Abstract
Enzymes (endonucleases) are coupled to constitutional dynamic networks to stimulate the selection of a constituent and cascaded emergence of a new network. This is exemplified with the EcoRI-dictated depletion of a network and selection of a constituent that activates the cascaded emergence of a new network. The new network is further depleted by HindIII to a selected constituent that can be coupled to the cascaded emergence of a dynamic network. In addition, upon subjecting a [3 × 3] constitutional dynamic network to endonucleases EcoRI and HindIII, the programmed hierarchical selection of [2 × 2] constitutional dynamic networks followed by the biocatalytic selection of a constituent for the subsequent emergence of new networks is demonstrated.
Keywords: nucleic acid, systems chemistry, endonuclease, DNAzyme, DNA nanotechnology
Constitutional dynamic networks (CDNs) play key functions in controlling intracellular transformations,1−6 and substantial research efforts are directed to assemble synthetic systems mimicking these processes.7−10 The simplest CDN consists of four interequilibrated constituents AA′, AB′, BA′, and BB′. The stabilization of one of the constituents, e.g., AA′, by an auxiliary trigger, results in the upregulation of the stabilized constituent at the expense of constituents AB′ and BA′ (as they share components with AA′), which are downregulated. The separation and downregulation of the constituents leads, however, to the concomitant upregulation of constituent BB′ (that does not share components with AA′). Extensive research efforts were directed to the assembly and reconfiguration of molecular11 or supramolecular12−15 CDNs using auxiliary triggers, such as temperature,16 light,17,18 pH,16 chemical agents,19,20 solvents,21 electric fields,22 and supramolecular H-bonds.23,24 Recently, we introduced nucleic acids as versatile modules to assemble and reconfigure CDNs.25−30 Different triggers, such as the formation and dissociation of T–A·T triplexes,25 the K+-ion assembly of G-quadruplexes and their separation by crown ethers,25 and the stabilization and destabilization of duplexes by means of photoisomerizable intercalators,26 were used to reversibly reconfigure CDN systems. These processes were applied to design nucleic-acid-based networks of enhanced complexity.27−30 Beyond the reported advances and specifically to mimic complex natural networks, major challenges are still ahead of us. For example, the selection of networks, and particularly the hierarchical selection of networks, is a key feature of natural systems.31−34 In the present paper, we introduce an unprecedented concept in nucleic-acid-based constitutional dynamic networks, whereby we couple enzymes (endonucleases) to constituent modules associated with CDNs as a means to control the hierarchical selection and cascaded emergence of networks. The study paves versatile means to select and to control over many different cascaded emergent networks by applying libraries of “dormant” nucleic acid modules and different “activating” enzymes.
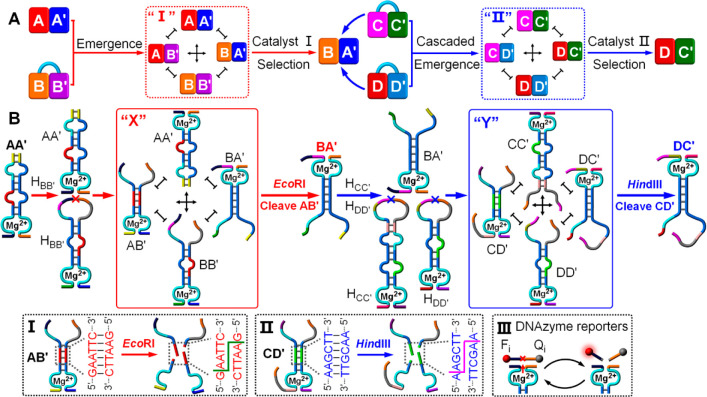
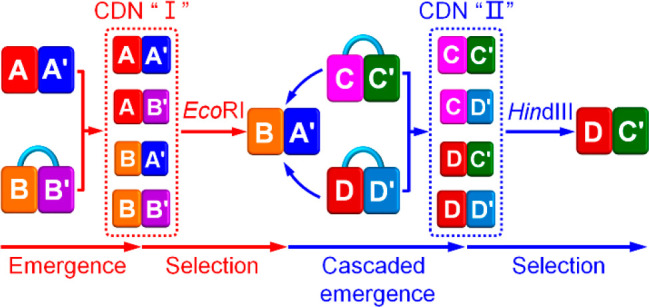
Figure 1A depicts schematically the emergence, selection, and cascaded catalytic emergence of networks. The process assumes the existence of a “pool” of modules. In the first step, the interaction of an active module, AA′, with a caged module B–B′ gives rise to the evolved emergence of a network, CDN “I”, consisting of the constituents AA′, AB′, BA′, and BB′. Subjecting the generated CDN “I” to catalyst (I) leads to the depletion of CDN “I” and the survival of constituent BA′. The selected constituent BA′ reacts with the caged modules C–C′ and D–D′, leading to the cascaded emergence of a new network, CDN “II”, composed of the constituents CC′, CD′, DC′, and DD′. The subsequent treatment of CDN “II” with catalyst (II) results in the catalyzed selection of constituent DC′, while depleting the other constituents associated with CDN “II”. That is, the scheme presents a general pathway to evolve a network to catalytically select a constituent, that activates the cascaded emergence of networks.
Figure 1.
(A) Schematic presentation of the evolved emergence, selection, and cascaded catalytic emergence of networks. (B) Schematic design of the compositions of the nucleic-acid-based constitutional dynamic networks coupled to two endonucleases that allow the biocatalytically triggered selection and cascaded emergence processes. Panels I and II - Schematic cleavage of the sequence-specific domains associated with constituents AB′ and CD′ by the endonucleases EcoRI and HindIII, respectively. Panel III - The cleavage of a fluorophore/quencher (Fi/Qi)-functionalized substrate by the Mg2+-ion-dependent DNAzyme reporter unit associated with the respective constituent.
Figure 1B shows the compositions of the nucleic-acid-based CDN systems that allow the selection and cascaded emergence processes, stimulated by two endonucleases, e.g., EcoRI and HindIII. The sequence-specific-stimulated cleavage of the duplex domains of the constituents by the respective endonucleases dictates the selection and the activation of the cascaded emergence of the CDN systems. Constituent AA′ includes a Mg2+-ion-dependent DNAzyme unit.35,36 Its interaction with a ribonucleobase-modified hairpin HBB′ that includes the caged components B and B′ leads to the cleavage of HBB′ to form constituent BB′. The equilibration of AA′ and BB′ evolves CDN “X” composed of the four constituents AA′, AB′, BA′, and BB′, where AB′ is, however, designed to be cleaved by endonuclease EcoRI,37Figure 1B, panel I. Subjecting CDN “X” to EcoRI leads to the cleavage of AB′, and thus the depletion of AB′ and the concomitant separation of constituents AA′ and BB′ that share components with AB′. The released components B and A′ from the separation of AA′ and BB′, however, enrich BA′. That is, EcoRI-stimulated cleavage of AB′ in CDN “X” results in the depletion of three constituents AA′, AB′, and BB′ and the survival and upregulation of constituent BA′. The selected BA′ is, then, subjected to the two ribonucleobase-modified hairpins HCC′ and HDD′. These hairpins include caged components in self-“dormant” configurations. Nonetheless, the cleavage of the two hairpins by BA′ yields constituents CC′ and DD′ that dynamically equilibrate into CDN “Y” consisting of CC′, DC′, CD′, and DD′. The formation of CDN “Y” represents a cascaded emergence process activated by the selected constituent BA′. The generated CDN includes, however, in constituent CD′, the encoded sequence that can be cleaved by endonuclease HindIII,38Figure 1B, panel II. Thus, in the presence of HindIII, the cleavage of CD′ depletes this constituent, and the process is accompanied by the depletion of CC′ and DD′ and the selection and upregulation of DC′, Figure 1B. To each of the constituents of the CDNs is conjugated a different Mg2+-ion-dependent DNAzyme reporter unit that cleaves a different fluorophore/quencher (Fi/Qi)-functionalized substrate to provide a fluorescence readout signal for the quantitative evaluation of the concentration of the constituent, Figure 1B, panel III. By determining the cleavage activities associated with the constituents and applying appropriate calibration curves that follow the cleavage rates of the substrates by the respective DNAzyme reporter units associated with the intact constituents at variable concentrations, the concentrations of the constituents of the CDNs can be evaluated. (It should be noted that, in principle, the dynamic transitions of the CDNs could be followed by the direct labeling of the CDN components with donor/acceptor FRET pairs, rather than employing the Mg2+-ion-dependent DNAzyme reporter units. We preferred, however, the latter read transduction means, mainly because of two reasons: (i) the donor/acceptor pairs may affect the equilibrium of the CDN system; (ii) the number of FRET pairs is limited to allow the resolved fluorescence readout of the [3 × 3] CDN system presented in the paper, vide infra.)
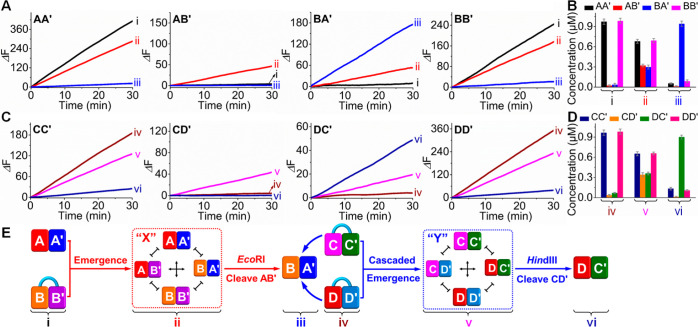
Figure 2A shows the time-dependent fluorescence changes generated by the cleavage of the Fi/Qi-functionalized substrates by the respective DNAzyme reporter units associated with the constituents, before the interaction of AA′ with HBB′ (t = 0 h, curves i), after the AA′-stimulated cleavage of HBB′ and the generation of CDN “X” (t = 24 h, curves ii), and after the treatment of the resulting CDN “X” with EcoRI and the selection of BA′ (curves iii). At time t = 0 h, AA′ and HBB′ reveal their inherent DNAzyme activities reflecting their initial concentrations, and AB′ and BA′ do not show any catalytic activities. These results are consistent with the fact that, at time t = 0 h, CDN “X” is basically nonexistent. At time t = 24 h, the formation of CDN “X” is demonstrated by the decrease of the catalytic activities associated with AA′ and BB′ and the emergence of catalytic activities associated with AB′ and BA′. That is, AB′ and BA′ are generated at the expense of AA′ and BB′ in the dynamically evolved CDN “X” (for the experimental time-dependent fluorescence spectra consistent with the time-dependent fluorescence changes shown in Figure 2A, see Figure S1). Upon subjecting CDN “X” to EcoRI, the catalytic activities corresponding to constituents AA′, AB′, and BB′ are almost zero, while the fluorescence signal of constituent BA′ is upregulated, implying the depletion of AA′, AB′, and BB′ and the selection and upregulation of BA′. (It should be noted that EcoRI reveals specific activity toward the cleavage of constituent AB′, and all other constituents are unaffected by the enzyme, Figure S2). By using the appropriate calibration curves (Figures S3 and S4), the contents of the constituents of CDN “X” before and after the formation of the CDN and after EcoRI-dictated selection of BA′ from the CDN were quantified, and the results are summarized in Table S1 and Figure 2B. The emergence of CDN “X” and the EcoRI-triggered selection of BA′ were further supported by quantitative gel electrophoresis, Figure S5. Using ImageJ software that compares the intensities of the separated bands to those of the individual structures at known concentrations (1 μM), we quantified the contents of the structures before and after the formation of CDN “X”, and after the treatment of the resulting CDN “X” with EcoRI to stimulate the selection of BA′ (Table S1 in brackets). The concentrations derived from the catalytic activities of the DNAzyme reporter units agree well with those derived by gel electrophoresis.
Figure 2.
(A) Time-dependent fluorescence changes generated by the Mg2+-ion-dependent DNAzyme reporter units associated with the constituents of CDN “X” undergoing the emergence and selection processes and (B) concentrations of the constituents of CDN “X” derived from the respective catalytic activities shown in part A, by using the appropriate calibration curves (Figures S3 and S4): (i) upon mixing constituent AA′ with hairpin HBB′ at time t = 0 h; (ii) in CDN “X” generated after the cleavage of HBB′ by AA′ (t = 24 h); (iii) after the treatment of evolved CDN “X” with EcoRI that induces the biocatalytic selection of BA′. (C) Time-dependent fluorescence changes generated by the Mg2+-ion-dependent DNAzyme reporter units associated with the constituents of CDN “Y” during the cascaded catalytic emergence and selection processes, and (D) concentrations of the constituents of CDN “Y” derived from the respective catalytic activities shown in part C, by applying the appropriate calibration curves (Figures S6 and S7): (iv) in the mixture of hairpins HCC′ and HDD′, in the presence of the survived BA′, at time t = 0 h; (v) in CDN “Y” emerged upon subjecting HCC′ and HDD′ to the survived BA′ for 24 h; (vi) after treatment of the resulting CDN “Y” with HindIII to stimulate the selection of DC′. The error bars in parts B and D were derived from N = 3 experiments. (E) Schematic emergence of CDN “X” and EcoRI-induced selection of constituent BA′ from CDN “X” followed by the BA′-dictated emergence of CDN Y and HindIII-guided survival of constituent DC′.
Figure 2C shows the time-dependent fluorescence changes generated by the reporter units associated with the constituents before the interaction of HCC′ and HDD′ with the selected BA′ (t = 0 h, curves iv), after the cascaded emergence of CDN “Y” (t = 24 h, curves v), and after the treatment of CDN “Y” with HindIII to induce the selection of DC′ (curves vi). At time t = 0, the reporter units associated with HCC′ and HDD′ yield effective fluorescence signals, whereas CD′ and DC′ are nonexistent. At time t = 24 h, the generation of equilibrated CDN “Y” occurs, as evident by the lower catalytic activities of CC′ and DD′ and the emergent catalytic activities associated with CD′ and DC′. This is consistent with the fact that CDN “Y” is formed at the expense of CC′ and DD′. After subjecting CDN “Y” to HindIII, the fluorescence signals generated by CC′, CD′, and DD′ are almost zero, while the catalytic activity associated with DC′ increases. These results reveal that constituents CC′, CD′, and DD′ are depleted and constituent DC′ is survived and upregulated. By applying appropriate calibration curves (Figures S6 and S7), the contents of the constituents before and after the formation of CDN “Y” and after the HindIII-driven selection of DC′ were quantified (Table S2 and Figure 2D). The cascaded emergence of CDN “Y” and the selection of DC′ from CDN “Y” were further supported by gel electrophoresis, Figure S8 and accompanying discussion. These results demonstrate that, by coupling endonucleases with constitutional dynamic networks, the very basic principles of the selection and cascaded emergence of the networks can be modeled, Figure 2E.
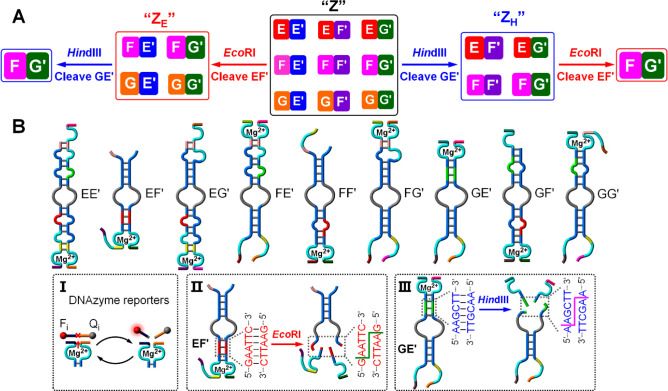
Beyond the endonuclease-induced selection of the dictated constituents from the [2 × 2] CDNs, one may use the biocatalytic reactions for the hierarchical selection of the complex networks. This is exemplified in Figure 3A with the biocatalyzed reduction of a [3 × 3] network to different [2 × 2] networks and the subsequent transition of the resulting networks into single selected constituents. The latter products may then act as initiators for the cascaded emergence of new networks (vide infra). The schematic structures of the nine constituents are provided in Figure 3B. Each of the constituents includes a different Mg2+-ion-dependent DNAzyme reporter unit, Figure 3B, panel I, and constituents EF′ and GE′ include sequence-specific domains to be cleaved by endonucleases EcoRI and HindIII, respectively, Figure 3B, panels II and III. Thus, treatment of CDN “Z” with EcoRI is anticipated to deplete constituent EF′, and hence, all other constituents that share components with EF′, e.g., EE′, EG′, FF′, and GF′, will be depleted, resulting in an upregulated survived [2 × 2] network, CDN “ZE”, which consists of FE′, FG′, GE′, and GG′. Subjecting CDN “ZE” to HindIII leads to the cleavage and the depletion of constituent GE′, resulting in the concomitant depletion of constituents GG′ and FE′, while constituent FG′ is survived and overexpressed. Similarly, treatment of CDN “Z” with HindIII results in the depletion of constituent GE′ and the concomitant depletion of the constituents sharing components with GE′, e.g., GF′, GG′, EE′, and FE′. This process leads to a new overexpressed [2 × 2] CDN “ZH” composed of EF′, EG′, FF′, and FG′, as the biocatalyically selected network. The sequential treatment of CDN “ZH” with EcoRI results in the depletion of constituent EF′ and a process that guides the concomitant depletion of constituents EG′ and FF′ and the overexpression of FG′ as the single selected constituent. The survived constituents originating from CDN “Z”/CDN “ZE” and CDN “Z”/CDN “ZH” can, then, lead to the subsequent cascaded emergence of new networks, in the presence of two sets of additionally engineered hairpins, demonstrating the variability and diversity of networks that can be evolved by the selection and cascaded emergence principles.
Figure 3.
(A) Schematic design of a [3 × 3] constitutional dynamic network, CDN “Z”, undergoing hierarchical biocatalytically guided selection processes. (B) The schematic structures of the nine constituents comprising CDN “Z”. The hierarchical control of the selection processes is shown in part A: treatment of CDN “Z” with EcoRI that cleaves constituent EF′ results in the selection of the [2 × 2] CDN “ZE”; interaction of the survived CDN “ZE” with HindIII that cleaves GE′ leads to the selection of constituent FG′; subjecting CDN “Z” to HindIII cleaves GE′, resulting in the selection of the [2 × 2] CDN “ZH”; treatment of the selected CDN “ZH” with EcoRI leads to the cleavage of EF′ and to the selection of FG′. Panel I - The cleavage of a fluorophore/quencher (Fi/Qi)-functionalized substrate by the Mg2+-ion-dependent DNAzyme reporter unit associated with the respective constituent. Panels II and III - Schematic cleavage of the sequence-specific domains associated with constituents EF′ and GE′ by the endonucleases EcoRI and HindIII, respectively.
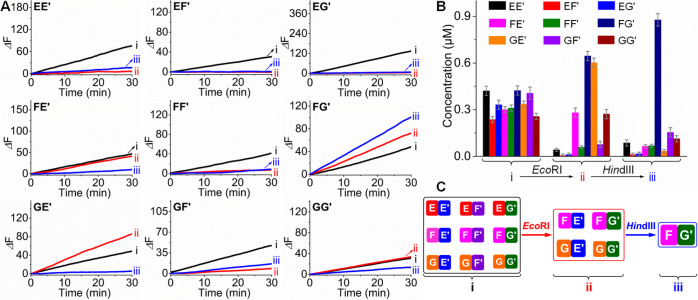
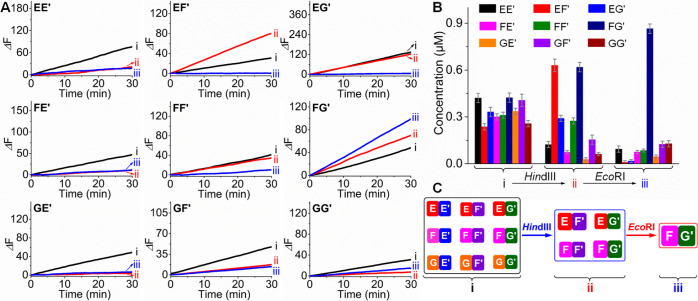
Figure 4A shows the time-dependent fluorescence changes generated by the Mg2+-ion-dependent DNAzyme reporter units associated with the nine constituents of CDN “Z” (curves i), upon the treatment of CDN “Z” with EcoRI and the selection of CDN ZE” (curves ii), and after the treatment of CDN “ZE” with HindIII and the selection of constituent FG′ (curves iii). In CDN “Z”, the reporter units associated with all constituents show catalytic activities, implying that all constituents exist in a dynamic equilibrium. After the treatment of CDN “Z” with EcoRI, the catalytic activities associated with EE′, EF′, EG′, FF′, and GF′ decrease to almost zero, consistent with the catalytical depletion of EF′ and concomitant depletion of the constituents sharing components with EF′. Interestingly, however, the catalytic activities generated by FG′ and GE′ are substantially higher than those in CDN “Z”, while the catalytic rates of the reporter units conjugated to FE′ and GG′ are very similar to those in CDN “Z”. Thus, the results demonstrate that the set of constituents FE′, FG′, GE′, and GG′ are indeed the dynamically equilibrated constituents in CDN “ZE”, where constituents FG′ and GE′ are upregulated in CDN “ZE” as compared to those in CDN “Z”. Treatment of CDN “ZE” with HindIII leads to the predominant increase in the catalytic activity associated with FG′, while all other constituents show very low catalytic activities. The results demonstrate that constituent FG′ is the only survived constituent upon the hierarchical selection process. By using the appropriate calibration curves (Figures S9 and S10), the concentrations of all constituents in CDN “Z” and after the sequential treatment of CDN “Z” with EcoRI and HindIII were quantified, and the results are summarized in Table S3 and Figure 4B. These results demonstrate the stepwise treatment of CDN “Z” with EcoRI and HindIII leads to the hierarchical selection of CDN “Z”, involving the selection of [2 × 2] CDN “ZE” from CDN “Z” and the selection of constituent FG′ from CDN “ZE”, Figure 4C.
Figure 4.
(A) Time-dependent fluorescence changes generated by the Mg2+-ion-dependent DNAzyme reporter units associated with the nine constituents and (B) concentrations of the constituents derived from the respective catalytic activities shown in part A, by using the appropriate calibration curves (Figures S9 and S10): (i) in CDN “Z”; (ii) in EcoRI-guided selection of CDN “ZE”; (iii) in HindIII-stimulated selection of constituent FG′ from the survived CDN “ZE”. The error bars in part B were derived from N = 3 experiments. (C) Schematic hierarchical selection of [3 × 3] CDN “Z” including EcoRI-induced selection of [2 × 2] CDN “ZE” from CDN “Z” and the subsequent HindIII-guided selection of constituent FG′ from CDN “ZE”.
In addition, Figure 5A shows the time-dependent fluorescence changes generated by the respective DNAzyme reporter units conjugated to the nine constituents in CDN “Z” (curves i), after subjecting CDN “Z” to HindIII and the selection of CDN “ZH” (curves ii), and after the treatment of the resulting CDN “ZH” with EcoRI and the selection of constituent FG′ (curves iii). Treatment of CDN “Z” with HindIII results in the depletion of the fluorescence signals associated with EE′, FE′, GE′, GF′, and GG′, accompanied by intensified fluorescence signals associated with EF′ and FG′ and almost unchanged catalytic rates associated with EG′ and FF′. The results are consistent with the depletion of EE′, FE′, GE′, GF′, and GG′ and the selection of CDN “ZH” composed of EF′, FG′, EG′, and FF′. Moreover, subjecting the resulting CDN “ZH” to EcoRI leads to the enhanced catalytic activity of FG′, while all other constituents show very low catalytic activities, implying the selection of FG′ from CDN “ZH”. By applying the appropriate calibration curves (Figures S9 and S10), the concentrations of the constituents before and after the sequential treatment of CDN “Z” with HindIII and EcoRI were evaluated, Table S3 and Figure 5B. The results confirm the endonuclease-induced hierarchical selection of CDN “Z”, including HindIII-guided selection of CDN “ZH” and the following EcoRI-driven selection of a single upregulated constituent FG′, Figure 5C.
Figure 5.
(A) Time-dependent fluorescence changes generated by the Mg2+-ion-dependent DNAzyme reporter units associated with the nine constituents and (B) concentrations of the constituents derived from the respective catalytic activities shown in part A, by applying the appropriate calibration curves (Figures S9 and S10): (i) in CDN “Z”; (ii) in HindIII-dictated selection of CDN “ZH”; (iii) in EcoRI-induced selection of constituent FG′ from the survived CDN “ZH”. The error bars in part B were derived from N = 3 experiments. (C) Schematic hierarchical selection of [3 × 3] CDN “Z” including HindIII-induced selection of [2 × 2] CDN “ZH” from CDN “Z” and the subsequent EcoRI-guided selection of constituent FG′ from CDN “ZH”.
In summary, the study has expanded the concept of constitutional dynamic networks to systems of enhanced complexity by integrating enzymes (endonucleases) as functional units for the operation of the networks. We introduced means to deplete a network by the biocatalytic selection of a constituent that is applied as a functional module for the emergence of a new network. By applying two different endonucleases on a multicomponent [3 × 3] constitutional dynamic network, the programmed hierarchical selection of different [2 × 2] networks followed by the selection of single functional constituents for the cascaded emergence of new diverse networks was demonstrated. The study introduces new dimensions to the field of “systems chemistry”39−41 by providing pathways for selective variation and emergence of networks.
Acknowledgments
This research was supported by the Israel Science Foundation and by the Minerva Center for Biohybrid Complex Systems.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c01939.
Additional experimental details such as materials, methods, calibration curves, and PAGE gel results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Jaenisch R.; Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Barabási A. L.; Oltvai Z. N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E.; Sattath S.; Kashtan N.; Itzkovitz S.; Milo R.; Pinter R. Y.; Alon U.; Margalit H. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (16), 5934–5939. 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M. M.; Luscombe N. M.; Aravind L.; Gerstein M.; Teichmann S. A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004, 14 (3), 283–291. 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Davidson E.; Levin M. Gene regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (14), 4935–4942. 10.1073/pnas.0502024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B.; Gompel N.; Carroll S. B. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (Suppl 1), 8605–8612. 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehn J.-M. From supramolecular chemistry towards constitutional dynamic chemistry and adaptive chemistry. Chem. Soc. Rev. 2007, 36 (2), 151–160. 10.1039/B616752G. [DOI] [PubMed] [Google Scholar]
- Rowan S. J.; Cantrill S. J.; Cousins G. R. L.; Sanders J. K. M.; Stoddart J. F. Dynamic covalent chemistry. Angew. Chem., Int. Ed. 2002, 41 (6), 898–952. . [DOI] [PubMed] [Google Scholar]
- Herrmann A. Dynamic combinatorial/covalent chemistry: A tool to read, generate and modulate the bioactivity of compounds and compound mixtures. Chem. Soc. Rev. 2014, 43 (6), 1899–1933. 10.1039/C3CS60336A. [DOI] [PubMed] [Google Scholar]
- Lehn J.-M. Perspectives in chemistry—aspects of adaptive chemistry and materials. Angew. Chem., Int. Ed. 2015, 54 (11), 3276–3289. 10.1002/anie.201409399. [DOI] [PubMed] [Google Scholar]
- Holub J.; Vantomme G.; Lehn J.-M. Training a constitutional dynamic network for effector recognition: Storage, recall, and erasing of information. J. Am. Chem. Soc. 2016, 138 (136), 11783–11791. 10.1021/jacs.6b05785. [DOI] [PubMed] [Google Scholar]
- Hasenknopf B.; Lehn J.-M.; Kneisel B. O.; Baum G.; Fenske D. Self-assembly of a circular double helicate. Angew. Chem., Int. Ed. Engl. 1996, 35 (16), 1838–1840. 10.1002/anie.199618381. [DOI] [Google Scholar]
- Hasenknopf B.; Lehn J.-M.; Boumediene N.; Dupont-Gervais A.; Dorsselaer A. V.; Kneisel B.; Fenske D. Self-assembly of tetra- and hexanuclear circular helicates. J. Am. Chem. Soc. 1997, 119 (45), 10956–10962. 10.1021/ja971204r. [DOI] [Google Scholar]
- Northrop B. H.; Aric F.; Tangchiavang N.; Badjić J. D.; Stoddart J. F. Template-directed synthesis of mechanically interlocked molecular bundles using dynamic covalent chemistry. Org. Lett. 2006, 8 (18), 3899–3902. 10.1021/ol061262u. [DOI] [PubMed] [Google Scholar]
- Valade A.; Urban D.; Beau J.-M. Target-assisted selection of galactosyltransferase binders from dynamic combinatorial libraries. An unexpected solution with restricted amounts of the enzyme. ChemBioChem 2006, 7 (7), 1023–1027. 10.1002/cbic.200600022. [DOI] [PubMed] [Google Scholar]
- Giuseppone N.; Lehn J.-M. Protonic and temperature modulation of constituent expression by component selection in a dynamic combinatorial library of imines. Chem. - Eur. J. 2006, 12 (6), 1715–1722. 10.1002/chem.200501038. [DOI] [PubMed] [Google Scholar]
- Ingerman L. A.; Waters M. L. Photoswitchable dynamic combinatorial libraries: Coupling azobenzene photoisomerization with hydrazone exchange. J. Org. Chem. 2009, 74 (1), 111–117. 10.1021/jo801783w. [DOI] [PubMed] [Google Scholar]
- Chaur M. N.; Collado D.; Lehn J.-M. Configurational and constitutional information storage: Multiple dynamics in systems based on pyridyl and acyl hydrazones. Chem. - Eur. J. 2011, 17 (1), 248–258. 10.1002/chem.201002308. [DOI] [PubMed] [Google Scholar]
- Giuseppone N.; Schmitt J.-L.; Lehn J.-M. Generation of dynamic constitutional diversity and driven evolution in helical molecular strands under Lewis acid catalyzed component exchange. Angew. Chem., Int. Ed. 2004, 43 (37), 4902–4906. 10.1002/anie.200460343. [DOI] [PubMed] [Google Scholar]
- Kovaříček P.; Lehn J.-M. Merging constitutional and motional covalent dynamics in reversible imine formation and exchange processes. J. Am. Chem. Soc. 2012, 134 (22), 9446–9455. 10.1021/ja302793c. [DOI] [PubMed] [Google Scholar]
- Ramírez J.; Stadler A.-M.; Kyritsakas N.; Lehn J.-M. Solvent-modulated reversible conversion of a [2 × 2]-grid into a pincer-like complex. Chem. Commun. 2007, (3), 237–239. 10.1039/B612222A. [DOI] [PubMed] [Google Scholar]
- Giuseppone N.; Lehn J.-M. Electric-field modulation of component exchange in constitutional dynamic liquid crystals. Angew. Chem., Int. Ed. 2006, 45 (28), 4619–4624. 10.1002/anie.200600430. [DOI] [PubMed] [Google Scholar]
- Berl V.; Huc I.; Lehn J.-M.; DeCian A.; Fischer J. Induced fit selection of a barbiturate receptor from a dynamic structural and conformational/configurational library. Eur. J. Org. Chem. 1999, 1999 (11), 3089–3094. . [DOI] [Google Scholar]
- Au-Yeung H. Y.; Cougnon F. B. L.; Otto S.; Pantoş G. D.; Sanders J. K. M. Exploiting donor-acceptor interactions in aqueous dynamic combinatorial libraries: Exploratory studies of simple systems. Chem. Sci. 2010, 1 (5), 567–574. 10.1039/c0sc00307g. [DOI] [Google Scholar]
- Wang S.; Yue L.; Shpilt Z.; Cecconello A.; Kahn J. S.; Lehn J.-M.; Willner I. Controlling the catalytic functions of DNAzymes within constitutional dynamic networks of DNA nanostructures. J. Am. Chem. Soc. 2017, 139 (28), 9662–9671. 10.1021/jacs.7b04531. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yue L.; Li Z.-Y.; Zhang J.; Tian H.; Willner I. Light-induced reversible reconfiguration of DNA-based constitutional dynamic networks: Application to switchable catalysis. Angew. Chem., Int. Ed. 2018, 57 (27), 8105–8109. 10.1002/anie.201803371. [DOI] [PubMed] [Google Scholar]
- Yue L.; Wang S.; Wulf V.; Lilienthal S.; Remacle F.; Levine R. D.; Willner I. Consecutive feedback-driven constitutional dynamic networks. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (8), 2843–2848. 10.1073/pnas.1816670116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L.; Wang S.; Lilienthal S.; Wulf V.; Remacle F.; Levine R. D.; Willner I. Intercommunication of DNA-based constitutional dynamic networks. J. Am. Chem. Soc. 2018, 140 (28), 8721–8731. 10.1021/jacs.8b03450. [DOI] [PubMed] [Google Scholar]
- Yue L.; Wang S.; Willner I. Triggered reversible substitution of adaptive constitutional dynamic networks dictates programmed catalytic functions. Sci. Adv. 2019, 5 (5), eaav5564 10.1126/sciadv.aav5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L.; Wulf V.; Wang S.; Willner I. Evolution of nucleic-acid-based constitutional dynamic networks revealing adaptive and emergent functions. Angew. Chem., Int. Ed. 2019, 58 (35), 12238–12245. 10.1002/anie.201905235. [DOI] [PubMed] [Google Scholar]
- Darwin C.; Wallace A. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. Proc. Linn. Soc. London 1858, 3 (9), 45–62. 10.1111/j.1096-3642.1858.tb02500.x. [DOI] [Google Scholar]
- Fernando C.; Rowe J. Natural selection in chemical evolution. J. Theor. Biol. 2007, 247 (1), 152–167. 10.1016/j.jtbi.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Wagner A.Gene networks and natural selection. In Evolutionary Genomics and Proteomics; Pagel M., Pomiankowski A., Eds.; Sinauer Associates: Sunderland, MA, 2008; pp 255–270. [Google Scholar]
- Danger G.; d’Hendecourt L. L. S.; Pascal R. On the conditions for mimicking natural selection in chemical systems. Nat. Rev. Chem. 2020, 4, 102–109. 10.1038/s41570-019-0155-6. [DOI] [PubMed] [Google Scholar]
- Kolpashchikov D. M. A binary deoxyribozyme for nucleic acid analysis. ChemBioChem 2007, 8 (17), 2039–2042. 10.1002/cbic.200700384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R.; Matamoros E.; Liu M.; Stefanovic D.; Stojanovic M. N. Training a molecular automaton to play a game. Nat. Nanotechnol. 2010, 5, 773–777. 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]
- Heitman J. How the EcoRI endonuclease recognizes and cleaves DNA. BioEssays 1992, 14 (7), 445–454. 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- Old R.; Murray K.; Roizes G. Recognition sequence of restriction endonuclease III from Hemophilus influenzae. J. Mol. Biol. 1975, 92 (2), 331–339. 10.1016/0022-2836(75)90232-6. [DOI] [PubMed] [Google Scholar]
- Ludlow R. F.; Otto S. Systems chemistry. Chem. Soc. Rev. 2008, 37 (1), 101–108. 10.1039/B611921M. [DOI] [PubMed] [Google Scholar]
- Mattia E.; Otto S. Supramolecular systems chemistry. Nat. Nanotechnol. 2015, 10, 111–119. 10.1038/nnano.2014.337. [DOI] [PubMed] [Google Scholar]
- Ashkenasy G.; Hermans T. M.; Otto S.; Taylor A. F. Systems chemistry. Chem. Soc. Rev. 2017, 46 (9), 2543–2554. 10.1039/C7CS00117G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.