Abstract
Deep convolutional neural networks are emerging as the state of the art method for supervised classification of images also in the context of taxonomic identification. Different morphologies and imaging technologies applied across organismal groups lead to highly specific image domains, which need customization of deep learning solutions. Here we provide an example using deep convolutional neural networks (CNNs) for taxonomic identification of the morphologically diverse microalgal group of diatoms. Using a combination of high-resolution slide scanning microscopy, web-based collaborative image annotation and diatom-tailored image analysis, we assembled a diatom image database from two Southern Ocean expeditions. We use these data to investigate the effect of CNN architecture, background masking, data set size and possible concept drift upon image classification performance. Surprisingly, VGG16, a relatively old network architecture, showed the best performance and generalizing ability on our images. Different from a previous study, we found that background masking slightly improved performance. In general, training only a classifier on top of convolutional layers pre-trained on extensive, but not domain-specific image data showed surprisingly high performance (F1 scores around 97%) with already relatively few (100–300) examples per class, indicating that domain adaptation to a novel taxonomic group can be feasible with a limited investment of effort.
Subject terms: Optical imaging, Marine biology, Software, Scientific data, Software, High-throughput screening
Introduction
Diatoms are microscopic algae possessing silicate shells called frustules1. They inhabit marine and freshwater environments as well as terrestrial habitats. Taxonomic composition of their assemblages is routinely assessed using light microscopy in ecological, bioindication and paleoclimate research2–4. Silicate frustules cleaned of organic material and embedded into high refractive index mountant on cover slips represent the most widely used type of microscopic preparations for such analyses5,6. Attempts to computerize parts or the whole of this workflow have been made repeatedly, starting with Cairns7, and in the most complete manner so far by the ADIAC project8, motivated by the desire to speed up the taxonomic enumeration process, to reduce its dependence on highly trained taxonomic experts, and to make identification results more reproducible and transparent. Recently, we described an updated re-implementation of most parts of this workflow, covering high throughput microscopic imaging, segmentation and outline shape feature extraction of diatom specimens9,10 which we since mainly applied in morphometric investigations11–14. The missing component in this workflow has been automated or computer-assisted taxonomic identification.
For this purpose, i.e. image classification in a taxonomic context, deep convolutional neural networks (CNNs) are currently becoming the state-of-the-art technique. Due to the broadening availability of high throughput, in part, in situ, imaging platforms15–18, and large publicly available image sets19, marine plankton has probably been addressed most commonly in such attempts in the aquatic realm20–23. The attention, however, recently also broadened to fossil foraminifera24, radiolarians25, as well as diatoms26,27. Due to the availability of deep learning software libraries28,29, well performing network architectures pre-trained on massive data sets like ImageNet30, and experiences accumulating related to transfer learning, i.e., application of pre-trained networks upon smaller data sets from a specialized image domain, the utilization of deep CNNs for a particular labelled image library is now within reach of taxonomic specialists of individual organismic groups.
In the case of diatom analyses, most studies thus far have addressed individual aspects of the taxonomic enumeration workflow in isolation, such as image acquisition by slide scanning31, diatom detection, segmentation and contour extraction9,32,33, or taxonomic identification26,34,35. Although all these aspects have been considered in detail by ADIAC8 and, with the exception of the final identification step, in our recent work10, a practicable end-to-end digital diatom analysis workflow has not emerged thus far. In this work, we introduce substantial further developments to these previously described workflows, now covering all aspects from imaging to deep learning-based classification, and apply it in a low diversity diatom habitat, the pelagic Southern Ocean, harbouring a unique and paleo-oceanographically and biogeochemically interesting diatom flora36–40.
The so far most extensive work on the application of deep learning on taxonomic diatom classification from brightfield micrographs26 tested only one CNN architecture to investigate the effects of training set size, histogram normalization and a coarse object segmentation that aimed more for a figure ground separation than for an exact segmentation.
We propose a procedure combining high resolution focus-enhanced light microscopic slide scanning, web-based taxonomic annotation of gigapixel-sized “virtual slides”, and highly customized and precise object segmentation, followed by CNN-based classification. In a transfer learning experiment employing a full factorial design varying CNN architecture, data set size, background masking and out-of-set testing (i.e. using data from different sampling campaigns for training and prediction), we address the questions (1) how well do different CNN architectures perform on the task of diatom classification; (2) to what extent does the increase in the size of training image sets improve transfer learning performance; (3) to what extent does a precise segmentation of diatom frustules influence classification performance; (4) to what extent is a CNN trained on one sample set (in this case, expedition) applicable to samples obtained from a different set.
Material and methods
Sampling and preparation
Samples were obtained by 20 µm mesh size plankton nets from ca. 15 to 0 m depth during two summer Polarstern expeditions ANT-XXVIII/2 (Dec. 2011–Jan. 2012, https://pangaea.de/?q=ANT-XXVIII%2F2) and PS103 (Dec. 2016–Jan. 2017, https://pangaea.de/?q=PS103). In both cases, a north to south transect from around the Subantarctic Front into the Eastern Weddell Sea was sampled, roughly following the Greenwich meridian, covering a range of Subantarctic and Antarctic surface water masses. To obtain clean siliceous diatom frustules, the samples were oxidized using hydrochloric acid and potassium permanganate after Simonsen41 and mounted on coverslips on standard microscopic slides in Naphrax resin (Morphisto GmbH, Frankfurt am Main, Germany).
Digitalization
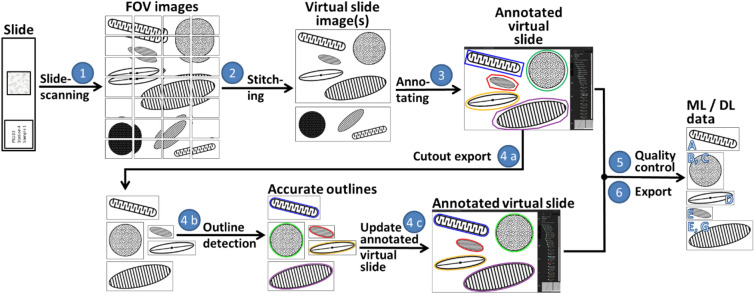
For converting these physical diatom samples into digital machine/deep learning data sets, we developed an integrated workflow consisting of the following steps (numbering refers to Fig. 1)
Slide scanning: Utilizing brightfield microscopy, we imaged a continuous rectangular region per slide, mostly ca. 5 × 5 mm2, in the form of several thousand overlapping field-of-view images (FOVs), where the scanned area usually contained hundreds to thousands of individual objects, mostly diatom frustules. For each FOV, at distances of one µm each, 80 focal planes were imaged and combined into one focus-enhanced image to overcome depth of field limitations. This technique is referred to as focus stacking and allows to observe a frustule's surface structure as well as its outline at the same time. Scanning and stacking were performed utilizing a Metafer slide scanning system (MetaSystems Hard & Software GmbH, Altlussheim, Germany) equipped with a CoolCube 1 m monochrome CCD camera (MetaSystems GmbH) and a high resolution/high magnification objective (Plan-APOCHROMAT 63x/1.4, Carl Zeiss AG, Oberkochen, Germany) with oil immersion (Immersol 518 F, Carl Zeiss AG, Oberkochen, Germany). This resulted in FOV images of 1,360 × 1,024 pixels at a resolution of 0.10 × 0.10 µm2/pixel. Device-dependent settings are detailed in10.
Slide stitching: The several thousand individual FOVs obtained during step 1 were combined into so-called virtual slides, gigapixel images capturing large portions of the scanned microscope slide at a resolution of ca. 0.1 × 0.1 µm2 per pixel. These were produced by a process called stitching, for which we used two different approaches. The Metafer VSlide Software (version 1.1.101) was applied to the PS103 scans, but produced misalignment artefacts (see Supplement I), frequently creating so-called ghosting (objects appeared doubled and shifted by a few pixels) and sometimes substantial displacement of FOVs, causing parts of the virtual slide image missing. As a consequence, during the course of the project we developed a method combining two ImageJ/FIJI plugins, MIST42 for exact alignment of FOV images and Grid/collection stitching43 for blending them into one large virtual slide image, which led to less stitching artefacts specifically for diatom slides. The scans from ANT-XXVIII/2 were processed using this stitching method.
Collaborative annotation: The virtual slide images were uploaded to the BIIGLE 2.0 web service44 for collaborative image annotation of objects of interest (OOIs). This term refers to all object categories used in the labelling process, in our case diatom frustules and valves (the frontal plates of a frustule) from various species and genera, diatom girdlebands (the radial parts of a frustule) and silicate skeletons or shells of non-diatom organisms like silicoflagellates or radiolarians. The OOIs were marked manually using BIIGLE’s annotation tool. Since following object boundaries precisely is very time-consuming, the OOI outlines were sketched only very roughly by a bounding box or a simple polygonal approximation. Objects distorted substantially by stitching artefacts (see step 2) were skipped. Predefined labels, mostly specifying names of Southern Ocean diatom taxa imported from WORMS45, were attached by four users, two of them (B.B., M.K.) being among the authors of this work. In some cases, multiple users annotated the same object, either agreeing or disagreeing on previously attached labels, where taxonomic disagreement is not uncommon11. This issue was resolved during data export (step 6).
Outline refinement: The roughly marked object outlines were refined to the exact object shape utilizing the semi-automatic segmentation feature of the diatom morphometry software SHERPA9. To this end, cut-outs depicting individual OOIs were produced from the virtual slide images by our software SHERPA2BIIGLE (step 4a). These cut-outs were processed with SHERPA for computing the actual object outlines, where faulty segmentations were refined manually (step 4b). Using another function of SHERPA2BIIGLE, these accurate segmentations were then uploaded into BIIGLE to replace the roughly marked object outlines (step 4c).
Quality control: The BIIGLE Largo46 feature (see Supplement II), as well as SHERPA2BIIGLE, were used to validate annotated labels. BIIGLE Largo enabled inspecting a large number of objects marked with a certain label simultaneously by displaying a series of scaled-down thumbnail images, whilst SHERPA enabled scrutinizing such objects one at a time at their original resolution/screen size. Erroneous label assignments were then corrected.
Data export: For each annotated OOI, a rectangular cut out was extracted with a minimum margin of 10 pixels, utilizing SHERPA2BIIGLE. Cut-outs were produced with and without background masking in order to study the effect on the classification (see below). If background masking was applied, the background (i.e. the area outside the annotated object) was replaced by the average background grey value, with a smooth transition close to the object boundary. Labels and metadata were exported in CSV format. Downstream processing was executed by R47 scripts (R version 3.6.1). The most important steps here were filtering annotations according to specific labels and defining the gold standard if multiple diverging labels had been attached to the same object, in which case the label attached by the senior expert (B.B.) was used.
Figure 1.
Workflow for generating annotated machine learning/deep learning data from physical slide specimens. Slides are scanned using a high resolution oil immersion objective as overlapping fields-of-view (1), those are stitched together to virtual slide images (2), which are uploaded to BIIGLE for annotating objects of interest (3), in our case diatom valves. The manually defined, rough object outlines can optionally be refined making use of SHERPA and SHERPA2BIIGLE (4a-c). After quality control using BIIGLE Largo or SHERPA2BIIGLE (5), cut-outs of annotated areas were exported along with label data (6) to assemble machine/deep learning data sets.
Data
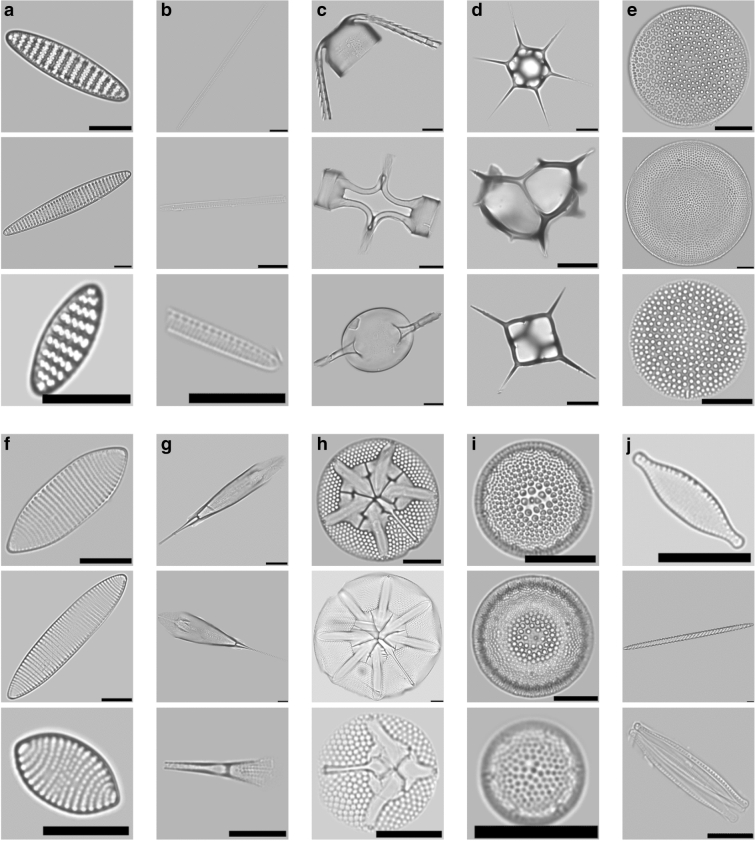
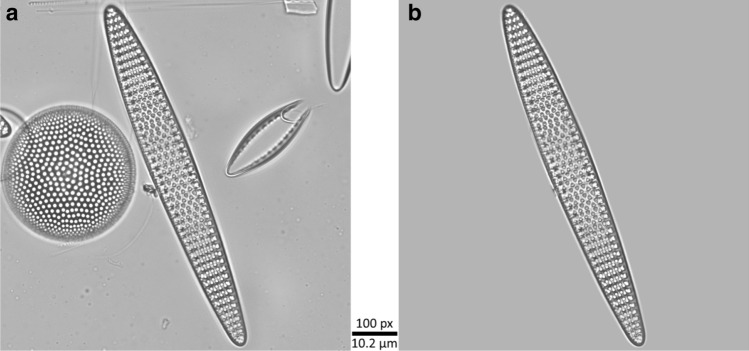
Using the protocol described above, we were able to collect annotations for nearly 10,000 OOIs of 51 different classes, originating from 26 virtual slides representing 19 physical slides. To allow for a sound comparison of the two expeditions ANT-XXVIII/2 and PS103, we limited this work to those 10 classes for which at least 40 specimens were present in each of the expeditions. Objects representing the diatom Fragilariopsis kerguelensis, which in the raw data accounted for nearly 40% of all specimens, were randomly subsampled to 660 specimens to reduce imbalance. This resulted in a total of 3,319 specimens from four diatom species, five diatom genera and the non-diatom taxon silicoflagellates (Table 1, Fig. 2). An example for a typical cut-out with and without background masking is given in Fig. 3.
Table 1.
Base data set composition.
| Class | ntotal | nANT-XXVIII/2 | nPS103 |
|---|---|---|---|
| Fragilariopsis kerguelensis | 660 | 418 | 242 |
| Pseudonitzschia | 520 | 173 | 347 |
| Chaetoceros | 434 | 82 | 352 |
| Silicoflagellate | 342 | 177 | 165 |
| Thalassiosira lentiginosa | 311 | 89 | 222 |
| Fragilariopsis rhombica | 272 | 57 | 215 |
| Rhizosolenia | 227 | 153 | 74 |
| Asteromphalus | 223 | 63 | 160 |
| Thalassiosira gracilis | 212 | 88 | 124 |
| Nitzschia | 118 | 76 | 42 |
| ∑ | 3,319 | 1,376 | 1,943 |
Figure 2.
Three typical representatives of each annotated class, illustrating their variability in size and morphology: (a) Fragilariopsis kerguelensis, (b) Pseudonitzschia, (c) Chaetoceros, (d) Silicoflagellate, (e) Thalassiosira lentiginosa, (f) Fragilariopsis rhombica, (g) Rhizosolenia, (h) Asteromphalus, (i) Thalassiosira gracilis, (j) Nitzschia. Black scale bars represent a width of 100 pixels or 10.2 µm, resp.
Figure 3.
Example of a typical cut-out without (a) and with background masking (b). The diatom in the centre of both cut-outs is Fragilariopsis kerguelensis.
This enabled us to generate data sets (Table 2, Fig. 2) to design a deep learning-based classification. The data sets A100,− (ANT-XXVIII/2, without background masking) and A100,+ (ANT-XXVIII/2, with background masking) contain 1,376 original cut-outs, the data sets B100,− (PS103, without background masking) and B100,+ (PS103, with background masking) contain 1,943 original cut-outs (Table 1; instructions on downloading the original data are given in Supplement III). For splitting the data into training/validation/test sets, sampling was performed class-wise to ensure an even split of each class between the sets.
Table 2.
Data set composition and denomination.
| Expedition | ANT-XXVIII/2 | PS103 |
|---|---|---|
| Base data set (both expeditions merged), without background masking | AB100,− | |
| Full data set, without background masking | A100,− | B100,− |
| Full data set, with background masking | A100,+ | B100,+ |
| Data subset 10%, without background masking | A10,− | B10,− |
| Data subset 10% with background masking | A10,+ | B10,+ |
Experiments
The machine learning community continuously proposes new deep learning network architectures. In this work, we have tested a set of nine convolutional neural network architectures (Table 3)48–53. These were applied to learn diatom classification from the datasets described above using the KERAS default application models54. For each model, the pre-defined convolutional base with frozen weights pre-trained on ImageNet data55 was used as a basis and a new fully connected classifier module with a final softmax classification was trained on top of it. This approach usually is referred to as simple implementation of transfer learning without fine-tuning. Weights were adapted using the Adam optimizer56. We conducted the experiments utilizing the R interface to KERAS V2.2.4.157. The input image intensity values were scaled to [0:1] and training data were augmented by rotation, shift, shear, zoom and flipping, using the functionality provided by the KERAS image data generator (see file “03-CNN functions.R” provided in Supplement III for details). The models were trained for 50 epochs, which for all investigated CNN architectures was sufficient to prohibit over- as well as underfitting. A batch size of 32 was used for the 100% data sets, and a batch size of 8 for the 10% data sets. All scripts were written in R and run on a Windows 10 system equipped with a nVidia Quadro P2000 GPU. R scripts are provided in Supplement III.
Table 3.
CNN architectures.
| Model | CNN convolutional basea | Classification layer(s) b | Input shape |
|---|---|---|---|
| VGG16_1FC | VGG16 | One 256 neuron dense layer | 224 × 224 |
| VGG16_2FC | VGG16 | Two 256 neuron dense layers | 224 × 224 |
| VGG19_1FC | VGG19 | One 256 neuron dense layer | 224 × 224 |
| VGG19_2FC | VGG19 | Two 256 neuron dense layers | 224 × 224 |
| Xception | Xception | Global average pooling 2d | 299 × 299 |
| DenseNet | DenseNet | Global average pooling 2d | 224 × 224 |
| InceptionResNetv2 | Inception-ResNet V2 | Global average pooling 2d | 299 × 299 |
| MobileNetV2 | MobileNet V2 | Global average pooling 2d | 224 × 224 |
| InceptionV3 | Inception V3 | Global average pooling 2d | 299 × 299 |
aFrozen, pre-trained on ImageNet data.
bTrained on our data for 50 epochs.
Classification performance was assessed by micro- and macro-averaged F1 scores according to
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
With = number of true positives, = number of false positives, = number of false negatives, = number of classes. Subindex “” refers to values per individual class, “” to micro-averaged values, “” to macro-averaged values.
If a class was not predicted (i.e. = 0), was set to 0 to allow for calculation of .
In order to address the questions raised in the introduction, we conducted 17 experiments (Table 4), one to test our data and setup and 16 to study the effects of (a) small/large number of training samples, (b) background masking and (c) possible concept drifts between data sets collected at similar geographic locations, but during different expeditions (i.e. in different years, Table 2, Fig. 4) and processed with different stitching methods. The individual data sets are referred to as shown in Table 2. The machine learning experiments and the results (as shown in Tables 4, 7, Fig. 4) are referred to as follows: the first part of the designation (left of “|”) denominates the data set used for training/validation, whilst the second part denotes the data set used for testing prediction performance. The test data were never used during the training and optimization of the network and thus are disjunct from the training/validation data, but were collected and prepared with identical parameters and conditions, with the exception of the expedition when indicated, which also implies application of a different stitching method.
Table 4.
Deep learning experiments.
| Row | Experiment | Training/validation data | Test data | Portion (%) | Background masking | Replication | Batch size |
|---|---|---|---|---|---|---|---|
| 1 | A100,−|A100,− | A | A | 100 | No | Fourfold cross validation | 32 |
| 2 | B100,−|B100,− | B | B | 100 | No | 32 | |
| 3 | A100,+|A100,+ | A | A | 100 | Yes | 32 | |
| 4 | B100,+|B100,+ | B | B | 100 | Yes | 32 | |
| 5 | A10,−|A10,− | A | A | 10 | No | 8 | |
| 6 | B10,−|B10,− | B | B | 10 | No | 8 | |
| 7 | A10,+|A10,+ | A | A | 10 | Yes | 8 | |
| 8 | B10,+|B10,+ | B | B | 10 | Yes | 8 | |
| 9 | A100,−|B100,− | A | B | 100 | No | 3 replicates | 32 |
| 10 | B100,−|A100,− | B | A | 100 | No | 32 | |
| 11 | A100,+|B100,+ | A | B | 100 | Yes | 32 | |
| 12 | B100,+|A100,+ | B | A | 100 | Yes | 32 | |
| 13 | A10,−|B10,− | A | B | 10 | No | 5 replicates | 8 |
| 14 | B10,−|A10,− | B | A | 10 | No | 8 | |
| 15 | A10,+|B10,+ | A | B | 10 | Yes | 8 | |
| 16 | B10,+|A10,+ | B | A | 10 | Yes | 8 |
Figure 4.
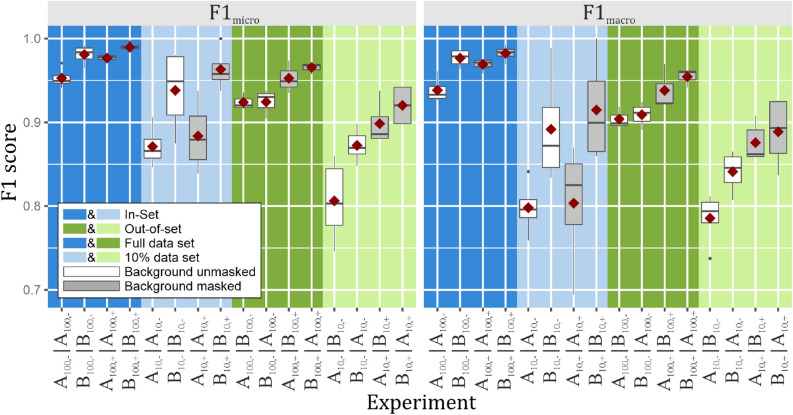
Boxplots comparing the classification performance for model “VGG16_1FC” experiments (Table 4). Mean values are indicated by red diamonds, black dots indicate outliers.
Table 7.
Deep learning experiments, results for the best performing model “VGG16_1FC”.
| Row | Experiment | ||
|---|---|---|---|
| 1 | A100,−|A100,− | 0.95 | 0.94 |
| 2 | B100,−|B100,− | 0.98 | 0.98 |
| 3 | A100,+|A100,+ | 0.98 | 0.97 |
| 4 | B100,+|B100,+ | 0.99 | 0.98 |
| 5 | A10,−|A10,− | 0.87 | 0.80 |
| 6 | B10,−|B10,− | 0.94 | 0.89 |
| 7 | A10,+|A10,+ | 0.88 | 0.80 |
| 8 | B10,+|B10,+ | 0.96 | 0.91 |
| 9 | A100,−|B100,− | 0.92 | 0.90 |
| 10 | B100,−|A100,− | 0.92 | 0.91 |
| 11 | A100,+|B100,+ | 0.95 | 0.94 |
| 12 | B100,+|A100,+ | 0.97 | 0.95 |
| 13 | A10,−|B10,− | 0.81 | 0.79 |
| 14 | B10,−|A10,− | 0.87 | 0.84 |
| 15 | A10,+|B10,+ | 0.90 | 0.88 |
| 16 | B10,+|A10,+ | 0.92 | 0.89 |
Experiments: X|Y corresponds to data set X used for training/validation and Y for testing, with “A” = expedition ANT-XXVIII/2, “B” = expedition PS103 and indices “100” = full data set, “10” = 10% subset, “−“ = background unmasked, “ + ” = background masked (cmp. Table 4). F1 scores were calculated according to Eqs. (6) and (7) and averaged over the cross validation folds or the replicates, respectively. Highest/lowest F1 scores are marked in bold font.
Initial validation experiment
For testing our data and setup, the initial experiment “AB100,−|AB100,−” was conducted. This experiment represents the most commonly reported scenario where all of the data were used, i.e. all data from both expeditions were merged, and no background masking was applied. These data were split into 72% training, 18% validation and 10% test data, and a batch size of 32 was used for training the network.
Experiments investigating influence of data set reduction, background masking and possible concept drift
Next, the two experiments “A100,−|A100,−” and “B100,−|B100,−” were conducted (Table 4 rows 1 and 2). To the data from each expedition without masking the image background (A100,−, B100,−), fourfold cross-validation was applied. For each of the four runs, the combined data from three folds (75% of the base data) were used to train the network. From these data, per class 80% were used for training and 20% for validating the training progress. The remaining fold (25% of the data) was used entirely to test classification performance.
Next, further intra-set experiments were conducted with modifications regarding background masking (Table 4 rows 3, 4, 7 and 8) and number of training data (rows 5–8). Data split and cross-validation were applied in the same way as in the first two experiments. The index “+” indicates that background masking was applied (cmp. Fig. 3), A10 (or B10) indicate that per class only 10% of the data were used for the entire experiment in order to simulate a significantly low number of training data. Accordingly, A10,+ refers to the experiment that uses only 10% of all data from ANT-XXVIII/2 to create training, validation and test set and where the background is masked in all the input images.
Subsequently, we investigated for the possible effect of transect-induced (expedition/year and stitching algorithm, respectively) concept drifts (Table 4 rows 9–16). Here, data from one expedition was used exclusively for training the network (split into 80% training and 20% validation data), and the trained network was applied to classify all cut-outs from data of the other expedition; we refer to this setup as “out-of-set” in the following. Experiments using the complete data sets (index “100”) were run with a batch size of 32 and three replications, for the reduced data sets (index “10”) a batch size of 8 was used with five replications Experiments.
Downstream processing
The results were evaluated with R scripts, provided in Supplement III. Experiments were compared by analysis of variance (ANOVA), investigating the effects of CNN architecture, data set reduction, background masking and out-of-set prediction.
Results
Performance of different CNN architectures
From the variety of CNN models we investigated, those based on VGG architectures clearly outperformed the other models with respect to and values (Table 5, Eqs. (6) and (7)). In the following discussion, we will focus on the best performing model “VGG16_1FC” (VGG16 convolutional base with one downstream fully connected 256 neuron layer and a softmax classification layer). Detailed results of this model are provided in Supplement IV, a comprehensive comparison of all models is provided in Supplement V.
Table 5.
Classification performance of different models, average over experiments 1–16, calculated by ANOVA.
| Model | ||
|---|---|---|
| VGG16_1FC | 0.92*** | 0.89*** |
| VGG16_2FC | − 0.01 | − 0.01 |
| VGG19_1FC | − 0.01 | − 0.01 |
| VGG19_2FC | − 0.02 | − 0.02 |
| Xception | − 0.09*** | − 0.12*** |
| DenseNet | − 0.18*** | − 0.19*** |
| InceptionResNetV2 | − 0.19*** | − 0.26*** |
| MobileNetV2 | − 0.22*** | − 0.31*** |
| InceptionV3 | − 0.23*** | − 0.31*** |
General classification performance of model “VGG16_1FC”
For our initial experiment “AB100,−|AB100,−”, which utilized the merged complete data from both expeditions without background masking, and values of 0.97 were achieved. The classification performance was below average only for classes where specimens were represented solely on the genus level (Table 6, F1 values marked in bold), thus containing a very wide range of morphologies (cmp. Fig. 2 b, c, g, j).
Table 6.
Classification performance per class for the initial experiment AB100,−|AB100,−.
| Class | ||||||
|---|---|---|---|---|---|---|
| Asteromphalus | 24 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| Chaetoceros | 43 | 4 | 2 | 0.91 | 0.96 | 0.93 |
| Fragilariopsis kerguelensis | 68 | 2 | 0 | 0.97 | 1.00 | 0.99 |
| Fragilariopsis rhombica | 28 | 0 | 1 | 1.00 | 0.97 | 0.98 |
| Nitzschia | 11 | 0 | 3 | 1.00 | 0.79 | 0.88 |
| Pseudonitzschia | 51 | 1 | 3 | 0.98 | 0.94 | 0.96 |
| Rhizosolenia | 24 | 2 | 0 | 0.92 | 1.00 | 0.96 |
| Silicoflagellate | 37 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| Thalassiosira gracilis | 23 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| Thalassiosira lentiginosa | 33 | 0 | 0 | 1.00 | 1.00 | 1.00 |
Influence of data set reduction
Intra-set experiments based on full data sets from the individual expeditions with background masking (A100,+|A100,+, B100,+|B100,+) in general achieved the best classification performance (Table 7 rows 3 and 4) and thus have been chosen as base-line for analyses of variance (ANOVA) investigating the effects of data set reduction, background masking and out-of-set prediction (Table 8). Here, reducing the data sets to 10% of the original size resulted in a substantial and significant decrease in classification performance ( − 0.06, − 0.12).
Table 8.
ANOVA results of F1 scores for model “VGG16_1FC” experiments (Table 4).
| Factor interactions | ||||
|---|---|---|---|---|
| Portion 10% | Background masked | Out-of-Set | ||
| ✗ | ✓ | ✗ | 0.98*** | 0.98*** |
| ✗ | ✗ | ✗ | − 0.02 | − 0.02 |
| ✓ | ✓ | ✓ | 0.01 | 0.05 |
| ✓ | ✓ | ✗ | − 0.06** | − 0.12*** |
| ✗ | ✓ | ✓ | − 0.02 | − 0.03 |
| ✓ | ✗ | ✓ | − 0.03 | − 0.03 |
| ✗ | ✗ | ✓ | − 0.02 | − 0.02 |
| ✓ | ✗ | ✗ | 0.00 | 0.00 |
Base-line values for experiments without factor interactions (i.e. average of “A100,+|A100,+” and “B100,+|B100,+”) are highlighted in bold font, effects of factor interactions are given relative to this base-line. Significance codes: ***p < 0.001, **p < 0.01.
Influence of background masking and possible concept drift
Interactions of other factors, i.e. background masking and out-of-set prediction, were obscured by the low sample sizes of the 10% data sets (Fig. 4 bright blue and green areas). As a consequence, some classes were represented with a very low number of examples in the test data. This resulted in a higher variance for the F1 scores. To overcome this impediment, we investigated the other factor interactions on experiments utilizing only the full data sets (Table 9). Here, out-of-set prediction as well as not executing background masking resulted in a significant decrease in classification performance (F1 scores ca. − 0.02).
Table 9.
ANOVA results of F1 scores for model “VGG16_1FC” experiments utilizing only the full data sets (Table 4 rows 1–4 and 9–12).
| Factor interactions | |||
|---|---|---|---|
| Background masked | Out-of-Set | ||
| ✓ | ✗ | 0.98*** | 0.98*** |
| ✗ | ✗ | − 0.02* | − 0.02· |
| ✓ | ✓ | − 0.02** | − 0.03** |
| ✗ | ✓ | − 0.02 | − 0.02 |
Base-line values for experiments without factor interactions (i.e. average of “A100,+|A100,+” and “B100,+|B100,+”) are highlighted in bold font, effects of factor interactions are given relative to this base-line. Significance codes: ***p < 0.001, **p < 0.01, *p < 0.05, · p < 0.1
Discussion
This work applied newly developed methods for producing annotated image data for investigating the influence of a range of factors on deep learning-based taxonomic classification of light microscopic diatom images. These methods and factors are discussed in the following:
Workflow
Our workflow covers the complete process of generating annotated image data from physical slide specimens in a user-friendly way. This is achieved by combining microscopic slide scanning, virtual slides, web-based (multi-)expert annotation and (semi-)automated image analysis. Scanning larger areas of microscopy slides instead of individual user-selected fields of view helps to avoid overlooking taxa at the object detection step and enables later re-analysis. Multi-user annotation, as implemented in BIIGLE, facilitates consensus-building and defining the gold standard in case of ambiguous labelling11.
Data quality
Our workflow (Fig. 1) produced cut-outs of a high visual quality, with a resolution close to the optical limit and enhanced focal depth (Fig. 2). This allowed to investigate the very fine and intricate structures of diatom frustules, which usually are essential for taxonomic identification. The specimens contained a variety of problematic but typical cases, for example incomplete frustules (e.g. Pseudonitzschia); ambiguous imaging situations where multiple, sometime overlapping objects are included in the same cut-out (see Fig. 3a for an example); large intra-class variability of object size, which for larger specimens distorts local features by scaling them to different sizes when the cut-out is downsized to the CNN’s input size (for most classes); different imaging angles causing substantial changes in the specimens’ appearance (e.g. Chaetoceros, Silicoflagellates Fig. 2c,d); broad morphological variability within one class (e.g. Asteromphalus, Silicoflagellates Fig. 2d,h); and pooling of morphologically diverse species into the same class (e.g. genus Nitzschia Fig. 2j). The latter problem will of course be solved with the accumulation of more images covering different species of genus, but the rest will in a large part remain characteristic of diatom image sets. In the full data sets (A100,−/+, B100,−/+), individual classes were covered by ca. 40–400 specimens each. This represents an amount of imbalance that is not uncommon in taxonomic classification.
Classification performance of different CNN architectures
For our experimental setup, which used transfer learning but re-trained only the classification layers, the relatively old VGG16 architecture48 clearly outperformed (Table 5) the newer CNNs Xception50, DenseNet51 Inception-ResNet V249, MobileNet V252 and Inception V353. The reason for this interesting observation may be that the large number of model parameters in the VGG16 CNNs allows learning of models that differ in a large number of single not strongly correlated details. We assume that the reason for not observing overfitting, even though classifier modules of CNN architectures of different complexity were trained for the same number of epochs, might be owed our augmentation scheme. The models were trained exclusively on augmented versions of the original data, and since we used 7 randomly parameterized augmentation features (rotation, width shift, height shift, shear, zoom, horizontal flip and vertical flip) the input seems to be distinct enough for each epoch to prohibit overfitting during 50 epochs.
Classification performance of VGG16_1FC
Using the most commonly reported scenario where all of the data were pooled (experiment AB100,−|AB100,−), the VGG16_1FC network achieved a classification success of 97% (F1 scores 0.97). This is slightly lower than the 99% accuracy reported by Pedraza et al.26 for their classification of 80 diatom classes. A possibly important difference between both data sets is the higher proportion of morphologically heterogeneous classes in our case. In terms of methods used, Pedraza et al. applied fine tuning of the feature extraction layers, a technique which was not tested in our experiments, because in our opinion a 97% classification success already is suitable for routine application, whilst re-training the convolutional base for further improvement would be very demanding in terms of computational costs. However, the CNN architectures used in both studies are different, so a direct comparison for drawing deeper conclusions at this point is difficult. It will be interesting to more systematically investigate the effects of these and other further factors on deep learning diatom classification in future studies. Classification performance of the other investigated VGG architectures, i.e. VGG16_2FC, VGG19_1FC and VGG19_2FC (Table 3), was slightly, but not significantly worse. Accordingly, we conclude that for only 10 classes, one 256 neuron fully connected layer is sufficient for processing the information from the convolutional base for the final softmax classification layer.
Influence of data set size
It is a common observation in machine learning that larger training sets result in better classification performance (condition to a good labelling quality). Nevertheless, using a tenfold of data increased the classification performance by only 6% () to 12% absolute (). From the factors we investigated, this is the most substantial improvement (Tables 8, 9), but it also comes at the highest costs. This once again underlines that the availability of training data usually is the most crucial prerequisite in deep learning. Nevertheless, in this study already sample sizes of mostly below 100 specimens per class resulted in 95% correct classifications (F1 scores ca. 0.95 for experiment A100,−|A100,−), an astonishingly good result underlining the value of using networks pre-trained on a different image domain in situations where the amount of annotated images is a bottleneck. Our observation is also in line with the results of Pedraza et al.26, indicating that slightly below 100 specimens (plus augmentation) per class might be taken as a desirable minimum number for future investigations.
Influence of masking
Background masking improved the classification performance by ca. 2% absolute (Table 9), but required substantial efforts for exact outline computation. The improvement probably results from avoiding ambiguities in cases where multiple objects of different classes are contained within the same cut-out (Fig. 3a) or where the OOI’s structures have an only weak contrast compared to debris in the background (Fig. 2b). Contrasting our findings, in Pedraza et al.26 background-segmentation impaired classification performance slightly. We assume this might be due to their hard masking of the image background in black, which might introduce structures that could be misinterpreted as significant features by the CNN, whereas we tried to avoid adding artificial structures by blending the OOI’s surroundings softly into the homogenized background. A second difference possibly contributing to explaining this difference might be that nearby objects might be depicted in the cut-outs generated by our workflow. Such situations were presumably avoided in26 where the objects were cropped manually by a human expert. Looking at it this way, it could be said that the accurate soft masking we applied (Fig. 3) more than compensates for the difficulties caused by the less selective automated imaging workflow. An additional benefit of the exact object outlines produced by our workflow is their potential use for training deep networks performing instance segmentation like Mask R-CNN58, Unet59 or Panoptic-DeepLab60.
Possible concept drift
We observed a decline in classification performance of ca. 2% absolute for out-of-set classification (Table 9). Though significant, this effect is minimal. This speaks for the efficiency of our standardization of sampling, imaging, processing (with the exception of the stitching method) and analysis. The still remaining small shift might represent either a slight residual methodological drift, or genuine biological signal, i.e. morphological variation due to changes in environmental conditions or sampled populations.
Conclusion
We revisited the challenge of automation of the light microscopic analysis of diatoms and propose a full workflow including high-resolution multi-focus slide scanning, collaborative web-based virtual slide annotation, and deep convolutional network-based image classification. We demonstrated that the workflow is practicable end to end, and that accurate classifications (in the range of 95% accuracy/F1 score) are attainable already with relatively small training sets containing around 100 specimens per class using transfer learning. Although more images, as well as more systematic testing of different network architectures, still have a potential to improve on these results, this accuracy is already in a range that a routine application of the workflow for floristic, ecological or monitoring applications now seems within near reach.
Supplementary information
Acknowledgements
We wish to thank Andrea Burfeid Castellanos and Barbara Glemser for annotating virtual slides and the physical oceanography team on RV Polarstern expeditions PS103 and ANT-XXVIII/2 for their support.
Author contributions
M.K. developed the proposed workflow and conducted the experiments, supervised by B.B. and T.W.N., who also designed the experimental setup. D.L. supported data management and verified the experimental results. M.Z. developed BIIGLE 2.0 and extended it to facilitate the proposed workflow. M.K., T.W.N. and B.B. authored this manuscript. All authors read and approved of the final manuscript.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the priority programme SPP 1991 Taxon-OMICS under grant nrs. BE4316/7-1 & NA 731/9-1. M.Z.’s contribution was supported by the German Federal Ministry of Education and Research (BMBF) project COSEMIO (FKZ 03F0812C), D.L.’s contribution was supported by the German Federal Ministry for Economic Affairs and Energy (BMWi) project ISYMOO (FKZ 0324254D). BIIGLE is supported by the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A537B, 031A533A, 031A538A, 031A533B, 031A535A, 031A537C, 031A534A, 031A532B). Sample collection was performed in the frame of RV Polarstern expeditions PS103 (grant nr. AWI_PS103_04) and ANT-XXVIII/2. Open Access funding provided by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71165-w.
References
- 1.Round FE, Crawford RM, Mann DG. Diatoms: Biology and Morphology of the Genera. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- 2.Seckbach J, Kociolek P. The Diatom World. Berlin: Springer; 2011. [Google Scholar]
- 3.Necchi JRO. River Algae. Berlin: Springer; 2016. p. 279. [Google Scholar]
- 4.Esper O, Gersonde R. Quaternary surface water temperature estimations: New diatom transfer functions for the Southern Ocean. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2014;414:1–19. doi: 10.1016/j.palaeo.2014.08.008. [DOI] [Google Scholar]
- 5.Hasle GR, Fryxell GA. Diatoms: Cleaning and mounting for light and electron microscopy. Trans. Am. Microsc. Soc. 1970;20:469–474. doi: 10.2307/3224555. [DOI] [Google Scholar]
- 6.Kelly M, et al. Recommendations for the routine sampling of diatoms for water quality assessments in Europe. J. Appl. Phycol. 1998;10:215. doi: 10.1023/A:1008033201227. [DOI] [Google Scholar]
- 7.Cairns J, Jr, et al. Determining the accuracy of coherent optical identification of diatoms. J. Am. Water Resour. Assoc. 1979;15:1770–1775. doi: 10.1111/j.1752-1688.1979.tb01187.x. [DOI] [Google Scholar]
- 8.du Buf H, Bayer MM. Automatic Diatom Identification. Singapore: World Scientific; 2002. [Google Scholar]
- 9.Kloster M, Kauer G, Beszteri B. SHERPA: An image segmentation and outline feature extraction tool for diatoms and other objects. BMC Bioinform. 2014;15:218. doi: 10.1186/1471-2105-15-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloster M, Esper O, Kauer G, Beszteri B. Large-scale permanent slide imaging and image analysis for diatom morphometrics. Appl. Sci. 2017;7:330. doi: 10.3390/app7040330. [DOI] [Google Scholar]
- 11.Beszteri B, et al. Quantitative comparison of taxa and taxon concepts in the diatom genus Fragilariopsis: A case study on using slide scanning, multi-expert image annotation and image analysis in taxonomy. J. Phycol. 2018 doi: 10.1111/jpy.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloster M, Kauer G, Esper O, Fuchs N, Beszteri B. Morphometry of the diatom Fragilariopsiskerguelensis from Southern Ocean sediment: High-throughput measurements show second morphotype occurring during glacials. Mar. Micropaleontol. 2018;143:70–79. doi: 10.1016/j.marmicro.2018.07.002. [DOI] [Google Scholar]
- 13.Glemser B, et al. Biogeographic differentiation between two morphotypes of the Southern Ocean diatom Fragilariopsiskerguelensis. Polar Biol. 2019;42:1369–1376. doi: 10.1007/s00300-019-02525-0. [DOI] [Google Scholar]
- 14.Kloster M, et al. Temporal changes in size distributions of the Southern Ocean diatom Fragilariopsiskerguelensis through high-throughput microscopy of sediment trap samples. Diatom. Res. 2019;34:133–147. doi: 10.1080/0269249X.2019.1626770. [DOI] [Google Scholar]
- 15.Olson RJ, Sosik HM. A submersible imaging-in-flow instrument to analyze nano-and microplankton: Imaging FlowCytobot. Limnol. Oceanogr. Methods. 2007;5:195–203. doi: 10.4319/lom.2007.5.195. [DOI] [Google Scholar]
- 16.Poulton NJ. FlowCam: Quantification and classification of phytoplankton by imaging flow cytometry. In: Barteneva NS, Vorobjev IA, editors. Imaging Flow Cytometry: Methods and Protocols. New York: Springer; 2016. pp. 237–247. [DOI] [PubMed] [Google Scholar]
- 17.Schulz J, et al. Imaging of plankton specimens with the lightframe on-sight keyspecies investigation (LOKI) system. J. Eur. Opt. Soc. Rapid Publ. 2010;5:20. doi: 10.2971/jeos.2010.10026. [DOI] [Google Scholar]
- 18.Cowen RK, Guigand CM. In situ ichthyoplankton imaging system (ISIIS): System design and preliminary results. Limnol. Oceanogr. Methods. 2008;6:126–132. doi: 10.4319/lom.2008.6.126. [DOI] [Google Scholar]
- 19.Orenstein, E. C., Beijbom, O., Peacock, E. E. & Sosik, H. M. Whoi-plankton-a large scale fine grained visual recognition benchmark dataset for plankton classification. https://arxiv.org/abs/1510.00745(arXiv preprint) (2015).
- 20.Cheng K, Cheng X, Wang Y, Bi H, Benfield MC. Enhanced convolutional neural network for plankton identification and enumeration. PLoS One. 2019;14:e0219570. doi: 10.1371/journal.pone.0219570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunker S, Boho D, Wäldchen J, Mäder P. Combining high-throughput imaging flow cytometry and deep learning for efficient species and life-cycle stage identification of phytoplankton. BMC Ecol. 2018;18:51. doi: 10.1186/s12898-018-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lumini A, Nanni L. Deep learning and transfer learning features for plankton classification. Ecol. Inform. 2019;51:33–43. doi: 10.1016/j.ecoinf.2019.02.007. [DOI] [Google Scholar]
- 23.Luo JY, et al. Automated plankton image analysis using convolutional neural networks. Limnol. Oceanogr. Methods. 2018;16:814–827. doi: 10.1002/lom3.10285. [DOI] [Google Scholar]
- 24.Mitra R, et al. Automated species-level identification of planktic foraminifera using convolutional neural networks, with comparison to human performance. Mar. Micropaleontol. 2019;147:16–24. doi: 10.1016/j.marmicro.2019.01.005. [DOI] [Google Scholar]
- 25.Keçeli AS, Kaya A, Keçeli SU. Classification of radiolarian images with hand-crafted and deep features. Comput. Geosci. 2017;109:67–74. doi: 10.1016/j.cageo.2017.08.011. [DOI] [Google Scholar]
- 26.Pedraza A, et al. Automated diatom classification (Part B): A deep learning approach. Appl. Sci. 2017;7:460. doi: 10.3390/app7050460. [DOI] [Google Scholar]
- 27.Zhou Y, et al. Digital whole-slide image analysis for automated diatom test in forensic cases of drowning using a convolutional neural network algorithm. Forensic Sci. Int. 2019;302:109922. doi: 10.1016/j.forsciint.2019.109922. [DOI] [PubMed] [Google Scholar]
- 28.Abadi, M. et al. Tensorflow: A system for large-scale machine learning. In 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), 265–283 (2016).
- 29.Chen, T. etal. Mxnet: A flexible and efficient machine learning library for heterogeneous distributed systems. https://arxiv.org/abs/1512.01274(arXiv preprint) (2015).
- 30.Russakovsky O, et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 31.Pech-Pacheco, J. L. & Cristóbal, G. Automatic slide scanning. In Automatic Diatom Identification 259–288 (World Scientific, Singapore, 2002).
- 32.Fischer, S., Shahabzkia, H. R. & Bunke, H. Contour extraction. In Automatic Diatom Identification 93–107 (World Scientific, Singapore, 2002).
- 33.Rojas Camacho O, Forero M, Menéndez J. A tuning method for diatom segmentation techniques. Appl. Sci. 2017;7:762. doi: 10.3390/app7080762. [DOI] [Google Scholar]
- 34.Bueno G, et al. Automated diatom classification (Part A): Handcrafted feature approaches. Appl. Sci. 2017;7:753. doi: 10.3390/app7080753. [DOI] [Google Scholar]
- 35.Sánchez, C., Vállez, N., Bueno, G. & Cristóbal, G. Diatom classification including morphological adaptations using CNNs. In Iberian Conference on Pattern Recognition and Image Analysis 317–328 (Springer, Berlin, 2019).
- 36.Crosta X. Holocene size variations in two diatom species off East Antarctica: Productivity vs environmental conditions. Deep Sea Res. Part I. 2009;56:1983–1993. doi: 10.1016/j.dsr.2009.06.009. [DOI] [Google Scholar]
- 37.Smetacek V, et al. Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature. 2012;487:313–319. doi: 10.1038/nature11229. [DOI] [PubMed] [Google Scholar]
- 38.Mock T, et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature. 2017;541:536–540. doi: 10.1038/nature20803. [DOI] [PubMed] [Google Scholar]
- 39.Assmy P, et al. Thick-shelled, grazer-protected diatoms decouple ocean carbon and silicon cycles in the iron-limited Antarctic Circumpolar Current. Proc. Natl. Acad. Sci. USA. 2013;110:20633–20638. doi: 10.1073/pnas.1309345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cárdenas P, et al. Biogeochemical proxies and diatoms in surface sediments across the Drake Passage reflect oceanic domains and frontal systems in the region. Prog. Oceanogr. 2019;174:72–88. doi: 10.1016/j.pocean.2018.10.004. [DOI] [Google Scholar]
- 41.Simonsen R. The Diatom Plankton of the Indian Ocean expedition of RV "Meteor" 1964–1965. Meteorology. 1974;66:25. [Google Scholar]
- 42.Chalfoun J, et al. MIST: Accurate and scalable microscopy image stitching tool with stage modeling and error minimization. Sci. Rep. 2017;7:4988. doi: 10.1038/s41598-017-04567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preibisch, S. Grid/CollectionStitchingPlugin—ImageJ. https://imagej.net/Grid/Collection_Stitching_Plugin.
- 44.Langenkämper D, Zurowietz M, Schoening T, Nattkemper TW. BIIGLE 2.0—browsing and annotating large marine image collections. Front. Mar. Sci. 2017;4:20. doi: 10.3389/fmars.2017.00083. [DOI] [Google Scholar]
- 45.Horton, T. et al.World Register of Marine Species (WoRMS). WoRMS Editorial Board (2020).
- 46.Schoening T, Osterloff J, Nattkemper TW. RecoMIA—recommendations for marine image annotation: Lessons learned and future directions. Front. Mar. Sci. 2016;3:59. doi: 10.3389/fmars.2016.00059. [DOI] [Google Scholar]
- 47.R Core Team. R:ALanguageandEnvironmentforStatisticalComputing. https://www.R-project.org (2015).
- 48.Simonyan, K. & Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. https://arxiv.org/abs/1409.1556(arXiv preprint) (2014).
- 49.Szegedy, C., Ioffe, S., Vanhoucke, V. & Alemi, A. A. Inception-v4, Inception-ResNet and the impact of residual connections on learning. In Thirty-First AAAI Conference on Artificial Intelligence (2017).
- 50.Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision And Pattern Recognition, 1251–1258 (2017).
- 51.Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4700–4708 (2017).
- 52.Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. & Chen, L.-C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4510–4520 (2018).
- 53.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2818–2826 (2016).
- 54.Chollet, F. et al.Keras. https://keras.io (2015).
- 55.Deng, J. et al. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition, 248–255 (2009).
- 56.Kingma, D. & Adam, B. J. A method for stochastic optimization. https://arxiv.org/abs/1412.6980(arXiv preprint) (2014).
- 57.Chollet, F., & Allaire, J. J., et al.R interface to Keras. https://github.com/rstudio/keras (2017).
- 58.He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. In 2017 IEEE International Conference on Computer Vision (ICCV), 2980–2988 (2017).
- 59.Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds Navab N. et al.) 234–241 (Springer International Publishing, Cham, 2015).
- 60.Cheng, B. et al.Panoptic-DeepLab: A simple, strong, and fast baseline for bottom-up panoptic segmentation. https://arxiv.org/abs/1911.10194(arXiv preprint) (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.