Abstract
Alpinia belongs to a large genus with many species found in Peninsular Malaysia. Several species of Alpinia exhibit important medicinal potential. However, progressive studies on the genus Alpinia were hampered by difficulties encountered in species identification. With the advancement achieved in genomic technology, more sensitive tools such as DNA barcoding were developed, which can be used for species identification. Internal Transcribe Spacer 2 (ITS2) is a DNA barcode which has proven to be a promising tool for species identification. The criterions of ITS2 efficacy namely universality and efficacy for species identification were tested on Alpinia species collected from Peninsular Malaysia. The results showed that a success rate of 96.97% was achieved using ITS2 for screening 11 species of Alpinia and an outgroup sample (Zingiber specatabile). Combined with 15 additional sequences from the Genbank for five Alpinia species, ITS2 demonstrated high species identification efficacy with 88.2% of species identified using phylogenetic and distance analysis. The analysis was further improved with the use of ITS2 secondary structure. The results of both criterions demonstrated the ability of ITS2 to successfully discriminate Alpinia species, which will help to improve species identification of Alpinia species in Peninsular Malaysia.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00868-1) contains supplementary material, which is available to authorized users.
Keywords: Alpinia, Peninsular Malaysia, DNA barcoding, ITS2, Molecular identification
Introduction
The genus Alpinia Roxb. is the largest genus in the Zingiberaceae (ginger) family with approximately 235 species (Govaerts et al. 2017). Several traits distinguish the genus from other genera in the Zingiberaceae family namely terminally borne inflorescence on the leafy stem, poorly developed or absence of lateral staminodes and typically attractive labellum. The genus has an extensive distribution range, which can be found growing in Japan, tropical and sub-tropical Asia, Australia and the Pacific Islands. There are currently 27 recognised Alpinia species in Peninsular Malaysia, 13 of which are endemic to the region (Govaerts et al. 2017). Alpinia species are small or large herbaceous plants, up to 2 m in height. Most species are found in understory of rainforest while some favour disturbed habitats (e.g. A. mutica and A. javanica) or wetlands (A. aquatica). Many species of Alpinia are rich in phytochemicals making them pharmaceutically important (Ghosh et al. 2013). Chemical constituents from species such as A. mutica, A. pahangensis and A. scabra exhibited cytotoxic activity towards cancerous cell (Phang et al. 2013; Reddy et al. 2013; Malek et al. 2011). Studies have demonstrated their cardomin property in treating inflammatory (Chow et al. 2012; Lee 2005). Extracted essential oil of A. pahangensis showed antimicrobial activity against five different Staphylococcus aureus strain (Awang et al. 2011).
Despite its potential applications, the studies on the genus Alpinia in Peninsular Malaysia were faced with several hurdles. As with many plant taxa, species identification for Alpinia is often difficult due to vague congeneric morphological characteristics especially in the rhizome form, phenotypic plasticity and short seasonal flowering periods (Shi et al. 2011; Vinitha et al. 2014). This could lead to counterfeit medicinal herbs which is a recurring problem in the pharmaceutical and food industry (Vinitha et al. 2014). Moreover, the population and distribution data on the genus are limited and remains unassessed for the International Union for Conservation of Nature Red List.
With deforestation being a looming threat in Peninsular Malaysia, many Alpinia species are at risk of extinction as many are only recorded in several restricted locations. As traditional morphological identification techniques might prove inadequate at times, DNA barcoding can be used to complement species identification for Alpinia species.
DNA barcoding utilizes short segments of DNA for rapid and standardized identification at the species level based on two criterions (Kress et al. 2005; Hollingsworth et al. 2011). A DNA barcode must remain conserved among taxa for species resolution by possessing universal primer sites while retaining sufficient genetic polymorphism to discriminate (Chase et al. 2007). In contrast to the success of mitochondrial Cytochrome Oxidase I (COI) in DNA barcoding efforts for animals, current DNA barcode for plants do not have the universality and resolving power equivalent to that of COI. Instead, evaluation for candidate DNA barcodes in plants identified several potential loci such as intron maturase K (matK) and RuBisCo (rbcL) or nuclear-encoded ribosomal internal transcribed spacer (ITS) or its shorter fragment ITS2 (Hollingsworth et al. 2009a). Each respective plant DNA barcode has their own strength and weaknesses resulting in varying levels of success for identification between taxa (Hollingsworth et al. 2009b). To identify plant based medicine, DNA barcode ITS2 is the preferred choice as it has good universality, small intraspecific variation, but high interspecific divergence, and a short fragment length (Chen et al. 2010).
Several studies have evaluated candidate DNA barcodes for Alpinia as part of a family wide study in China (Shi et al. 2011) and India (Vinitha et al. 2014), each with varying results. Shi et al. (2011) reported that the use of DNA barcode ITS2 had the highest species identification efficacy among six chosen barcode candidates for Zingiberaceae. Conversely, Vinitha et al. (2014) suggested the combination of matK and rbcL to be the better suited candidate for Zingiberaceae. As different results were observed, DNA barcode ITS2 must first be evaluated prior to establishing a DNA barcode library to aid in taxonomic identification of Alpinia species in Peninsular Malaysia. Thus, in this study, criterions of universality and efficacy of species identification were assessed to determine the suitability of ITS2 as a candidate DNA barcode for the genus Alpinia in Peninsular Malaysia.
Materials and methods
Plant sample collection and preparation
A total of 33 samples were collected from living specimens either at or in conjunction with Suriana Botanical Gardens, Penang as listed in Supplementary Table 1. For each species, leaves were handpicked from three individual plant to serve as biological replicates. However, Alpinia oxymitra and Alpinia suriana were sampled once as only a single individual plant was available. Leaves were dried in silica gel and stored at − 80 °C.
DNA extraction and PCR amplification
Genomic DNA was extracted using GeneAll® plant sv mini extraction kit (GeneAll, Seoul, South Korea) from the plant tissue with slight modifications to the protocol i.e. plant material was incubated at 65 °C for 15 min while periodically shaken every 5 min.
Primer design and annealing temperature for plant barcode ITS2 were adapted from Vinitha et al. (2014). The sequence for forward primer is 5′- AATTGCAGAATCCCGTGAAC-3′ and reverse primer is 5′-CTCGCCGTTACTAGGGGAAT-3′. Polymerase chain reaction (PCR) assays were performed in 25-μl reactions containing: 1 × Green GoTaq® Flexi Buffer (Promega Corporation, Madison, USA), 0.08 M of dNTP mix, 4 mM of MgCl2, 1 μM of both primers, 1.25 U of Taq polymerase (Promega Corporation, Madison, USA) and 50 ng of DNA. The PCR reaction cycle was as follows: initial denaturation at 95 °C for 5 min, 40 cycles of denaturation at 95 °C for 30 s, primer annealing at 58 °C for 40 s and DNA strand extension at 72 °C for 1 min with a final extension at 72 °C for 7 min. PCR products were purified using GeneAll® Expin™ PCR centrifugation protocol (GeneAll, Seoul, South Korea).
Sequencing and data analysis
Purified PCR products were sequenced in both directions by the commercial sequencing service company (First BASE Laboratories Sdn Bhd. Seri Kembangan, Malaysia). To improve species coverage of Alpinia in Peninsular Malaysia, additional ITS2 sequences of published Alpinia species were retrieved from GenBank. Species were only selected if there are at least three distinct accessions present in the GenBank as listed in Supplementary Table 1.
Nucleotide sequences were assembled and trimmed using Sequencher 5.4.6 (Gene Codes Corporation, Ann Arbor, Michigan, USA). The trimmed sequences were annotated for the true ITS2 region based on a Hidden Markov model (Keller et al. 2009) using the annotate tool on Internal Transcribed Spacer 2 Ribosomal RNA database. The DNA sequences of accessions were uploaded to GenBank and Barcode of Life data system. Edited DNA sequences were then aligned with MUSCLE (Edgar 2004) and manually improved with Jalview (Waterhouse et al. 2009). Sequence characteristics were computed using MEGA 7.0.
Genetic distances
Six parameters of genetic distances of ITS2 sequences were calculated based on Kimura 2-Parameter (K2P) model using MEGA7.0 (Kumar et al. 2016). The average inter-specific divergence, minimum inter-specific divergence and average theta prime were calculated based on K2P model to evaluate inter-specific divergence (Meyer and Paulay 2005). The average intra-specific distance, coalescent depth, and theta were calculated to determine the intra-specific variation based on the K2P model (Meyer and Paulay 2005). Differences between interspecific and intraspecific divergence were evaluated using Wilcoxon two-sample tests. Wilcoxon 2-sample test is a commonly non-parametric equivalent paired Student’s T test for simple statistical analysis.
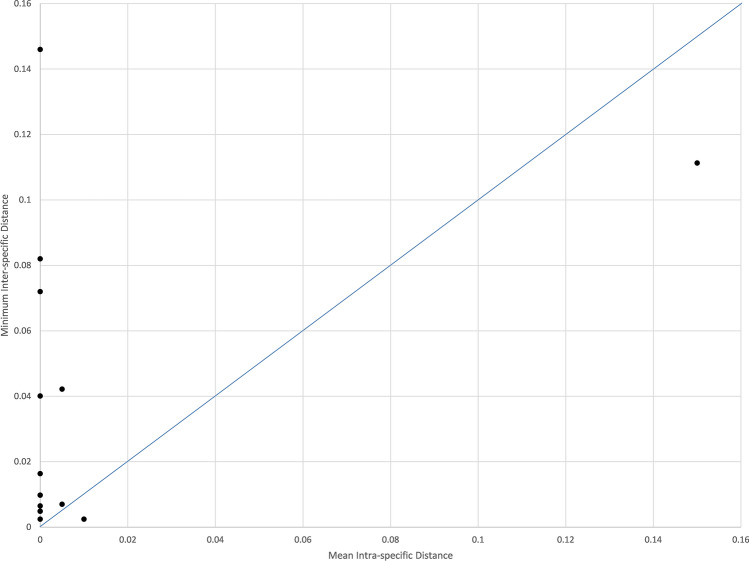
As recommended by Collins and Cruickshank (2013), barcode gap was analysed using a dot plot in which the maximum intraspecific distance (x-axis) was plotted against the nearest neighbour distance (y-axis). A barcode gap is present in a species if the dot is positioned above the 1:1 slope emerging from the origin of the graph.
Efficacy of species delineation
To determine the species identification efficacy of ITS2 as a DNA barcode for the genus Alpinia of Peninsular Malaysia, two different strategies were used namely tree-based and distance methods.
The tree-based approach was used to resolve the taxonomy of the sampled Alpinia species using different phylogenetic analysis strategies on all accessions of Alpinia along with outgroup sample Z. spectabile. A species is considered identified if it is recovered monophyletically, in which all accession of the same species is clustered. Phylogenetic trees were constructed using neighbour joining (NJ), and maximum likelihood (ML) using MEGA 7.0. Bootstrap support values were calculated by running 1000 bootstrap replicates of the data.
For the distance method, two parameters from TaxonDNA namely ‘best match’ and ‘best close match’ based on K2P model were computed using Species Identifier 1.8 software (Meier et al. 2006). Each parameter is defined below:
‘Best match’ module is used to determine the closest barcode for each query. An identification is deemed successful if both the sequences are of the same species; misidentification occurs when both sequences are mismatched; and scored ambiguous if multiple equally good best matches from different species were matched to the queried sequence.
‘Best close match’ module, a threshold below 95% of all intraspecific distances was computed from relative frequency of intraspecific distances. Queries will be scored unidentified if the match’s value exceeds the computed threshold. The remaining categories are like ‘best match’ module.
Secondary structures prediction
The ITS2 sequences were folded into its secondary structures using the predict tool in Internal Transcribed Spacer 2 Ribosomal RNA database (Wolf et al. 2005) with the default value of E-value cutoff (E < 1e−16).
Results
Amplification and sequencing
The universality of DNA barcode ITS2 was determined by the results of polymerase chain reaction (PCR) and sequencing. In this study, a total 32 new ITS2 sequences representing 11 species were successfully recovered. High PCR success (100%, n = 33) was observed for loci ITS2 using a single primer pair indicating good primer universality. The amplicon size produced was in congruent with the recovered size of 350 bp in Zingiberaceae (Vinitha et al. 2014) and the estimated size of ITS2 in monocots which ranges from a 100 to 480 bp (Yao et al. 2010). Conversely, sequencing success was slightly lower (96.97%, n = 33). The average sequence length of ITS2 produced from sampled accessions is 364 bp (n = 32) with the length of sequences ranging from 263 bp to 430 bp. To further improve the coverage of Alpinia species in Peninsular Malaysia, a total of 15 accessions representing 5 additional species were retrieved from GenBank as listed in Supplementary Table 1. Once annotated, aligned and trimmed, the sequences were 251 bp in length. The number of variable sequence sites found for ITS2 was 33.86%. The average GC content was 59.45% ranging from 56.33 to 63.52%.
Intra and inter-specific genetic divergence analyses and barcode gap
The genetic-divergence of the genus Alpinia from Peninsular Malaysia were estimated using MEGA 7.0 which were represented by six different parameters (average inter-specific distance, minimum inter-specific distance, theta prime, average intra-specific distance, coalescent depth, and theta) based on the K2P model as shown in Table 1. The calculated average intraspecific distance (0.002) is relatively lower than average interspecific distance (0.094). The differences between interspecific and intraspecific divergence were significant (p value = 0.00) according to the Wilcoxon two-sample test.
Table 1.
Analyses of inter-specific and intra-specific divergence of ITS2 sequences in 17 Alpinia species
| Measurement | K2P distance |
|---|---|
| Average inter-specific distance | 0.094 ± 0.037 |
| Theta Prime | 0.094 ± 0.037 |
| Minimum inter-specific distance | 0.000 ± 0.000 |
| Average intra-specific distance | 0.002 ± 0.003 |
| Theta | 0.003 ± 0.005 |
| Coalescent depth | 0.015 ± 0.008 |
An ideal barcode must display a ‘barcoding gap’ between inter- versus intra-specific divergences. The dot plot method recommended by Collins and Cruickshank (2013) was used to determine the presence of a ‘barcoding gap’. Dots positioned above the 1:1 slope indicates the presence of the barcode gap, while dots positioned below the 1:1 slope do not have a barcode gap. In this study, the barcode gap was present in 88% of the species as shown in Fig. 1.
Fig. 1.
DNA barcoding gap plotted through dot plot graph with minimum inter-specific distance against mean intra-specific distance. DNA barcoding gap is present for species above the 1:1 line
Efficacy of DNA barcode ITS2 in identifying Alpinia species
The efficacy of DNA barcode ITS2 in identifying Alpinia species from Peninsular Malaysia were evaluated using two different approaches namely tree and distance-based approach.
For the tree-based approach, species are considered successfully identified if sequences from the species are clustered monophyletically. Figures 2 and 3 show that both neighbour-joining and maximum likelihood tree were identical with 88.2% of species recovered monophyletically. However, the phylogenetic arrangement of species were different in both analysis. An ambiguous identification was observed for A. rafflesiana and A. suriana in both analysis as individuals from these species were clustered together.
Fig. 2.
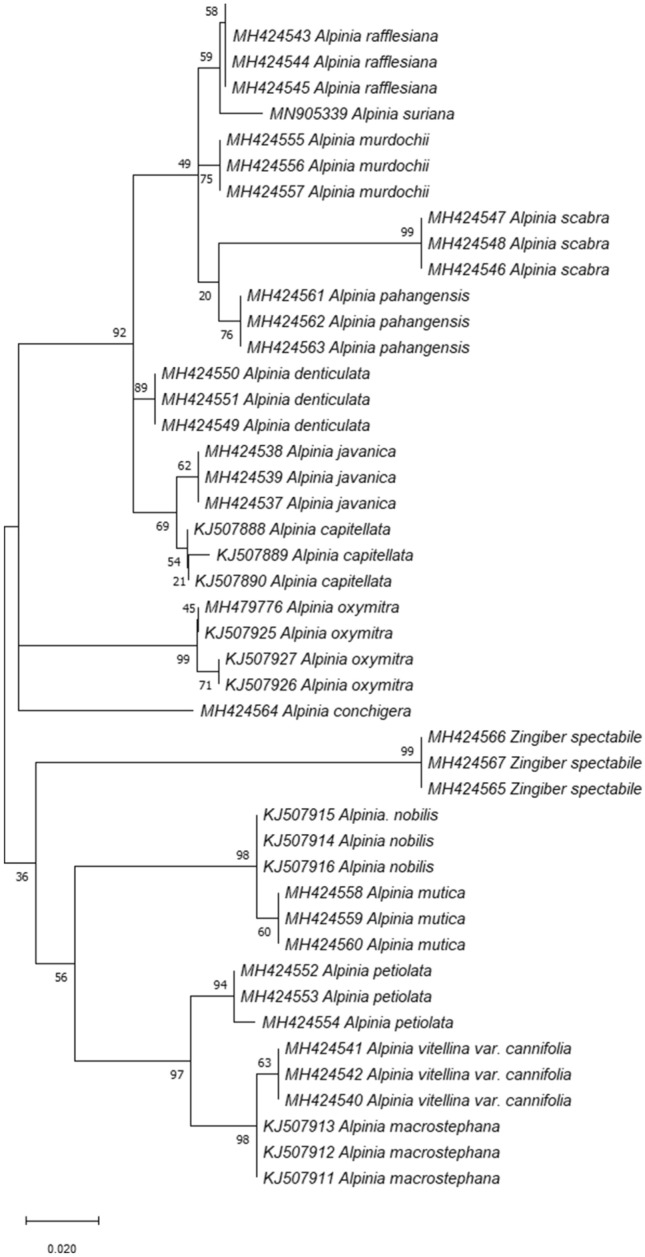
Neighbor-Joining (NJ) Tree of Alpinia species in Peninsular Malaysia constructed with ITS2. The bootstrap score (1000 replicates) are shown for each branch
Fig. 3.
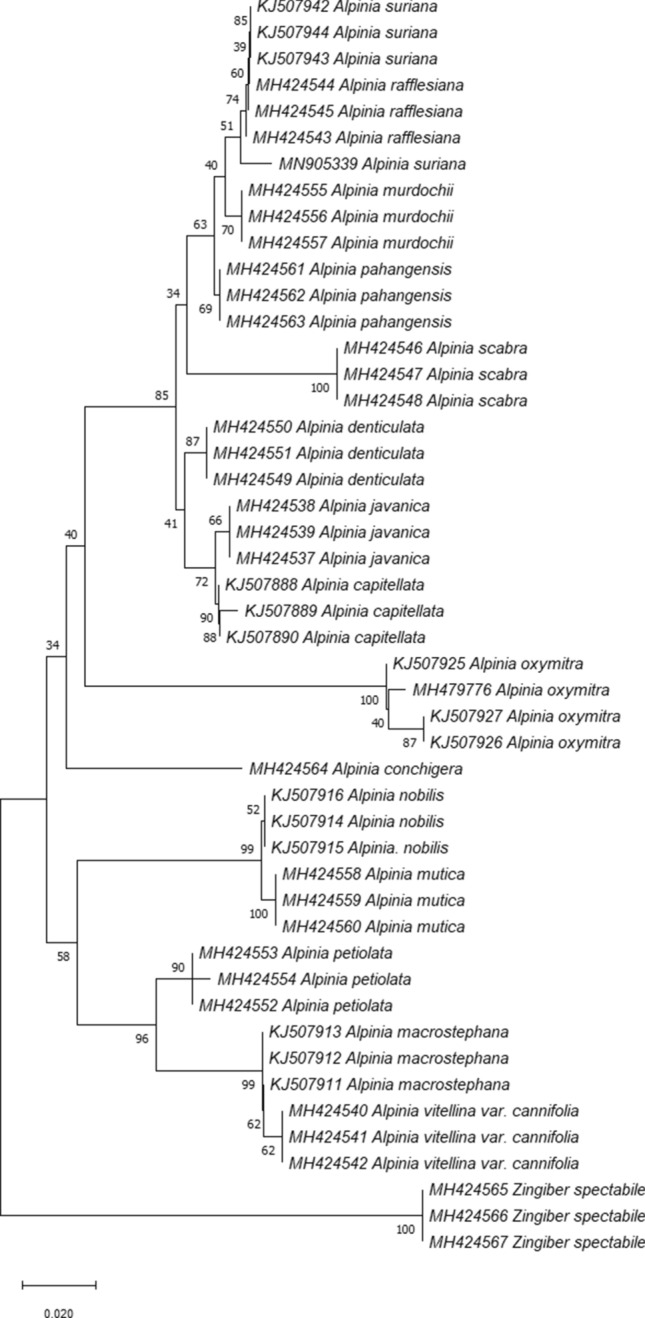
Maximum Likelihood (ML) Tree of Alpinia species in Peninsular Malaysia constructed with ITS2. The bootstrap score (1000 replicates) are shown for each branch
The accuracy of barcode assignment was evaluated using the distance-based approach with two parameters from TaxonDNA namely ‘best match’ and ‘best close match’. In both parameters, sequences will be assigned against others in the dataset simulating a species search. The results from both parameters were similar to the tree-based methods with 88.2% of species successfully identified. Moreover, the successfully identified speices were also similar, with A. rafflesiana and A. suriana remain ambiguously identified.
Secondary structure of ITS2 regions
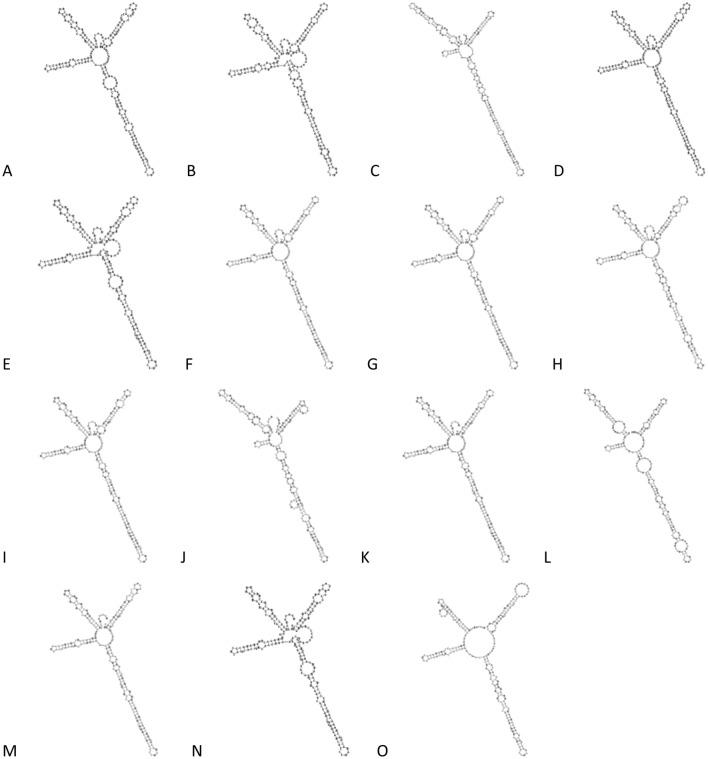
Besides using the DNA sequences of ITS2 for species identification, the variation found in the secondary structure of ITS2 can also be used. The ITS2 secondary structure for 14 Alpinia species and one Zingiber species were predicted and shown in Fig. 4. The secondary structures of ITS2 sequences for A. javanica and A. oxymitra could not be displayed as no reference models were found for them. All ITS2 secondary structures possess a central ring and four similar helices: Helix I, II, III, and IV. The central ring was different in size between the ITS2 secondary structure of Z. spectabile and Alpinia species. Variations in helices regions in terms of loop number, size, position and screw angle were observed between 12 Alpinia species. However, identical ITS2 secondary structures were predicted for A. macrostephana and A. vitellina var. cannifolia. From the data observed, ITS2 secondary structures predicted were able to delineate Alpinia species relatively well.
Fig. 4.
The ITS2 secondary structures of Alpinia species a A. aquatica, b A. capitellata, c A. conchigera, d A. denticulate, e A. macrostephana, f A. murdochii, g A. mutica, h A. nobilis, i A. pahangensis, j A. petiolata, k A. rafflesiana, l A. scabra, m A. suriana, n A. vitellina var cannifolia and o Z. spectabile
Discussion
As the pharmaceutical significance of Alpinia species continues to gain recognition, an improved method of species identification is needed as many Alpinia species are difficult to differentiate morphologically. The introduction of DNA barcoding to complement descriptive taxonomy is a viable solution as demonstrated by studies involving other taxa in Zingiberaceae (Vinitha et al. 2014; Shi et al. 2011). This is first accomplished by creating a DNA barcode database for example BOLD. Through this database, query sequences can be assigned to a list of known species for identification. Though there are several loci used in DNA barcoding, ITS2 is favored in the field of plant based traditional medicine due to its favorable characteristics namely good universality, small intraspecific but high interspecific variation, and a small fragment length (Chen et al. 2010; Yao et al. 2010). Previous studies have demonstrated the success of DNA barcode ITS2 in species identification for Zingiberaceae (Shi et al. 2011). To our current knowledge, the efficacy of DNA barcoding has not been tested on Alpinia species from Peninsular Malaysia. Based on the two criterions of universality and efficacy of species identification, DNA barcode ITS2 was found to be efficient and therefore appropriate to be used for identifying Alpinia species from Peninsular Malaysia.
Universality is an important characteristic of DNA barcoding as the recovery of the target loci must be simple, efficient and consistent to perform (Michel et al. 2016). This is determined by the success of PCR and quality of sequence produced. In this study, DNA barcode ITS2 demonstrated good universality with PCR being successful (100%) at recovering ITS2 amplicons from all samples using a single primer pair developed by Vinitha et al. (2014). The sequencing of ITS2 was also relatively successful (96.97%). High levels of universality were also observed in several plant taxa (Gu et al. 2013; Feng et al. 2016; Michel et al. 2016; Guo et al. 2017; Yu et al. 2017; Zhu et al. 2017). The presence of conserved flanking regions 5.8S and 28S around ITS2 promotes universal amplification in numerous plant taxa (Shi et al. 2011). Though good universality was observed in this study, the failed sequencing of two A. conchigera samples can be due to the presence of intragenomic heterogeneity of ITS2. This outcome is attributed to the hybrid origin or polyploidy nature of certain species of Zingiberaceae (Chen et al. 2015; Vinitha et al. 2014). By testing via a taxon-wise manner, the problem of intragenomic heterogeneity does not appear to be prevalent among Alpinia species in the dataset. Improvement in amplification and sequencing success was observed when a subsection of ITS namely ITS2 was used (Hollingsworth et al. 2011).
The success of species identification is determined by the selected locus having sufficient genetic variation and the method used for assessing the efficacy used (Collins and Cruickshank 2013). Sufficient genetic variability was found in ITS2 as both types of analysis were consistently successful in resolving 88.2% of the species sampled. Similar levels of species resolution were observed in other studies on Zingiberaceae (Shi et al. 2011; Vinitha et al. 2014) and plant taxa (Michel et al. 2016; Zhu et al. 2017; Yu et al. 2017; Guo et al. 2017). The ITS2 region was shown to have high genetic variability due to its high mutation rate allowing it to resolve a large range of species (Chen et al. 2010; Yao et al. 2010). In congruent with other studies, the use of ITS2 secondary structure improves species delineation (Yao et al. 2010; Feng et al. 2016). For example, distinct ITS2 secondary structures of A. rafflesiana and A. suriana allows for both species to be differentiated.
Though ITS2 performed well as a DNA barcode for resolving Alpinia species of Peninsular Malaysia, other DNA barcodes have yet to be tested on Alpinia species in this region. Differences in DNA barcode performance occur between plant species. In a study by Shi et al. (2011), ITS2 was an effective DNA barcode for Zingiberaceae but found to have poor universality when tested on Indian Zingiberaceae (Vinitha et al. 2014) and curcuma species from Myanmar and China (Chen et al. 2015). This is due to the presence of intragenomic heterogeneity in ITS2 which can be overcome by cloning, an unideal step in DNA barcoding (Hollingsworth et al. 2011). Although our study showed that intragenomic heterogeneity in ITS2 was not a prevalent issue in Alpinia species of Peninsular Malaysia, it would be beneficial for future studies to incorporate other DNA barcodes such as matK and rbcL for Alpinia species in Peninsular Malaysia.
Conclusion
In this study, efficacy of DNA barcode ITS2 was tested on Alpinia species from Peninsular Malaysia based on two criterions namely universality and species resolution. DNA barcode ITS2 was found to possess good levels of universality after screening through 11 species using a single primer pair designed by Vinitha et al. (2014). Good levels of species identification were observed using ITS2 with an 88.2% success rate using two different methods of analysis. This is further improved when ITS2 secondary structures were used for species delineation. Based on the results observed, ITS2 appears as a suitable candidate for use in the species identification of Alpinia species in Peninsular Malaysia. This work will allow the establishment of an ITS2 library to aid species identification for Alpinia species in Peninsular Malaysia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1: Ascension of Alpinia species in this study. (DOCX 16 kb)
Acknowledgements
We would like to thank Datuk Seri Dr Lim Chong Keat from the Suriana Botanical Gardens, Penang, Malaysia, for kindly provided us with the Alpinia plant samples. This work was supported by the University of Nottingham Malaysia campus internal grant. Many thanks to volunteers, Boey Jian Sheng, Stephane Arul Mariampilai, Kartik Kanasarmurthy, Kong Wei Yang and Henrik Seyersted for participating in the field sampling.
Authors contribution
CCF conceptualized and designed the experiments. WHT performed the experiments with assistance from LCC. WHT, LCC and CCF analysed the data. WHT and CCF wrote and edited the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Awang K, Ibrahim H, Rosmy Syamsir D, et al. Chemical constituents and antimicrobial activity of the leaf and rhizome oils of Alpinia pahangensis Ridl., an endemic wild ginger from Peninsular Malaysia. Chem Biodivers. 2011;8:668–673. doi: 10.1002/cbdv.201000225. [DOI] [PubMed] [Google Scholar]
- Chase MW, Cowan RS, Hollingsworth PM, et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. doi: 10.1002/tax.562004. [DOI] [Google Scholar]
- Chen S, Yao H, Han J, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:1–8. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao J, Erickson DL, et al. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol Ecol Resour. 2015;15:337–348. doi: 10.1111/1755-0998.12319. [DOI] [PubMed] [Google Scholar]
- Chow YL, Lee KH, Vidyadaran S, et al. Cardamonin from Alpinia rafflesiana inhibits inflammatory responses in IFN-γ/LPS-stimulated BV2 microglia via NF-κB signalling pathway. Int Immunopharmacol. 2012;12:657–665. doi: 10.1016/j.intimp.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Collins RA, Cruickshank RH. The seven deadly sins of DNA barcoding. Mol Ecol Resour. 2013;13:969–975. doi: 10.1111/1755-0998.12046. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jiang M, Shi Y, et al. Application of the ribosomal DNA ITS2 region of physalis (Solanaceae): DNA barcoding and phylogenetic study. Front Plant Sci. 2016;7:1–11. doi: 10.3389/fpls.2016.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Indukuri K, Bondalapati S, et al. Unveiling the mode of action of antibacterial labdane diterpenes from Alpinia nigra (Gaertn.) B. L. Burtt seeds. Eur J Med Chem. 2013;66:101–105. doi: 10.1007/s13205-012-0089-x. [DOI] [PubMed] [Google Scholar]
- Govaerts, R., Newman M., Lock JM (2017) World checklist of Zingiberaceae. In: Bot R (ed) Gard. Kew. http://wcsp.science.kew.org/advsearch.do. Accessed 21 Dec 2017
- Gu W, Song J, Cao Y, et al. Application of the ITS2 region for barcoding medicinal plants of selaginellaceae in pteridophyta. PLoS ONE. 2013;8:2–9. doi: 10.1371/journal.pone.0067818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ren L, Pang X. Inspecting the true identity of herbal materials from Cynanchum using ITS2 barcode. Front Plant Sci. 2017;8:1–10. doi: 10.3389/fpls.2017.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth ML, Andra Clark A, Forrest LL, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009;9:439–457. doi: 10.1111/j.1755-0998.2008.02439.x. [DOI] [PubMed] [Google Scholar]
- Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim K-J, Kress WJ, Schneider H, van AlphenStahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacon J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim Y-D, Lahaye R, Lee H-L, Long DG, Madrinan S, Maurin O, Meusnier I, Newmaster SG, Park C-W, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi D-K, Little DP. A DNA barcode for land plants. Proc Natl Acad Sci. 2009;106(31):12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS ONE. 2011;6(5):e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Schleicher T, Schultz J, et al. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, et al. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7.0: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H. Blockade of nuclear factor-B signaling pathway and anti-inflammatory activity of cardamomin, a chalcone analog from Alpinia conchigera. J Pharmacol Exp Ther. 2005;316:271–278. doi: 10.1124/jpet.105.092486. [DOI] [PubMed] [Google Scholar]
- Malek SNA, Phang CW, Ibrahim H, et al. Phytochemical and cytotoxic investigations of Alpinia mutica rhizomes. Molecules. 2011;16:583–589. doi: 10.3390/molecules16010583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Shiyang K, Vaidya G, Ng PKL. DNA barcoding and taxonomy in diptera: a tale of high intraspecific variability and low identification. Syst Biol. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:1–10. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel CI, Meyer RS, Taveras Y, Molina J. The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J Appl Res Med Aromat Plants. 2016;3:94–100. doi: 10.1016/j.jarmap.2016.02.002. [DOI] [Google Scholar]
- Phang C, Nurestri S, Malek A, Ibrahim H. Antioxidant potential, cytotoxic activity and total phenolic content of Alpinia pahangensis rhizomes. BMC Complement Altern Med. 2013;13:1. doi: 10.1186/1472-6882-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Abd Malek SN, Ibrahim H, Sim KS. Cytotoxic effect of Alpinia scabra (Blume) Náves extracts on human breast and ovarian cancer cells. BMC Complement Altern Med. 2013;13:314. doi: 10.1186/1472-6882-13-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LC, Zhang J, Han JP, et al. Testing the potential of proposed DNA barcodes for species identification of Zingiberaceae. J Syst Evol. 2011;49:261–266. doi: 10.1111/j.1759-6831.2011.00133.x. [DOI] [Google Scholar]
- Vinitha MR, Kumar US, Aishwarya K, et al. Prospects for discriminating Zingiberaceae species in India using DNA barcodes. J Integr Plant Biol. 2014;56:760–773. doi: 10.1111/jipb.12189. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Achtziger M, Schultz J, Dandekar T, Müller T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA. 2005;11:1616–1623. doi: 10.1261/rna.2144205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Song J, Liu C, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010 doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Wei YL, Zhang X, et al. Barcode ITS2: a useful tool for identifying Trachelospermum jasminoides and a good monitor for medicine market. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-04674-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R-W, Li Y-C, Zhong D-L, Zhang J-Q. Establishment of the most comprehensive ITS2 barcode database to date of the traditional medicinal plant Rhodiola (Crassulacaee) Sci Rep. 2017;7:10051. doi: 10.1038/s41598-017-09769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Ascension of Alpinia species in this study. (DOCX 16 kb)