Abstract
In higher plants, chloroplast ATP synthase has a unique redox switch on its γ subunit that modulates enzyme activity to limit ATP hydrolysis at night. To understand the molecular details of the redox modulation, we used single-particle cryo-EM to determine the structures of spinach chloroplast ATP synthase in both reduced and oxidized states. The disulfide linkage of the oxidized γ subunit introduces a torsional constraint to stabilize the two β hairpin structures. Once reduced, free cysteines alleviate this constraint, resulting in a concerted motion of the enzyme complex and a smooth transition between rotary states to facilitate the ATP synthesis. We added an uncompetitive inhibitor, tentoxin, in the reduced sample to limit the flexibility of the enzyme and obtained high-resolution details. Our cryo-EM structures provide mechanistic insight into the redox modulation of the energy regulation activity of chloroplast ATP synthase.
Subject terms: Bioenergetics, Cryoelectron microscopy
Jay-How Yang et al. use single-particle cryo-EM to determine the structures of spinach chloroplast ATP synthase in reduced and oxidized states. They report a torsional constraint in the oxidized γ subunit that is alleviated by free cysteines in the reduced state. Their work provides mechanistic insights into the redox modulation of the ATP synthesis by the chloroplast ATP synthase.
Introduction
ATP synthase is a molecular motor that converts energy from a membrane electrochemical potential into the high energy phosphate bonds in the ATP molecule, which is utilized throughout the cell to sustain its life by hydrolyzing ATP to ADP. ATP synthase is present in all forms of life, ranging from bacteria to animals and plants1,2. In a healthy living cell, mitochondrial and bacterial membranes are always energized for maintaining the routine activities of the cell, whereas photosynthetic membranes are de-energized during the night when no light energy is available to facilitate energy production via the photosynthetic electron transport chain. The ATP synthase motor can rotate reversibly in the opposite direction, if there is no regulatory control, to consume ATP molecules that are generated during the day3. Thus, to minimize energy waste, photosynthetic organisms have developed a fast, light-dependent mechanism to prevent the ATP synthase from hydrolyzing ATP molecules at night4,5.
Chloroplast ATP synthase (CF1FO) from higher plants has a unique redox switch that serves to modulate the ATP synthesis activity4,6,7. The CF1FO enzyme actively synthesizes ATP in the reduced state, whereas the oxidized form has a low activity6,8. At sunrise, in the thylakoid membrane, the photosynthetic electron transport chain, consisting of photosystem II, cytochrome b6f complex, and photosystem I, performs a light-induced charge separation, which energizes the membrane and creates an electrochemical gradient9. This gradient activates the CF1FO, which releases a tightly bound ATP and enters an active but still oxidized state, synthesizing ATP molecules at a slower rate10–12. Meanwhile, PSI changes the redox state of the chloroplasts by reducing ferredoxin protein, which docks to the ferredoxin NADP reductase and enables the reduction of NADP+ to NADPH. Ferredoxin also acts as a reductant messenger to reduce thioredoxin, which in turn reduces and activates the CF1FO13,14, leading to the synthesis of ATP at a full rate of more than 200 ATP molecules per second4. At night, the plant needs to prevent the enzyme from wasting energy in the absence of light, since the CF1FO can also hydrolyze ATP by rotating in the opposite direction15. A general hypothesis is that the oxidization of the CF1FO limits this rotary action, thereby hindering unnecessary ATP hydrolysis13,14.
In recent years, single-particle electron cryogenic microscopy (cryo-EM) has become a powerful tool in the study of ATP synthase structures16. In vitro biochemical assays and mutagenesis have shown that the redox state modulates the CF1FO activities4–6,8,17, and cryo-EM imaging of the CF1FO in an autoinhibited and oxidized state has shown a unique disulfide linkage in the γ subunit (γCys240–γCys246)18, which stabilizes the local structure of the two β hairpin motifs of the γ subunit. Although the cryo-EM structure of the oxidized CF1FO has been generated, structural information of the reduced form is still missing. To understand the molecular mechanism of the redox modulation of CF1FO, a more complete structural view is required in order to provide a fundamental framework of energy regulation in plants.
To investigate the structure of CF1FO in different redox states, we isolated and purified the full enzyme complex from spinach leaves (Spinacia oleracea). We modulated the redox state of the enzyme using external redox agents, dithiothreitol (DTT) and iodosobenzoate (IBZ), and characterized the activities of ATP synthesis19–23. Single-particle cryo-EM was utilized to determine the CF1FO structures, with a total of nine full complex structures (including the control) at resolutions in the range of 3.4–7.9 Å. Among the individual redox states, the particle images could be categorized into three distinct rotary states. We further focused the density refinement on the F1 domains, which have improved map resolutions from 3.0 to 4.4 Å. These cryo-EM density maps allowed us to build atomic models of CF1FO under the redox states in question, giving insights into the mechanisms of redox modulation on the enzyme activity.
Results
Characterization of the CF1FO redox states
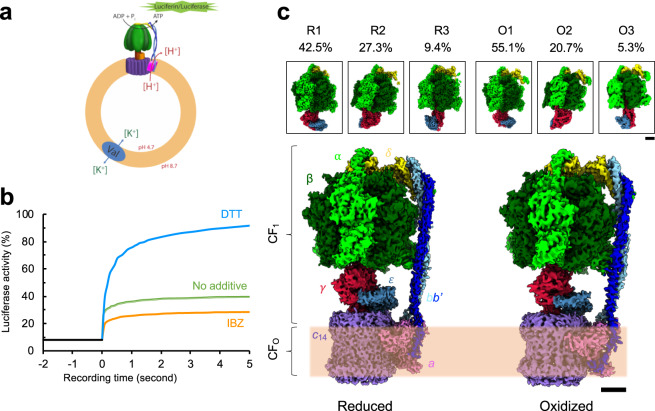
It has been shown that the reduced form of the plant CF1FO is more active in producing ATP molecules than the oxidized form, and the rate of the ATP synthesis of the reduced form is much faster than that of the oxidized form4. To produce the CF1FO in the various redox states, we first isolated the full CF1FO complex from spinach leaves (Supplementary Fig. 1a, b) and reconstituted it in the membrane bilayer of liposomes with a pH gradient established across the membrane using the ΔpH-step jump method. We then applied DTT or IBZ as the reducing and oxidizing agents, respectively, to mix with the reconstituted liposomes, generating different redox states of the CF1FO. One sample without DTT or IBZ treatment was taken as the control sample. ADP was supplied to initiate the ATP synthesis reaction, and the generated ATP molecules were detected using a luciferin–luciferase assay19–23 (Fig. 1a, b). Using the curve of the control sample as a reference, luciferase activity of the reduced sample was higher than that of the oxidized sample (Fig. 1b). This supports the conclusion that the CF1FO redox state can be modulated using these external redox agents, thus changing the enzymatic activity accordingly.
Fig. 1. Chloroplast ATP synthase (CF1FO) of Spinacia oleracea in two different redox states.
a Schematics of the experimental design for measuring CF1FO function. Purified CF1FO was reconstituted into a liposome (orange) mixed with lipids of phosphatidylcholine and phosphatidic acid. The generated pH gradient across the membrane drove the reconstituted CF1FO to synthesize ATP molecules, which were detected using a luciferin/luciferase assay (green). Val indicates valinomycin. b Profile of the CF1FO activity measurements of the CF1FO in different redox states. Blue curve represents the sample with dithiothreitol (DTT) (reduced), orange the sample with iodosobenzoate (IBZ) (oxidized), and green the sample with no redox agent added (control). c Cryo-EM density maps of the oxidized and reduced forms of the CF1FO. Percentages of the particle images used are listed for individual rotary states. Scale bars indicate 25 nm. Color codes: α (light green), β (dark green), δ (yellow), bb' (blue and light blue), γ (crimson), ε (indigo), a (light pink), and c ring (purple). R indicates a reduced state, and O indicates an oxidized state. Membrane bilayer is indicated with the light orange band. The three-dimensional (3D) reconstructions are categorized into three different rotary states (states 1, 2, and 3). Upper insets are the density maps of the F1 domains.
Single-particle cryo-EM of the CF1FO
Different redox states were generated by the external reagents and imaged by cryo-EM for three-dimensional (3D) reconstruction. As prepared for the functional measurements, three different protein samples in detergent micelles were imaged: reduced (DTT), oxidized (IBZ), and control enzyme complexes. The electron micrographs of individual groups showed a homogeneous distribution of the purified CF1FO enzyme complex (Supplementary Figs. 2a–e, 3a–e, 4a–e). After iterative 3D classification procedures, the reconstructions of three different rotary states were categorized and determined in individual groups. Comparison of the three different redox states showed that rotary state 1 was the most populated, while rotary state 3 was the least populated (Supplementary Figs. 2b, 3b, 4b). The percentage of the particle images of the reduced enzyme consisting of rotary state 2 (27.3%) and 3 (9.44%) are higher than in the oxidized enzyme (rotary state 2: 20.7%; rotary state 3: 5.26%) (Supplementary Figs. 2b, 3b, 4b). In each sample, about ~20% of the total particle images could not be unambiguously categorized into any of the three rotary states; these uncategorized particle images were possibly the 2D projections with the particles in the intermediate rotary states or ones lacking discernible features. All the resulting 3D density maps unambiguously feature the structural elements of the extracellular F1 domain and the membrane FO part. By increasing the map contours, the inside of the detergent-bound region can be distinguished from the extracellular domains, and the 14 hairpin structures of the c14 ring rotor were clearly resolved (Supplementary Figs. 2e, 3e, 4e). The resolutions among the density maps of the full complexes ranged from 3.4 to 7.9 Å (CF1 3.0 to 4.4 Å) (Fig. 1c; Supplementary Figs. 2c, 3c, 4c, 5). Although the density maps generated from the control dataset could be categorized into three different rotary states (Supplementary Fig. 4), they were generated at relatively low resolutions (5.2–7.9 Å). One explanation could be that the control dataset was composed of multiple states, conferring a greater conformational variability in the population and thus flattening out the high-resolution details24–26. The statistics of data processing are shown in Tables 1, 2, 3.
Table 1.
Statistics of the single-particle cryo-EM structure of the reduced chloroplast ATP synthase of Spinacia oleracea.
| Chloroplast ATP synthase of Spinacia oleracea in dithiothreitol (DTT) |
|||
|---|---|---|---|
| R1 | R2 | R3 | |
| (EMD-21270, EMD-21271) | (EMD-21268, EMD-21269) | (EMD-21266, EMD-21267) | |
| (PDB codes: 6VON, 6VOO) | (PDB codes: 6VOL, 6VOM) | (PDB codes: 6VOJ, 6VOK) | |
| Data collection and processing | |||
| Magnification | 130,000× | ||
| Voltage (kV) | 300 | ||
| Electron exposure (e−/Å2) | 49 | ||
| Defocus range (μm) | −1.0 to −2.8 | ||
| Pixel size (Å) | 1.053 | ||
| Symmetry imposed | C1 | ||
| Initial particle images (no.) | 108,691 | ||
| Final particle images (no.) | 46,180 | 29,667 | 10,264 |
| Map resolution (Å) | 3.35 (3.05) | 4.06 (3.60) | 4.34 (3.85) |
| FSC threshold | |||
| Refinement | |||
| Initial model used (PDB code) | 6FKF | 6FKH | 6FKI |
| Model resolution (Å) | 3.3 (3.1) | 3.9 (3.6) | 4.2 (3.7) |
| FSC threshold of 0.143 | |||
| Cross correlation | |||
| Masked | 0.718 (0.760) | 0.705 (0.702) | 0.668 (0.708) |
| Volume | 0.704 (0.747) | 0.718 (0.702) | 0.676 (0.713) |
| Map sharpening b factor (Å2) | −50.65 (−34.10) | −64.42 (−61.15) | −76.98 (−60.57) |
| Model composition | |||
| Non-hydrogen atoms | 39,163 (27,551) | 38,991 (27,492) | 39,155 (27,512) |
| Protein residues | 5186 (3574) | 5165 (3566) | 5185 (3569) |
| Ligands | |||
| ATP | 4 | 4 | 4 |
| ADP | 2 | 2 | 2 |
| Tentoxin | 1 | 1 | 1 |
| B factors (Å2) | |||
| Protein | 52.25 (58.02) | 33.99 (45.03) | 63.72 (40.88) |
| Ligands | 26.06 (44.20) | 22.88 (34.80) | 30.79 (26.45) |
| RMS deviations | |||
| Bond lengths (Å) | 0.005 (0.006) | 0.005 (0.005) | 0.005 (0.007) |
| Bond angles (°) | 0.861 (0.787) | 0.848 (0.862) | 0.899 (0.974) |
| Validation | |||
| MolProbity score | 1.85 (1.90) | 1.98 (1.98) | 1.92 (2.03) |
| Clashscore | 8.30 (8.99) | 12.48 (11.60) | 9.88 (10.75) |
| Poor rotamers (%) | 0.05 (0.03) | 0.12 (0.10) | 0.34 (0.51) |
| Ramachandran plot | |||
| Favored (%) | 94.08 (93.64) | 94.66 (93.88) | 94.04 (92.31) |
| Allowed (%) | 5.82 (6.21) | 5.20 (5.95) | 5.88 (7.58) |
| Disallowed (%) | 0.10 (0.14) | 0.14 (0.17) | 0.08 (0.11) |
Values in the parentheses are the refinement statistics for the F1 region.
Table 2.
Statistics of the single-particle cryo-EM structure of the oxidized chloroplast ATP synthase of Spinacia oleracea.
| Chloroplast ATP synthase of Spinacia oleracea in iodosobenzoate (IBZ) | |||
|---|---|---|---|
| O1 | O2 | O3 | |
| (EMD-21264, EMD-21265) | (EMD-21262, EMD-21263) | (EMD-21241) | |
| (PDB codes: 6VOH, 6VOI) | (PDB codes: 6VOF, 6VOG) | (PDB codes: 6VMG) | |
| Data collection and processing | |||
| Magnification | 48,077× | ||
| Voltage (kV) | 300 | ||
| Electron exposure (e−/Å2) | 44.4 | ||
| Defocus range (μm) | −1.5 to −4.0 | ||
| Pixel size (Å) | 1.04 | ||
| Symmetry imposed | C1 | ||
| Initial particle images (no.) | 552,893 | ||
| Final particle images (no.) | 304,879 | 114,542 | 29,090 |
| Map resolution (Å) | 4.16 (4.03) | 4.51 (4.35) | 6.46 |
| FSC threshold | |||
| Refinement | |||
| Initial model used (PDB code) | 6FKF | 6FKH | 6FKI |
| Model resolution (Å) | 4.1 (3.9) | 4.4 (4.3) | 6.6 |
| FSC threshold of 0.143 | |||
| Cross correlation | |||
| Masked | 0.728 (0.752) | 0.774 (0.824) | 0.711 |
| Volume | 0.712 (0.734) | 0.773 (0.821) | 0.696 |
| Map sharpening b factor (Å2) | −211.95 (−203.09) | −179.39 (−232.12) | −304.57 |
| Model composition | |||
| Non-hydrogen atoms | 39,159 (27,497) | 35,182 (27,137) | 25,375 |
| Protein residues | 5193 (3575) | 5175 (3563) | 5170 |
| Ligands | |||
| ATP | 4 | 4 | 0 |
| ADP | 1 | 1 | 0 |
| B factors (Å2) | |||
| Protein | 84.67 (64.35) | 144.23 (158.45) | 222.15 |
| Ligands | 68.15 (55.87) | 119.07 (142.64) | |
| RMS deviations | |||
| Bond lengths (Å) | 0.005 (0.005) | 0.007 (0.006) | 0.006 |
| Bond angles (°) | 0.941 (0.881) | 0.983 (1.025) | 0.970 |
| Validation | |||
| MolProbity score | 2.14 (2.12) | 2.19 (2.20) | 2.05 |
| Clashscore | 14.08 (13.35) | 15.13 (15.24) | 9.65 |
| Poor rotamers (%) | 0.49 (0.24) | 0.76 (0.49) | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 92.02 (91.96) | 91.63 (91.11) | 90.31 |
| Allowed (%) | 7.68 (7.73) | 8.32 (8.80) | 9.60 |
| Disallowed (%) | 0.29 (0.31) | 0.06 (0.08) | 0.10 |
Values in the parentheses are the refinement statistics for the F1 region.
Table 3.
Statistics of the single-particle cryo-EM structure of the control chloroplast ATP synthase of Spinacia oleracea.
| Chloroplast ATP synthase of Spinacia oleracea | |||
|---|---|---|---|
| C1 | C2 | C3 | |
| (EMD-21239, EMD-21240) | (EMD-21238) | (EMD-21235) | |
| (PDB codes: 6VMB, 6VMD) | (PDB codes: 6VM4) | (PDB codes: 6VM1) | |
| Data collection and processing | |||
| Magnification | 48,077× | ||
| Voltage (kV) | 300 | ||
| Electron exposure (e−/Å2) | 43.5 | ||
| Defocus range (μm) | −1.5 to −4.0 | ||
| Pixel size (Å) | 1.04 | ||
| Symmetry imposed | C1 | ||
| Initial particle images (no.) | 208,371 | ||
| Final particle images (no.) | 127,760 | 26,291 | 13,947 |
| Map resolution (Å) | 5.23 (4.53) | 7.08 | 7.90 |
| FSC threshold | |||
| Refinement | |||
| Initial model used (PDB code) | 6FKF | 6FKH | 6FKI |
| Model resolution (Å) | 6.5 (4.5) | 7.2 | 7.9 |
| FSC threshold of 0.143 | |||
| Cross correlation | |||
| Masked | 0.764 (0.841) | 0.787 | 0.786 |
| Volume | 0.753 (0.836) | 0.764 | 0.768 |
| Map sharpening b factor (Å2) | −187.09 (−195.21) | −269.31 | −507.69 |
| Model composition | |||
| Non-hydrogen atoms | 35,401 (27,501) | 25,445 | 25,331 |
| Protein residues | 5194 (3576) | 5184 | 5161 |
| Ligands | |||
| ATP | 4 | 0 | 0 |
| ADP | 1 | 0 | 0 |
| B factors (Å2) | |||
| Protein | 306.88 (130.00) | 376.31 | 286.00 |
| Ligands | 233.70 (101.89) | ||
| RMS deviations | |||
| Bond lengths (Å) | 0.006 (0.008) | 0.007 | 0.007 |
| Bond angles (°) | 1.064 (1.097) | 0.948 | 1.054 |
| Validation | |||
| MolProbity score | 2.01 (2.18) | 1.98 | 2.27 |
| Clashscore | 10.54 (12.90) | 7.94 | 12.03 |
| Poor rotamers (%) | 0.51 (0.82) | 0.00 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 92.59 (89.74) | 90.02 | 84.46 |
| Allowed (%) | 7.23 (10.12) | 9.90 | 15.42 |
| Disallowed (%) | 0.18 (0.14) | 0.08 | 0.12 |
Values in the parentheses are the refinement statistics for the F1 region.
Atomic models that were built along with the resulting cryo-EM density maps (Supplementary Figs. 6, 7, 8) showed that the overall architectures of the three different rotary states in both redox groups. The F1 domain is composed of a catalytic α3β3 hexamer, a δ stator, a γ-ε central shaft, and the extracellular domain of a heterodimeric bb′ stator. Our structure of the oxidized state is in good agreement with previous structures of the oxidized state of the CF1FO, while significant differences are revealed when the structure of the inactive oxidized state was compared to our first revealed structure of the active, reduced state of the CF1FO. The packing of the catalytic unit with the δ and bb′ stators is the same as in both oxidized and reduced forms (Supplementary Fig. 9a, b). The interactions between the catalytic head and the peripheral stalk are essential to stabilize the F1 domain during the process of ATP synthesis occurring at the αβ interfaces and triggered by the rotation of the central shaft via the binding change mechanism27,28.
Nucleotide-binding states in α3β3 catalytic unit
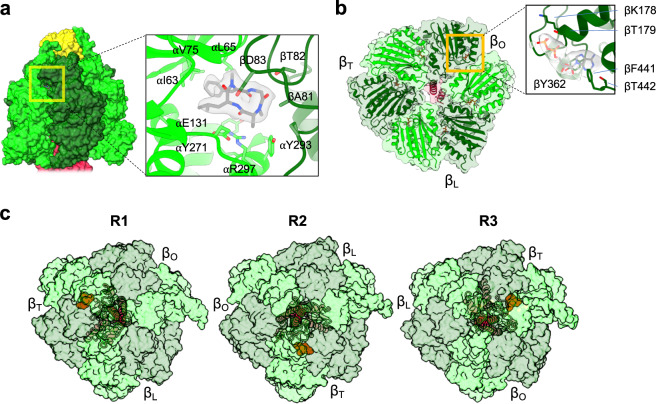
According to the binding change mechanism, the asymmetric α3β3 hexamer with the γ-ε central shaft alternates the three nucleotide-binding sites on the αβ interfaces for ATP synthesis27,29. These three sites are identified as loose (partially open, ADP and phosphate bound, βL), tight (closed, ATP bound, βT), and open (empty, βO) sites27. Note that the γ-ε central shaft faces the βO open site30. In our cryo-EM densities of both the reduced and oxidized forms, three ATP molecules were modeled into the clearly defined densities in the α subunits (Supplementary Fig. 10a, b). Our cryo-EM density maps allow us to identify one ADP located in the loose βL catalytic site and one ATP located in the tight βT site (Supplementary Fig. 10a). This finding fully agrees with the binding change mechanism proposed by Boyer27. The energy generated from the proton gradient then changes the binding affinity and conformation of the F1 domain, leading to the release of ATP molecule. We were now for the first time able to identify an ATP in the tight binding site of the CF1FO, in contrast to the previous structures of the inactive, oxidized chloroplast ATP synthase (PDB codes: 6FKF, 6FKH, and 6FKI), which showed the ADPs occupying both the βL and βT sites18. We show the interactions of the bound nucleotides in these binding sites are conserved: the aromatic side chains of the βTyr362 and βPhe441 stack with the adenine of the nucleotide and βLys178, βThr179, and αArg366 (arginine finger) interact with the phosphate group (Supplementary Fig. 10a, b). The βO open site does not have any density for a nucleotide. Thus, the locations of the nucleotide binding in our structures are consistent with previous findings28. In contrast to Hahn et al.18, our oxidized CF1FO structures match the nucleotide-binding occupancy proposed in the binding change mechanism27.
We had initially pursued single-particle cryo-EM of the reduced CF1FO with the reducing agent DTT, but the resolution of the reconstructions was not high enough for us to model the atomic coordinates. This may be a result of the enzyme being highly flexible in its active, reduced form, leading to a low-resolution density map. To test this idea, we aimed to fix the CF1FO in its active, reduced state in the native membrane by adding tentoxin after the enzyme was reduced before the enzyme was extracted from the membrane. The goal was to block the enzyme “in action” by restriction of the rotary action thereby limiting the mobility of the complex. Tentoxin, a cyclic peptide produced from the fungus Alternaria fungi, was reported to influence the CF1FO activity in a concentration-dependent manner31–33. It blocks rotation and thereby multisite catalysis, but still allows for activation of single site catalysis34. Although the crystal structure of the F1 head complexed with tentoxin were determined previously, our cryo-EM structure is to our knowledge the first structure showing the intact enzyme in complex with tentoxin33,35. Our resulting cryo-EM densities imaged after the addition of tentoxin were overall well-resolved, corroborating the highly mobile nature of the reduced CF1FO. Our density maps also showed one consistent tentoxin-binding site, at the same site identified in previous crystal structure (PDB code: 1KMH)33 (Fig. 2a). Tentoxin binds to the interface between the α and β subunit (with the βT binding site). Its cyclic ring interacts with the charged or polar residues (βAsp83, βThr82, αGlu131, αArg297, and αTyr271), and its isobutyl and phenyl moieties interact with the hydrophobic residues (αIle63, αLeu65, and αVal75) (Fig. 2a, c). Our structure of the reduced CF1FO reveals that the tentoxin binds specifically to the α-βT interface thereby blocking the most critical and rate limiting step in the rotary mechanism: the opening of the βT site and the release of the ATP molecule. All three rotary states of the active, reduced CF1FO showed the density of the bound tentoxin ligand, and its binding site alternates with the rotary state (Fig. 2c). Thus, in the three rotary states, the tentoxin-binding site is alternated together with the βT subunit, which indicates that tentoxin-binding indeed fixed each of the rotary state of the active, reduced enzyme.
Fig. 2. Structure of the reduced CF1 domain.
a Tentoxin-binding site. Surface overview of the F1 domain is shown on the left, with an enlarged view of the tentoxin-binding site shown on the right. Light green, dark green, and yellow are the α, β, and δ subunits, respectively. Tentoxin and its cryo-EM density are shown in gray. b Cross section of the α3β3 catalytic unit from the reduced CF1FO. Central coiled coil is the central shaft γ subunit (crimson). ADP (gray stick) binds onto the βO site in the enlarged view on the right. c Different rotary states bound with tentoxin. The tentoxin binds to the αβ interface, where the β subunit associates with a βT nucleotide-binding site. The αβ hexamers (light and dark green) are shown in surface representation and the central γ subunit (crimson) is shown in cartoon representation. Tentoxin molecules are shown as orange balls.
In addition to the identification of the tentoxin-binding site, the density of one additional ADP molecule was found in the β open site (βO) (Fig. 2b), which did not appear in the oxidized or control structures, nor in the previous structure of the oxidized CF1FO18. The density of this extra ADP molecule was clearly resolved, but we did not identify any additional density for the phosphate (Fig. 2b). This ADP is bound to the β subunit via interactions with the residues of βLys178, βThr179, βTyr362, and βPhe441 (Fig. 2b). Because the βO binding site is away from the α subunit, the ADP has no interaction with the arginine finger (αArg366) of the adjacent α subunit. According to the binding change mechanism27, the βO open site is both the entry site for the ADP and phosphate and the exit site for the ATP during the process of ATP synthesis27. It has also been shown that tentoxin inhibits the step of the ADP release from the βO open site in ATP hydrolysis32,36. Because ATP synthesis and hydrolysis are reversible processes for CF1FO, our tentoxin-bound and reduced CF1FO structure is to our knowledge very likely the first visualization of the entry step of the ADP molecule for the subsequent ATP synthesis.
γ-ε central shaft and the redox switch
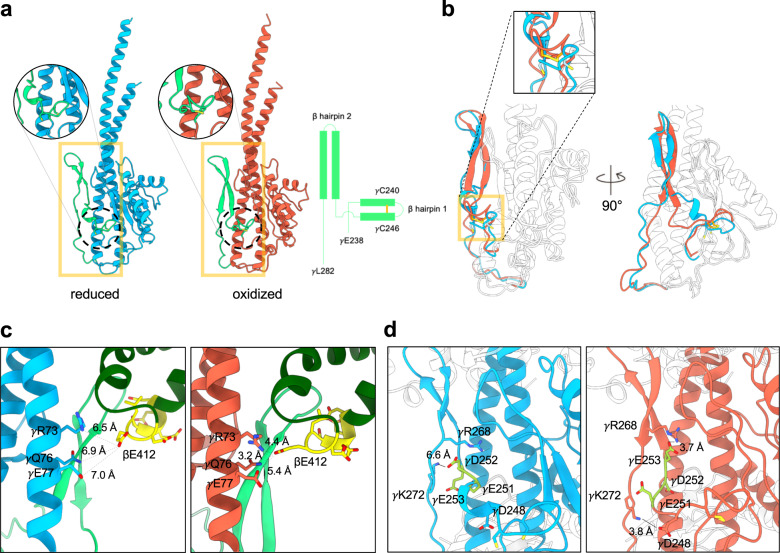
The two cysteines, γCys240 and γCys246, comprise the redox switch in the γ subunit of the CF1FO4,18. They are located on a short β hairpin loop motif (β hairpin 1), connected to a long anti-parallel β hairpin motif (β hairpin 2) at its C-terminus (Fig. 3a, b). Our oxidized γ subunits are similar to the previous structures (PDB code: 6FKF; RMSD of the γ subunits: 0.877 Å)18. Superposition of the reduced and oxidized γ subunits showed the same architecture of the secondary structures (RMSD 1.016 Å), but the local structures of the β hairpins 1 and 2 are different (γGlu238 – γLeu282) (Fig. 3b). The oxidized γ subunit structure shows that the β hairpin 1 is formed by the disulfide bond formation and the β hairpin 2 is stabilized by two long anti-parallel β strands with extensive hydrogen bond pairs between their N–H and C = O groups on the peptide bonds (Fig. 3a, b). However, the reduced γ subunit does not maintain the β hairpin 1 structure due to the disconnection of the disulfide bond, and the β strands of the β hairpin 2 are shorter than those of the oxidized form. The shorter β strands have less hydrogen bonding between the two long loops to stabilize the hairpin structure, and as such the disconnected cysteines are more likely to destabilize the structures of the two β hairpins. The reduced cysteines may release the torsional constraint on the β hairpin 1, forming a one-turn helix, uncoupling the β strands of the β hairpin 2, and destabilizing the loop structures of the γ subunit (Fig. 3a, b).
Fig. 3. Structures of the reduced and oxidized γ subunits.
a Structures of the reduced (light blue) and oxidized (orange) γ subunits. Two β hairpin structures (from γGlu238 to γLeu282) are shown in light green, and the two cysteines of the redox switch are shown in yellow in circular enlarged views. Diagram on right shows the topology of the two β hairpin structures. b Superposition of the reduced and oxidized γ subunits (RMSD 1.016 Å). The two β hairpins are shown in light blue and orange for the reduced and oxidized forms, respectively. Other regions are shown in white. c Interaction networks of the β hairpin 2 and βDELSEED motif. Left and right panels are the reduced (γ subunit in light blue) and oxidized (γ subunit in orange) forms. Light green represents the β hairpin 2, dark green for the β subunit, and yellow for the βDELSEED motif. The distances connecting the residues of the γ coiled coil (γArg73, γGln76, and γGlu77) with the βGlu412 are labeled. d Interaction of the EDE motif with the γ subunit. The EDE motif (yellow) does not interact with any part of the reduced γ subunit but forms an extensive interaction network with its neighborhood when the γ subunit is oxidized.
One motif with a helix-turn-helix structure in the β subunit has a highly conserved DELSEED sequence (βDELSEED motif) and features multiple negatively charged residues37. The conformation of the βDELSEED motif correlates with the nucleotide-binding state of the β subunit37 (Supplementary Fig. 11a). Generally, the interactions between the βDELSEED and the β hairpin 2 motifs do not seem to have a significant change in the two redox states (Supplementary Fig. 11a). Of the three reduced rotary states, the β hairpin 2 loop in the γ subunit has extensive interactions with the βDELSEED motif on the β subunit with the βO site mainly via polar-polar (βSer414, βGlu416, βAsp417, γThr258, γThr259, γLys260, and γGlu267) and hydrophobic (βLeu408, βLeu413, γLeu257, and γLeu264) interactions (Fig. 3c; Supplementary Fig. 11b). Secondly, within the γ subunit, the aromatic side chain of the γPhe255 stacks with γPhe217 and buries within hydrophobic residues in the coiled coil of the γ subunit (γVal72, γVal79, γAla313, and γAla317)18. These interactions also contribute to stabilizing the interaction between the loop and βDELSEED motif, which are similar to the corresponding interactions in the oxidized state (Fig. 3c; Supplementary Fig. 11b).
The βGlu412 interacts with the γ central coiled coil in the oxidized state, but not in the reduced state (Fig. 3c). In the oxidized state, the side chain of the βGlu412 is very likely to form hydrogen bonds with the side chains of the γGln76 and γGlu77 (Fig. 3c). While in the reduced state, the βGlu412 is far from the γ coiled coil, which has less interaction with the central coiled coil and may result in less restriction for the central shaft rotation (Fig. 3c). On the other hand, the lower loop of the β hairpin 2 (γIle271 – γGlu285) has a low signal level (1.2 σ) in our cryo-EM density map of the reduced form, implying a high level of mobility. This entropic gain may also lead to less resistance for the central shaft rotation.
A previous mutagenesis study on the γ subunit showed that the mutant with the deletion of the three negatively charged residues, γGlu251, γAsp252, and γGlu253, was insensitive to redox regulation38. These three residues are located between the β hairpin 1 and 2, in the so-called EDE motif. In the reduced form, the EDE motif does not seem to interact with the adjacent residues, allowing the residues between γVal247 and γAla250 to form a short one-turn helix (Fig. 3d). In contrast, in the oxidized form, the EDE motif interacts with the adjacent residues and stabilizes the two β hairpin structures (Fig. 3d). Disulfide bond formation also introduces torsional stress to break the one-turn helix and form an anti-parallel β hairpin 2 (Fig. 3b). Thus, the oxidized form has an organized structure that provides a stable interaction network, and as a result, the oxidized γ subunit hampers the rotary action and has a slow rotation speed4.
The ε subunit binds peripherally and rotates together with the γ subunit. For bacterial F-type of ATP synthases, the ε subunit acts as a molecular switch to the catalysis by exerting a conformational transition of its C-terminal dual helices, which make up the so-called C-terminal domain (εCTD)39–43. However, a conformational change of the ε subunit was not observed in the CF1FO structures. This may suggest a different role of the ε subunit in regulating CF1FO given the low sequence similarity to the ε subunit of the E. coli and mitochondrial ATP synthases44,45.
Membrane FO domain
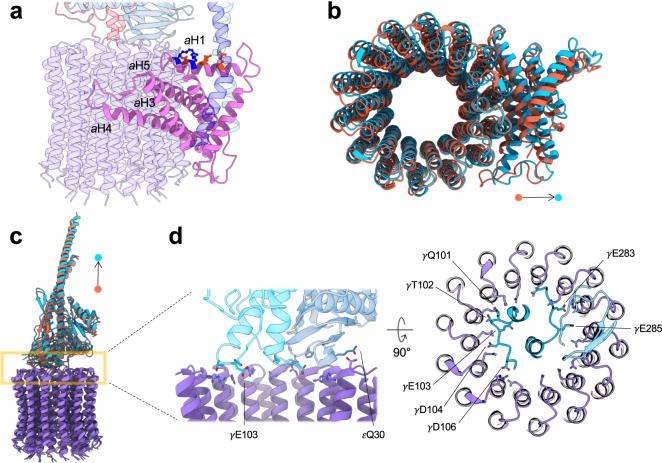
The membrane FO domain includes the c14 ring, subunit a, and the membrane part of the bb′ stator. Both the oxidized and reduced forms show the common overall FO domain structure found in all F-type ATP synthase, which is consistent with the previously determined CF1FO structure (PDB code: 6FKF, 6FKH, and 6FKI)18 (Fig. 1c; Supplementary Figs. 2, 3, 4). Subunit a is embedded in the membrane and sandwiched between the bb′ stator and c ring (Fig. 4a). The subunit a is composed of two half-channels responsible for proton translocation46, driving one full rotation of the c-ring rotor coupled to the synthesis of three ATP molecules47. The subunit a features a characteristic four-helix bundle from aH2 to aH5, binding peripherally to the c ring. The helix aH1 is parallel to the membrane plane with one side consisting of negatively charged residues (aGlu73, aGlu77, and aAsp81) facing the c ring and the peripheral bb′ stator, and the other side consisting of positively charged residues (aArg80 and aLys84) facing the membrane lipids (Fig. 4a). As the FO domains of the reduced and oxidized structures are similar, we conclude that the redox modulation process does not affect the overall structure of the proton half-channels in the a subunit or the c14 membrane rotor ring. The previously reported crystallographic structures of the isolated c14 ring from the spinach CF1FO showed a symmetric arrangement48,49. However, it is conceivable that the membrane ring may take on some level of flexibility to interact with the subunit a and the γ-ε central shaft. Our cryo-EM structures (Fig. 4d) showed that the residues on the top of the c ring are not symmetric, which may be caused by the electrostatic interactions between these conserved residues (cArg41, cGln42, and cPro43) and the polar or acidic residues on the bottom of the γ-ε central shaft (γGln101, γThr102, γGlu103, γAsp104, γAsp106, γGlu283, γGlu285, and εGln30)18,50,51 (Fig. 4d).
Fig. 4. Interaction of the γ-ε central shaft with the membrane FO domain.
a Spatial arrangement of the subunit a in the membrane. Color codes: subunit a (light pink), c14 ring (purple), bb′ stator (blue and light blue), γ subunit (crimson), and ε subunit (steel blue). Charged residues on the aH1 are shown as a ball-and-stick model. Negatively (aGlu73, aGlu77, and aAsp81) and positively (aArg80 and aLys84) charged residues are shown in orange and blue, respectively. b Alignment of the membrane c-ring rotors of the reduced and oxidized states. Superposition shows the subunit a of the reduced CF1FO is slightly away from the membrane c ring. c As in b, the superposition of the γ-ε central shafts of the reduced (light blue) and oxidized (orange) forms shows a slight translational movement of their centers of mass. d Interaction between the central shaft and the membrane c ring. Light blue and indigo represent the reduced γ subunit and ε subunit, respectively. cArg41 are shown as sticks with their side chains mostly pointing to the ring center. In the γ and ε subunits, the negatively charged and polar residues that interact with the top of the c ring are shown as sticks.
To analyze the movements between the two redox states, we superimposed the membrane c ring and analyzed the spatial arrangements of other subunits in different redox and rotary states (Fig. 4c). Among the three rotary states, the membrane c rings of the reduced and oxidized forms were superimposed (Fig. 4b). An exciting finding is that the peripheral subunit a slightly moves away from the c14 ring when the enzyme is active, reduced state in all three rotary states (Fig. 4b and Supplementary Fig. 12). This implicates that the reduction of the γ subunit may have an influence in facilitating the rotation of the membrane c14 ring, thereby generating ATP molecules at a higher turnover speed (Fig. 1b).
The peripheral stators bb′ has a large tilt and shift in its ectodomain, and the α3β3 catalytic unit with the δ stator moves along with the bb′ stator (Supplementary Fig. 12). The tilting angles of the F1 catalytic domains around the axis perpendicular to the membrane plane are 1.74°, 5.89°, and 3.25° in the rotary state 1, 2, and 3, respectively (Supplementary Fig. 12). Thus, even in the same rotary state, the reduced and oxidized forms do not have the same spatial arrangements between the subunit components. Moreover, the reduced γ-ε central shaft is slightly further away from the top of the c14 ring than the oxidized form (rotary state 1: 1.75 Å; rotary state 2: 2.55 Å; rotary state 3: 1.02 Å) (Fig. 4c). Rather than resulting from one single structural change, these concerted movements may synergistically contribute to the differences in efficiency, resulting in a change in the rotation transmission from the membrane rotor to the central shaft in the reduced and oxidized enzymes. Thus, the conformational changes between different redox γ subunits are likely to collaborate with other subunits to influence the rotation of the ATP synthase motor. However, because the movements between these subunits are small, these findings will need further investigation on higher resolution structures to define the trajectory of the movements. Due to its highly flexible nature, this may require further technical development to derive high-resolution structures in the future.
Discussion
What makes the chloroplast ATP synthase unique from other ATP synthases is the ability to modulate its enzymatic activity through different redox states6,52. In this work, we performed a redox titration on the spinach CF1FO enzyme, investigating the structures of different CF1FO redox forms using single-particle cryo-EM. Single-particle cryo-EM enables direct imaging of the molecular motors, and the subsequent image classification allows the data to be sorted into different conformations. Our cryo-EM reconstructions showed a complete view of the enzyme complexes, composed of α3β3δγεabb′c14 subunits (Fig. 1c), in different redox and rotary states. The overall architecture, especially the oxidized form, was consistent with the previous CF1FO structural data18. The resolutions of our 3D densities were anisotropic, which implies that either multiple metastable states co-exist or the enzyme features structural differences in the mobility of domains.
Previous cryo-EM studies and our structures of the ATP synthase showed different proportions of the three rotary states: rotary state 1 is the most populated, and rotary state 3 is the least populated18,42,53. This uneven proportion of the particle distribution implies that different energy levels are populated in individual rotary states while the motor rotates. As such, in the oxidized and control samples, the rotary state 1 is more energetically stable than the rotary state 3. While the disulfide bond was reduced, the proportions of the rotary state 2 and 3 were increased and that of the rotary state 1 was decreased (Supplementary Figs. 2b, 3b). Assuming that the proportion of the rotary state follows the Boltzmann distribution, our analysis may implicate that the energy differences for the transitions between the rotary states of the reduced form are lower than those of the oxidized form, allowing a smooth transition in the rotary action and thereby a fast rotation. On the other hand, single-molecule measurement of the CF1FO showed that the oxidized rotor can frequently take long pauses during rotation54. The oxidized state is very likely to stabilize certain rotary states, leading to an uneven rotation. The stabilization may result from the interaction network of the β hairpin structures of the γ subunit with its neighbor motifs.
Superposition of the reduced and oxidized γ subunit showed that the two hairpin structures may be the key to modulating the ATP synthase activity (Fig. 3b). The torsional restraints given by the disulfide linkage may stabilize the structure, strengthening the interactions between the γ and β subunits. When the γ subunit is reduced, the breakdown of the disulfide bond and instability of this short-hairpin loop probably confers an entropic gain to relax the mechanical torsion. On the other hand, the concerted movements of the CF1FO caused by the γ redox change modulate the properties of the transmission between the rotors. The γ subunit may act as a molecular clutch to control the rotation of the CF1FO motor. Understanding the mechanism in more detail will be required for further confirmation from biochemical studies or high-resolution structures.
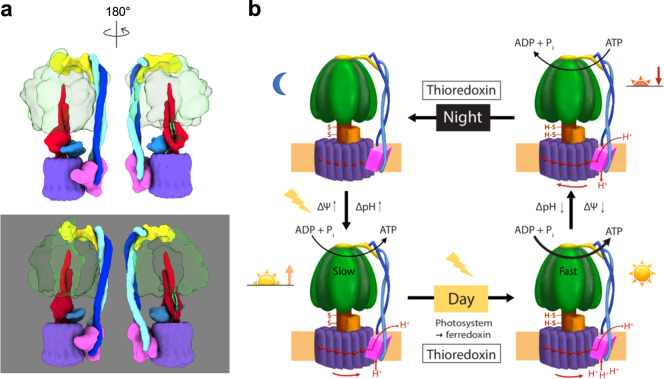
From our results summarized above, a mechanistic view of the molecular redox modulation of CF1FO can be deduced. When the plant leaves are in the darkness, the γ subunit of the CF1FO is oxidized to form a disulfide linkage, which introduces a torsional constraint on the structures of β hairpins 1 and 2. The hairpin structures also stabilize the interactions between the βDELSEED motif and the coiled coil of the γ subunit. In addition, the membrane-embedded subunit a and membrane c ring closely interact with the central shaft. These may limit the rotary actions to prevent unnecessary ATP hydrolysis and minimize the loss of ATP in the darkness (Fig. 5b). At sunrise, when the photosynthetic electron transport chain of the plant is activated, PSI transfers the electron to ferredoxin, which reduces thioredoxin and subsequently reduces the γ subunit of CF1FO (Fig. 5). The disconnected cysteines alleviate the torsion on the β hairpin 1 and remove the constraints of the β hairpin 2 of the γ subunit. This leads to multiple concerted movements within the CF1FO assembly to transition into efficient ATP synthesis. At the same time, the electron transport from the photosystem generates the proton gradient across the thylakoid membranes, which drives the c-ring rotation with less friction and synthesizes the ATP molecules. These synergistic actions would then keep the ATP synthesis in the activated state at full speed during the day.
Fig. 5. Proposed working model for the light regulatory mechanism.
a Cartoon schematics of the redox modulation. Upper and lower models are the reduced and oxidized states, respectively. Color codes are the same as in Fig. 1c and the β hairpin structures of the γ subunit are shown in light green. The two redox states are aligned in the same view. b At night, no energy input from light is available for the photosynthetic electron transport chain, and thus, no electrochemical potential (ΔΨ) and proton gradient (ΔpH) are generated. The oxidized γ subunit prevents CF1FO from hydrolyzing ATP. During the day, light induces charge separation to generate an electrochemical potential across the membrane. Although the CF1FO begins to synthesize ATP molecules, the γ subunit is still oxidized while ΔΨ is small. The rate of ATP synthesis is not at its maximum. At sunrise, thioredoxin subsequently reduces the γ subunit, fully activating CF1FO. The molecular motor, consisting of the γ-ε central shaft and the c14-ring, is free to rotate at full speed to maximize its ATP synthesis activity. Three ATP molecules per rotation of the c14 ring are produced. At sunset, the membrane becomes de-energized, leading to small ΔΨ and ΔpH, and the ATP hydrolysis starts to take place. To prevent ATP loss from excess ATP hydrolysis, the γ subunit is then oxidized again. This process of light regulation and redox modulation on the CF1FO will cyclize daily.
In summary, we determined the first structure of the active, reduced chloroplast ATP synthase to our knowledge. Our cryo-EM structures of the chloroplast ATP synthase revealed significant differences of the reduced and oxidized enzyme triggered by distinct redox states of the γ subunit. These results provide a fundamental framework of how CF1FO is regulated by the presence or absence of light. Our first structure of the active, reduced CF1FO brings together the experimental observations with the lines of structural evidence of the redox modulation that was investigated over multiple decades from researchers in the field into one consistent picture. This reveals the structural basis for light-dependent regulation of ATP synthesis in chloroplasts as a robust model for redox-dependent molecular switching.
Methods
Purification of the chloroplast ATP synthase complex
Samples of the chloroplast ATP synthase complexes were purified from baby spinach, Spinacia oleracea, as previously described for Heliobacterium modesticaldum55. Five kilograms of fresh baby spinach leaves (Fresh Express, Chiquita Brands International Inc.) were stored at 4 °C in the darkness for at least 3 days. Selected leaf tissues were placed in ice-cold water and subsequently homogenized in 100 mM Tricine (pH 8.0), 400 mM sucrose, and 2 mM MgCl2. The tissue lysate was centrifuged at 16,000 × g at 4 °C for 25 min.
Cell lysis was performed at 4 °C in the darkness using osmotic shock, followed by centrifugation at 16,000 × g for 30 min at 4 °C. The resuspended pellet was homogenized to reach a final concentration of 5 mM chlorophyll in 50 mM Tricine (pH 8.0), 400 mM sucrose, and 4 mM MgCl2. The membrane part was then solubilized in a buffer containing 10 mM Tricine-NaOH (pH 8.0), 100 mM sucrose, 2.5 mM MgCl2, 2.5 mM KCl, 2 mM ATP, 10% (w/v) NH4SO4, 50 mM dithiothreitol (DTT), 12.5 mM sodium cholate, and 30 mM β-D-octylglucoside (OG; Glycon Biochemicals, Luckenwalde, Germany). The supernatant was collected using centrifugation at 208,000 × g for 60 min at 4 °C. The purified complexes were precipitated using ammonium sulfate at the cut-off saturated concentrations of 32.5 to 45% (v/v). The precipitates were collected by centrifugation at 12,000 × g for 15 min at 4 °C and resuspended in 30 mM NaH2PO4 (pH 7.2), 200 mM sucrose, 2 mM MgCl2, 0.5 mM EDTA, and 4 mM n-dodecyl-β-D-maltoside (β-DDM; Glycon Biochemicals, Luckenwalde, Germany). The protein samples were further purified by discontinuous sucrose gradient centrifugation with the steps of 20, 28, 36, 44, 52, and 60% (w/v) sucrose in the gradient buffer consisting of 30 mM NaH2PO4 (pH 7.2), 2 mM MgCl2, 0.5 mM EDTA, 1 mg/ml asolectin, and 8 mM β-DDM. The bands at the interface between 44 and 52% sucrose interface were collected and loaded onto a POROS 20 HQ anion-exchange column (Thermo Fisher Scientific/Life Technologies, Waltham, MA) in 10 mM Tris-Cl (pH 8.0), 10 mM MgCl2, and 0.4 mM β-DDM with an elution gradient generated by 500 mM NaCl. The collected peak fraction was further purified using size-exclusion chromatography (Superdex 200 10/300 column, GE Healthcare, Chicago, IL) with 10 mM Tris-Cl (pH 8.0), 2 mM MgCl2, 500 µM ATP, and 0.4 mM β-DDM. The peak fraction was characterized using SDS-PAGE electrophoresis with silver staining and negative-stain electron microscopy (EM).
Luciferin/luciferase ATP synthesis assay
Functional assays were followed by the previous methods with modifications23,56. Liposomes were prepared by mixing L-α-phosphatidylcholine (Egg PC, Avanti) and L-α-phosphatidic acid (Egg PA, Avanti) at a lipid-to-protein ratio (LPR) of 19:1 (w/w) in 10 mM Tricine (pH 8.0), 100 μM EDTA, 500 µM DTT, 7.2 mg/ml sodium cholate, and 3.6 mg/ml sodium desoxycholate. The mixture was sonicated to form an emulsion and dialyzed at 30 °C for 5 h against 15 L of 10 mM Tricine (pH 8.0), 0.2 mM EDTA, 0.25 mM DTT, and 2.5 mM MgCl2. The purified proteins were reconstituted into liposomes by detergent removal with Bio-Beads (Bio-Rad SM2). The pH gradient was generated using a pH-pump step method23,56. Valinomycin was used to generate the potassium gradient across the membrane, leading to an additional chemiosmotic potential, Δφ, across the membrane when the buffers of high (pH 4.7) and low (pH 8.7) proton concentrations were mixed. The luminescence generated with luciferin and luciferase was detected using an LKB WALLAC-1250 luminometer (Vienna, Austria).
Electron microscopy of the enzyme complex
The purified complexes were negatively stained with 0.75% (w/v) uranyl formate. Electron images were recorded using a Philips CM12 transmission electron microscope (TEM) equipped with a side-mounted CCD camera (Model 791, Gatan, Pleasanton, CA). TEM imaging was operated at an acceleration voltage of 80 keV with the settings of a calibrated magnification of ×18,680, corresponding to a pixel size of 8.03 Å/pixel at the specimen level, and a defocus setting of about −1.6 μm.
For cryo-EM specimen preparation, 5 µl of 10 mg/ml protein sample was applied onto a glow-discharged C-flat holey carbon grid (1.2/1.3–4C or 2/1–4C, Protochips, Morrisville, NC). The grid specimen was plunge-frozen into liquid ethane using a Thermo Fisher/FEI Vitrobot Mark IV (Thermo Fisher/FEI, Hillsborough, OR) with a blotting time of 6 s and at a humidity of 100% at 22 °C. The frozen grid specimens were stored in the liquid-nitrogen dewar before imaging.
The protein samples were mixed with 100 mM DTT or 20 mM iodosobenzoate (IBZ; Sigma-Aldrich, Burlington, MA) to generate the reduced or oxidized samples, respectively. The protein samples were incubated for 10 min before plunge freezing. For the reduced sample, 0.05 mM tentoxin (Cayman Chemical, Ann Arbor, MI) was also added.
The data were collected in the Eyring Materials Center (EMC) at Arizona State University (ASU) and the European Synchrotron Radiation Facility (ESRF)57. The samples were imaged at the EMC using a Thermo Fisher/FEI Titan Krios TEM (Thermo Fisher/FEI, Hillsborough, OR) at an accelerating voltage of 300 kV, and the dose-fragmented frames were recorded on a Gatan K2 Summit direct electron detector (DED) camera in super-resolution mode (Gatan, Pleasanton, CA). The imaging was performed in the nanoprobe mode with a C2 aperture diameter of 50 µm. The low-dose procedure was applied with a defocus setting ranging from −1.5 to −4.0 µm and a nominal magnification of ×48,077, which corresponds to a physical pixel size of 1.04 Å/pixel at the specimen level. The beam intensity was adjusted to a counting rate of 2 counts/sub-pixel/s on the camera. The exposure time was 6 s with a subframe rate of 200 ms for each movie, accumulating to a total dosage of 43.5 e−/Å2. For the images of oxidized sample, the beam-image shift was applied to promote the data acquisition speed58. The data collection was automated using SerialEM program (version 3.7) with customized macros59.
The electron movies collected at the ESRF were recorded on a Thermo Fisher/FEI Titan Krios TEM equipped with a Quantum LS energy filter and a Gatan K2 Summit DED camera. The image data was recorded in the counting mode at a nominal magnification of 130,000 X, corresponding to a pixel size of 1.053 Å/pixel at the specimen level. The defocus range was set from −1.0 to −2.8 µm. The exposure time was 7 s for each movie, accumulating to a total dosage of 49 e−/Å2. The beam-image shift was applied during data collection for multiple images per foil-hole. The data collection was automated using EPU software (version 1.11; Thermo Fisher/FEI, Hillsborough, OR).
Image processing
3935, 2064, and 1637 movies were collected for the oxidized, reduced, and control samples, respectively. For the dataset of the oxidized and control samples, the IMOD program (version 4.9) “clip” was used to unpack and gain-normalize the movie data60. The movie frames were motion corrected and dose weighted using the MotionCor2 program (version 1.2.1)61, and the final frame average was performed with a Fourier cropping at the spatial frequency of 2 and 1.5 times physical Nyquist frequency, resulting in a pixel size of 1.04 and 0.788 Å/pixel for the movies of the oxidized and control samples, respectively. The processing workflow was generally followed the RELION program (version 3.1-beta-commit-da823c)62. The defocus and astigmatism of individual image was estimated by CTFFIND4 (version 4.1.13)63. The particle images were automatically selected using a template-based approach that was implemented in Relion software. The false positives were removed by iterative two-dimensional (2D) classification procedures and selection of 2D class averages. The final particle numbers used for 3D reconstruction and classification were 552,893, 108,691, and 208,371 for oxidized, reduced, and control samples, respectively.
The 3D initial densities were built using stochastic gradient descent method that was implemented in Relion software, except the initial density for the control sample was built using “sxviper.py” (SPARX) supplied with 2D class averages64. The initial densities were then refined against the particle images using regularized likelihood optimization in Relion. The density maps of different states and samples were generated by iterative 3D classification and refinement. To further improve the densities of the F1 region, the signal subtraction and focused refinement was applied65. The final density map was sharpened by an estimated b factor using Guinier plot66 and the modulation transfer function of the DED camera at 300 keV. The gold-standard Fourier-shell correlation (FSC) was used to estimate the overall resolution of the density map67. Local resolution was estimated using Relion software.
Modeling
The initial templates used for modeling were the structures of the spinach chloroplast ATP synthase (PDB codes: 6FKF, 6FKH, and 6FKI)18. The initial rigid-body fitting of the templates was performed against one of the half maps using “Fit in the Volume” function implemented in the UCSF Chimera (version 1.14)68. The density maps that were not sufficient to identify the side chains were only used to model the coordinates of the main chain. The atomic coordinates were rebuilt and fit to the density map using Coot (version 0.9-pre). The residue assignment was based upon the densities of bulky side chains, such as aromatic residues. The ligand configuration of the tentoxin coordinate was optimized using AM1 (Austin Model 1)69 quantum-mechanical method in eLBOW program70. The rebuilt model was refined against the density map using “phenix.real_space_refine” program in the PHENIX suite (version: 1.16–3546 or 1.17.1–3660)71. The representation of the atomic models was made by the UCSF Chimera or ChimeraX (version 0.91)68,72.
Statistics and reproducibility
Individual cryo-EM density maps are representative from over 10,000 particle images and the observations were described by quantitative data (Supplementary Figs. 2, 3, 4). The resolution of the reconstruction was estimated by randomly dividing dataset into two independent groups and determined using the gold-standard FSC method67.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The establishment of the isolation procedure for chloroplast ATP synthase (originally aimed for structure determination by X-ray crystallography) was supported by the National Institute of Health grant (R01GM081490) to P.F. We would like to acknowledge the use of the Titan Krios TEM in the EMC at ASU with the support funding for the instrumentation by NSF MRI 1531991. We thank David Lowry in the EMC at ASU for technical assistance. We also acknowledge the ESRF for the provision of cryo-EM time on CM01. The computation was partly supported by the NVIDIA GPU Grant Program to P.-L.C. The cryo-EM studies were partly supported by the ASU Biodesign Center for Applied Structural Discovery (CASD) and the ASU start-up fund to P.-L.C. We thank Yuval Mazor, Kuang-Lei Tsai, and Debra Hansen for proofreading the article. We thank Shangji Zhang and David Flesher for their assistance in some steps of the protein purification as well as Karie Robertson for designing and producing the graphic art for Figure 5b of the manuscript.
Author contributions
J.-H.Y. purified the protein samples and performed the functional assays. D.W., E.K., and P.-L.C. collected single-particle cryo-EM movie data. P.-L.C. performed single-particle image analysis and atomic modeling. P.F. and P.-L.C. conceived the ideas of the project and J.-H.Y., P.F., and P.-L.C. wrote the paper.
Data availability
Cryo-EM density maps (MRC format) were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-21270 (R1), EMD-21271 (R1-F1), EMD-21268 (R2), EMD-21269 (R2-F1), EMD-21266 (R3), EMD-21267 (R3-F1), EMD-21264 (O1), EMD-21265 (O1-F1), EMD-21262 (O2), EMD-21263 (O2-F1), EMD-21241 (O3), EMD-21239 (C1), EMD-21240 (C1-F1), EMD-21238 (C2), and EMD-21235 (C3). Model coordinates were deposited in the Worldwide Protein Data Bank (wwPDB) under accession numbers 6VON (R1), 6VOO (R1-F1), 6VOL (R2), 6VOM (R2-F1), 6VOJ (R3), 6VOK (R3-F1), 6VOH (O1), 6VOI (O1-F1), 6VOF (O2), 6VOG (O2-F1), 6VMG (O3), 6VMB (C1), 6VMD (C1-F1), 6VM4 (C2), and 6VM1 (C3). All the data are available in the EMDB and wwPDB database or from the corresponding author upon request. Data of the luminescence measurements for the reconstituted CF1FO function in different redox states are presented in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Petra Fromme, Email: Petra.Fromme@asu.edu.
Po-Lin Chiu, Email: plchiu@asu.edu.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-020-01221-8.
References
- 1.Boyer PD. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 3.Okuno D, Iino R, Noji H. Rotation and structure of FOF1-ATP synthase. J. Biochem. 2011;149:655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- 4.Nalin CM, McCarty RE. Role of a disulfide bond in the γ subunit in activation of the ATPase of chloroplast coupling factor 1. J. Biol. Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- 5.Mills JD, Mitchell P, Schürmann P. Modulation of coupling factor ATPase activity in intact chloroplasts. FEBS Lett. 1980;112:173–177. [Google Scholar]
- 6.Hisabori T, Sunamura E-I, Kim Y, Konno H. The chloroplast ATP synthase features the characteristic redox regulation machinery. Antioxid. Redox Signal. 2013;19:1846–1854. doi: 10.1089/ars.2012.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hisabori T, Ueoka-Nakanishi H, Konno H, Koyama F. Molecular evolution of the modulator of chloroplast ATP synthase: origin of the conformational change dependent regulation. FEBS Lett. 2003;545:71–75. doi: 10.1016/s0014-5793(03)00395-8. [DOI] [PubMed] [Google Scholar]
- 8.Junesch U, Gräber P. Influence of the redox state and the activation of the chloroplast ATP synthase on proton-transport-coupled ATP synthesis/hydrolysis. Biochim. Biophys. Acta Bioenerg. 1987;893:275–288. [Google Scholar]
- 9.Nelson N, Ben-Shem A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004;5:971–982. doi: 10.1038/nrm1525. [DOI] [PubMed] [Google Scholar]
- 10.Fromme P, Gräber P. Activation/inactivation and uni-site catalysis by the reconstituted ATP-synthase from chloroplasts. Biochim. Biophys. Acta. 1990;1016:29–42. doi: 10.1016/0005-2728(90)90003-m. [DOI] [PubMed] [Google Scholar]
- 11.Fromme P, Gräber P. Uni-site catalysis in thylakoids. The influence of membrane energization on ATP hydrolysis and ATP-Pi exchange. FEBS Lett. 1990;269:247–251. doi: 10.1016/0014-5793(90)81165-k. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 13.Schumann J, Richter ML, McCarty RE. Partial proteolysis as a probe of the conformation of the γ subunit in activated soluble and membrane-bound chloroplast coupling factor 1. J. Biol. Chem. 1985;260:11817–11823. [PubMed] [Google Scholar]
- 14.Ort DR, Oxborough K. In situ regulation of chloroplast coupling factor activity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:23. [Google Scholar]
- 15.Itoh H, et al. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Rubinstein JL. Cryo-EM of ATP synthases. Curr. Opin. Struct. Biol. 2018;52:71–79. doi: 10.1016/j.sbi.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Buchert F, Bailleul B, Hisabori T. A γ-subunit point mutation in Chlamydomonas reinhardtii chloroplast F1FO-ATP synthase confers tolerance to reactive oxygen species. Biochim. Biophys. Acta Bioenerg. 2017;1858:966–974. doi: 10.1016/j.bbabio.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, A., Vonck, J., Mills, D. J., Meier, T. & Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science360, 10.1126/science.aat4318 (2018). [DOI] [PMC free article] [PubMed]
- 19.Turina P, Samoray D, Gräber P. H+/ATP ratio of proton transport‐coupled ATP synthesis and hydrolysis catalysed by CFOF1—liposomes. EMBO J. 2003;22:418–426. doi: 10.1093/emboj/cdg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagendorf AT, Uribe E. ATP formation caused by acid-base transition of spinach chloroplasts. Proc. Natl Acad. Sci. U.S.A. 1966;55:170–177. doi: 10.1073/pnas.55.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard P, Rigaud JL, Gräber P. Reconstitution of CFOF1 into liposomes using a new reconstitution procedure. Eur. J. Biochem. 1990;193:921–925. doi: 10.1111/j.1432-1033.1990.tb19418.x. [DOI] [PubMed] [Google Scholar]
- 22.Fischer S, et al. ATP synthesis catalyzed by the ATP synthase of Escherichia coli reconstituted into liposomes. Eur. J. Biochem. 1994;225:167–172. doi: 10.1111/j.1432-1033.1994.00167.x. [DOI] [PubMed] [Google Scholar]
- 23.Fischer S, Gräber P. Comparison of ΔpH- and Δφ-driven ATP synthesis catalyzed by the H+-ATPases from Escherichia coli or chloroplasts reconstituted into liposomes. FEBS Lett. 1999;457:327–332. doi: 10.1016/s0014-5793(99)01060-1. [DOI] [PubMed] [Google Scholar]
- 24.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 2015;4:e10180. doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 27.Boyer PD. The binding change mechanism for ATP synthase–some probabilities and possibilities. Biochim. Biophys. Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 28.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 29.Boyer PD. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 1989;3:2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki K, et al. Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor. Nat. Commun. 2016;7:13235. doi: 10.1038/ncomms13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele JA, Uchytil TF, Durbin RD, Bhatnagar P, Rich DH. Chloroplast coupling factor 1: A species-specific receptor for tentoxin. Proc. Natl Acad. Sci. U.S.A. 1976;73:2245–2248. doi: 10.1073/pnas.73.7.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meiss E, Konno H, Groth G, Hisabori T. Molecular processes of inhibition and stimulation of ATP synthase caused by the phytotoxin tentoxin. J. Biol. Chem. 2008;283:24594–24599. doi: 10.1074/jbc.M802574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groth G. Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc. Natl Acad. Sci. U.S.A. 2002;99:3464–3468. doi: 10.1073/pnas.052546099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromme P, Dahse I, Gräber P. Effect of tentoxin on the activation and on the catalytic reaction of reconstituted H+-ATPase from chloroplasts. Z. für Naturforschung C. 1992;47:239–244. [Google Scholar]
- 35.Groth G, Pohl E. The structure of the chloroplast F1-ATPase at 3.2 Å resolution. J. Biol. Chem. 2001;276:1345–1352. doi: 10.1074/jbc.M008015200. [DOI] [PubMed] [Google Scholar]
- 36.Hu N, Mills DA, Huchzermeyer B, Richter ML. Inhibition by tentoxin of cooperativity among nucleotide binding sites on chloroplast coupling factor 1. J. Biol. Chem. 1993;268:8536–8540. [PubMed] [Google Scholar]
- 37.Mnatsakanyan N, Krishnakumar AM, Suzuki T, Weber J. The role of the βDELSEED-loop of ATP synthase. J. Biol. Chem. 2009;284:11336–11345. doi: 10.1074/jbc.M900374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konno H, et al. Inverse regulation of F1-ATPase activity by a mutation at the regulatory region on the γ subunit of chloroplast ATP synthase. Biochem. J. 2000;352:783–788. [PMC free article] [PubMed] [Google Scholar]
- 39.Capaldi RA, Aggeler R, Gogol EP, Wilkens S. Structure of the Escherichia coli ATP synthase and role of the γ and ε subunits in coupling catalytic site and proton channeling functions. J. Bioenerg. Biomembr. 1992;24:435–439. doi: 10.1007/BF00762359. [DOI] [PubMed] [Google Scholar]
- 40.Yagi H, et al. Structures of the thermophilic F1-ATPase ε subunit suggesting ATP-regulated arm motion of its C-terminal domain in F1. Proc. Natl Acad. Sci. U.S.A. 2007;104:11233–11238. doi: 10.1073/pnas.0701045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobti, M. et al. Cryo-EM reveals distinct conformations of E. coli ATP synthase on exposure to ATP. eLife8, 10.7554/eLife.43864 (2019). [DOI] [PMC free article] [PubMed]
- 43.Suzuki T, et al. FOF1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of ε subunit in response to proton motive force and ADP/ATP balance. J. Biol. Chem. 2003;278:46840–46846. doi: 10.1074/jbc.M307165200. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi-Matsui M, Sekiya M, Futai M. ATP synthase from Escherichia coli: Mechanism of rotational catalysis, and inhibition with the ε subunit and phytopolyphenols. Biochim. Biophys. Acta. 2016;1857:129–140. doi: 10.1016/j.bbabio.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Feniouk BA, Suzuki T, Yoshida M. The role of subunit ε in the catalysis and regulation of FOF1-ATP synthase. Biochim. Biophys. Acta. 2006;1757:326–338. doi: 10.1016/j.bbabio.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 46.Vik SB, Antonio BJ. A mechanism of proton translocation by F1FO ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 47.Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK. The rotary mechanism of the ATP synthase. Arch. Biochem. Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vollmar M, Schlieper D, Winn M, Büchner C, Groth G. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J. Biol. Chem. 2009;284:18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vlasov AV, et al. Unusual features of the c-ring of F1FO ATP synthases. Sci. Rep. 2019;9:18547. doi: 10.1038/s41598-019-55092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allegretti M, Mills DJ, McMullan G, Kühlbrandt W, Vonck J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. eLife. 2014;3:e01963. doi: 10.7554/eLife.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonck J, et al. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 2002;321:307–316. doi: 10.1016/s0022-2836(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 52.Kohzuma K, et al. The role of light-dark regulation of the chloroplast ATP synthase. Front. Plant Sci. 2017;8:1248. doi: 10.3389/fpls.2017.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobti, M. et al. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. eLife5, 10.7554/eLife.21598 (2016). [DOI] [PMC free article] [PubMed]
- 54.Bald D, Noji H, Yoshida M, Hirono-Hara Y, Hisabori T. Redox regulation of the rotation of F1-ATP synthase. J. Biol. Chem. 2001;276:39505–39507. doi: 10.1074/jbc.C100436200. [DOI] [PubMed] [Google Scholar]
- 55.Yang JH, et al. Purification and biochemical characterization of the ATP synthase from Heliobacterium modesticaldum. Protein Expr. Purif. 2015;114:1–8. doi: 10.1016/j.pep.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turina P, Samoray D, Gräber P. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CFOF1-liposomes. EMBO J. 2003;22:418–426. doi: 10.1093/emboj/cdg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kandiah E, et al. CM01: a facility for cryo-electron microscopy at the European Synchrotron. Acta Crystallogr. D. Struct. Biol. 2019;75:528–535. doi: 10.1107/S2059798319006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng A, et al. High resolution single particle cryo-electron microscopy using beam-image shift. J. Struct. Biol. 2018;204:270–275. doi: 10.1016/j.jsb.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 61.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Bai, X.-C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S.H.W. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife4, 10.7554/eLife.11182 (2015). [DOI] [PMC free article] [PubMed]
- 66.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. Development and use of quantum mechanical molecular models. 76. AM1: a new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985;107:3902–3909. [Google Scholar]
- 70.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Cryo-EM density maps (MRC format) were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-21270 (R1), EMD-21271 (R1-F1), EMD-21268 (R2), EMD-21269 (R2-F1), EMD-21266 (R3), EMD-21267 (R3-F1), EMD-21264 (O1), EMD-21265 (O1-F1), EMD-21262 (O2), EMD-21263 (O2-F1), EMD-21241 (O3), EMD-21239 (C1), EMD-21240 (C1-F1), EMD-21238 (C2), and EMD-21235 (C3). Model coordinates were deposited in the Worldwide Protein Data Bank (wwPDB) under accession numbers 6VON (R1), 6VOO (R1-F1), 6VOL (R2), 6VOM (R2-F1), 6VOJ (R3), 6VOK (R3-F1), 6VOH (O1), 6VOI (O1-F1), 6VOF (O2), 6VOG (O2-F1), 6VMG (O3), 6VMB (C1), 6VMD (C1-F1), 6VM4 (C2), and 6VM1 (C3). All the data are available in the EMDB and wwPDB database or from the corresponding author upon request. Data of the luminescence measurements for the reconstituted CF1FO function in different redox states are presented in Supplementary Data 1.