Abstract
GLI1 expression is broadly accepted as a marker of Hedgehog pathway activation in tumors. Efficacy of Hedgehog inhibitors is essentially limited to tumors bearing activating mutations of the pathway. GLI2, a critical Hedgehog effector, is necessary for GLI1 expression and is a direct transcriptional target of TGF-β/SMAD signaling. We examined the expression correlations of GLI1/2 with TGFB and HH genes in 152 distinct transcriptome datasets totaling over 23,500 patients and representing 37 types of neoplasms. Their prognostic value was measured in over 15,000 clinically annotated tumor samples from 26 tumor types. In most tumor types, GLI1 and GLI2 follow a similar pattern of expression and are equally correlated with HH and TGFB genes. However, GLI1/2 broadly share prognostic value with TGFB genes and a mesenchymal/EMT signature, not with HH genes. Our results provide a likely explanation for the frequent failure of anti-Hedgehog therapies in tumors, as they suggest a key role for TGF-β, not Hedgehog, ligands, in tumors with elevated GLI1/2-expression.
Subject terms: Cancer, Computational biology and bioinformatics, Risk factors
Introduction
Elevated expression and nuclear localization of GLI1, reminiscent of Hedgehog (HH) pathway activation, has been reported in a wide variety of tumor types1. Although extensive experimental evidence exists for a pro-tumorigenic and pro-metastatic role of GLI1, efficacy of HH inhibitors is restricted to a handful of cancers with genetic activation of upstream components of the pathway2,3. The sole FDA-approved indication for HH inhibitors is advanced cutaneous basal cell carcinoma4. Understanding their lack of efficacy in other tumors despite high GLI1 expression remains a challenge5,6.
Members of the HH family of growth factors, Sonic (SHH), Indian (IHH) and Desert (DHH) control tissue patterning, limb and skeletal polarity during embryonic life, and broadly contribute to tissue homeostasis and repair processes during adulthood, by controlling cell proliferation, migration, as well as stem cell maintenance and self-renewal1,7. Ligand binding to the 12-transmembrane receptor PATCHED-1 (PTCH1) allows activation of the 7-transmembrane G-coupled receptor Smoothened (SMO), and HH signal transduction proceeds towards activation and nuclear accumulation of GLI transcription factors with activator or repressor functions dependent upon proteolytic cleavage8. GLI1 is a prototypic HH target gene and its expression is widely considered a read-out of HH pathway activation. GLI2 is the primary substrate and effector of the pathway that largely contributes to GLI1 induction by HH, as well as that of other HH target genes in cooperation with GLI19,10. GLI3 has weak transcriptional activity and is considered an inhibitor of HH activity8.
Ligand-independent HH pathway activation as a result of mutations in genes encoding upstream pathway components, such as loss-of-function mutations in PTCH1 or SUFU (suppressor of fused) and activating mutations of SMO are rare in cancers and only found in cutaneous basal cell carcinoma, rhabdomyosarcoma, and medulloblastoma, cancers which exhibit notable therapeutic response to HH inhibitors11.
Clinical studies on HH inhibitors have often relied on increased expression or nuclear localization of GLI1 as a marker of HH pathway activation. It may be argued that in a number of cases, HH activation was stated empirically, with no direct evidence of active upstream HH signaling. Similarly, studies targeting GLI1/2 expression or function in tumor cells in vitro or in mouse models of cancer have shown remarkable anti-tumor efficacy and concluded to a pathogenic role for the HH pathway, although most studies targeted its main downstream effectors, not the pathway itself. Thus, while there is little doubt that GLI transcription factors contribute substantially to cancer progression, direct evidence that would link GLI activity in a given tumor setting to HH ligands activating their receptors is often missing. This may explain the overall lack of anti-tumor therapeutic efficacy of HH inhibitors, most of which target SMO activity1.
TGF-β is secreted abundantly by both tumor and stromal cells, allowing tumor evasion from immune surveillance, peri-tumoral angiogenesis and EMT, processes that all contribute to tumor progression2,3. TGF-β signals via ubiquitously expressed membrane-bound heteromeric serine-threonine kinase receptor complexes2. We identified TGF-β as a powerful inducer of GLI2 and GLI1 expression as well as GLI-dependent transcription, independent from SMO activity12,13. We established a role for both TGF-β signaling and GLI2 in driving melanoma invasion and metastasis, that could be targeted with TGF-β receptor inhibitors, the latter inhibiting GLI2 expression in tumor cells, or by knocking down GLI2 expression14–16. High GLI2 expression in invasive melanoma cells depends largely upon autocrine TGF-β signaling and is associated with a mesenchymal transition and loss of E-cadherin expression, events associated with enhanced cell motility and capacity to metastasize15,17. Similar observations have since been reported for other tumor types, including ovarian and oral squamous cell carcinomas, that link GLI2 and TGF-β expression to tumor aggressiveness via various mechanisms such as induction of PTHrP, leading to enhanced osteolytic bone metastases, or that of a stemness-like phenotype that also promotes metastatic progression18–20.
Herein, we hypothesized that the lack of efficacy of SMO antagonists in numerous tumors occurs because high GLI1 expression and activity may not be linked to HH, but rather to TGF-β, ligand expression, taken as surrogates for HH and TGF-β signaling in tumors, irrespective of the cellular compartment. We compiled data from publicly available gene expression datasets from over 23,500 cancer patients, of which over 15,000 with survival annotations, well above those available from The Cancer Genome Atlas (TCGA). While GLI1/2 expression is correlated with both HH and TGFB expression, their prognostic value is tightly correlated with that of TGFB, not HH. High GLI1/2 and TGFB expression, associated with a mesenchymal/EMT signature, often represent parallel markers of poor clinical outcome. Inversely, high expression of HH is mostly associated with increased survival.
Results
Pan cancer correlation between GLI1, GLI2, TGFB, and HH genes
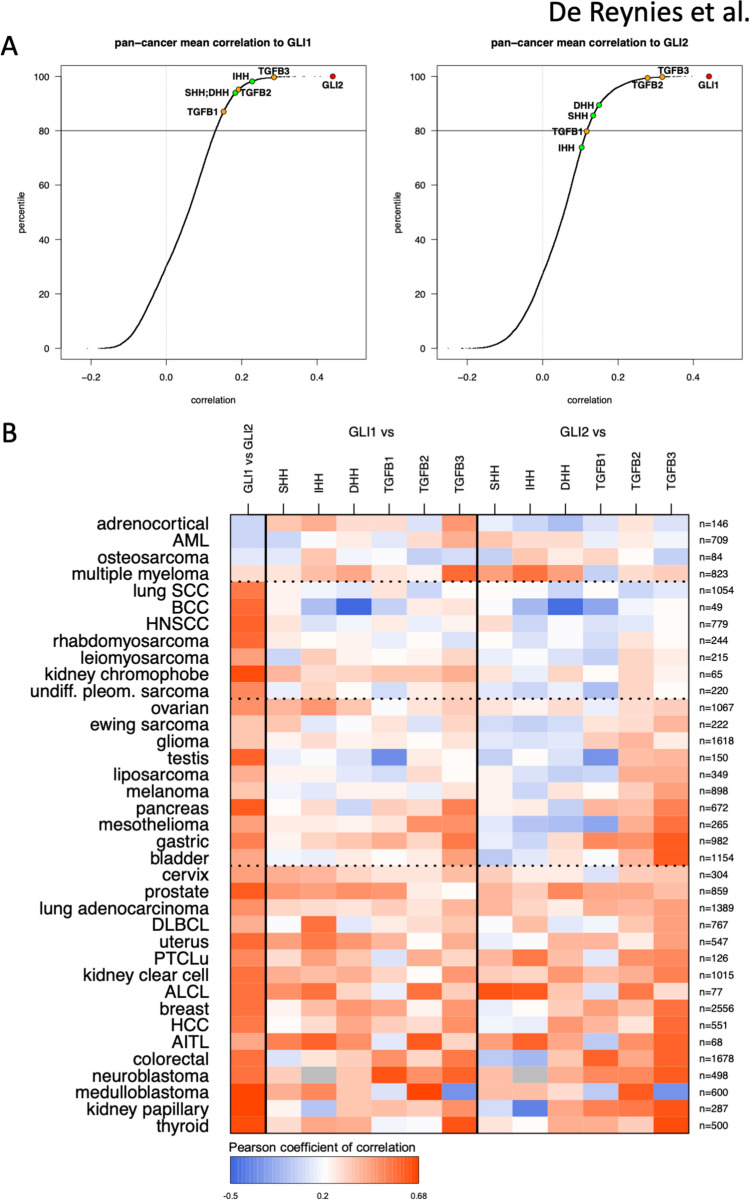
We hypothesized that the correlation between GLI1 and GLI2 expression with HH (SHH, IHH, DHH) and TGF-β ligands (TGFB1, TGFB2, TGFB3) transcript levels represent adequate surrogates for the respective pathogenic implication of HH and TGF-β ligands in GLI1/2 expression and activity in tumors. Pan-cancer analysis of the correlation of GLI1 and GLI2 with 19,540 genes expressed in 30 tumor types revealed that GLI1 and GLI2 are mutually the most correlated genes (Fig. 1A), and that all HH and TGFB genes are in the top 20 percentiles of most correlated genes with GLI1 and GLI2, with one exception. Expression of at least one of the TGFB genes was more closely related than that of any HH genes to both GLI1 and GLI2 expression.
Figure 1.
Pan-cancer expression correlations with GLI1 and GLI2. (A) Data from 152 expression public datasets from 37 cancer types, spanning over 23,500 patients were sorted and the correlation of 19,540 genes (expressed in at least 30 tumor types) with that of GLI1 (left panel) and GLI2 (right panel) was calculated. The respective position of HH and TGFB genes is indicated. (B) Heatmap representation of the expression correlation between GLI1 and GLI2 with each other and between HH and TGFB genes in 37 cancer types. Corresponding numerical values are provided in Supplementary Table 2. The number of patients for each neoplasm is indicated.
Positive correlation (arbitrary threshold: r > 0.25) between the GLI1 and GLI2 genes was observed in 33/37 tumor types (Fig. 1B), representing 92.5% (21,825/23,587) of patients. Most tumor types exhibited similar correlation values between GLI1 expression and that of at least one of either HH or TGFB genes (Fig. 1B). The correlation pattern between HH and TGFB genes with GLI2 (Fig. 1B) was similar to that with GLI1, yet tumors with high GLI2/TGFB correlation could be discriminated into two subgroups: one exhibiting low GLI2/HH correlation (tumor types from ovarian down to bladder), the other exhibiting high GLI2/HH correlation (tumor types from cervix down to thyroid). We did not identify a single neoplasm for which GLI1/2 expression was correlated with that of HH genes without a simultaneous correlation with that of at least one of the TGFB genes.
Expression of GLI1, GLI2, HH and TGFB genes differentially associates with key oncogenic signatures
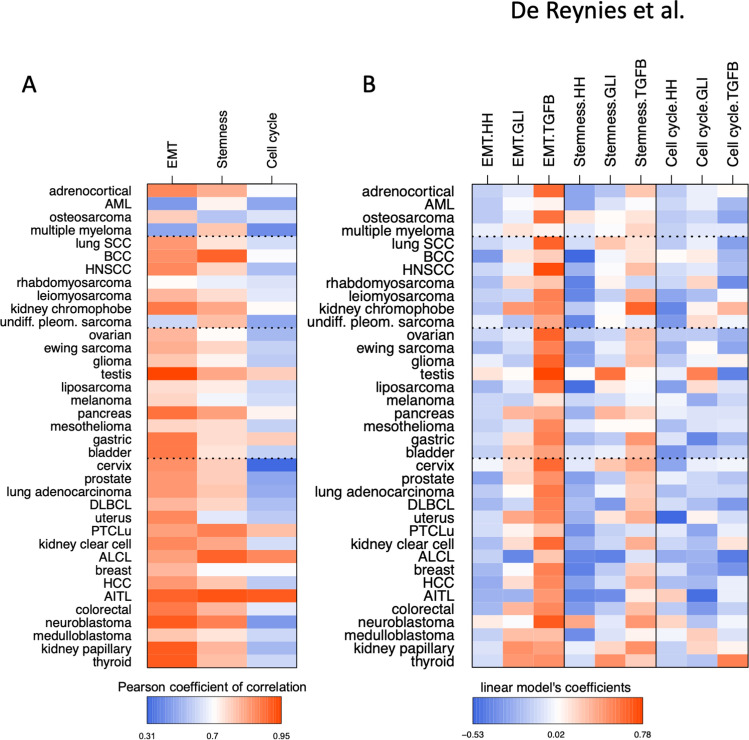
Cell cycle progression, acquisition of a mesenchymal phenotype through epithelial-to-mesenchymal transition (EMT), and cell stemness are cellular traits considered hallmarks of cancer progression21, to which both the HH and TGF-β pathways are linked2,3. For each of the 37 tumor types, we generated a multivariate linear model based on the expression of the eight genes of interest taken together (three HH and three TGFB genes, GLI1 and GLI2) to determine whether it may be predictive of these metagenes. To assess the goodness-of-fit of these models, correlations between the predicted values and observed values for each metagene were calculated in each tumor type. As shown in Fig. 2A, strong correlations were observed in most tumor types for the mesenchymal/EMT and cancer cell stemness metagenes albeit to a lesser extent, while it was seldom observed with the cell cycle metagene.
Figure 2.
Multivariate linear prediction models of metagene signatures for select major oncogenic traits (mesenchymal/EMT, stemness, cell cycle). (A) Heatmap representation of the correlations between predicted and observed values of metagenes in 37 tumor types, taking GLI1/GLI2, the three HH and three TGFB genes together. Corresponding numerical values are provided in Supplementary Table S4B. (B) Heatmap representation of the coefficients from the linear models for each of the 3 metagenes in each tumor type, based on the combined expression of either GLI1 and GLI2, or the three HH or TGFB genes. Corresponding numerical values are provided in Supplementary Table S4B.
A simplified multivariate linear model using compounded expression of either GLI1/2, the HH or TGFB genes was next calculated for each metagene. Coefficients for these three predictive variables within each model, presented in Fig. 2B, demonstrate the dominant role of TGFB gene expression, followed by that of GLI1/2, not HH, in predicting mesenchymal and cancer cell stemness metagene expression in a broad array of tumor types. None of them was associated with the cell cycle metagene. Data for each GLI, HH and TGFB gene taken individually in each model are provided in Supplementary Figure S1. It should be noted that the higher correlation observed with the mesenchymal/EMT metagene is consistent with the fact that the latter comprises a number of known TGF-β/SMAD target genes.
Pan-cancer prognostic values associated with GLI1/2, HH and TGFB transcript levels and select oncogenic signatures
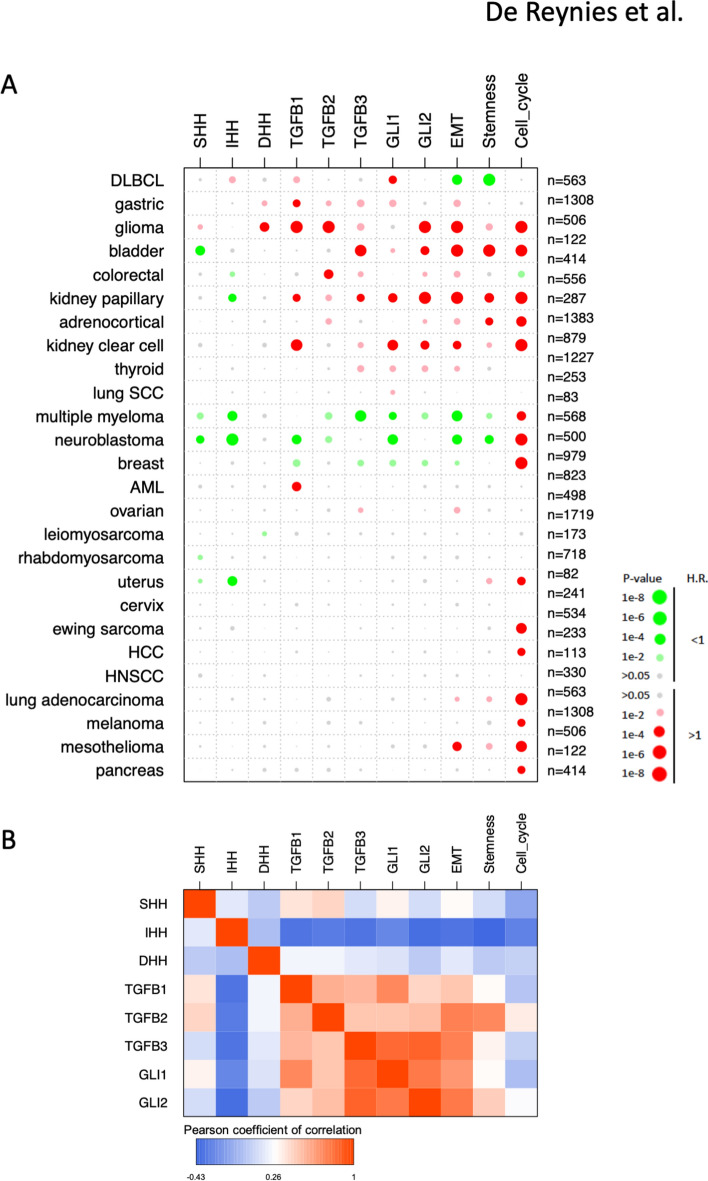
Univariate Cox survival analysis from over 15,000 clinically annotated tumor samples brought critical information (Fig. 3A). At odds with a generalized assumption in the clinical setting that HH signaling is deleterious, expression of all HH genes in tumors was associated with good prognosis (H.R. < 1, green color). Not a single tumor type was found for which expression of any HH gene was associated with morbidity (H.R. > 1, red color) without a parallel pejorative prognostic value for at least one TGFB gene and either GLI1, or GLI2. Also, for each occurrence when high GLI1 or GLI2 expression was of bad prognosis, the same held true for at least one of the TGFB genes. On the other hand, when either GLI1 and GLI2 expression were of good prognosis, the same applied for at least one of the TGFB genes. Noticeably, in bladder, colorectal and kidney papillary carcinoma, GLI2 (and GLI1) expression shared bad prognostic value with that of TGFB genes, while high HH expression was associated with a positive outcome. As expected, the three metagenes were largely of bad prognosis, with mesenchymal/EMT and cancer cell stemness metagenes exhibiting a largely overlapping, yet cancer type-specific pattern of prognostic significance, while the cell cycle metagene was almost universally associated with poor outcome.
Figure 3.
Prognostic value associated with GLI(1/2), HH(S/I/D) and TGFB(1/2/3) genes and the mesenchymal/EMT, cell stemness and cell cycle metagenes in human cancers. (A) Data were derived from a meta-analysis for the univariate prognostic value for overall survival in 26 types of human cancers for which sufficient events were available. Bigger circles represent lower p values. Marked colors represent p value below 0.001, dull colors represent p values below 0.05. Green and red colors represent H.R. below and above 1, respectively (see scale bar). (B) Correlations between pan-cancer profiles of prognostic scores of GLI(1/2), HH(S/I/D) and TGFB(1/2/3) genes and the mesenchymal/EMT, cell stemness and cell cycle metagenes. Prognostic scores are defined as the log2(H.R.) if the related p-value is below 0.05, or 0 otherwise.
In breast cancer, HH expression had no prognostic value while GLI1/2 and TGFB expression were associated with better survival, together with the mesenchymal/EMT and cell stemness metagenes. These results are at odds with most neoplasms where GLI1/2 and TGFB genes share pejorative prognostic value. To understand this discrepancy, a large cohort of breast cancer patients’ data22 was further analyzed. Intra-dataset z-score GLI(1/2), TGFB(1/2/3) and HH(S/I/D) and metagene expression values were sorted according to increasing GLI2 expression, then aligned to the molecular subtypes. High GLI1/2/TGFB and mesenchymal/EMT metagene expression was associated with the normal-like subgroup of tumors with better prognosis, and inversely correlated with luminal-type tumors of poor prognosis (Supplementary Figure S2). A hypothesis may be that the mesenchymal/EMT metagene, which follows GLI2 and TGFB expression pattern, may not be representative of actual EMT in normal-like breast tumors, but rather represents that of fibroblastic heterogeneity found in breast tumors23.
For each cancer type, we next defined a prognostic score as the log2(H.R.) if the related p-value was below 0.05, or 0 otherwise. Correlations between the prognostic scores for GLI(1/2), TGFB(1/2/3), (S/I/D)HH and oncogenic function metagenes were calculated. The results, presented in Fig. 3B, overwhelmingly demonstrate that GLI(1/2), TGFB(1/2/3), Mesenchymal/EMT and stemness metagenes have highly correlated pan-cancer prognostic profiles, with no or modest correlation to that of HH(S/I/D). Little or no correlation between GLI(1/2), TGFB(1/2/3), (S/I/D)HH was found with the cell cycle metagene H.R., consistent with the expression correlation data from Fig. 2.
Discussion
HH inhibitors have failed to fulfill expectations placed on them as powerful anti-cancer drugs2. Despite widespread expression of GLI1 in tumors, considered to be a marker of HH activation, HH inhibitors have solely been granted FDA approval for the treatment of advanced basal cell carcinoma of the skin, a type of tumor that bears activating mutations of the HH pathway25. Clinical trials on other solid (or non-solid) tumors have overall failed2. Based on our earlier work that identified TGF-β as a potent transcriptional inducer of GLI2, consequently leading to SMO-independent induction of GLI112,13, we hypothesized that GLI1 expression in tumors may be driven by TGF-β, not HH, which would explain the lack of efficacy of anti-HH approaches. Large-scale datamining from gene expression datasets representing 23,500 patients and 37 types of neoplasms addressed the issue of whether GLI1/2 expression in tumors warrants therapeutic approaches targeting upstream HH signaling. While significant correlation was found between the expression of GLI1/2 and HH genes in tumors, correlation with that of TGFB genes was as strong. Thus, not only GLI1 is not a relevant marker to predict HH pathway activity, but neither GLI1 nor GLI2 expression in tumors discriminate between HH and TGF-β ligands as potential upstream inducers of their expression.
Strikingly, the prognostic value of GLI1/2 expression in tumors was mostly at odds with that of HH genes, while paralleling that of one or more TGFB genes, as well as that of mesenchymal/EMT and cell stemness metagenes. Contrary to the pejorative prognosis associated with high GLI1/2/TGFB gene expression, high HH expression was mostly associated with good prognosis. In the rare occurrences when HH expression was associated with poor outcome, at least one TGFB gene shared the prognosis. This is schematized in Fig. 4.
Figure 4.
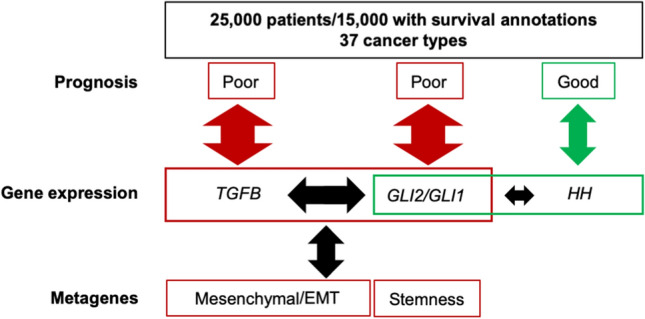
Schematic summary. Pan cancer analysis of gene expression and survival data demonstrates shared poor prognostic value for TGFB, GLI1/2, together with mesenchymal/EMT and Stemness signatures, not HH. Arrow size illustrates the relative correlations between genes (TGFB, GLI2/GLI1 or HH), prognosis and metagene signatures.
Our broad pan-cancer analysis herein indicates that GLI1/2 functions parallel or match those of TGF-β ligands, not HH, as identified both in linear models of oncogenic functions and in prognostic analyses of tumors. Multivariate analysis demonstrated the dominant role of TGFB, followed by that of GLI1/2, not HH, in predicting mesenchymal/EMT and cell stemness metagene expression in a broad array of tumor types. Noteworthy, these analyses were performed using gene expression data from whole tumors, that do not discriminate between tumor and stromal cells. These data fit a mechanistic model whereby TGF-β, acts in a paracrine or autocrine manner to control GLI2 expression which, in turn and depending upon context, allows for GLI1 expression in either a HH-dependent or -independent fashion, as proposed previously24.
Our data hint that HH ligand-driven signaling in tumors leading to GLI1 expression without an overlap and contribution of the TGF-β pathway is not only a rare event but is also unlikely to be pathogenic. The divergence between the prognostic value of GLI1/2 expression and that of HH ligands indicates that there is no sensible justification for targeting HH for cancer treatment if GLI1 expression is used as a prognostic variable. We speculate that patients’ selection based upon an inadequate marker of HH pathway activation may therefore contribute to the lack of clinical efficacy of SMO antagonists in various neoplasms. atients’ selection based upon an inadequate marker of HH pathway activation may therefore contribute to the lack of clinical efficacy of SMO antagonists in various neoplasms. The uniqueness of the cell cycle metagene prognostic value is independent from GLI1/2, TGFB and HH gene signatures. Together with its broad pan-cancer pejorative prognostic value distinct from that of TGFB/GLI1/2/EMT/Stemness, suggests a potential therapeutic benefit for the combination of cytostatic drugs together with anti TGF-β/GLI inhibitors.
Limitations
Both tumor and stromal cells can express TGF-β ligand with is able to act in a paracrine or autocrine manner. This is highly context-specific and highly variable. Since the data originate from non-dissected tumors, we have to assume that the cellular origin of ligands does not affect the outcome of our analyses.
Driver mutations in either HH or TGF-β signaling are likely to influence GLI1/2 expression. Yet, we would like to contend that the outcome of HH signaling from driver mutations would not modify the conclusions drawn from our analyses, as those driver mutations are rare and restricted to a limited number of cancer types.
While a number of published mechanistic studies fit with our model, it would be interesting for our analyses to be functionally validated at the protein level in patients’ samples.
Material and methods
Transcriptome series
A set of 135 transcriptome series related to 37 cancer types was collected from public repositories (ArrayExpress, GEO, TCGA). Series that included multiple cancer types were split accordingly, yielding to a total of 152 distinct transcriptome datasets representing over 23,500 patients. Details and accession numbers are provided in Supplementary Table S1.
Pre-treatment, normalization
Datasets based on Affymetrix microarrays were normalized independently using the justRMA function from the Affy R package (with default parameter). For TCGA RNA-Seq datasets, raw counts were normalized using the upper quartile method26. Datasets from other sources were used as furnished, after log2 transformation of expression values. All probe sets data were aggregated by HUGO Gene Symbol.
Correlation analyses
Transcripts with available measures in at least 30 cancer types were selected for further analyses (n = 19,540). The correlation between their expression and that of GLI1 and GLI2 was calculated independently in each of the 152 datasets, yielding 152 correlation matrices (dimension: 19,540 × 2). Correlations were then averaged across all 152 dataset matrices, yielding a unique (19,540 × 2) matrix used to plot Fig. 1A. The 152 matrices were also averaged per cancer type, yielding 37 sub-matrices that were reduced to the 8 genes of interest (GLI1, GLI2, TGFB(1/2/3) and HH(S/I/D), Supplementary Table S2) and used to plot Fig. 1B.
Metagene calculation
Three published gene signatures corresponding to critically important oncogenic activities were selected: (i) mesenchymal-EMT27, (ii) Stemness28; (iii): Cell Cycle: https://www.genome.jp/dbget-bin/www_bget?pathway+hsa04110. Gene content for each signature is listed in Supplementary Table S3. For each of these signatures and each dataset, the average zero-centered expression of the genes, both measured and included in the signature, was calculated for each sample.
Linear models
Within each dataset, based upon the metagene values for each of the three oncogenic signatures, the lm function from the stats R package was used to perform a linear regression of the metagene variable, using GLI1, GLI2, SHH, IHH, DHH, TGFB1, TGFB2 and TGFB3 expression as predictive variables. To allow for inter-datasets and inter-variables comparisons, all variables were z-scored within each dataset before linear modeling (common unit = standard deviation). For each model in each dataset, correlations between predicted values and observed metagene values (z-scores) was recorded and averaged across datasets by cancer type, then represented as heatmaps. For each linear model in each dataset, coefficients of the predictive variables were recorded and averaged across datasets by cancer type and metagene. Numerical values are provided per series and per cancer type (Supplementary Tables S4A and S4B, respectively).
Survival analyses
Univariate Cox models of overall survival in 26 tumor types with cohorts of more than 50 patients and at least 10 death events were calculated using the survival R package. Aggregation of Hazard Ratios (H.R.) and related confidence intervals across datasets of a given cancer type for a given variable were calculated using the meta R package. All genes and metagenes were z-scored intra-dataset prior to modeling, to allow for inter-datasets comparisons (common unit = standard deviation). Numerical values are provided per series and per cancer type (Supplementary Tables S5A and S5B, respectively).
Supplementary information
Acknowledgements
This work was supported by the French national program Cartes d’Identité des Tumeurs (CIT) funded and developed by the Ligue Nationale Contre le Cancer, grants from Ligue Nationale Contre le Cancer (Equipe Labellisée LIGUE-2016), Institut National du Cancer (INCa MELA13-002) and Donation Henriette et Emile Goutière (to A.M), Fondation ARC (to D.J.), and institutional funding from Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Institut Curie, and Université Paris-Sud.
Author contributions
A.R., D.J. and A.M. conceptualized the project, performed the bioinformatics analyses, and wrote the manuscript. N.E. formatted several of the datasets used in this study. V.M. and C.G. provided help in presenting the data and writing the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aurélien de Reyniès, Email: Aurelien.DeReynies@ligue-cancer.net.
Alain Mauviel, Email: alain.mauviel@curie.fr.
Supplementary information
is available for this paper at 10.1038/s41598-020-71559-w.
References
- 1.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog signaling: from basic biology to cancer therapy. Cell Chem. Biol. 2017;24:252–280. doi: 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrot CY, Javelaud D, Mauviel A. Overlapping activities of TGF-beta and Hedgehog signaling in cancer: therapeutic targets for cancer treatment. Pharmacol. Ther. 2013;137:183–199. doi: 10.1016/j.pharmthera.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Javelaud D, Pierrat MJ, Mauviel A. Crosstalk between TGF-beta and hedgehog signaling in cancer. FEBS Lett. 2012;586:2016–2025. doi: 10.1016/j.febslet.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the sonic Hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel) 2016 doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laukkanen MO, Castellone MD. Hijacking the Hedgehog pathway in cancer therapy. Anticancer Agents Med. Chem. 2016;16:309–317. doi: 10.2174/1871520615666151007160439. [DOI] [PubMed] [Google Scholar]
- 6.Justilien V, Fields AP. Molecular pathways: novel approaches for improved therapeutic targeting of Hedgehog signaling in cancer stem cells. Clin. Cancer Res. 2015;21:505–513. doi: 10.1158/1078-0432.CCR-14-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pak E, Segal RA. Hedgehog signal transduction: key players, oncogenic drivers, and cancer therapy. Dev. Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. J. Biol. Chem. 2012;287:17905–17913. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- 10.Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- 11.Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim. Biophys. Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Dennler S, Andre J, Alexaki I, Li A, Magnaldo T, ten Dijke P, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 13.Dennler S, Andre J, Verrecchia F, Mauviel A. Cloning of the human GLI2 Promoter: transcriptional activation by transforming growth factor-beta via SMAD3/beta-catenin cooperation. J. Biol. Chem. 2009;284:31523–31531. doi: 10.1074/jbc.M109.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, et al. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexaki VI, Javelaud D, Van Kempen LC, Mohammad KS, Dennler S, Luciani F, et al. GLI2-mediated melanoma invasion and metastasis. J. Natl. Cancer Inst. 2010;102:1148–1159. doi: 10.1093/jnci/djq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Newolna M, et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 17.Perrot CY, Gilbert C, Marsaud V, Postigo A, Javelaud D, Mauviel A. GLI2 cooperates with ZEB1 for transcriptional repression of CDH1 expression in melanoma cells. Pigment Cell Melanoma Res. 2013;26:861–873. doi: 10.1111/pcmr.12149. [DOI] [PubMed] [Google Scholar]
- 18.Steg AD, Bevis KS, Katre AA, Ziebarth A, Dobbin ZC, Alvarez RD, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin. Cancer Res. 2012;18:869–881. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merker AR, Oyajobi BO, et al. TGF-beta promotion of Gli2 induced PTHrP expression is independent of canonical Hedgehog signaling. Cancer Res. 2011;71:822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannonier SA, Gonzales CB, Ely K, Guelcher SA, Sterling JA. Hedgehog and TGFbeta signaling converge on Gli2 to control bony invasion and bone destruction in oral squamous cell carcinoma. Oncotarget. 2016;7:76062–76075. doi: 10.18632/oncotarget.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A. TGF-beta/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res. 2011;71:5606–5610. doi: 10.1158/0008-5472.CAN-11-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Reynies A, Jaurand MC, Renier A, Couchy G, Hysi I, Elarouci N, et al. Molecular classification of malignant pleural mesothelioma: identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin. Cancer Res. 2014;20:1323–1334. doi: 10.1158/1078-0432.CCR-13-2429. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Porath I, Thompson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.