantly is, however, the miRNA-stimulated selectivity of the enhanced release of DOX-D/DOX or CPT-CMC/CPT. Namely, the release of DOX-D by CDN “Y” generated in the presence of miRNA-124 is almost depleted, and the release of CPT-CMC in the presence of CDN “X”, formed upon subjecting CDN “S” to miRNA-155, is almost depleted. Such control over the miRNA-driven release of a specific drug from a mixture of drug-loaded carriers might be important for personalized medicine treatment. A major point that needs to be addressed relates, however, to the need to control the release of the drugs by means of the CDNs. One may argue that the microcapsules would be engineered in such a way that the miRNAs (miRNA-155 or miRNA-124) directly unlock the carrier for the respective release processes. Nonetheless, one should realize that the concentrations of miRNA are low, and thus, the direct triggering of the carriers would be inefficient. The miRNA-triggered activation of the CDNs provides an amplification path for the unlocking of the carriers. The DNAzyme associated with the constituents BB′ (in CDN “X”) and BA′ (in CDN “Y”) catalyzes the cleavage of the hairpins H1 and H2 thus providing amplified strands for unlocking the carriers. That is, the catalytic cleavage of hairpins H1 and H2 by the respective CDN constituents provides a mechanism to amplify the unlocking of the respective microcapsule and to release the drugs. This is exemplified in Figure 6, where MC-1 and MC-2 were subjected to different concentrations of the respective miRNA-155 and miRNA-124, in the presence of H1 and H2, in the absence and presence of CDN “S”. Evidently, in the absence of the CDN no release of DOX-D from MC-1 or of CPT-CMC from MC-2 can be detected within this concentration range of miRNA, (curves i and ii). In turn, successful release of DOX-D from MC-1 or of CPT-CMC from MC-2 is observed within the entire range of miRNA concentration in the presence of the CDN (curves iv and vi). As the concentrations of miRNA increase, the release rates of the respective drugs are enhanced. It should be noted that MC-1, engineered to be directly uncaged by miRNA-155, to release DOX-D, or MC-2 engineered to be directly, uncaged by miRNA-124 to release CPT-CMC, did not show any release properties at miRNAs concentrations corresponding to 5–10 nM, in contrast to the release of the drugs observed in the presence of the CDNs. These results emphasize the significance of the CDNs in amplifying the release processes. (For experimental details and the structures of the respective microcapsules, see the Supporting Information, Figures S21 and S22, and the accompanying discussion.) To further emphasize and explain the selectivity and amplification features introduced by the CDNs-stimulated release of the two drugs from the mixture of the two-drug loaded microcapsules, we note the following: (i) As stated, one may design microcapsules that are directly unlocked by the respective miRNA. As shown in Figure S21, such microcapsules are not being unlocked at miRNAs concentrations that correspond to 5 and 10 nM. The constituents of the CDNs unlock the respective microcapsules at these concentrations. This is due to the miRNA-stimulated stabilization of DNAzymes that catalyze the amplified formation of strands p′ and q′ that unlock the microcapsules. (ii) One might argue that we could apply the pure AA′-T1 DNAzyme and AB′-T2 DNAzyme to unlock the microcapsules without the CDNs systems. Indeed, such a procedure would lead to the amplified unlocking and release of the drugs. Nonetheless, such a procedure will lead to the loss of the selectivity principle introduced by the CDNs. Namely, the formation of CDN “X” inhibits the release of CPT, and the formation of CDN “Y” decreases the release of DOX. Saying that, the CDNs introduce selectivity/amplification paths for the release of the two drugs.
Figure 6.
Time-dependent release of the DOX from miRNA-155-responsive MC-1 microcapsule (panel I) and from miRNA-124-responsive MC-2 microcapsule (panel II) driven by (i) 10 nM of miRNA in the presence of H1 and in the absence of CDN “S”, (ii) 100 nM of miRNA in the presence of H1 and in the absence of CDN “S”, (iii) 1 μM of miRNA in the presence of H1 and in the absence of CDN “S”, (iv) 10 nM of miRNA in the presence of H1 and in the presence of CDN “S”, (v) 50 nM of miRNA in the presence of H1 and in the presence of CDN “S”, (vi) 500 nM of miRNA in the presence of H1 and in the presence of CDN “S”, and (vii) 1 μM of miRNA in the presence of H1 and in the presence of CDN “S”. Error bars derived from N = 3 experiments. It should be noted that the solid lines (i–vii) correspond to a fit of the experimental results to a first-order drug release profile (r = 0.9804 for MC-1 and r = 0.9882 for MC-2).
Experimental Section
The sequences of all nucleic acids used in this paper are listed as follows (from 5′ to 3′):
A: GATATCAGCGATACGATACAAACATTAGCATTAACGTGCCTTAA
A′: ACCCCTATCACGGTTTGTATCGTCACCCATGTTCGTCA
B: CTGCTCAGCGATACGATACAAACAATCCTTAA
B′: GCATTCACCGGTTTGTATCGTCACCCATGTTACTCT
T1-microRNA-155: UUAAUGCUAAUCGUGAUAGGGGU
T2-microRNA-124: UUAAGGCACGCGGUGAAUGC
A-MOD: GATATCAGCGATACGATACAAACATTAGCATTAACGTGCCTTAACACACACACA
A′-MOD: TCTCTCTCTCACCCCTATCACGGTTTGTATCGTCACCCATGTTCGTCA
Sub 1 (AB′): FAM-AGAGTATrAGGATATC-BHQ1
Sub 2 (BA′): ROX-TGACGATrAGGAGCAG-BHQ2
Sub 3 (BB′): CY5-AGAGTATrAGGAGCAG-BHQ2
Sub 4 (AA′): CY5.5-TGACGATrAGGATATC-IBRQ
Sub1-noFQ (AB′): AGAGTATrAGGATATC
Sub2-noFQ (BA′): TGACGATrAGGAGCAG
Sub3-noFQ (BB′): AGAGTATrAGGAGCAG
Sub4-noFQ (AA′): TGACGATrAGGATATC
H1: CCGGAGTATTGCGGTTCCAGAGTATrAGGAGCAGCCTTCCTGGCGGACCGCAATACTCCGGGCTGCCCCCAGG
H2: TTCCAGACAAGAGTGCCATTGACGATrAGGAGCAGTACCCTGTAGCGGCACTCTTGTCTGGACCGGGACGGAA
p′/L1, (9): GGAGCAGCCTTCCTGGCGGACCGCAATACTCCGGGCTGCCCCCAGG
q′/L2, (10): GGAGCAGTACCCTGTAGCGGCACTCTTGTCTGGACCGGGACGGAA
(1): GAATCGATTACCCCCAGG
(2)/p: TCGATTCCCTGGGGGCAGCCCGGAGTATTGCGGTCCGCCAGGAAGGTAGTTGGT
(3): CCTTCCTTTTACCAACTA
(4): CTACTATAATGACGGCC
(5)/q: ATAGTAGGGCCGTCCCGGTCCAGACAAGAGTGCCGCTACAGGGTATAAGAAT
(6): TACCCTGTAAATTCTTA
(7): NH2-(CH2)6-GGAGCAGCCTTCCTGGCGGACCGCAATACTCCGGGCTGC
(7′): CCTGGGGGCAGCCCGGAGTATTGCGGTCCGCCAGGAAGGCTGCTCC
(8): NH2-(CH2)6-GGAGCAGTACCCTGTAGCGGCACTCTTGTCTGGACCGG
(8′): TTCCGTCCCGGTCCAGACAAGAGTGCCGCTACAGGGTACTGCTCC
Additionally, the ribonucleobase cleavage site of rA in all the substrates of the different Mg2+-dependent DNAzymes is marked in bold; the sequences of corresponding Mg2+-dependent DNAzyme are indicated with underline.
Generation of CDNs
The [2 × 2] CDN “S” depicted in Figure 1, consisting of the constituents AA′, AB′, BA′, and BB′, was prepared as follows. A mixture of A, A′, B, and B′ (2 μM each) in HEPES buffer (10 mM, 20 mM MgCl2, pH = 7.2) was annealed at 65 °C for 15 min, subsequently cooled down to 25 °C with the rate of 0.33 °C/min, and eventually allowed to equilibrate at 25 °C for 2 h, yielding CDN “S”. For the conversion of CDN “S” to CDN “X” or CDN “Y”, the mixture of AA′, AB′, BA′, and BB′ (CDN “S”, 1 μM) was mixed with the corresponding trigger miRNA-155 or miRNA-124 (1.3 μM) and was allowed to equilibrate at 33 °C for 12 h, to generate CDN “X” or CDN “Y”, respectively.
Controlling the Release of Loads from Microcapsules or NMOFs Using Auxiliary CDNs
The equilibrated CDN “S”, “X”, or “Y” (10 μL, 2 μM) was subjected to the hairpins H1 or H2 (5 μL, 2 μM), and to the prepared solution of microcapsules or NMOFs (100 μL for each). The resulting mixture was divided into different batches; each batch was incubated at room temperature for different time intervals. After incubation, the respective batches were centrifuged at 500 rpm for 20 min to precipitate the residual capsules, and the fluorescence intensity of the released drugs in the supernatant solution was monitored. Notably, all the hairpins were annealed at 95 °C and slowly cooled down to room temperature before using.
Acknowledgments
This paper was supported by the Minerva Center for Biohybrid Complex Systems.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.9b06047.
Materials and instrumentation, measurements, calibration curves, gel electrophoresis, schematic description of NMOFs preparation and CDNs-guided release of loads from NMOFs, and additional results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Jaenisch R.; Bird A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Barabaśi A. L.; Oltvai Z. N. Network Biology: Understanding the Cell’s Function Organization. Nat. Rev. Genet. 2004, 5, 101–113. 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Vogel C.; Marcotte E. M. Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 2012, 13, 227–232. 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.; Levin M. Gene Regulatory Networks. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 4935–4942. 10.1073/pnas.0502024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehn J.-M. Perspectives in Chemistry-Aspects of Adaptive Chemistry and Materials. Angew. Chem., Int. Ed. 2015, 54, 3276–3289. 10.1002/anie.201409399. [DOI] [PubMed] [Google Scholar]
- Giuseppone N.; Lehn J.-M. Protonic and Temperature Modulation of Constituent Expression by Component Selection in a Dynamic Combinatorial Library of Imines. Chem. - Eur. J. 2006, 12, 1715–1722. 10.1002/chem.200501038. [DOI] [PubMed] [Google Scholar]
- Vantomme G.; Jiang S.; Lehn J.-M. Adaptation in Constitutional Dynamic Libraries and Networks, Switching between Orthogonal Metalloselection and Photoselection Processes. J. Am. Chem. Soc. 2014, 136, 9509–9518. 10.1021/ja504813r. [DOI] [PubMed] [Google Scholar]
- Giuseppone N.; Schmitt J.-L.; Lehn J.-M. Generation of Dynamic Constitutional Diversity and Driven Evolution in Helical Molecular Strands under Lewis Acid Catalyzed Component Exchange. Angew. Chem., Int. Ed. 2004, 43, 4902–4906. 10.1002/anie.200460343. [DOI] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. A DNA Enzyme that Cleaves RNA. Chem. Biol. 1994, 1, 223–229. 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. Directed Evolution of Nucleic Acid Enzymes. Annu. Rev. Biochem. 2004, 73, 791–836. 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Willner I.; Shlyahovsky B.; Zayats M.; Willner DNAzymes for Sensing, Nanobiotechnology and Logic Gate Applications. Chem. Soc. Rev. 2008, 37, 1153–1165. 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- Golub E.; Albada H. B.; Liao W.-C.; Biniuri Y.; Willner I. Nucleoapzymes: Hemin/G-Quadruplex DNAzyme–Aptamer Binding Site Conjugates with Superior Enzyme-Like Catalytic Functions. J. Am. Chem. Soc. 2016, 138, 164–172. 10.1021/jacs.5b09457. [DOI] [PubMed] [Google Scholar]
- Osborne S. E.; Matsumura I.; Ellington A. D. Aptamers as Therapeutic and Diagnostic Reagents: Problems and Prospects. Curr. Opin. Chem. Biol. 1997, 1, 5–9. 10.1016/S1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Mayer G. Aptamer Modules as Sensors and Detectors. Acc. Chem. Res. 2011, 44, 1349–1358. 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. DNA Nanotechnology: Novel DNA Constructions. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 225–248. 10.1146/annurev.biophys.27.1.225. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. An Overview of Structural DNA Nanotechnology. Mol. Biotechnol. 2007, 37, 246–257. 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.-H.; Willner B.; Willner I. DNA Nanotechnology: From Sensing and DNA Machines to Drug-Delivery Systems. ACS Nano 2013, 7, 8320–8332. 10.1021/nn404613v. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu X.; Willner I. DNA Switches: From Principles to Applications. Angew. Chem., Int. Ed. 2015, 54, 1098–1129. 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- Teller C.; Willner I. Functional Nucleic Acid Nanostructures and DNA Machines. Curr. Opin. Biotechnol. 2010, 21, 376–391. 10.1016/j.copbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Goodman R. P.; Heilemann M.; Doose S.; Erben C. M.; Kapanidis A. N.; Turberfield A. J. Reconfigurable, Braced, Three-Dimensional DNA Nanostructures. Nat. Nanotechnol. 2008, 3, 93–96. 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. From Genes to Machines: DNA Nanomechanical Devices. Trends Biochem. Sci. 2005, 30, 119–125. 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.; Willner I. pH-Stimulated Reconfiguration and Structural Isomerization of Origami Dimer and Trimer Systems. Nano Lett. 2016, 16, 6650–6655. 10.1021/acs.nanolett.6b03418. [DOI] [PubMed] [Google Scholar]
- Wang J. B.; Yue L.; Wang S.; Willner I. Triggered Reversible Reconfiguration of G-Quadruplex-Bridged “Domino”-Type Origami Dimers: Application of the Systems for Programmed Catalysis. ACS Nano 2018, 12, 12324–12336. 10.1021/acsnano.8b06191. [DOI] [PubMed] [Google Scholar]
- Lu C.-H.; Guo W.; Hu Y.; Qi X.-J.; Willner I. Multitriggered Shape-Memory Acrylamide-DNA Hydrogels. J. Am. Chem. Soc. 2015, 137, 15723–15731. 10.1021/jacs.5b06510. [DOI] [PubMed] [Google Scholar]
- Guo W.; Lu C. H.; Orbach R.; Wang F.; Qi X. J.; Cecconello A.; Seliktar D.; Willner I. pH-Stimulated DNA Hydrogels Exhibiting Shape-Memory Properties. Adv. Mater. 2015, 27, 73–78. 10.1002/adma.201403702. [DOI] [PubMed] [Google Scholar]
- Wang C.; Liu X.; Wulf V.; Vaźquez-Gonzaĺez M.; Fadeev M.; Willner I. DNA-Based Hydrogels Loaded with Au Nanoparticles or Au Nanorods: Thermoresponsive Plasmonic Matrices for Shape-Memory, Self-Healing, Controlled Release, and Mechanical Applications. ACS Nano 2019, 13, 3424–3433. 10.1021/acsnano.8b09470. [DOI] [PubMed] [Google Scholar]
- Kahn J. S.; Hu Y. W.; Willner I. Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications. Acc. Chem. Res. 2017, 50, 680–690. 10.1021/acs.accounts.6b00542. [DOI] [PubMed] [Google Scholar]
- Lu C.-H.; Willner I. Stimuli-Responsive DNA-Functionalized Nano-/Microcontainers for Switchable and Controlled Release. Angew. Chem., Int. Ed. 2015, 54, 12212–12235. 10.1002/anie.201503054. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Luo G.-F.; Sohn Y. S.; Nechushtai R.; Willner I. miRNA-Specific Unlocking of Drug-Loaded Metal-Organic Framework Nanoparticles: Targeted Cytotoxicity toward Cancer Cells. Small 2019, 15, 1900935. 10.1002/smll.201900935. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Yu X.; Cecconello A.; Sohn Y. S.; Nechushtai R.; Willner I. Stimuli-Responsive Nucleic Acid-Functionalized Metal-Organic Framework Nanoparticles using pH- and Metal-Ion-Dependent DNAzymes as Locks. Chem. Sci. 2017, 8, 5769–5780. 10.1039/C7SC01765K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-H.; Liao W.-C.; Sohn Y. S.; Fadeev M.; Cecconello A.; Nechushtai R.; Willner I. Stimuli-Responsive Nucleic Acid-Based Polyacrylamide Hydrogel-Coated Metal-Organic Framework Nanoparticles for Controlled Drug Release. Adv. Funct. Mater. 2018, 28, 1705137. 10.1002/adfm.201705137. [DOI] [Google Scholar]
- Liao W.-C.; Lu C.-H.; Hartmann R.; Wang F. A.; Sohn Y. S.; Parak W. J.; Willner I. Adenosine Triphosphate-Triggered Release of Macromolecular and Nanoparticle Loads from Aptamer/DNA-Cross-Linked Microcapsules. ACS Nano 2015, 9, 9078–9086. 10.1021/acsnano.5b03223. [DOI] [PubMed] [Google Scholar]
- Liao W.-C.; Sohn Y. S.; Riutin M.; Cecconello A.; Parak W. J.; Nechushtai R.; Willner I. The Application of Stimuli-Responsive VEGF- and ATP-Aptamer-Based Microcapsules for the Controlled Release of an Anticancer Drug, and the Selective Targeted Cytotoxicity toward Cancer Cells. Adv. Funct. Mater. 2016, 26, 4262–4273. 10.1002/adfm.201600069. [DOI] [Google Scholar]
- Liao W.-C.; Lilienthal S.; Kahn J. S.; Riutin M.; Sohn Y. S.; Nechushtai R.; Willner I. pH- and Ligand-Induced Release of Loads from DNA-Acrylamide Hydrogel Microcapsules. Chem. Sci. 2017, 8, 3362–3373. 10.1039/C6SC04770J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Yue L.; Shpilt Z.; Cecconello A.; Kahn J. S.; Lehn J.-M.; Willner I. Controlling the Catalytic Functions of DNAzymes within Constitutional Dynamic Networks of DNA Nanostructures. J. Am. Chem. Soc. 2017, 139, 9662–9671. 10.1021/jacs.7b04531. [DOI] [PubMed] [Google Scholar]
- Zhou Z. X.; Yue L.; Wang S.; Lehn J.-M.; Willner I. DNA-Based Multiconstituent Dynamic Networks: Hierarchical Adaptive Control over the Composition and Cooperative Catalytic Functions of the Systems. J. Am. Chem. Soc. 2018, 140, 12077–12089. 10.1021/jacs.8b06546. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yue L.; Li Z.-Y.; Zhang J. J.; Tian H.; Willner I. Light-Induced Reversible Reconfiguration of DNA-Based Constitutional Dynamic Networks: Application to Switchable Catalysis. Angew. Chem. 2018, 130, 8237–8241. 10.1002/ange.201803371. [DOI] [PubMed] [Google Scholar]
- Yue L.; Wang S.; Wulfa V.; Lilienthal S.; Remacle F.; Levine R. D.; Willner I. Consecutive Feedback-Driven Constitutional Dynamic Networks. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 2843–2848. 10.1073/pnas.1816670116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L.; Wang S.; Lilienthal S.; Wulf V.; Remacle F.; Levine R. D.; Willner I. Intercommunication of DNA-Based Constitutional Dynamic Networks. J. Am. Chem. Soc. 2018, 140, 8721–8731. 10.1021/jacs.8b03450. [DOI] [PubMed] [Google Scholar]
- Zhou Z. X.; Liu X.; Yue L.; Willner I. Controlling the Catalytic and Optical Properties of Aggregated Nanoparticles or Semiconductor Quantum Dots Using DNA-Based Constitutional Dynamic Networks. ACS Nano 2018, 12, 10725–10735. 10.1021/acsnano.8b05452. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Yu X.; Liao W.-C.; Sohn Y. S.; Cecconello A.; Kozell A.; Nechushtai R.; Willner I. ATP-Responsive Aptamer-Based Metal-Organic Framework Nanoparticles (NMOFs) for the Controlled Release of Loads and Drugs. Adv. Funct. Mater. 2017, 27, 1702102. 10.1002/adfm.201702102. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.