Abstract

Over the past decade, DNA nanotechnology has spawned a broad variety of functional nanostructures tailored toward the enabled state at which applications are coming increasingly in view. One of the branches of these applications is in synthetic biology, where the intrinsic programmability of the DNA nanostructures may pave the way for smart task-specific molecular robotics. In brief, the synthesis of the user-defined artificial DNA nano-objects is based on employing DNA molecules with custom lengths and sequences as building materials that predictably assemble together by obeying Watson–Crick base pairing rules. The general workflow of creating DNA nanoshapes is getting more and more straightforward, and some objects can be designed automatically from the top down. The versatile DNA nano-objects can serve as synthetic tools at the interface with biology, for example, in therapeutics and diagnostics as dynamic logic-gated nanopills, light-, pH-, and thermally driven devices. Such diverse apparatuses can also serve as optical polarizers, sensors and capsules, autonomous cargo-sorting robots, rotary machines, precision measurement tools, as well as electric and magnetic-field directed robotic arms. In this review, we summarize the recent progress in robotic DNA nanostructures, mechanics, and their various implementations.
Keywords: dynamic DNA nanotechnology, DNA origami, biomedicine, autonomous devices, photonics
From the initially static objects to rudimentary switching of states and finally multistage movement, environmental triggering, and information relay, the field of DNA nanotechnology1,2 has evolved to the increasingly dynamic, enabled state3 it is today. Remarkable development in building functional nanostructures using DNA molecules as fundamental building blocks4 has elicited various stimuli-responsive mechanisms and elementary functions that can be integrated into solving sophisticated tasks in a preprogrammed manner. In other words, DNA nanotechnology is encroaching on the realm of robotics.
There is an incredible amount of digital information that can be encoded into and read from DNA as demonstrated by Church and colleagues.5,6 Despite this ingenious and far-reaching approach to use DNA as a digital information storage, Richard Feynman once famously postulated7 that the key feature in biology is not just about writing the information, but rather what we could do with it. The information in DNA, i.e., the sequence of nucleobases, directs the construction of countless nanoscopic atomically precise molecular machines that can perform intricate tasks in a dynamic and reliable manner. On the other hand, this protein-dominating machinery has evolved to meet its primary function, and therefore, it is remarkably demanding—yet possible through de novo protein design8,9—to revamp the machines for any other purpose. Thus, one way to manufacture artificial and accurate nanoscale tools for performing similarly complex, but now user-defined, functions is to harness DNA molecules in additive, bottom-up fabrication. DNA molecules and their derivatives, DNA nanostructures, composed of a few to dozens of custom-length and -sequence DNA strands and whose assembled shapes are therefore also encoded in the sequences of the building blocks,4 can change their conformations or behavior in response to a plethora of physical and chemical inputs, including heat, presence of certain molecules, pH, and light.10,11
Due to the modularity and high addressability of DNA nanostructures, the responses of these tiny nanomachines can be engineered toward specific applications. Furthermore, while DNA nanostructures are based on molecular self-assembly, an intrinsically additive (bottom-up) fabrication process, there exists both “bottom-up” and “top-down” approaches for designing them. Features can be either gradually added to a design from nothing (bottom-up), or a user can directly create a 3D model and then systematically route a DNA-strand frame around it, sculpting the model’s likeness with DNA (top-down). Through these design paradigms DNA nanostructures are respectively afforded excellent customizability, addressability, and freedom of form. Thanks to effortless automated design12,13 and powerful simulation tools,14 programmable DNA-based nanostructures such as scaffolded DNA origami15,16 and various top-down fabricated wireframe structures17 have enjoyed widely spread interest as drug carriers,18,19 measurement and sensing tools,20 photonic instruments,21,22 molecular templates,23,24 spectroscopic rulers,25,26 and eventually the dynamic nanoscale devices discussed in depth in this review.
The first major steps in creating dynamic DNA-based systems were taken around two decades ago.27 In the very end of the 1990s it was reported how manipulating solution conditions could induce simple rotary28 and linear29 motion in primitive DNA structures via conformational changes of helices. Soon after, Yurke et al. presented the first DNA-based molecular tweezers30,31 that could reversibly switch between open and closed states by using additional DNA strands as fuel. Next, Yan et al.(32) employed a similarly reversible fuel-based strategy to change the hybridization topology of a DNA system, causing it to partially rotate between two topologies. These mechanisms of controlled movement eventually led to the creation of the first DNA walkers,33,34 that could travel step-by-step along a molecular track by consuming fuel. These reversible mechanisms were based on so-called toehold-regions. Toeholds are short single-stranded DNA (ssDNA) extensions that serve as the docking sites for the reactant ssDNA at the end of the dsDNA domains. In other words, they are essentially short ssDNA extensions that could initiate the strand displacement reaction and energetically favor one of the two sides in this reaction. Around the same time, also other kinds of DNA motors were devised,34 similarly expending various molecular fuel sources35,36 to induce a rudimentary cycle of motion. The mechanisms have since evolved into more complex ones,37 and alternative regulation methods like external fields,38 weak base stacking interactions,39 and logic gates40 have recently opened completely new possibilities for building dynamic systems. Currently, we are approaching the level of advanced and feasible DNA-based nanorobotics.
To probe the mechanical movements of individual DNA nanostructures, several single-molecule techniques could be employed. The label-free techniques like transmission electron microscopy (TEM) and atomic force microscopy (AFM) are commonly applied for acquiring static configurations of DNA nanorobots with subnanometer spatial resolution. With the recent developments of high-speed AFM (HS-AFM), even dynamic events could be recorded with a temporal resolution down to hundreds of milliseconds per frame. On the other hand, DNA nanostructures labeled with fluorophores could be characterized by the single-molecule fluorescence resonance energy transfer (smFRET) to resolve their dynamics with a much higher temporal resolution. Fluorescence-based super-resolution imaging, e.g., DNA points accumulation for imaging in nanoscale topography (DNA-PAINT)25 is natively compatible with DNA nanostructures for high spatial resolution imaging in aqueous environments.
Two interesting research areas that deserve to be mentioned but are not touched here in detail are DNA computing and protein–DNA hybrid structures. DNA computing is based on dynamic DNA interactions combined with the programmable nature of the DNA function. The programmability enables for example creation of neural-networks using DNA molecules and thus realization of the neural-computing at the molecular scale and scaled up digital circuit computation by DNA strand-displacement cascades.41,42 Meanwhile, in the hybrids one could combine the addressability and modularity of DNA nanostructures with the precision and functionality of proteins.43 For example, in the work of Derr et al.(44) the dynein and kinesin motor proteins—that have also inspired many versions of DNA walkers—were attached to a DNA origami strut, thereby facilitating a molecular scale tug-of-war between different protein populations. While the goals of protein–DNA hybrids are similar as for purely DNA-based machines,45,46 the approach is fundamentally different as functionality stems more from the proteins instead of the DNA components. The topic is thus discussed elsewhere.45−48 Here, we describe the progress in user-defined robotic DNA nanostructures by summarizing the recent achievements of device development for a variety of applications. In the context of this review, we define nanorobots as dynamic nanoscale devices that display sophisticated functions such as multistage and repeatable movement, interconnectivity, or autonomous regulation. In particular, we mainly focus on the structurally complex DNA nanodevices, i.e., the newly emerged ones that usually consist of dozens of DNA strands. First, we introduce the mechanisms of the DNA nanostructure movement and provide examples of DNA nanostructure-based force spectroscopy, thus laying the foundation for all the other emerging implementations. We describe regulatory and information relay systems, nanomedical robots for imaging, diagnostics and drug delivery, dynamic photonic and plasmonic structures, as well as DNA-based instruments driven by external fields. Finally, we discuss advanced robotics that are capable of autonomous operation.
Principles of Mechanical Movement and Characterization of Molecular Scale Forces
The fundamental core of DNA structures is highly specific and based on ordered Watson–Crick base pairs.49 However, DNA is not brittle, but instead pliable and robust. Mechanically it behaves as an entropic spring, i.e., when the DNA molecule is stretched, the entropy decreases (fewer available conformations) thus resulting in a restoring force. A DNA helix can be elastically stretched and bent by external forces and in proper conditions it possesses the ability to reconstitute itself in case part of it unravels due to excessive stress.50 This malleability makes DNA an excellent building material for durable moving parts.51 DNA retains most of these spring-like properties when assembling into larger 3D structures such as DNA origami, the mechanical properties of which have been documented by Kauert et al.(52) with direct force measurements. Importantly, the architecture of the assembly (crossovers, etc.) affects the bending and torsional rigidity of the object. Thus, the mechanical properties can be tuned from beamlike stiffness to wire-like flexibility by intelligent design of the structures, using only DNA as material. A nice demonstration of this was made by Liedl and co-workers,53 where they crafted prestressed tensegrity structures from DNA origami.
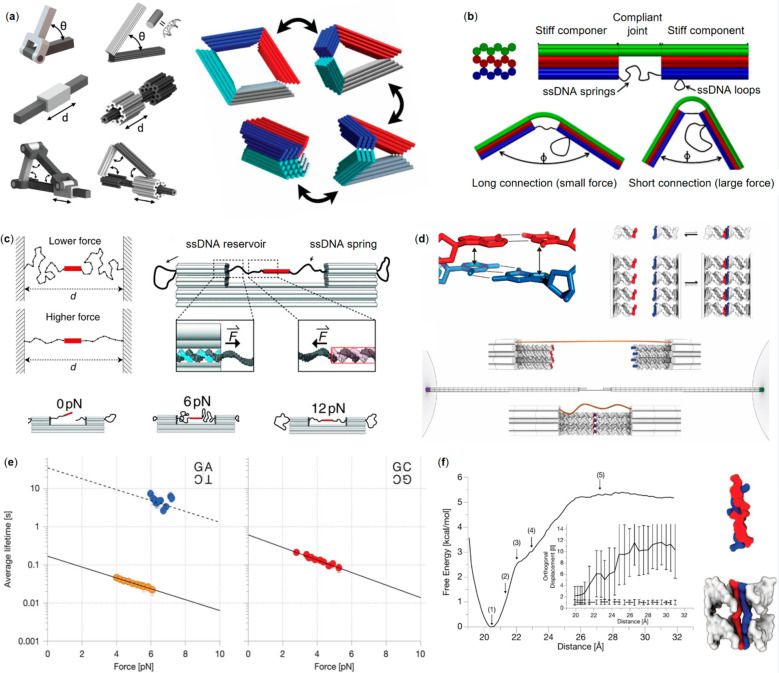
Stiff beams can be connected by short single-strands to constitute hinges and facilitate restricted, mechanical movement. Marras et al.(54) used this strategy to create DNA origami analogues of a hinge for angular motion, a slider for linear motion, a crank-slider that links angular and linear motion (Figure 1a, left panel), and finally a Bennet linkage capable of a reversible, 3D movement cycle (Figure 1a, right panel). Their hinge was made of two beams connected from their ends with short ssDNA wires, enabling angular opening and closing of the beams. The slider was built by positioning a mobile DNA origami ring around a beam with a stopper ring and then anchoring it from the both ends with ssDNA. Three hinges and the slider were then combined to constitute the crank-slider. The opening and closing cycle of the Bennet linkage could be initiated by addition of single-stranded DNA inputs that “zipper” the sides of the linkage together. The linkage could then be opened via strand displacement of the input strands. After the “zippering” was undone, electrostatic repulsion forced the structure to its original, open shape.
Figure 1.
DNA nanostructure mechanics and force spectroscopy. (a) Mechanic DNA nanostructures. Left panel from top to bottom: a hinge for angular motion, a slider for linear motion, and a crank-slider that executes angular and linear movement. Right panel: a Bennett linkage.54 (b) Compliant DNA origami structures.55 (c) A DNA origami force clamp created using ssDNA as an entropic spring (force adjustment demonstrated in the left panel).57 (d) Measurement setup for unraveling the base stacking (top left) forces. The opposite DNA origami pylons with blunt-ended dsDNA variants (top right panel) are linked together through a ssDNA tether and the pylons are attached to the optical tweezer beads for force spectroscopy (bottom panel).58 (e) Average lifetimes of select base stacking interactions as a function of force, as measured with the setup depicted in (d). Six, four, and two stack arrays are depicted as blue, orange, and red data points, respectively. The solid line represents an exponential fit of the data. Meanwhile, the dashed line is a visual guide for combinations with particularly long lifetimes, that were difficult to dissect with the optical trap.58 (f) A free energy diagram of the dissociation of two GC:CG stacks as a function of distance. The energy minimum highlights relaxed, fully formed stacks (∼20 Å) and a plateau is accordingly reached at full dissociation of the stacking (>26 Å). In addition, a sharp increase in average orthogonal displacement respective to the helical axis can be observed as the stacks are pulled apart by the measurement rig.58 (a) Reprinted with permission from ref (54). Published by 2015 National Academy of Sciences. (b) Reprinted with permission from ref (55). Copyright 2013 American Chemical Society. (c) Reprinted with permission from ref (57). Copyright 2016 The American Association for the Advancement of Science. (d–f) Reprinted with permission from ref (58). Copyright 2016 The American Association for the Advancement of Science.
In terms of control, the inherent flaw with floppy ssDNA hinges is that they are subject to random, thermal motion. However, the locally addressable flexibility also provides the basis for making other kinds of joints that offer more direct control. A bit earlier, in 2014, Zhou et al.(55) showed how stiffer and more compliant origami components could be combined to form a joint-like structure. The stiff regions, 3-fold stacks of layered helices, were connected by a singular layer from one end at the middle, while the other side was connected by a lone DNA strand acting as a spring (Figure 1b). By changing the length of the spring, varying forces were exerted, and the joint complied accordingly. In this way, the bending force could actually be controlled very carefully.
The following year, Zhou and his co-workers56 exploited this when they developed a dynamic, compliant DNA origami mechanism that displayed two rationally designed stable configurations separated by an energetic barrier. The device was an assembly of four connected bars that had two possible resting positions where no deformation was present. Changing configuration required bending the bars, thus creating an energy barrier to overcome. They then displayed how such a mechanism could be actuated and released by DNA strand inputs and strand displacement, respectively.
Employing a ssDNA as a roughly ideal entropic spring, Nickels et al.(57) were able to craft a DNA origami-based nanoscopic force clamp (Figure 1c). In the clamp, a sample molecule could be propped between stiff DNA origami towers by ssDNA springs. The force applied to the target molecule could be controlled incrementally by changing the length of the ssDNA spring strands base by base. With the additional help from Förster resonance energy transfer (FRET) pairs, the authors used the clamp to study conformational changes in a Holliday junction and the bending that a TATA-binding protein causes to a DNA helix. A slightly similar approach was taken by Kilchherr and co-workers58 to study weak DNA base stacking forces in a time-resolvable manner. They attached stiff honeycomb-packed DNA origami pylons to micrometer-sized beads that could then be manipulated with optical tweezers (Figure 1d). The pylons were connected to each other via a ssDNA tether and the blunt-ended double helices protruded from the ends of the pylons. Owing to the optical tweezers, they were able to record forces and associated lifetimes for the blunt-ended base stacking interactions, resolving the exact moments when stacked bases disconnected upon applying a set force. The use of such a novel molecular scale tool provided unique insight into the mechanics and dynamics of stacking bonds, for example differentiating various nucleotide combinations by their lifetimes (Figure 1e) and allowing observation into the energetics of stacking bond formation and dissociation (Figure 1f).
Finally, nanoscale force calipers have also been constructed from DNA by the research groups of Carlos E. Castro and Hendrik Dietz. In a design visually like the hinge in Figure 1a, the opening angle of the hinge is controlled, thereby controlling also the tensile force applied to a sample tethered at the ends of the caliper. These have proven especially useful precise measurement tools for molecular scale features that may otherwise be impossible to characterize. These include various nucleosome properties, such as the forces between nucleosomes,59 nucleosome unwrapping,60 and the stability of nucleosomes.61
It has become clear from above that the structural evolution of dynamic objects and the requirements of the delicate molecular scale measurements have further increased the importance of a powerful software for simulating structural details of the DNA nanoshapes and their movement. Here, our main focus is on the different robotic mechanisms and their intriguing applications, so the readers are suggested to refer to other sources for multiple DNA design and simulation software options. Both modeling and validation methods have been recently reviewed in ref (62), and in more detail, for the design of the DNA nanostructures, see, e.g., ref (14) (various approaches) or ref (17) (wireframe constructions), and for the simulation software, see, e.g., rigid-beam models in finite element frameworks (software CanDo),63,64 atomistic molecular-dynamics simulations,65 or coarse-grained modeling (software oxDNA).66−68
Information Relay
In order to be able to handle complex tasks, it may be necessary for the components of a system to communicate with each other and to propagate information. By triggering one component with the chosen input, it can undergo a change and release an output that will in turn act as the input for the next component, eventually translating into a predetermined function. With these kinds of cascaded selective triggering mechanisms and precise internal interactions, system modularity, dynamics and automation can be achieved. To this end, several interaction cascade and information relay strategies have been devised for DNA-based systems.
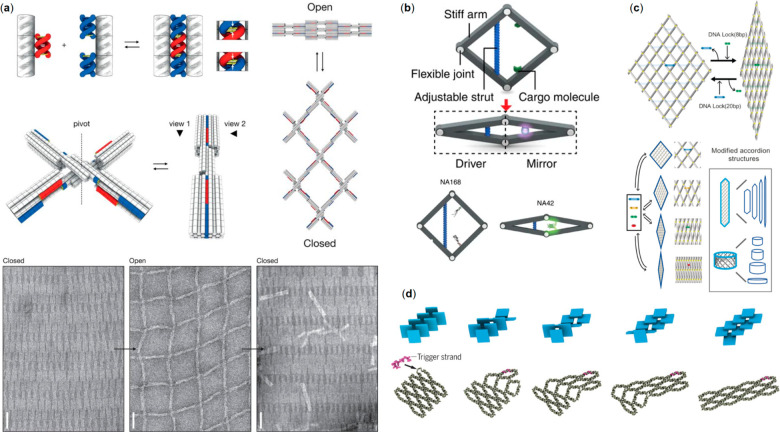
In 2015 Gerling et al.(39) published a strategy for manipulating connections between ready-assembled 3D DNA structures by the weak base stacking interactions between the components, first introduced by Woo and Rothemund using 2D tile stacking69 (Figure 2a). The authors created shape-complementary features on the lateral surfaces of DNA structures, enhancing the weak interactions at these interfaces and effectively docking DNA bundles together in a shape-specific manner. The attractive force between the nanostructures could be overcome by electrostatic repulsion forces by changing the prevalent solution conditions such as salt concentration or temperature. Thus, the DNA structures could be reversibly disassembled and recombined as necessary, in a time frame of seconds, making them toggleable and forming a basis for dynamic functions. Furthermore, combined bundles could then constitute a new shape-complementary interface in a manner similar to a 3D jigsaw puzzle, that could allow the next structure to dock to the previously combined ones. Finally, Gerling et al. also showed how singular elements of the system could be selectively disabled and enabled by using single-stranded DNA loops and toehold-mediated mechanisms. By including ssDNA loops at a shape-complementary interface and then adding a loop-specific and complementary strand to the system, that interface would deform and become unusable for assembly. The reverse could then be realized by removing the complementary strand via the toehold-mediated hybridization of a so-called recovery strand. In this way, the authors presented one option for making complex DNA nanostructure systems, where components can reversibly interact in a dynamic, cascading manner, and where individual elements can be activated and deactivated at will. One example is a large-scale reconfigurable lattice based on non-base-pairing interactions (Figure 2a). The individual dynamic cross-like switches (Figure 2a, middle panel) can be polymerized into a network (Figure 2a, right panel) that reversibly changes between the closed (high magnesium level) and open state (low magnesium level) depending on the prevalent cation concentration (Figure 2a, bottom panel).
Figure 2.
DNA nanostructures for information relay and regulation. (a) Top left panel: a dsDNA protrusion (red) fits tightly to the designed dsDNA recession (blue) thus stacking the counterparts through non-base-pairing interaction. Middle left panel: A reversible DNA switch with designed protrusions and recessions. Middle right panel: Working principle of a reconfigurable large-scale network obtained by polymerization of multiple switches. Bottom panel: TEM images verifying reversible switching of the network (closed state: high magnesium concentration; open state: low magnesium concentration).39 (b) A rhombus-shape DNA origami nanoactuator; movement on the driver side (left) is mirrored to the right side. The lower panel depicts the regaining of eGFP fluorescence by bringing its two halves in close proximity using the actuator.70 (c) DNA accordion rack that can adapt different geometries relying on DNA lock strand positioning.71 (d) Long-range step-by-step information relay process in DNA “domino” nanoarrays, launched by the hybridization of a trigger strand to a single unit.72 (a) Reprinted with permission from ref (39). Copyright 2015 The American Association for the Advancement of Science. (b) Reprinted with permission from ref (70). Copyright 2016 Springer Nature Ltd. (c) Reprinted with permission from ref (71). Copyright 2018 John Wiley & Sons. (d) Reprinted with permission from ref (72). Copyright 2017 The American Association for the Advancement of Science.
A year later Ke and co-workers70 introduced a rhombus-shape DNA origami nanoactuator (Figure 2b). The device was composed of a driver and a mirror, i.e., four stiff rod-like arms assembled into a rhombus shape using flexible ssDNA scaffolds in every corner. The conformation of the device (opening angle) was controlled via ssDNA “strut-locking” strands of different lengths embedded on driver-side arms. The mirror-side contained two capturing strands for cargo attachment. Tunable fluorescent behavior via long-range allosteric regulation was demonstrated by attaching two halves of enhanced green fluorescent protein (eGFP) to the capturing strands in open conformation. Closing of the device with short locking strands regained the strong fluorescence of eGFP (Figure 2b, bottom panel).
Choi et al.(71) expanded the actuator concept by constructing a reconfigurable DNA accordion rack using a network of long DNA beams joined together via multiple flexible joints. Conformational switching in 2D DNA 6 × 6 rhombus meshes was controlled by adding DNA lock strands at predefined positions in the structure (Figure 2c). Multistep lattice reconfigurations were generated using a toehold-mediated strand-displacement process by repeating the locking process after detaching the prior DNA locks. Moreover, controlled multistep lattice reconfigurations of the accordion rack and the wrapped up 2D DNA configuration resulting in a 3D DNA nanotubular structure with various dimensions was demonstrated.
One step further in the information propagation systems, i.e., transformations with programmable initiation, propagation, and regulation was presented by Song et al.(72) who employed base-stacking interactions as a driving force to perform long-range information relay on dynamic DNA molecular arrays. The modular system was constructed from dynamic multi-interconnected trapezoidal DNA “antijunction units” that could switch between two stable conformations via an open intermediate conformation. The array transformation analogous to molecular domino array was launched by adding trigger strands at the predefined “antijunction” unit. This resulted in a conformational switch in an adjacent unit, which then migrated through the system without the need of additional strands (Figure 2d). This is due to declined base-stacking and higher energy at the interface between the units. The reconfiguration process is reversible since the array can be transformed to its initial conformation by releasing the old trigger strands with new ones. Such artificial systems allow the study of complex dynamic behaviors and allosteric mechanisms observed in biological systems.
Robots for Nanomedicine
Biobased nanorobots are typically operating at the subcellular level and consist of stimuli-responsive biomaterials with excellent precision and efficiency and these nanorobots have recently attracted increasing attention particularly in medical applications.73 With delicate design, nanorobots can outperform conventional techniques through a combination of several unique properties, such as cargo-loading, degradation resistance, site-targeting, tissue penetration and stimuli responsiveness. The multifunctionality makes them ideal candidates for drug delivery and imaging. For instance, light,74 ultrasound,74 and magnetic fields75,76 have been utilized to transport or trigger the release of loaded drugs or imaging agents. Additionally, they may reach targets in the human body that are otherwise inaccessible to conventional robotics.77,78
As one of the most important biomacromolecules in all life forms, DNA possesses advantages over other smart materials in the construction of such robots. First of all, DNA is intrinsically biocompatible and biodegradable, which lightens the concerns on its possible adverse effects. On top of this, the tremendous development in automated DNA synthesis has enabled production of DNA strands in large quantities, yet with site-specific addressability, which may lead to integrated functionality, such as drug-loading, site-targeting and stimuli-responsiveness within one rationally designed architecture.79 There are several reviews that have well summarized the design and construction of DNA nanostructures, as well as their potential in drug delivery applications,19,80−82 while their bioimaging possibilities83 and biological responses84 have been addressed in others. For an in-depth discussion on the various drug payloads and their delivery mechanisms, interested readers are respectively referred to ref (85). In this section, we will focus on the advances in the emerging field of logic-gated dynamic DNA nanorobots in nanomedicine, which includes two main categories: imaging/diagnostics and delivery/computing.
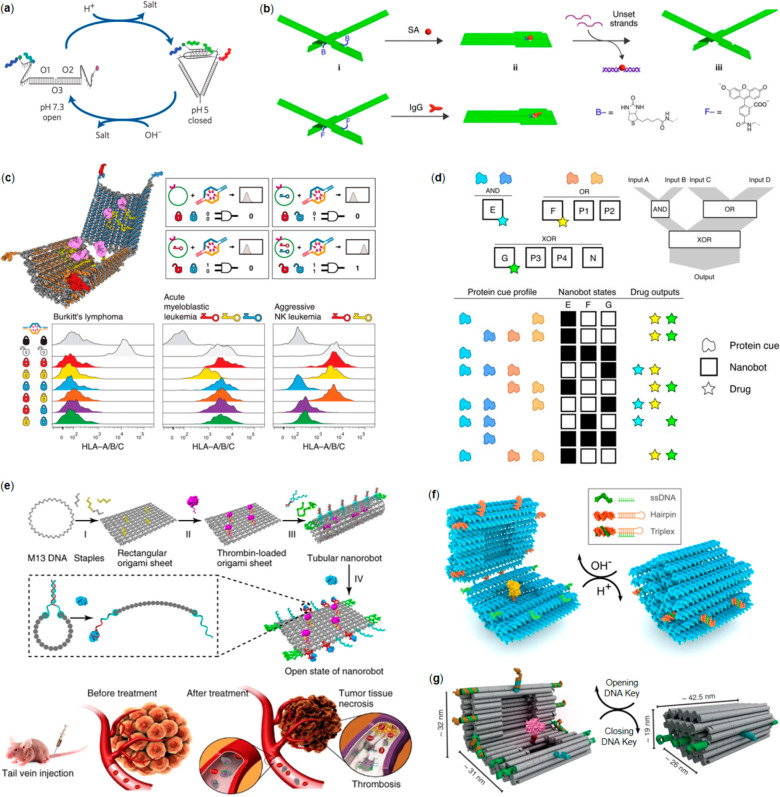
Yamuna Krishnan’s research group has performed seminal work on in situ imaging using several smart DNA nanorobot designs. In their early versions, the nanorobot was constructed with several fluorescent dye-modified oligonucleotides that underwent a conformational transition of an i-tetraplex in response to protons, which was used for pH-mapping (Figure 3a).86 The spatiotemporal pH-mapping based on the so-called I-switch was demonstrated to be efficient in living cells using FRET. Later, the same design was applied for pH-mapping in a living organism using nematode Caenorhabditis elegans as a model.87In-vivo study revealed that the internalization of I-switch to coelomocytes followed a receptor-mediated endocytosis pathway and that the nanorobot functioned as efficiently in vivo as in vitro. By using two nanorobot designs that were modulated with distinct endocytic pathway-trafficking proteins, they managed to simultaneously map pH gradients through intersecting cellular entry pathways in the same cell.88 They also extended the design, termed as 2-IM, which could simultaneously detect protons and chloride ions in living cells.89 The 2-IM enters lysosomes through the process mediated by a lysosome-targeting tag. Following the entrance, a H+-induced conformational change of an i-tetraplex triggers FRET between a fluorophore pair while a Cl–-sensitive dye reports Cl– gradient. It was found that 2-IM is able to report the pH and chloride ions within one lysosome quantitatively and simultaneously. As a demonstration of this concept, 2-IM was employed to image fibroblasts derived from skin biopsies of both healthy individuals and patients with Niemann–Pick disease resulting in highly distinct lysosome subpopulation profiles, thus demonstrating the efficiency of 2-IM in disease diagnosis.
Figure 3.
DNA nanorobotics for biomedicine. (a) An i-tetraplex for pH-mapping.86 (b) A nanomechanical DNA origami plier for detecting target molecules (observed through the plier configuration change).94 (c) A logic-gated DNA nanorobot for encapsulation and display of cargo. Top left panel: the open robot with loaded antibodies. Top right panel: Boolean AND gating with two-lock (two aptamer locks for robot) and two-key (antigens at the cell surface) combinations. Bottom panel: Logic-gating in action; different lock-key combinations against selected cancer cells.40 (d) Universal computing with DNA nanorobots; a generalized version of the robots shown in (c) with multiple different gate options. The logic-gated DNA nanorobots can adopt distinct states, and depending on the interaction with the protein cue profile, the interaction can result in different drug output.101 (e) Top panel: Working principle of the thrombin-loaded DNA nanorobot that is wrapped into a tubular shape using strands with nucleolin aptamers (robot interaction with nucleolin opens the robot and displays the cargo). Bottom panel: In vivo mouse model for inhibition of tumor growth and tumor necrosis.103 (f) A reconfigurable DNA origami nanocapsule with “pH-latches” for encapsulation and display of cargo. The capsule can reversibly open (dsDNA and ssDNA do not form a triplex) and close (dsDNA and ssDNA form a triplex) upon the pH change.107 (g) A similar device as in (f), but this type of DNA origami nanovault closes and opens through strand displacement reactions (DNA keys).108 (a) Reprinted with permission from ref (86). Copyright 2009 Springer Nature Ltd. (b) Reprinted with permission from ref (94). Copyright 2011 Springer Nature Ltd. (c) Reprinted with permission from ref (40). Copyright 2012 The American Association for the Advancement of Science. (d) Reprinted with permission from ref (101). Copyright 2014 Springer Nature Ltd. (e) Reprinted with permission from ref (103). Copyright 2018 Springer Nature Ltd. (f) Reprinted with permission from ref (107). Copyright 2019 American Chemical Society. Further permissions related to the material excerpted should be directed to the American Chemical Society. (g) Reprinted with permission from ref (108). Published by 2017 Springer Nature Ltd.
Other than the simple i-motif structure, a DNA icosahedra was also designed and used for cargo-loading, which was further employed in cell imaging.90−93 For example, they introduced three different endocytic ligand (folic acid (FA), galectin-3 (Gal3), and the Shiga toxin B-subunit (STxB)) to the quantum dot (QD)-loaded DNA icosahedra respectively through site-specific modification and demonstrated the feasibility to characterize the dynamics of endocytic pathways facilitated by Gal3 and STxB.91 Hence, the QD-loaded DNA icosahedra exhibited an excellent platform in real-time endocytic pathway mapping at single-particle level. They also loaded the DNA icosahedra with photoresponsive polymers and realized targeted spatiotemporal imaging of single endosomes in specific cells in Caenorhabditis elegans.92
Kuzuya et al. developed a nanomechanical DNA origami plier-like device that consists of two DNA lever domains joined together by a Holliday junction.94 With this design, target molecules of various sizes can be detected at a single-molecule level with three independent mechanisms: “pinching”, “zipping”, or “unzipping”. For example, by attaching two ligands to the concavities of DNA arms, the DNA plier could cooperatively capture a single target molecule, transforming the origami from a cross or antiparallel configuration into a parallel closed form (Figure 3b). The shape transition can then be detected by AFM, spectroscopic analysis and agarose gel electrophoresis. Later on, they realized a more sophisticated system with precise control over the transitions between three structural conformations, thus showing potential in molecular computing.95
In addition to the previous examples, Francesco Ricci’s research group has also shown remarkable progress in developing a variety of antibody sensors using smart DNA robots.96 Omitting often complex and large DNA nanostructures, they have designed sensors that consist of only several functionalized DNA sequences and realized sensitive antibody probing based on the structural transition of their devices.97,98 Other than sensors, it is worth noting that with the help of antibodies the group has also produced DNA robotic vectors for controlled release of molecular payloads.99,100
Through deliberate design, Douglas et al. constructed a capsule-like DNA robot that can open and close in response to a binding antigen (Figure 3c).40 In the open state, the nanocapsule can be loaded with designated cargos such as nanoparticles and proteins through complementary DNA hybridization (Figure 3c, left panel). The loaded capsule can then be closed by two DNA aptamer “locks” and stabilized by the guiding staples. Upon interaction with the antigens expressed on the human leukocyte surface, the DNA aptamers change conformation, which leads to the open state and subsequent display of encapsulated cargo. Owing to the modularity of the system, the authors used two distinct aptamer “locks” for two different targets/keys to “unlock” the capsule, which is equivalent to a logical AND gate (shown in Figure 3c top right panel and bottom panel).
By utilizing similar devices as above, Amir et al. developed the universal computing system that was operational in a living animal.101 The barrel-shaped DNA nanorobot was conjugated with DNA aptamer gates that targeted different protein cues, platelet derived growth factor (PDGF) and vascular endothelial growth factor (VEGF). Together with the interaction between nanorobots that triggered molecular payloads on or off, they managed to realize various logic gates (AND, OR, XOR, NAND, NOT, CNOT and a half adder) in a living animal model, cockroach (Blaberus discoidalis). Possible outcomes of the robot interactions, i.e., drug outputs, depended on different protein cues and the prevalent robot states (Figure 3d). In a more recent report, they also showed that the DNA nanorobot in a living Blaberus discoidalis could be controlled by human brain activity.102 An electromagnetic field was regulated by an online algorithm which recognized electroencephalography (EEG) patterns. The thought-induced switch in the field caused the heating of metal nanoparticles that were attached to the gate, which led to the opening of nanorobots and subsequent display of payloads.
Li et al. designed an intelligent DNA robot and brought the test subject from cultured cells to tumor-bearing mouse models, demonstrating a promising strategy for cancer treatment in mammals.103 Their tubular DNA nanorobot was loaded with blood coagulation protease, thrombin, and fastened along the periphery by targeting DNA, a DNA aptamer (AS1411) that targets nucleolin that is specifically expressed in tumor-associated endothelial cells (Figure 3e). Upon intravenous injection in tumor-bearing mouse models, it was found that the DNA nanorobots explicitly accumulated on tumor vessels where the encapsulated thrombin was released, finally leading to inhibition of tumor growth and tumor necrosis. The therapeutic effect was confirmed in mouse models bearing melanoma, lung cancer, breast cancer, and ovarian cancer, while the safety and immunological inertia was proved in mice and Bama miniature pigs. Besides the above-mentioned examples, other stimuli have also been demonstrated to be feasible in triggering the cargo release from DNA nanorobotics, such as temperature,104 light,105 and mRNA.106
Using a modified capsule design, Ijäs et al. changed the trigger to a more general subject, pH, by replacing the aptamer “lock” with Hoogsteen triplexes, dubbed pH-latches (Figure 3f).107 The Hoogsteen triplexes form at low pH, resulting in a closed state of the nanocapsule. When the pH was elevated, the Hoogsteen triplexes dehybridized and thus rapidly opened the nanocapsule revealing the encapsulated cargo. In this study, AuNPs and horseradish peroxidase (HRP) were successfully loaded and displayed selectively in response to the specified external trigger. Similar process has also been demonstrated by Grossi et al.,108 but in their approach strand-displacement reactions were employed in achieving successive opening and closing of the “nanovault” (Figure 3g).
In summary, the genetic information-bearing DNA and its derivative nanorobots hold tremendous potential for the preparation of personalized medicine,109 especially so with the foreseeable simple and cost-effective mass production of DNA nanostructures.110 As the key limitation, however, detailed elucidation on the in vivo features such as the pharmacokinetics, structure-performance relationships and circulating half-lives is still missing. As of today, no clinical trials have yet been reported,85 although an increasing amount of efforts have been invested in the research on DNA nanostructure stability and behavior in physiologically relevant conditions.111−113 Currently, covalent cross-linking,114−116 intelligent crossover design,117 and structure-dependent digestion118,119 have yielded promising results in increasing resistance against enzyme digestion, denaturation, and low-magnesium environments.
Plasmonic and Photonic Robots as Sensors and Walkers
Photons serve as ideal readout signals for nanorobotics as they can be detected remotely and instantaneously. To detect the state of a DNA nanodevice by optical means, it often requires a mechanism to translate the spatial configuration to a measurable optical quantity. Two most common mechanisms that have been employed are chiral plasmonics120 and FRET. Gold nanorods (AuNRs), which have distinct longitudinal and transversal plasmon modes, can couple with each other in different chiral modes depending on their relative spatial configurations. This chirality can thus be measured by circular dichroism (CD) spectroscopy. FRET, for one, is a well-established method to understand the distance between molecules by measuring the energy-transfer efficiency from donor fluorophore to acceptor molecule. In this section, recent robotic DNA structures relying on these two mechanisms will be discussed, and these nanodevices can be roughly categorized into sensors and walkers.
DNA nanostructures are actively studied and employed as sensing units in DNA nanotechnology. The input from DNA sensing strands can be translated to a measurable CD or fluorescent output via mechanical reconfiguration of the nanorobots. Based on this operational principle, a variety of sensing devices have been built.
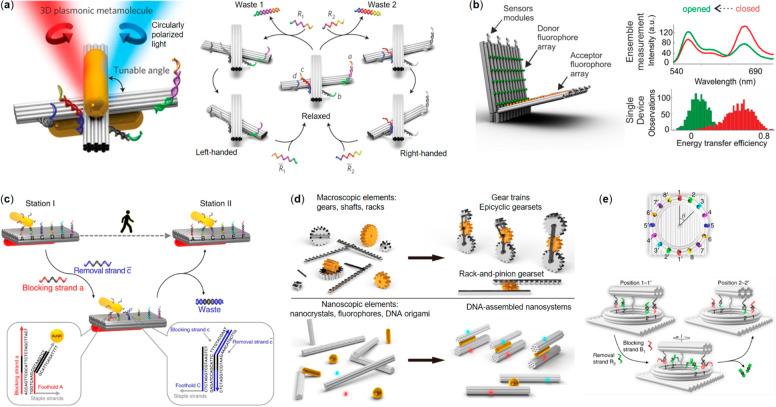
Kuzyk et al. have developed a reconfigurable chiral plasmonic nanomachine or “metamolecule” that can be modified to convert various input signals to a shift in its CD spectroscopy.121−125 The framework of the metamolecule consists of two DNA origami beams connected via a single Holliday junction that enables the free rotation of the beams with respect to each other. One oligonucleotide-functionalized AuNR is attached to each beam in a parallel fashion so that the AuNRs move following the rotation of the DNA beam (Figure 4a left panel). The metamolecule can switch between a relaxed configuration and a fixed configuration by the sensing strands attached at the end or edge of the system. By employing different sensing strands, the nanomachine can be used to detect ssDNA121 and RNA124 with specific sequence (Figure 4a right panel shows the switching in action), as well as pH changes,123 light irradiation,122 and aptamer-binding molecules.125
Figure 4.
Dynamic plasmonic and photonic DNA nanodevices. (a) Left panel: a reconfigurable plasmonic metamolecule made from AuNRs and a DNA origami with lock strands (colorful strands) on both sides. Right panel: operational principle of the switching between relaxed state and left-handed or right-handed state via strand displacement.121 (b) Left panel: a hinge-like fluorescence beacon with multiple donor–acceptor pairs and sensor modules. Right panel: comparisons of fluorescent spectra and energy transfer efficiency between opened and closed state.128 (c) Working principle of AuNR walking on predefined tracks on a DNA origami plate.129 (d) Macroscopic mechanical elements and their corresponding nanoscale counterparts in different DNA nanosystems.131 (e) A rotary nanoclock; a DNA origami hand rotates along a ring track via strand displacement.132 (a) Reprinted with permission from ref (121). Copyright 2014 Springer Nature Ltd. (b) Reprinted with permission from ref (128). Copyright 2018 American Chemical Society. (c) Reprinted with permission from ref (129). Published by 2015 Springer Nature Ltd. (d) Reprinted with permission from ref (131). Published by 2019 The American Association for the Advancement of Science. (e) Reprinted with permission from ref (132). Published by 2019 Springer Nature Ltd.
A similar system has been developed by Zhou et al.(126) Instead of using two beams, they employed a rectangular platform with a beam able to rotate in a plane parallel to the plate. The involvement of the plate provided more addressable space for attaching several sets of sensing strands, thus allowing the systems to be regulated by multiple aptamer-targets at the same time. Wang et al.(127) have taken the approach one step further by combining up to four AuNRs together into diastereomers with the help of multiple beam-and-plate platforms.
Besides these chiral plasmonic approaches, FRET has also been used as an output of DNA nanorobots. Selnihhin et al.(128) designed a hinge-like DNA origami beacon equipped with donor and acceptor fluorophore arrays (Figure 4b). The nanosystem can switch between an opened and a closed state by the sensor modules attached at the edges opposite of the rotation axis. Because of the high number of FRET pairs, the beacon can detect a target ssDNA with specific sequence at the concentration down to 100 pM and therefore demonstrates its potential as a point-of-care diagnostic device.
Besides a plasmonic signal reporter, the AuNR can also serve as a walker itself. Zhou et al.(129) designed a system that allows a AuNR walker to move along a predefined track on a 2D or 3D DNA origami platform. With a stator AuNR equipped on the opposite side of the platform, the location of the walker is reflected in the CD spectroscopy. The walking behavior of the AuNR can be precisely controlled in a reversible stepwise manner by fueling the system with specific sets of blocking strands and removal strands for toehold-mediated strand displacements. The oligonucleotides attached to the surface of the AuNR act as feet during the walking, and a series of ssDNA extrusions on the platform serve as the footholds thus defining the track of the motion. The walker moves forward as the blocking strands cause the feet on the backside of AuNR to dissociate from the footholds, while the removal strands simultaneously free the footholds in the walking direction. This “release and capture” process is demonstrated in Figure 4c. The authors have also shown that the walker can walk on a 3D uneven platform with prescriptive tracks. Such a dynamic DNA nanodevice is not only able to report its own structural dynamics, but it also enables fine-tuning its plasmonic properties in situ for optical applications.
To expand the toolbox of DNA-based dynamic nanomachines for accomplishing even more diverse tasks, other types of mechanical movement have also been explored. For example, sliding is one important basic motion required for many nanofactories. Urban et al.(130) implemented the same principle of programmable walking as above but translated the coordinated walking of two AuNPs to a mechanical sliding of two antiparallel DNA origami filaments. However, instead of the reporting function of chiral arrangement of AuNRs, the sliding was characterized in situ by a FRET pair located at one end of the beams. Powered by similar DNA fuels as in ref (129), the sliding motion can also be controlled in a stepwise and reversible manner. When the two beams slide away from each other, the donor and acceptor pairs will be spatially separated with increasing distances, and therefore, the progress of the sliding could be monitored by fluorescence spectroscopy. In addition, in the attempt to investigate the system in more detail, the authors found an intriguing fact that the sliding could occur even when the structures were confined by DNA sidelocks. However, the underlying mechanism was not clear and needs further investigation.
Zhan et al.(131) further increased the complexity and motion types of nanomachinery by hierarchically assembling multiple DNA origami with AuNP/AuNR and fluorophores. These sophisticated assemblies can perform a variety of highly regulated and coordinated mechanical movements, which include an independent revolving, a synchronized revolving and a complex joint motion. These DNA nanomachines and their macroscopic analogues were depicted in Figure 4d. The authors demonstrated that two DNA origami beams equipped with footholds composed of different sequences can be independently controlled to revolve around the same AuNR by feeding the system with corresponding instructions via toehold-mediated reactions. When the footholds contained the same sequence, such motions were synchronized. Furthermore, a joint motion which combines simultaneous sliding and revolving were also realized by placing the footholds in a shifted pattern. It is worth mentioning that all the motions in this work are monitored in situ via the changes of fluorescence intensity owing to the distance-dependent interactions between fluorophores and AuNRs.
Another intriguing implementation of the “release and capture” type of walker is a nanoclock designed by Xin et al.(132) (as shown in Figure 4e). The clock consists of three DNA origami parts: a rectangular plate foundation, a ring-shaped track, and a rotary beam. A rotor-stator AuNR pair was equipped with the beam and foundation for reporting the structural dynamics. Both the track and rotor were connected to the foundation, the only difference being that the track was immobilized, while the rotor was linked to the foundation via two adjacent scaffold crossovers with 30 unpaired bases. The long scaffold linkers provide sufficient flexibility for the beam to freely rotate along the track. In total, 16 footholds were placed on the track, allowing a 360° rotation with 22.5° intervals. Periodic CD intensity changes as a function of rotation angles were observed for both the clockwise and counterclockwise rotations, which were in good agreement with theoretical simulations. While the stepwise rotation was still controlled by external inputs, the authors realized an autonomous clock by adding DNAzyme sequences as feet and replacing footholds with RNA molecules. After activating the DNAzyme feet and releasing the rotor, the DNAzyme cut the RNA substrates into shorter segments one after another in a time span of a few hours. In other words, the autonomous nanoclock was powered by RNA hydrolysis with a burnt-bridge mechanism.
Robots Guided and Driven by External Fields
The plasmonic and nanophotonic mechanisms mentioned in the previous subsection may provide instantaneous output information about the structural dynamics of DNA nanorobots, but the actuation of the structural reconfiguration is still relatively slow when, for example, diffusion-limited strand-displacement reactions133 are employed. However, charged DNA nanostructures can be manipulated by electric fields based on (di)electrophoretic motion,134−136 and therefore, DNA nanomachines with considerably swift responsiveness and well-defined temporal and spatial control driven by external electromagnetic fields have been realized.
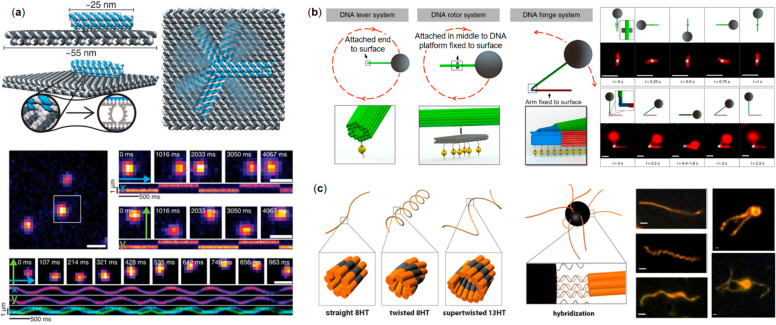
Kopperger et al.(38) fabricated a dynamic nanoelectromechanical robotic arm composed of a rigid rectangular DNA origami tile equipped with a 6HB arm that was joined to the tile via ssDNA scaffold crossovers (Figure 5a, top panel). The flexible joint enabled stochastic switching of the arm due to transient binding, detected from FRET signals generated by the donor fluorophore (Alexa Fluor 488) on the tip of the arm and two acceptor dyes (ATTO 565 and ATTO 647N) connected on the opposite sides of the origami tile. Electrically driven movement of the robot arm was measured when the system was seated at the center of a cross-shaped electrophoretic chamber having two perpendicular fluid channels. The electric field was applied by two pairs of platinum electrodes inserted in the reservoirs at the ends of the channels that resulted in a movement of the pointers, detected with an electron-multiplying charge-coupled device camera using total internal reflection fluorescence (TIRF) microscopy (Figure 5a, bottom panel). Moreover, the arm enabled electrically driven transport of cargo which is useful for the control of photonic and plasmonic processes. This DNA robotic arm was reported to be at least 5 orders of magnitude faster than previous DNA motor systems and in the same class as adenosine triphosphatase-driven biohybrid motors.137
Figure 5.
Electromagnetic field-driven DNA nanorobotics. (a) Top panel: The design and the working principle of the electrically driven movement of a 6HB robot arm that enables stochastic switching. Bottom panel: TIRF images showing the movement of the arm under the electric field (x- and y-coordinates follow sinusoidal behavior).38 (b) Left panel: DNA origami–superparamagnetic particle configurations to realize lever, rotor, and hinge systems. Right panel: TIRF images showing rotor and hinge system in operation.138 (c) Left panel: Different helix tube designs acting as flagella for magnetic beads. Right panel: Micrographs of the tubes (left) and two magnetic beads–DNA origami hybrids (twisted and supertwisted tubes, right).141 (a) Reprinted with permission from ref (38). Copyright 2018 The American Association for the Advancement of Science. (b) Reprinted with permission from ref (138). Published by 2018 Springer Nature Ltd. (c) Reprinted with permission from ref (141). Copyright 2016 American Chemical Society.
Unlike electric fields that can directly exert forces on the negatively charged DNA robotic arms, magnetic fields need magnetic particles (MPs) as media to apply their forces. It is challenging to scale down the MPs to nanoscale to match DNA nanodevices while still creating sufficient forces to overcome thermal fluctuations. Therefore, instead of scaling down the MPs, Lauback et al.(138) designed stiff DNA levers with persistence length over 20 μm to match the sizes of micron-scale superparamagnetic particles. These levers could be integrated into DNA nanorotors and nanohinges to actuate their motions. To facilitate the assembly of these nanomachines, one segment of the rigid lever was first connected to the nanoscale system, and then it served as a seed to extend the microscale lever arm with a NP attached at the end. During the experiments, either the lever itself or the stationary part of the rotor or hinge was immobilized on the surface via biotin–streptavidin affinity (Figure 5b, left panel). Several fluorophores were attached to the lever so its movement can be visualized by TIRF, in Figure 5b the right panel shows the rotor and hinge in action. The authors demonstrated the direct manipulation of DNA origami nanodevices with subsecond response times and torques ranging from ∼20 to 80 pN nm rad–1 at magnetic fields of ∼10–100 Oe. Moreover, compared to other systems which usually have a few discrete states, a continuous rotation of the rotor was established in this magnetic field driven system, exhibiting a high level of spatial control over the DNA nanodevices.
Yang et al.(139) composed a light-driven nanomachine based on a DNA walker that was fueled by 350 nm wavelength photoirradiation instead of strand-displacement or enzymatic reactions. Walking strands comprising two pyrene moieties moved on a rectangular 2D DNA tile along four disulfide-modified “stator” strands aligned linearly. Hopping from the cleaved stator to the adjacent one took place via duplex formation. Stepwise movement of the walker was time-dependent and visualized using HS-AFM.
Most of the DNA nanorobots presented in this review so far are focused on actuation of a local structural reconfiguration. In fact, the locomotion of the entire nanorobot for traveling a longer distance is also an essential aspect for many applications, e.g., in targeted biomedicine delivery. Recently, fundamental research along this line has been carried out, including proof-of-concept experiments on the movement of mechanically interlocked DNA nanostructures on DNA filaments140 or long-range swimmers propelled by flagella made from DNA nanostructures (see below).
Maier et al.(141) have built a micron-scale swimmer driven by a rotating external magnetic field. The hybrid microswimmer consists of a magnetic bead of 1 μm diameter and several artificial flagella assembled from tile-based DNA n-helix tubes (nHT) having various degrees of twist, which were linked together via biotin–streptavidin coupling (Figure 5c). The magnetic bead functions as a rotating power-driven hub that follows the homogeneous external field rotating perpendicular to the swimming direction. While swimming, the DNA flagella formed a hydrodynamically assembled corkscrew-like bundle of several micrometers long. The swimmer can propel with a speed up to 0.6 μm/s when it was driven by a 3 Hz magnetic field. In addition, by manipulating the magnetic field, complex maneuvers like moving in a curved path have been demonstrated by fluorescent microscopy.
Another plausible concept to fabricate microswimmers is thermophoresis. Herms et al.(142) and Heerwig et al.(143) proposed janus-type hybrid nanoparticles made from AuNP and DNA origami as self-thermophoretic swimmers. The authors predicted that the temperature gradient created by the asymmetric particles when the AuNPs absorb energy from laser irradiation could generate a slip flow to propel the swimmer forward. However, although these nanostructures have been assembled and their thermal stability has been studied, experimental work showing the swimming behavior has not been reported at the time of this review.
Advanced Autonomous Robots
Arguably, one of the most intriguing classes of nanomachinery in synthetic biology is autonomous robotics. These robots may be seamlessly integrated into biological systems and they could mimic for example naturally occurring motor protein functions or execute completely unnatural tasks with high efficiency and precision. A number of different DNA nanostructure based autonomous robots with defined moving pathways have been introduced.
One of the earliest examples taking advantage of the programmability and addressability of DNA origami was presented by Lund et al.(144) They created an autonomous DNA robot able to carry out sequences of actions such as start, follow, turn, and stop on a DNA platform. Previously engineered “molecular spiders”145 that consist of a streptavidin “body” and three catalytic deoxyribozyme “legs” (adapted from the 8–17 DNA enzyme) were set to autonomously move along the predefined paths coded on a rectangular DNA origami canvas. Movement was based on Brownian sensing and modifying tracks of substrate molecules by catalytic DNA binding and degradation of oligodeoxynucleotide substrates via hydrolysis. Real time movement of single spider particles was tracked by super-resolution total-internal-reflection fluorescence video microscopy.
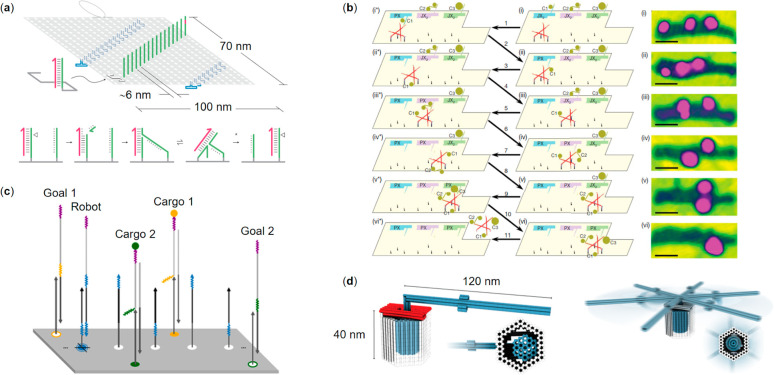
Wickham and co-workers146 employed a 100 nm long track on DNA origami rectangle to demonstrate stepwise directional movement of ssDNA motor through precisely aligned “stators” (1D array of complementary single-stranded attachment sites), including a distinct start and stop “stators”. A multistep autonomous movement at constant speed through stators was initiated by a thermal strand displacement reaction to form a motor-stator duplex (Figure 6a). Hydrolysis by restriction enzyme drives the movement of the motor to the adjacent intact stator via branch migration. The terminal stator has a built-in mismatch in the motor–stator duplex, which shuts down the motor and shields it from the nicking enzyme. Transport of individual motor molecules was monitored with a HS-AFM imaging system. Extension of the concept demonstrated integrated long-range transport and information processing. Authors established that the path of a ssDNA motor through a network of tracks containing alternative routes can be programmed using instructions that are added externally (87% operational yield) or embedded internally to the motor (71% operational yield). Programmable motion will pave the way, e.g., to the trajectory of computing networks and the evolution of self- and cargo-sorting molecular systems and assembly lines.147
Figure 6.
Autonomous and semiautonomous DNA origami nanorobotics. (a) Top panel: a stepwise directional movement of a ssDNA motor through aligned “stators” (green) (the hairpin loops are marked in blue). Bottom panel: a nicking restriction enzyme cuts the motor-bound stator revealing a toehold at the 3′ end of the motor (magenta) that drives the motor to the adjacent intact stator by branch migration (a “burnt bridges” mechanism).146 (b) Left panel: a proximity-based programmable DNA nanoscale assembly line and its stepwise operation. Right panel: atomic force micrographs of the system corresponding to the process steps sketched as states (i–vi) in the molecular assembly line.150 (c) A cargo-sorting DNA robot that employs an irreversible strand displacement reaction for picking up and delivering selected cargo to a goal on top of a 2D DNA origami platform.152 (d) A DNA origami rotary device fabricated from tight-fitting components showing closed brackets and a docked rotor (left) and an undocked, mobile rotor (right). Color code: blue = rotor unit, gray = clamp units, red = shape-complementary sockets on the rotor and the clamps, respectively.157 (a) Reprinted with permission from ref (146). Copyright 2011 Springer Nature Ltd. (b) Reprinted with permission from ref (150). Copyright 2010 Springer Nature Ltd. (c) Reprinted with permission from ref (152). Copyright 2017 The American Association for the Advancement of Science. (d) Reprinted with permission from ref (157). Published by 2016 The American Association for the Advancement of Science.
Inspired by a motion of kinesin, Liber et al.(148) demonstrated dynamic to and fro movement of the bipedal motor that can be extended over two DNA origami tiles (60 × 90 nm each) that were dimerized using bridging strands. Successive deployment of fuel and antifuel strands drives the bipedal motor from tile A to B back to A-tile through five footholds moving twice across the junction with an overall track distance of 64 nm. The uniformity of double-tiles and high operational yield was proven by smFRET and alternating laser excitation techniques (ALEX)149 and assisted by AFM.
Gu and colleagues150 created a proximity-based programmable DNA nanoscale assembly line constructed of three distinct components: a tensegrity-triangle walker possessing four ssDNA legs and three hands, a DNA origami platform that provide an automobile-style assembly line, and programmable cargo-donating cassettes mounted as stations. Three cassettes, each containing a robotic arm,151 can be independently switched between two states. The ON state (PX) allows transfer and loading of the cargo to the walker, whereas for the OFF state (JX2) the loading is disabled. All locomotion, orientation of the walker, and proximity-based attachment events of different Au-NPs is mediated by toehold-binding/branch-migration methods,30 which facilitate programming of eight assembly pathways and thus, eight different products. Schematic of the molecular assembly line steps and AFM images showing only the states of Au-NP cargo is depicted in Figure 6b.
A step toward a cargo-sorting DNA robot was introduced by Thubagere et al.(152) who developed a sophisticated algorithm capable of sorting two types of cargo and their destinations on a 2D DNA origami platform (Figure 6c). The DNA robot assembled from three modular domains was equipped to pick up and release the cargo at the specified location. The robot was designed to perform a random walk which requires no supplemental energy. Thus, a single robot was able to perform ca. 300 steps during the cargo sorting (on average), which is a significant enhancement on previously reported bipedal DNA walkers that performed tasks while moving.153−156
Ketterer and co-workers157 demonstrated the assembly of complex nanomachines from multiple DNA origami elements by taking advantage of DNA–DNA interactions. The group fabricated a miniature rotary apparatus that was inspired by the F1F0-adenosine triphosphate (ATP) synthase (Figure 6d). The device was constructed from tight-fitting DNA origami components, including a rotor, a clamp, and a socket, by steering the self-assembly with specific DNA hybridization events or base-stacking interactions. The rotating devices were not driven, but they were shown to exhibit random Brownian motion, as observed via single particle fluorescence microscopy recordings. Such a prototype equipped with external triggers and control of the movement may facilitate a basis toward creating complex and sophisticated DNA-based nanomachines.
As a fascinating example of how the different DNA robots can interact with each other, Kaminka et al.(158) used similar two-state (open-close) devices as explained in Amir et al.(101) This time the authors created mixtures of interacting robots that were able to recreate biochemistry professor and science fiction author Isaac Asimov’s infamous “Runaround” scenario159 based on his “Three Laws of Robotics” L1–L3 (see ref (159)). While the story is about a fictional robot and it is often regarded as impractical, some of its implementations may be rather thought-provoking when considering safety, autonomy and decision making of robotics in real life.
In Asimov’s book a robot gets stuck at equilibrium between the laws L2 and L3 during its mission, and is finally freed from this condition by overriding both L2 and L3 by the law L1. In this experimental setup the populations of logic-gated DNA nanorobots (L1, L2, and L3) were chosen in such a way that L1 and L2 formed a logical NOT gate (i.e., without a damage signal, L1 is closed and L2 is open and active, but when the damage signal is present, L2 is closed through binding to L1), and a microRNA molecule (a human miR-16 analogue) was employed as a damage signal. However, this gate function depends on the L1:L2 ratio, as the excess of L2 robots leads to exaggerated activity, and on the other hand, the excess of L1 robots halts the system. Therefore, the third robot L3 works as a sensor for the L1:L2 ratio and provides the functionality of the system, in other words, the necessary L1:L2 ratio is analogous to the robotic system protecting itself (L3). Importantly, as the proportion of robots required to exert action is relatively small, some L1 and L2 robots in the system may serve as ratio indicators, while others interact on accessible L3 robots in order to physically exert the effects. In practice, the inputs for the L3 robot are DNA strands attached to L1 and L2 robots. The scenario starts with L2 dominating, carrying out its defined task (open), followed by a conflict between L2 and L3. Experimentally it is reached by closing L3 robots by DNA strands, which mimics a skewed L1:L2 ratio thus resulting in a lower activity equilibrium of L2. This achieved equilibrium is then terminated by introducing a miR-16 damage, which leads to L1 override and thus a near-baseline activity of L2.
Conclusions and Future Directions
In this review we have introduced multiple DNA nanomachines that are able to execute tasks in a dynamic manner and described their main working principles ranging from the fast-moving external field driven devices to fully autonomous robots. The classification of the most important DNA devices, their sensing/actuation mechanisms, characterization techniques, and potential applications are listed in Table 1. All of these devices are based on programmable DNA nanostructures that are loaded with various (bio)chemical, physical, and logical features. During the past ∼40 years, the structural DNA nanotechnology has reached a state at which the design, fabrication, and cost of intricate DNA objects are no more obstacles on the road to real-life implementations. However, to date the great number of all presented DNA constructions have been static (usually on purpose), and therefore, the coming of age of the mechanically moving more complex DNA nanostructures has only recently been witnessed.160 That also means there are a great deal of challenges ahead in creating dynamic DNA devices, such as their stability under application-specific conditions.
Table 1. Selected Robot Types, Their Mechanisms of Action, Characterization Techniques, and Possible Applications.
| classification/robot type | mechanism of action | characterization/imaging | application |
|---|---|---|---|
| Mechanical tools | |||
| Force clamp57 | Entropic DNA springs | FRET | Resolving, e.g., DNA conformational changes |
| Pylons58 | DNA base stacking | Optical tweezers | Resolving DNA base stacking interaction |
| Calipers59−61 | DNA hinge + interaction between the investigated species | TEM, FRET, cryo-EM | Measuring, e.g., forces between nucleosomes and nucleosome unwrapping |
| Information relay | |||
| Networks39 | Base stacking (depends on ionic strength) | TEM | Large-scale movement |
| Nanoactuator70/accordion rack71 | DNA hybridization | AFM, TEM | Molecular regulation |
| Domino arrays72 | Base stacking | AFM | Long-distance step-by-step movement |
| Nanomedicine | |||
| Imaging tools86−93/antibody sensors97,98 | Various conformational (e.g., i-tetraplex) and structural transitions, DNA transient binding | FRET, fluorescence microscopy, DNA-PAINT | Diagnostics, studying pathway dynamics, payload delivery, super-resolution imaging, pH-mapping (also in vivo) |
| Pliers94,95 | Target molecule binding | AFM, spectroscopy | Diagnostics, molecular computing |
| Nanorobots40,96,99−103 | (Logic-gated) aptamer-protein interaction | TEM, AFM, flow cytometry | Targeted and programmable drug delivery, computing (also in vivo) |
| Capsules107,108/cages104−106 | Strand displacement/pH-sensitive DNA strands/light/temperature/mRNA | TEM, FRET, fluorescence microscopy, enzyme kinetics | Selective and controlled display/release of molecular cargo |
| Photonics/plasmonics | |||
| Metamolecules121−127/beacon128 | Strand displacement/pH-sensitive DNA strands/azobenzene-modified strands/aptamer-binding | CD, FRET, TEM | Sensors, diagnostics |
| AuNR walkers129−131/nanoclock132 | Strand displacement/DNAzyme | CD, FRET, TEM | Complex nanomachinery |
| External-field driven | |||
| Robotic arms38 | Electric field | FRET | Nanomachines with rapid and controlled movement |
| Nanohinge/nanorotor138 | Magnetic field | TIRF | Nanomachines with rapid and controlled movement |
| Swimmers141−143 | Magnetic field, thermophoresis | Fluorescence microscopy | Guided drug delivery |
| Autonomous robots | |||
| Walkers/motors/robots144,146−156 | Strand displacement/toeholds/restriction enzyme driven | AFM, HS-AFM, smFRET, ALEX | Nanoscale assembly lines, cargo-sorting, computing |
| Rotary apparatus157 | Controlled DNA base stacking + Brownian motion | Single-particle fluorescence microscopy | Toward biomimicking nanomachines |
| Interacting dynamic robot populations158 | Binding through hybridization/toeholds, detection of signals such as miR | Flow cytometry | Toward safe, decision-making robotics |
Currently, the DNA robots show considerable promise especially in biomedicine for rather obvious reasons, and thus prototypes of sophisticated delivery vehicles and point-of-care diagnostic molecular devices are coming increasingly in view. To just produce more advantageous robots may not sound too fancy, but the long-term goal in synthetic biology could be for example in the reprogramming of our immune systems using active biocompatible DNA machines. This, for one, would indeed have far-reaching outcomes. To use DNA nanorobots for realizing such scenarios as Isaac Asimov’s Runaround are arguably intriguing; however, we still remain miles away from the arenas often depicted in science fiction. This is due to the fact that it is extremely hard to maintain the autonomy of the device, while trying to increase the complexity of its functions. The present-day autonomous devices are still operating at the very fundamental level, but looking back into the recent rapid development of DNA nanotechnology may give us an impression of how swiftly the DNA robots could revolutionize synthetic biology and simultaneously open entirely new avenues for scientific, medical, and technological exploration.
Acknowledgments
Financial support by the Academy of Finland, Emil Aaltonen Foundation, the Sigrid Jusélius Foundation, the Jane and Aatos Erkko Foundation, and the Vilho, Yrjö, and Kalle Väisälä Foundation of the Finnish Academy of Science and Letters is gratefully acknowledged. This work was carried out under the Academy of Finland Centers of Excellence Programme (2014–2019).
The authors declare no competing financial interest.
References
- Bathe M.; Rothemund P. W. K. (2017) DNA Nanotechnology: A Foundation for Programmable Nanoscale Materials. MRS Bull. 42, 882–888. 10.1557/mrs.2017.279. [DOI] [Google Scholar]
- Seeman N. C.; Sleiman H. F. (2018) DNA Nanotechnology. Nat. Rev. Mater. 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Linko V.; Dietz H. (2013) The Enabled State of DNA Nanotechnology. Curr. Opin. Biotechnol. 24, 555–561. 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Jones M. R.; Seeman N. C.; Mirkin C. A. (2015) Programmable Materials and the Nature of the DNA Bond. Science 347, 1260901. 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- Church G. M.; Gao Y.; Kosuri S. (2012) Next-Generation Digital Information Storage in DNA. Science 337, 1628. 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- Lee H., Wiegand D. J., Griswold K., Punthambaker S., Chun H., Kohman R. E., and Church G. M. (2020) Photon-directed Multiplexed Enzymatic DNA Synthesis for Molecular Digital Data Storage. bioRxiv, Feb 20, 2020, 10.1101/2020.02.19.956888 (accessed 2020-04-30). [DOI] [PMC free article] [PubMed]
- Feynman R. P. (1960) There’s Plenty of Room at the Bottom. Eng. Sci. 23, 22–36. [Google Scholar]
- King N. P.; Sheffler W.; Sawaya M. R.; Vollmar B. S.; Sumida J. P.; Andre I.; Gonen T.; Yeates T. O.; Baker D. (2012) Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy. Science 336, 1171–1174. 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S.; Boyken S. E.; Baker D. (2016) The Coming of Age of De Novo Protein Design. Nature 537, 320–327. 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Ijäs H.; Nummelin S.; Shen B.; Kostiainen M. A.; Linko V. (2018) Dynamic DNA Origami Devices: From Strand-Displacement Reactions to External-Stimuli Responsive Systems. Int. J. Mol. Sci. 19, 2114. 10.3390/ijms19072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Pan V.; Li X.; Yang X.; Li H.; Wang P.; Ke Y. (2019) Dynamic DNA Structures. Small 15, 1900228. 10.1002/smll.201900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Kostiainen M. A. (2016) Automated Design of DNA Origami. Nat. Biotechnol. 34, 826–827. 10.1038/nbt.3647. [DOI] [PubMed] [Google Scholar]
- Veneziano R.; Ratanalert S.; Zhang K.; Zhang F.; Yan H.; Chiu W.; Bathe M. (2016) Designer Nanoscale DNA Assemblies Programmed from the Top Down. Science 352, 1534. 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nummelin S.; Kommeri J.; Kostiainen M. A.; Linko V. (2018) Evolution of Structural DNA Nanotechnology. Adv. Mater. 30, 1703721. 10.1002/adma.201703721. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. (2006) Folding DNA to Create Nanoscale Shapes and Patterns. Nature 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. (2009) Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskunen P.; Nummelin S.; Shen B.; Kostiainen M. A.; Linko V. (2020) Increasing Complexity in Wireframe DNA Nanostructures. Molecules 25, 1823. 10.3390/molecules25081823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Ora A.; Kostiainen M. A. (2015) DNA Nanostructures as Smart Drug-Delivery Vehicles and Molecular Devices. Trends Biotechnol. 33, 586–594. 10.1016/j.tibtech.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Jiang Q.; Zhao S.; Liu J.; Song L.; Wang Z.; Ding B. (2019) Rationally Designed DNA-Based Nanocarriers. Adv. Drug Delivery Rev. 147, 2–21. 10.1016/j.addr.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Castro C. E.; Dietz H.; Högberg B. (2017) DNA Origami Devices for Molecular-Scale Precision Measurements. MRS Bull. 42, 925–929. 10.1557/mrs.2017.273. [DOI] [Google Scholar]
- Shen B.; Kostiainen M. A.; Linko V. (2018) DNA Origami Nanophotonics and Plasmonics at Interfaces. Langmuir 34, 14911–14920. 10.1021/acs.langmuir.8b01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. (2019) DNA Nanotechnology for Building Artificial Dynamic Systems. MRS Bull. 44, 576–581. 10.1557/mrs.2019.155. [DOI] [Google Scholar]
- Maune H. T.; Han S.-P.; Barish R. D.; Bockrath M.; Goddard W. A. III; Rothemund P. W. K.; Winfree E. (2010) Self-Assembly of Carbon Nanotubes into Two-Dimensional Geometries Using DNA Origami Templates. Nat. Nanotechnol. 5, 61–66. 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- Hung A. M.; Micheel C. M.; Bozano L. D.; Osterbur L. W.; Wallraff G. M.; Cha J. N. (2010) Large-Area Spatially Ordered Arrays of Gold Nanoparticles Directed by Lithographically Confined DNA Origami. Nat. Nanotechnol. 5, 121–126. 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. (2014) Multiplexed 3D Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graugnard E.; Hughes W. L.; Jungmann R.; Kostiainen M. A.; Linko V. (2017) Nanometrology and Super-Resolution Imaging with DNA. MRS Bull. 42, 951–959. 10.1557/mrs.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. (2003) DNA in a Material World. Nature 421, 427–431. 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- Mao C.; Sun W.; Shen Z.; Seeman N. C. (1999) A Nanomechanical Device Based on the B-Z Transition of DNA. Nature 397, 144–146. 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- Yang X.; Vologodskii A. V.; Liu B.; Kemper B.; Seeman N. C. (1998) Torsional control of double-stranded DNA branch migration. Biopolymers 45, 69–83. . [DOI] [PubMed] [Google Scholar]
- Yurke B.; Turberfield A. J.; Mills A. P. Jr.; Simmel F. C.; Neumann J. L. (2000) A DNA-Fuelled Molecular Machine Made of DNA. Nature 406, 605–608. 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Simmel F. C.; Yurke B. (2002) A DNA-Based Molecular Device Switchable Between Three Distinct Mechanical States. Appl. Phys. Lett. 80, 883–885. 10.1063/1.1447008. [DOI] [Google Scholar]
- Yan H.; Zhang X.; Shen Z.; Seeman N. C. (2002) A Robust DNA Mechanical Device Controlled by Hybridization Topology. Nature 415, 62–65. 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- Shin J. S.; Pierce N. A. (2004) A Synthetic DNA Walker for Molecular Transport. J. Am. Chem. Soc. 126, 10834–10835. 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- Bath J.; Turberfield A. J. (2007) DNA Nanomachines. Nat. Nanotechnol. 2, 275–284. 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Wang M.; Mao C. (2004) An Autonomous DNA Nanomotor Powered by a DNA Enzyme. Angew. Chem., Int. Ed. 43, 3554–3557. 10.1002/anie.200453779. [DOI] [PubMed] [Google Scholar]
- Yin P.; Yan H.; Daniell X. G.; Turberfield A. J.; Reif J. H. (2004) A Unidirectional DNA Walker That Moves Autonomously Along a DNA Track. Angew. Chem., Int. Ed. 43, 4906–4911. 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- Pan J.; Li F.; Cha T. G.; Chen H.; Choi J. H. (2015) Recent Progress on DNA Based Walkers. Curr. Opin. Biotechnol. 34, 56–64. 10.1016/j.copbio.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Kopperger E.; List J.; Madhira S.; Rothfischer F.; Lamb D. C.; Simmel F. C. (2018) A Self-Assembled Nanoscale Robotic Arm Controlled by Electric Fields. Science 359, 296–301. 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- Gerling T.; Wagenbauer K. F.; Neuner A. M.; Dietz H. (2015) Dynamic DNA Devices and Assemblies Formed by Shape-Complementary, Non-Base Pairing 3D Components. Science 347, 1446–1452. 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. (2012) A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Qian L.; Winfree E.; Bruck J. (2011) Neural Network Computation with DNA Strand Displacement Cascades. Nature 475, 368–372. 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- Qian L.; Winfree E. (2011) Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 332, 1196–1201. 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- Hamdi M.; Ferreira A. (2009) Multiscale Design and Modeling of Protein-Based Nanomechanisms for Nanorobotics. Int. J. Robot. Res. 28, 436–449. 10.1177/0278364908099888. [DOI] [Google Scholar]
- Derr N. D.; Goodman B. S.; Jungmann R.; Leschziner A. E.; Shih W. M.; Reck-Peterson S. L. (2012) Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold. Science 338, 662–665. 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Nummelin S.; Aarnos L.; Tapio K.; Toppari J. J.; Kostiainen M. A. (2016) DNA-Based Enzyme Reactions and Systems. Nanomaterials 6, 139. 10.3390/nano6080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaekel A.; Stegemann P.; Saccà B. (2019) Manipulating Enzymes Properties with DNA Nanostructures. Molecules 24, 3694. 10.3390/molecules24203694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K.; Zhou Y.; Pan V.; Wang Q.; Ke Y. (2020) Programming Dynamic Assembly of Viral Proteins with DNA Origami. J. Am. Chem. Soc. 142, 5929–5932. 10.1021/jacs.9b13773. [DOI] [PubMed] [Google Scholar]
- Zhao D.; Kong Y.; Zhao S.; Xing H. (2020) Engineering Functional DNA-Protein Conjugates for Biosensing, Biomedical, and Nanoassembly Applications. Top. Curr. Chem. 378, 41. 10.1007/s41061-020-00305-7. [DOI] [PubMed] [Google Scholar]
- Watson J. D.; Crick F. H. C. (1953) Molecular Structure of Nucleic Acids. Nature 171, 737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Smith S. B.; Cui Y.; Bustamante C. (1996) Overstretching B-DNA: The Elastic Response of Individual Double-Stranded and Single-Stranded DNA Molecules. Science 271, 795–799. 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Castro C. E.; Su H. J.; Marras A. E.; Zhou L.; Johnson J. (2015) Mechanical Design of DNA Nanostructures. Nanoscale 7, 5913–5921. 10.1039/C4NR07153K. [DOI] [PubMed] [Google Scholar]
- Kauert D. J.; Kurth T.; Liedl T.; Seidel R. (2011) Direct Mechanical Measurements Reveal the Materials Properties of Three-Dimensional DNA Origami. Nano Lett. 11, 5558–5563. 10.1021/nl203503s. [DOI] [PubMed] [Google Scholar]
- Liedl T.; Högberg B.; Tytell J.; Ingber D. E.; Shih W. M. (2010) Self-Assembly of Three-Dimensional Prestressed Tensegrity Structures from DNA. Nat. Nanotechnol. 5, 520–524. 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras A. E.; Zhou L.; Su H.-J.; Castro C. E. (2015) Programmable Motion of DNA Origami Mechanics. Proc. Natl. Acad. Sci. U. S. A. 112, 713–718. 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.; Marras A. E.; Su H.-J.; Castro C. E. (2014) DNA Origami Compliant Nanostructures with Tunable Mechanical Properties. ACS Nano 8, 27–34. 10.1021/nn405408g. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Marras A. E.; Su H.-J.; Castro C. E. (2015) Direct Design of an Energy Landscape with Bistable DNA Origami Mechanisms. Nano Lett. 15, 1815–1821. 10.1021/nl5045633. [DOI] [PubMed] [Google Scholar]
- Nickels P. C.; Wünsch B.; Holzmeister P.; Bae W.; Kneer L. M.; Grohmann D.; Tinnefeld P.; Liedl T. (2016) Molecular Force Spectroscopy with a DNA Origami-Based Nanoscopic Force Clamp. Science 354, 305–307. 10.1126/science.aah5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchherr F.; Wachauf C.; Pelz B.; Rief M.; Zacharias M.; Dietz H. (2016) Single-Molecule Dissection of Stacking Forces in DNA. Science 353, aaf5508 10.1126/science.aaf5508. [DOI] [PubMed] [Google Scholar]
- Funke J. J.; Ketterer P.; Lieleg C.; Schunter S.; Korber P.; Dietz H. (2016) Uncovering the Forces Between Nucleosomes Using DNA Origami. Sci. Adv. 2, e1600974 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke J. J.; Ketterer P.; Lieleg C.; Korber P.; Dietz H. (2016) Exploring Nucleosome Unwrapping Using DNA Origami. Nano Lett. 16, 7891–7898. 10.1021/acs.nanolett.6b04169. [DOI] [PubMed] [Google Scholar]
- Le J. V.; Luo Y.; Darcy M. A.; Lucas C. R.; Goodwin M. F.; Poirier M. G.; Castro C. E. (2016) Probing Nucleosome Stability with a DNA Origami Nanocaliper. ACS Nano 10, 7073–7084. 10.1021/acsnano.6b03218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekic T.; Barisic I. (2020) In Silico Modelling of DNA Nanostructures. Comput. Struct. Biotechnol. J. 18, 1191–1201. 10.1016/j.csbj.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C. E.; Kilchherr F.; Kim D.-N.; Shiao E. L.; Wauer T.; Wortmann P.; Bathe M.; Dietz H. (2011) A Primer to Scaffolded DNA Origami. Nat. Methods 8, 221–229. 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- Kim D.-N.; Kilchherr F.; Dietz H.; Bathe M. (2012) Quantitative Prediction of 3D Solution Shape and Flexibility of Nucleic Acid Nanostructures. Nucleic Acids Res. 40, 2862–2868. 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffeo C.; Yoo J.; Aksimentiev A. (2016) De Novo Reconstruction of DNA Origami Structures Through Atomistic Molecular Dynamics Simulation. Nucleic Acids Res. 44, 3013–3019. 10.1093/nar/gkw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šulc P.; Romano F.; Ouldridge T. E.; Rovigatti L.; Doye J. P.; Louis A. A. (2012) Sequence-Dependent Thermodynamics of a Coarse-Grained DNA Model. J. Chem. Phys. 137, 135101. 10.1063/1.4754132. [DOI] [PubMed] [Google Scholar]
- Shi Z.; Castro C. E.; Arya G. (2017) Conformational Dynamics of Mechanically Compliant DNA Nanostructures from Coarse-Grained Molecular Dynamics Simulations. ACS Nano 11, 4617–4630. 10.1021/acsnano.7b00242. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Schreck J. S.; Romano F.; Louis A. A.; Doye J. P. K. (2017) Characterizing the Motion of Jointed DNA Nanostructures Using a Coarse-Grained Model. ACS Nano 11, 12426–12435. 10.1021/acsnano.7b06470. [DOI] [PubMed] [Google Scholar]
- Woo S.; Rothemund P. W. K. (2011) Programmable Molecular Recognition Based on the Geometry of DNA Nanostructures. Nat. Chem. 3, 620–627. 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Meyer T.; Shih W. M.; Bellot G. (2016) Regulation at a Distance of Biomolecular Interactions Using a DNA Origami Nanoactuator. Nat. Commun. 7, 10935. 10.1038/ncomms10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.; Choi H.; Lee A. C.; Lee H.; Kwon S. (2018) A Reconfigurable DNA Accordion Rack. Angew. Chem., Int. Ed. 57, 2811–2815. 10.1002/anie.201709362. [DOI] [PubMed] [Google Scholar]
- Song J.; Li Z.; Wang P.; Meyer T.; Mao C.; Ke Y. (2017) Reconfiguration of DNA Molecular Arrays Driven by Information Relay. Science 357, eaan3377 10.1126/science.aan3377. [DOI] [PubMed] [Google Scholar]
- Li J.; Esteban-Fernandez de Avila B.; Gao W.; Zhang L.; Wang J. (2017) Micro/Nanorobots for Biomedicine: Delivery, Surgery, Sensing, and Detoxification. Sci. Robot. 2, eaam6431 10.1126/scirobotics.aam6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gradilla V.; Sattayasamitsathit S.; Soto F.; Kuralay E.; Yardımcı C.; Wiitala D.; Galarnyk M.; Wang J. (2014) Ultrasound-Propelled Nanoporous Gold Wire for Efficient Drug Loading and Release. Small 10, 4154–4159. 10.1002/smll.201401013. [DOI] [PubMed] [Google Scholar]
- Walker D.; Käsdorf B. T.; Jeong H.-H.; Lieleg O.; Fischer P. (2015) Enzymatically Active Biomimetic Micropropellers for the Penetration of Mucin Gels. Sci. Adv. 1, e1500501 10.1126/sciadv.1500501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-Z.; Hoop M.; Shamsudhin N.; Huang T.; Özkale B.; Li Q.; Siringil E.; Mushtaq F.; Tizio L. D.; Nelson B. J.; Pané S. (2017) Hybrid Magnetoelectric Nanowires for Nanorobotic Applications: Fabrication, Magnetoelectric Coupling, and Magnetically Assisted In Vitro Targeted Drug Delivery. Adv. Mater. 29, 1605458. 10.1002/adma.201605458. [DOI] [PubMed] [Google Scholar]
- Nelson B. J.; Kaliakatsos I. K.; Abbott J. J. (2010) Microrobots for Minimally Invasive Medicine. Annu. Rev. Biomed. Eng. 12, 55–85. 10.1146/annurev-bioeng-010510-103409. [DOI] [PubMed] [Google Scholar]
- Wang J.; Gao W. (2012) Nano/Microscale Motors: Biomedical Opportunities and Challenges. ACS Nano 6, 5745–5751. 10.1021/nn3028997. [DOI] [PubMed] [Google Scholar]
- Kosuri S.; Church G. M. (2014) Large-Scale de Novo DNA Synthesis: Technologies and Applications. Nat. Methods 11, 499–507. 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M.; Sugiyama H. (2018) DNA Origami Nanomachines. Molecules 23, 1766. 10.3390/molecules23071766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.; Li H.; Wang L.; Gu H.; Fan C. (2018) DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 119, 6459–6506. 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- Keller A.; Linko V. (2020) Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem., Int. Ed. 10.1002/anie.201916390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K.; Veetil A. T.; Jaffrey S. R.; Krishnan Y. (2016) Nucleic Acid-Based Nanodevices in Biological Imaging. Annu. Rev. Biochem. 85, 349–376. 10.1146/annurev-biochem-060815-014244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana S.; Shenoy A. R.; Krishnan Y. (2015) Designing DNA Nanodevices for Compatibility with the Immune System of Higher Organisms. Nat. Nanotechnol. 10, 741–747. 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold K. E.; Lacroix A.; Sleiman H. F. (2018) DNA Nanostructures at the Interface with Biology. Chem. 4, 495–521. 10.1016/j.chempr.2018.02.005. [DOI] [Google Scholar]
- Modi S.; Swetha M. G.; Goswami D.; Gupta G. D.; Mayor S.; Krishnan Y. (2009) A DNA Nanomachine That Maps Spatial and Temporal pH Changes Inside Living Cells. Nat. Nanotechnol. 4, 325–330. 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- Surana S.; Bhat J. M.; Koushika S. P.; Krishnan Y. (2011) An Autonomous DNA Nanomachine Maps Spatiotemporal pH Changes in a Multicellular Living Organism. Nat. Commun. 2, 340. 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- Modi S.; Nizak C.; Surana S.; Halder S.; Krishnan Y. (2013) Two DNA Nanomachines Map pH Changes Along Intersecting Endocytic Pathways Inside the Same Cell. Nat. Nanotechnol. 8, 459–467. 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- Leung K.; Chakraborty K.; Saminathan A.; Krishnan Y. (2019) A DNA Nanomachine Chemically Resolves Lysosomes in Live Cells. Nat. Nanotechnol. 14, 176–183. 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia D.; Surana S.; Chakraborty S.; Koushika S. P.; Krishnan Y. (2011) A Synthetic Icosahedral DNA-Based Host-Cargo Complex for Functional In Vivo Imaging. Nat. Commun. 2, 339. 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- Bhatia D.; Arumugam S.; Nasilowski M.; Joshi H.; Wunder C.; Chambon V.; Prakash V.; Grazon C.; Nadal B.; Maiti P. K.; Johannes L.; Dubertret B.; Krishnan Y. (2016) Quantum Dot-Loaded Monofunctionalized DNA Icosahedra for Single-Particle Tracking of Endocytic Pathways. Nat. Nanotechnol. 11, 1112–1119. 10.1038/nnano.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veetil A. T.; Chakraborty K.; Xiao K.; Minter M. R.; Sisodia S. S.; Krishnan Y. (2017) Cell-Targetable DNA Nanocapsules for Spatiotemporal Release of Caged Bioactive Small Molecules. Nat. Nanotechnol. 12, 1183–1189. 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- Veetil A. T.; Jani M. S.; Krishnan Y. (2018) Chemical Control over Membrane-Initiated Steroid Signaling with a DNA Nanocapsule. Proc. Natl. Acad. Sci. U. S. A. 115, 9432–9437. 10.1073/pnas.1712792115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuya A.; Sakai Y.; Yamazaki T.; Xu Y.; Komiyama M. (2011) Nanomechanical DNA Origami “Single-Molecule Beacons” Directly Imaged by Atomic Force Microscopy. Nat. Commun. 2, 449. 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuya A.; Watanabe R.; Hashizume M.; Kaino M.; Minamida S.; Kameda K.; Ohya Y. (2014) Precise Structure Control of Three-State Nanomechanical DNA Origami Devices. Methods 67, 250–255. 10.1016/j.ymeth.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Ranallo S.; Porchetta A.; Ricci F. (2019) DNA-Based Scaffolds for Sensing Applications. Anal. Chem. 91, 44–59. 10.1021/acs.analchem.8b05009. [DOI] [PubMed] [Google Scholar]
- Ranallo S.; Rossetti M.; Plaxco K. W.; Vallée-Bélisle A.; Ricci F. (2015) A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins. Angew. Chem., Int. Ed. 54, 13214–13218. 10.1002/anie.201505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchetta A.; Ippodrino R.; Marini B.; Caruso A.; Caccuri F.; Ricci F. (2018) Programmable Nucleic Acid Nanoswitches for the Rapid, Single-Step Detection of Antibodies in Bodily Fluids. J. Am. Chem. Soc. 140, 947–953. 10.1021/jacs.7b09347. [DOI] [PubMed] [Google Scholar]
- Ranallo S.; Prévost-Tremblay C.; Idili A.; Vallée-Bélisle A.; Ricci F. (2017) Antibody-Powered Nucleic Acid Release Using a DNA-Based Nanomachine. Nat. Commun. 8, 15150. 10.1038/ncomms15150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti M.; Ranallo S.; Idili A.; Palleschi G.; Porchetta A.; Ricci F. (2017) Allosteric DNA Nanoswitches for Controlled Release of a Molecular Cargo Triggered by Biological Inputs. Chem. Sci. 8, 914–920. 10.1039/C6SC03404G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir Y.; Ben-Ishay E.; Levner D.; Ittah S.; Abu-Horowitz A.; Bachelet I. (2014) Universal Computing by DNA Origami Robots in a Living Animal. Nat. Nanotechnol. 9, 353–357. 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon S.; Dahan N.; Koren A.; Radiano O.; Ronen M.; Yannay T.; Giron J.; Ben-Ami L.; Amir Y.; Hel-Or Y.; Friedman D.; Bachelet I. (2016) Thought-Controlled Nanoscale Robots in a Living Host. PLoS One 11, e0161227. 10.1371/journal.pone.0161227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Jiang Q.; Liu S.; Zhang Y.; Tian Y.; Song C.; Wang J.; Zou Y.; Anderson G. J.; Han J. Y.; Chang Y.; Liu Y.; Zhang C.; Chen L.; Zhou G.; Nie G.; Yan H.; Ding B.; Zhao Y. (2018) A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger In Vivo. Nat. Biotechnol. 36, 258–264. 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- Juul S.; Iacovelli F.; Falconi M.; Kragh S. L.; Christensen B.; Frøhlich R.; Franch O.; Kristoffersen E. L.; Stougaard M.; Leong K. W.; Ho Y.-P.; Sørensen E. S.; Birkedal V.; Desideri A.; Knudsen B. R. (2013) Temperature-Controlled Encapsulation and Release of an Active Enzyme in the Cavity of a Self-Assembled DNA Nanocage. ACS Nano 7, 9724–9734. 10.1021/nn4030543. [DOI] [PubMed] [Google Scholar]
- Kohman R. E.; Cha S. S.; Man H. Y.; Han X. (2016) Light-Triggered Release of Bioactive Molecules from DNA Nanostructures. Nano Lett. 16, 2781–2785. 10.1021/acs.nanolett.6b00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold K. E.; Hsu J. C. C.; Sleiman H. F. (2016) Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of siRNA. J. Am. Chem. Soc. 138, 14030–14038. 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- Ijäs H.; Hakaste I.; Shen B.; Kostiainen M. A.; Linko V. (2019) Reconfigurable DNA Origami Nanocapsule for pH-Controlled Encapsulation and Display of Cargo. ACS Nano 13, 5959–5967. 10.1021/acsnano.9b01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi G.; Dalgaard Ebbesen Jepsen M.; Kjems J.; Andersen E. S. (2017) Control of Enzyme Reactions by a Reconfigurable DNA Nanovault. Nat. Commun. 8, 992. 10.1038/s41467-017-01072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J. H.; Moffitt R. A.; Stokes T. H.; Liu J.; Young A. N.; Nie S.; Wang M. D. (2009) Convergence of Biomarkers, Bioinformatics and Nanotechnology for Individualized Cancer Treatment. Trends Biotechnol. 27, 350–358. 10.1016/j.tibtech.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius F.; Kick B.; Behler K. L.; Honemann M. N.; Weuster-Botz D.; Dietz H. (2017) Biotechnological Mass Production of DNA Origami. Nature 552, 84–87. 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- Kielar C.; Xin Y.; Shen B.; Kostiainen M. A.; Grundmeier G.; Linko V.; Keller A. (2018) On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem., Int. Ed. 57, 9470–9474. 10.1002/anie.201802890. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S.; Ijäs H.; Linko V.; Keller A. (2018) Structural Stability of DNA Origami Nanostructures Under Application-Specific Conditions. Comput. Struct. Biotechnol. J. 16, 342–349. 10.1016/j.csbj.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bila H.; Kurisinkal E. E.; Bastings M. M. C. (2019) Engineering a Stable Future for DNA-Origami as a Biomaterial. Biomater. Sci. 7, 532–541. 10.1039/C8BM01249K. [DOI] [PubMed] [Google Scholar]
- Anastassacos F. M.; Zhao Z.; Zeng Y.; Shih W. M. (2020) Glutaraldehyde Cross-Linking of Oligolysines Coating DNA Origami Greatly Reduces Susceptibility to Nuclease Degradation. J. Am. Chem. Soc. 142, 3311–3315. 10.1021/jacs.9b11698. [DOI] [PubMed] [Google Scholar]
- Gerling T.; Kube M.; Kick B.; Dietz H. (2018) Sequence-Programmable Covalent Bonding of Designed DNA Assemblies. Sci. Adv. 4, eaau1157 10.1126/sciadv.aau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran A.; Endo M.; Katsuda Y.; Hidaka K.; Sugiyama H. (2011) Photo-Cross-Linking-Assisted Thermal Stability of DNA Origami Structures and Its Application for Higher-Temperature Self-Assembly. J. Am. Chem. Soc. 133, 14488–14491. 10.1021/ja204546h. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran A. R.; Vilcapoma J.; Dey P.; Wong-Deyrup S. W.; Dey B. K.; Halvorsen K. (2020) Exceptional Nuclease Resistance of Paranemic Crossover (PX) DNA and Crossover-Dependent Biostability of DNA Motifs. J. Am. Chem. Soc. 142, 6814–6821. 10.1021/jacs.0c02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopar A.; Coral L.; Di Giacomo S.; Adedeji A. F.; Castronovo M. (2018) Binary Control of Enzymatic Cleavage of DNA Origami by Structural Antideterminants. Nucleic Acids Res. 46, 995–1006. 10.1093/nar/gkx1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S.; Shen B.; Kostiainen M. A.; Grundmeier G.; Keller A.; Linko V. (2019) Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 20, 2818–2823. 10.1002/cbic.201900369. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Duan X.; Liu N. (2017) DNA-Nanotechnology-Enabled Chiral Plasmonics: From Static to Dynamic. Acc. Chem. Res. 50, 2906–2914. 10.1021/acs.accounts.7b00389. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Schreiber R.; Zhang H.; Govorov A.; Liedl T.; Liu N. (2014) Reconfigurable 3D Plasmonic Metamolecules. Nat. Mater. 13, 862–868. 10.1038/nmat4031. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Yang Y.; Duan X.; Stoll S.; Govorov A. O.; Sugiyama H.; Endo M.; Liu N. (2016) A Light-Driven Three-Dimensional Plasmonic Nanosystem That Translates Molecular Motion Into Reversible Chiroptical Function. Nat. Commun. 7, 10591. 10.1038/ncomms10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Urban M. J.; Idili A.; Ricci F.; Liu N. (2017) Selective Control of Reconfigurable Chiral Plasmonic Metamolecules. Sci. Adv. 3, e1602803 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck T.; Nicoli F.; Kuzyk A.; Liedl T. (2018) Sensing Picomolar Concentrations of RNA Using Switchable Plasmonic Chirality. Angew. Chem., Int. Ed. 57, 13495–13498. 10.1002/anie.201807029. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Nguyen M.; Natarajan A. K.; Nguyen V. H.; Kuzyk A. (2018) A DNA Origami-Based Chiral Plasmonic Sensing Device. ACS Appl. Mater. Interfaces 10, 44221–44225. 10.1021/acsami.8b19153. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Xin L.; Duan X.; Urban M. J.; Liu N. (2018) Dynamic Plasmonic System That Responds to Thermal and Aptamer-Target Regulations. Nano Lett. 18, 7395–7399. 10.1021/acs.nanolett.8b03807. [DOI] [PubMed] [Google Scholar]
- Wang M.; Dong J.; Zhou C.; Xie H.; Ni W.; Wang S.; Jin H.; Wang Q. (2019) Reconfigurable Plasmonic Diastereomers Assembled by DNA Origami. ACS Nano 13, 13702–13708. 10.1021/acsnano.9b06734. [DOI] [PubMed] [Google Scholar]
- Selnihhin D.; Sparvath S. M.; Preus S.; Birkedal V.; Andersen E. S. (2018) Multifluorophore DNA Origami Beacon as a Biosensing Platform. ACS Nano 12, 5699–5708. 10.1021/acsnano.8b01510. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Duan X.; Liu N. (2015) A Plasmonic Nanorod That Walks on DNA Origami. Nat. Commun. 6, 8102. 10.1038/ncomms9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M.; Both S.; Zhou C.; Kuzyk A.; Lindfors K.; Weiss T.; Liu N. (2018) Gold Nanocrystal-Mediated Sliding of Doublet DNA Origami Filaments. Nat. Commun. 9, 1454. 10.1038/s41467-018-03882-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P.; Urban M. J.; Both S.; Duan X.; Kuzyk A.; Weiss T.; Liu N. (2019) DNA-Assembled Nanoarchitectures with Multiple Components in Regulated and Coordinated Motion. Sci. Adv. 5, eaax6023 10.1126/sciadv.aax6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L.; Zhou C.; Duan X.; Liu N. (2019) A Rotary Plasmonic Nanoclock. Nat. Commun. 10, 5394. 10.1038/s41467-019-13444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Y.; Seelig G. (2011) Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat. Chem. 3, 103–113. 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Yurke B.; Toppari J. J.; Linko V.; Törmä P. (2008) Dielectrophoretic Trapping of DNA Origami. Small 4, 447–450. 10.1002/smll.200701320. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Dietz H.; Toppari J. J. (2015) Dielectrophoretic Trapping of Multilayer DNA Origami Nanostructures and DNA Origami-Induced Local Destruction of Silicon Dioxide. Electrophoresis 36, 255–262. 10.1002/elps.201400323. [DOI] [PubMed] [Google Scholar]
- Kroener F.; Traxler L.; Heerwig A.; Rant U.; Mertig M. (2019) Magnesium-Dependent Electrical Actuation and Stability of DNA Origami Rods. ACS Appl. Mater. Interfaces 11, 2295–2301. 10.1021/acsami.8b18611. [DOI] [PubMed] [Google Scholar]
- Soong R. K.; Bachand G. D.; Neves H. P.; Olkhovets A. G.; Craighead H. G.; Montemagno C. D. (2000) Powering an Inorganic Nanodevice with a Biomolecular Motor. Science 290, 1555–1558. 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- Lauback S.; Mattioli K. R.; Marras A. E.; Armstrong M.; Rudibaugh D. P.; Sooryakumar R.; Castro C. E. (2018) Real-Time Magnetic Actuation of DNA Nanodevices via Modular Integration with Stiff Micro-Levers. Nat. Commun. 9, 1446. 10.1038/s41467-018-03601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Goetzfried M. A.; Hidaka K.; You M.; Tan W.; Sugiyama H.; Endo M. (2015) Direct Visualization of Walking Motions of Photocontrolled Nanomachine on the DNA Nanostructure. Nano Lett. 15, 6672–6676. 10.1021/acs.nanolett.5b02502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List J.; Falgenhauer E.; Kopperger E.; Pardatscher G.; Simmel F. C. (2016) Long-Range Movement of Large Mechanically Interlocked DNA Nanostructures. Nat. Commun. 7, 12414. 10.1038/ncomms12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A. M.; Weig C.; Oswald P.; Frey E.; Fischer P.; Liedl T. (2016) Magnetic Propulsion of Microswimmers with DNA-Based Flagellar Bundles. Nano Lett. 16, 906–910. 10.1021/acs.nanolett.5b03716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A.; Günther K.; Sperling E.; Heerwig A.; Kick A.; Cichos F.; Mertig M. (2017) Concept, Synthesis, and Structural Characterization of DNA Origami Based Self-Thermophoretic Nanoswimmers. Phys. Status Solidi A 214, 1600957. 10.1002/pssa.201600957. [DOI] [Google Scholar]
- Heerwig A.; Schubel M.; Schirmer C.; Herms A.; Cichos F.; Mertig M. (2019) DNA Origami Ring Structures as Construction Element of Self-Thermophoretic Swimmers. Phys. Status Solidi A 216, 1970040. 10.1002/pssa.201970040. [DOI] [Google Scholar]
- Lund K.; Manzo A. J.; Dabby N.; Michelotti N.; Johnson-Buck A.; Nangreave J.; Taylor S.; Pei R.; Stojanovic M. N.; Walter N. G.; Winfree E.; Yan H. (2010) Molecular Robots Guided by Prescriptive Landscapes. Nature 465, 206–210. 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R.; Taylor S. K.; Stefanovic D.; Rudchenko S.; Mitchell T. E.; Stojanovic M. N. (2006) Behavior of Polycatalytic Assemblies in a Substrate-Displaying Matrix. J. Am. Chem. Soc. 128, 12693–12699. 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- Wickham S. F.; Endo M.; Katsuda Y.; Hidaka K.; Bath J.; Sugiyama H.; Turberfield A. J. (2011) Direct Observation of Stepwise Movement of a Synthetic Molecular Transporter. Nat. Nanotechnol. 6, 166–169. 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- Wickham S. F.; Bath J.; Katsuda Y.; Endo M.; Hidaka K.; Sugiyama H.; Turberfield A. J. (2012) A DNA-Based Molecular Motor That Can Navigate a Network of Tracks. Nat. Nanotechnol. 7, 169–173. 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- Liber A.; Tomov T. E.; Tsukanov R.; Berger Y.; Nir E. (2015) A Bipedal DNA Motor That Travels Back and Forth Between Two DNA Origami Tiles. Small 11, 568–575. 10.1002/smll.201402028. [DOI] [PubMed] [Google Scholar]
- Tomov T. E.; Tsukanov R.; Liber M.; Masoud R.; Plavner N.; Nir E. (2013) Rational Design of DNA Motors: Fuel Optimization Through Single-Molecule Fluorescence. J. Am. Chem. Soc. 135, 11935–11941. 10.1021/ja4048416. [DOI] [PubMed] [Google Scholar]
- Gu H.; Chao J.; Xiao S.-J.; Seeman N. C. (2010) A Proximity-Based Programmable DNA Nanoscale Assembly Line. Nature 465, 202–205. 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B.; Seeman N. C. (2006) Operation of a DNA Robot Arm Inserted into a 2D DNA Crystalline Substrate. Science 314, 1583–1585. 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thubagere A. J.; Li W.; Johnson R. F.; Chen Z.; Doroudi S.; Lee Y. L.; Izatt G.; Wittman S.; Srinivas N.; Woods D.; Winfree E.; Qian L. (2017) A Cargo-Sorting DNA Robot. Science 357, eaan6558 10.1126/science.aan6558. [DOI] [PubMed] [Google Scholar]
- Sherman W. B.; Seeman N. C. (2004) A Precisely Controlled DNA Biped Walking Device. Nano Lett. 4, 1203–1207. 10.1021/nl049527q. [DOI] [Google Scholar]
- Venkataraman S.; Dirks R. M.; Rothemund P. W. K.; Winfree E.; Pierce N. A. (2007) An Autonomous Polymerization Motor Powered by DNA Hybridization. Nat. Nanotechnol. 2, 490–494. 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- Yin P.; Choi H. M.; Calvert C. R.; Pierce N. A. (2008) Programming Biomolecular Self-Assembly Pathways. Nature 451, 318–322. 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Omabegho T.; Sha R.; Seeman N. C. (2009) A Bipedal DNA Brownian Motor with Coordinated Legs. Science 324, 67–71. 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterer P.; Willner E. M.; Dietz H. (2016) Nanoscale Rotary Apparatus Formed from Tight-Fitting 3D DNA Components. Sci. Adv. 2, e1501209 10.1126/sciadv.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminka G. A.; Spokoini-Stern R.; Amir Y.; Agmon N.; Bachelet I. (2017) Molecular Robots Obeying Asimov’s Three Laws of Robotics. Artif. Life 23, 343–350. 10.1162/ARTL_a_00235. [DOI] [PubMed] [Google Scholar]
- Asimov’s Three Laws of Robotics. L1: A robot may not injure a human being or, through inaction, allow a human being to come to harm; L2: A robot must obey the orders given it by human beings except where such orders would conflict with the First Law; L3: A robot must protect its own existence as long as such protection does not conflict with the First or Second Laws. See:; Asimov I. (2004) I, Robot, Bantam Books, New York, NY. [Google Scholar]
- DeLuca M.; Shi Z.; Castro C. E.; Arya G. (2020) Dynamic DNA Nanotechnology: Toward Functional Nanoscale Devices. Nanoscale Horiz. 5, 182–201. 10.1039/C9NH00529C. [DOI] [Google Scholar]