Abstract
Growing evidences suggest that the fibroblast growth factor/FGF receptor (FGF/FGFR) signaling has crucial roles in a multitude of processes during embryonic development and adult homeostasis by regulating cellular lineage commitment, differentiation, proliferation, and apoptosis of various types of cells. In this review, we provide a comprehensive overview of the current understanding of FGF signaling and its roles in organ development, injury repair, and the pathophysiology of spectrum of diseases, which is a consequence of FGF signaling dysregulation, including cancers and chronic kidney disease (CKD). In this context, the agonists and antagonists for FGF-FGFRs might have therapeutic benefits in multiple systems.
Subject terms: Diseases, Developmental biology
Introduction of the FGF/FGFR signaling
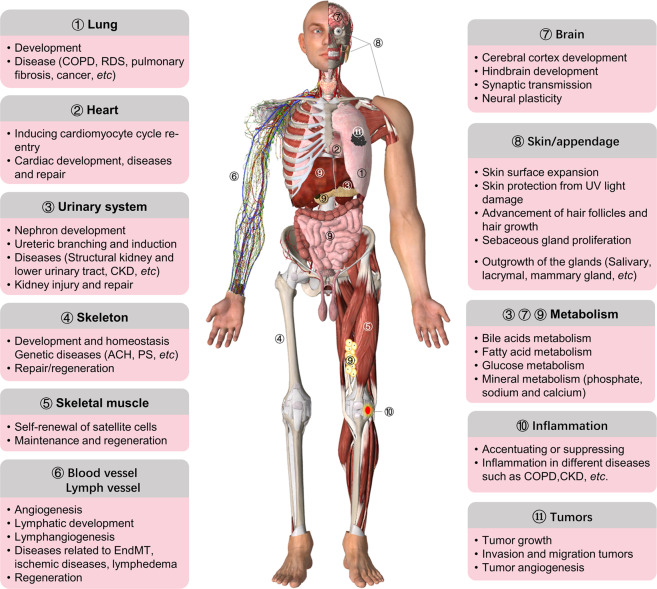
Fibroblast growth factors (FGFs) are broad-spectrum mitogens and regulate a wide range of cellular functions, including migration, proliferation, differentiation, and survival. It is well documented that FGF signaling plays essential roles in development, metabolism, and tissue homeostasis. The malfunction of FGF/FGF receptor (FGFR) signaling axis is observed in a variety of human diseases, such as congenital craniosynostosis and dwarfism syndromes, as well as chronic kidney disease (CKD), obesity, insulin resistance, and various tumors (Fig. 1).
Fig. 1.
Summary of the main roles of FGF/FGFR signaling in organ development, metabolism, and disease. FGF/FGFR signaling participates in the development of almost all organ such as lung, heart, urinary system, brain, skeleton, muscle, and skin/appendage, as well as angiogenesis and lymphangiogenesis. FGFs/FGFRs also have important effects on tissue repair, regeneration, and inflammation. Furthermore, endocrine FGFs play critical roles in metabolism by regulating kidney, liver, brain, intestine, and adipose tissue. The malfunctions of FGF/FGFR signaling lead to multiple kinds of diseases, such as genetic diseases, cancer, COPD, and CKD. The roles of FGF signaling in appendage development, such as epidermis, hair, and glands, and so on, is not mentioned in this review. ACH achondroplasia, CKD chronic kidney disease, COPD chronic obstructive pulmonary disease, PS Pfeiffer syndrome, RDS respiratory distress syndrome, EndMT endothelial-to-mesenchymal transition
FGF family is one of the most diverse growth factor groups in vertebrates. In mice and humans, 22 FGF ligands have been identified. Based on sequence homology and phylogeny, the 18 canonical mammalian FGFs are divided into six subfamilies, including five paracrine subfamilies and one endocrine subfamily.1 Five paracrine subfamilies contain the FGF1 subfamily (FGF1 and FGF2), the FGF4 subfamily (FGF4, FGF5, and FGF6), the FGF7 subfamily (FGF3, FGF7, FGF10, and FGF22), the FGF8 subfamily (FGF8, FGF17, and FGF18), and the FGF9 subfamily (FGF9, FGF16, and FGF20). The FGF19 subfamily (FGF19, FGF21, and FGF23) signals in an endocrine manner.1
FGFs exert their pleiotropic effects by binding and activating high-affinity tyrosine kinase receptors that are coded by four genes (FGFR1, FGFR2, FGFR3, and FGFR4) and FGFRL1, a truncated FGFR without intracellular domain,2 in mammals. FGFRs are single-pass transmembrane proteins containing an extracellular domain, a transmembrane domain (TMD), and an intracellular tyrosine kinase domain. Among them, the extracellular domain is composed of three immunoglobulin (Ig)-like domains (D1–D3), an acidic region, a heparin-binding motif for FGFs, heparan cofactors, and partner proteins. The TMD anchors the receptors in cell membrane and facilitates its dimerization. In the cytosol, the juxtamembrane region of FGFRs is involved in receptor dimerization, while the split kinase domains are required for the transmitting of FGF-related signaling.3
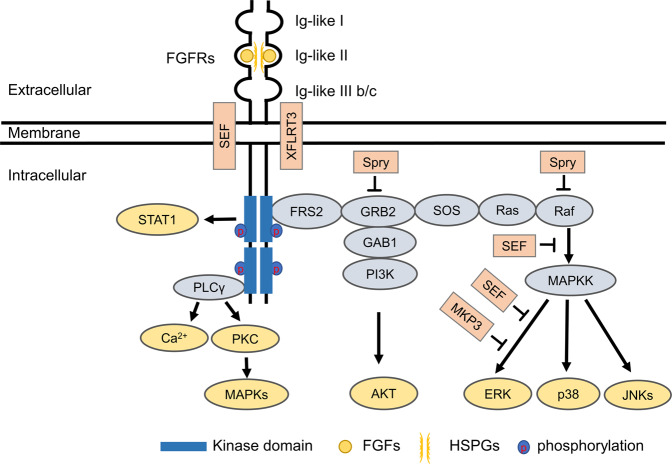
The binding of FGFs to the inactive monomeric FGFRs will trigger the conformational changes of FGFRs, resulting in dimerization and activation of the cytosolic tyrosine kinases by phosphorylating the tyrosine residues within the cytosolic tail of FGFRs.4 Then, the phosphorylated tyrosine residues serve as the docking sites for downstream signaling molecules, such as FGFR substrate 2α, which is localized on the plasma membrane.5 FGFRs also recruit and phosphorylate SH2 domain-containing substrate phospholipase Cγ (PLCγ) by formatting an allosteric 2:1 FGFR–PLCγ complex, indicating that FGFR dimerization plays an obligatory role in substrate phosphorylation.6 Depending on the cellular content in distinct cells and tissues, the classical FGF/FGFR downstream signaling pathways include Ras/Raf-MEK-MAPKs (mitogen-activated protein kinases), phosphatidylinositol-3 kinase/protein kinase B (PI3K/AKT), PLCγ, and signal transducer and activator of transcription (STAT).1,7 Additionally, several proteins belonging to FGF synexpression group have been identified, such as Sprouty (Spry),8,9 XFLRT3,10 SEF,11,12 MKP3,13,14 and so forth. These proteins are themselves regulated by FGF signaling and are tightly co-expressed with FGFs. Most of them inhibit FGF/FGFR signaling by establishing negative feedback loops15 (Fig. 2).
Fig. 2.
The classical FGF/FGFR pathways. Binding of appropriate growth factors to receptors triggers the conformational changes of FGFRs, resulting in dimerization and activation of FGFRs. Activated FGFRs phosphorylate FRS2a and FRS2a binds to SH2 domain-containing adaptor Grb2. Grb2 will subsequently bind to SOS, GAB1, and Cbl through its SH3 domain to activate Ras/Raf/MAPKs, including ERK MAPK, p38 MAPK, and JNK MAPK. The activated FGFRs also activate phosphatidylinositol (PI)-3 kinase and STAT. FGFRs recruit and phosphorylate PLCγ. Among the members of the FGF synexpression group, SEF and XFLRT3 are transmembrane proteins and can interact directly with FGFRs. SEF functions as a negative regulator by affecting the phosphorylation of the MAPK ERK cascade. XFLRT3 forms a complex with FGF receptors and enhances FGF/FGFR signaling. Spry acts at the level of Grb2 and/or the level of Raf to attenuate FGF/FGFR signaling. MKP3 negatively regulates FGF/FGFR signaling by dephosphorylating the activated ERK. FRS2α FGFR substrate 2α, GAB1 GRB2 associated binding protein 1, GRB2 growth factor receptor-bound 2, PKC protein kinase C, SOS son of sevenless
The diversified functions of FGF/FGFR signaling indicate the complex regulation of the signaling cascades. FGF/FGFR signaling can be modified at several levels, including ligand–receptor binding specificity,16 expressions1 and alternative splicing,17 and the crosstalk between FGFs/FGFRs and other signaling cascades,18 such as BMP (bone morphogenetic protein)19 and Wnt signalings.20,21 FGF/FGFR binding specificity/promiscuity combined with ligand-dependent differences in receptor orientation is the main mechanisms for the precise regulation of FGF-induced signaling.16 FGF/FGFR signaling is tightly regulated by the spatial and temporal expressions of FGFs, FGFRs, and heparan sulfate cofactors.15,22 Diversified tissue distribution and different expression levels of signaling components, which influences the function of FGF/FGFR signaling, eventually affect the tissue development, maintenance, and disease pathogenesis.1 Alternative splicing and translational initiation generate multiple isoforms of FGFs/FGFRs and regulate their expression levels.23 For example, the tissue-specific alternative splicing in D3 of FGFR1, FGFR2, and FGFR3 can generate b and c isoforms, and thus determines the binding specificity/promiscuity for individual FGFs at diverse cells and tissues.24 Furthermore, it is well documented that epigenetic mechanisms,25 the posttranslational modifications, such as phosphorylation,26 glycosylation,27 ubiquitination,28 and cellular trafficking of FGFs/FGFRs29,30 are also involved in the regulation of the expressions of FGF/FGFR signaling components and the signal specificity, intensity, and timing.
During the past decades, repaid progresses have been made about the modulation of FGF/FGFR signaling cascades; these studies not only deepen our understanding of the unique properties of FGF/FGFR signaling, but also raise the opportunity for developing new therapies targeting causative FGF/FGFR signaling.
Coreceptors of FGFs/FGFRs
Usually, specific ligands require assembly of the ternary complexes composed of ligand, receptor, and coreceptor at the cell surface to initiate signal transduction. The coreceptors of FGF/FGFR cascade include heparan sulfate proteoglycans (HSPGs) (for paracrine FGFs) and Klotho (for endocrine FGFs).
HSPGs
HSPGs are glycoproteins, containing one or more covalently attached heparan sulfate (HS) chains. According to their location, the HSPGs are grouped into three groups: membrane HSPGs, such as syndecans and glycosylphosphatidylinositol-anchored proteoglycans (glypicans), the secreted extracellular matrix HSPGs (agrin, perlecan, type XVIII collagen), and the secretory vesicle proteoglycan, serglycin.31 HSPGs is a mandatory cofactor in paracrine FGF signaling. Paracrine FGFs have moderate to high affinity for HSPGs, which shortens FGF diffusion distance away from their secretion cells. The interaction also provides a depot of regulatory factors that can be released by selective degradation of the HS chains facilitating the formation of FGF gradients essential for cell specification during development and regeneration.22
Structural studies have revealed that the HSPG binding site of FGFs contains the β1–β2 loop and the extended β10–β12 region, and each FGF ligand has discrete affinity for HSPGs.32 HSPG-mediated FGF-specific morphogenetic gradients contribute to the distinct function of FGFs. Importantly, endocrine FGFs such as FGF19 and FGF23 lack the paracrine-conserved glycine box and the truncated β10–β12 region in the potential HS binding region, reducing the binding affinity between HSPGs and the endocrine FGFs (FGF19 subfamily), which allows these FGF ligands to permeate through the HSPG-rich extracellular matrix (ECM) and subsequently enter the blood circulation.33
Detailed crystal studies reveal that HSPGs promote the formation of a 2:2:2 dimer between FGF, FGFR, and HSPGs.34 By engaging ligand and receptors in the dimer, HSPGs promote the kinetics and thermodynamics of FGF-FGFR binding and dimerization, which is required for the transmission of a sustained and robust intracellular signals.34
Klotho
Klotho are coreceptors for endocrine FGF signaling. As single-pass transmembrane proteins, Klotho consists of tandem KL domains, and are homologous to β-glucosidases. Modeling studies showed that the endocrine FGFs (FGF19, FGF21, and FGF23) exhibit a negligible HSPGs binding affinity and poor affinity for their cognate FGFRs, resulting in ineffective endocrine FGF/FGFR binding and dimerization.33 It is well established that α/β Klotho coreceptors are required for these ligands to initiate respective signaling activity.33 The Klotho coreceptors associate constitutively with the c-splice isoforms of FGFR1-3 and FGFR4 to promote their binding of FGFs and dimerization, reinforcing FGF/FGFR signaling specificity. For example, FGF23 can bind and activate FGFR1c-α-Klotho, FGFR3c-α-Klotho, and FGFR4-α-Klotho. A recent atomic structure study showed that α-Klotho simultaneously binds FGFR1c and FGF23, and dimerization of the stabilized ternary complexes and receptor activation depend on the binding of HS.35 FGF19 activates FGFR1c-β-Klotho (KLB) and FGFR4-KLB, whereas FGF21 mainly activates the FGFR1c-KLB complex.36
Endocrine FGF/FGFR signaling rely on the interaction between FGFRs and Klothos. Biochemical studies revealed that α-Klotho combines with FGFR1c to create a de novo site for the FGF23 carboxy tail, whereas KLB uses two distinct sites to independently bind FGFR and the carboxy tail of FGF19 or FGF21.37,38 The proteolytically cleaved FGF23 carboxy tail can competitively inhibit the binding of native FGF23 to the FGFR1c-α-Klotho complex and thus downregulate FGF23 signaling.39 In patients with autosomal-dominant hypophosphatemic rickets (ADHR), the mutations in the RXXR motif located in the carboxy tail abrogate the proteolytic cleavage of FGF23 and thus elevate the serum levels of full-length bioactive FGF23, which accelerates the excretion of phosphate from the kidney.40,41 Mutations in D3 hydrophobic groove of FGFRc isoforms and FGFR4 residues abolishes Klotho binding, indicating the overlapping between FGFs and Klotho binding sites on FGFRs.38 The association of FGFRs with the Klotho coreceptor decreases the ability of these receptors to respond to paracrine FGFs, such as FGF8, supporting the notion that endocrine and paracrine FGF signaling affect each other.38
Modulators of FGF/FGFR signaling
Cell adhesion molecules (CAMs)
CAMs are typically single-pass transmembrane receptors and include four major groups: cadherins, integrins, the Ig superfamily of CAMs (IgCAMs), and the superfamily of C-type of lectin-like domains proteins.42 A growing body of data reveals that various CAMs can act as FGFR binding partners, participating in the modulating of FGF/FGFR signaling and are strongly implicated in cell fate determination of different cell lineages.43
Cadherins play an essential role in the formation and adaptive reinforcement of adherens junctions, and modulation of the dynamics of actin cytoskeleton.44 Different members of the cadherin family are expressed in a cell type-specific manner, and most of the cell types express multiple cadherins, including VE-, N-, and T-cadherin. N-cadherin is associated with FGFRs through their acidic box-mediated activation of FGFRs and their downstream signaling in numerous cells.45 In breast cancer cells, formation of N-cadherin complexes with FGFR1 can decrease the internalization and lysosomal degradation of FGFR1, and thus sustain the receptor signaling via MAPKs, whereas silencing of N-cadherin results in the accelerated FGFR1 degradation. Thus, N-cadherin stabilizes FGFR1 and simultaneously enhances FGF2-induced proliferation and differentiation of epiblast stem cells.46 In addition, cadherin-11–FGFR1 interaction occurs through their extracellular domains. Cadherin-11 initiates intracellular signaling pathways via FGFR1 and recruits FGFR1 into the cell–cell contact area. The cadherin-11-induced FGFR1 signaling stimulates neurite outgrowth.47
The FGFR/neural CAM (NCAM) complexes have been observed in multiple cell types.48 The FN3 domains of NCAMs mediate its interaction with the Ig2–Ig3 region of FGFRs.49 NCAMs bind to FGFR1–FGFR3 to activate the receptor and initiation of signaling cascades and inhibit FGFR K27- and K29-linked polyubiquitination and lysosomal degradation.50 Interestingly, NCAMs can affect the cellular trafficking of FGFRs.51 In contrast to FGF-induced activation and lysosomal degradation of endocytic FGFR1, NCAM can promote the stabilization of FGFR1, which is recycled from endosomes to the cell surface through a Rab11 and Src-dependent manner.51
Integrins act as the receptors for extracellular matrix molecules, playing a key role in regulating intercellular contact and intracellular signaling. Eighteen α-subunits and eight β-subunits assemble into 24 functional integrins that vary in terms of ligand specificity and cellular function.52 Each α–β combination can bind to unique matrix components. Increasing evidences showed that integrins modify FGF/FGFR signaling.53 For example, the fibronectin-binding α5β1-integrin dimer upregulates FGF2 expression, while secreted FGF2 directly binds to αvβ3 integrin.54,55 FGF1, FGFR1 and integrin αvβ3 can be assembled into a ternary complex, in which FGF1 acts as a bridging molecule, to maintain sustained activation of FGFR1-dependent kinases ERK1/2.56
NCAM is a member of IgCAMs containing Ig-like and fibronectin type III (FNIII) domains. NCAM plays a critical role in neurite outgrowth as binding partners affecting the signaling process. A peptide derived from the NCAM FNIII region binds to FGFR1 directly to stimulate FGFR1 phosphorylation in primary rat neurons.51 In PC12 cells, NCAM requires FGFRs to promote neurite growth.57 Specifically, the NCAM-FGFR interaction activates PLCγ and diacylglycerol lipase to generate arachidonic acid, elevating intracellular calcium levels and activating Ca2+-dependent protein kinase C (PKCs).58 NCAM has been found to form a complex with FGFR4. This complex can lead to β1-integrin-mediated cell–matrix adhesion, and also decrease the mobility of pancreatic tumor cells by stimulating FGFR4 kinase activity.59
G protein-coupled receptors
G protein-coupled receptors (GPCRs) constitute the largest groups of receptors that mainly transmit various signals across cell membranes through binding and activating heterotrimeric G proteins. Structurally, GPCRs are composed of an N-terminal extracellular domain, seven-transmembrane helices, and a C-terminal region.60 A growing number of studies have revealed that various members of GPCRs and receptor tyrosine kinase (RTKs) can form heterocomplexes together and trigger different intracellular signaling and cellular response.61,62 The GPCRs can transactivate multiple RTKs,63 including epidermal growth factor receptor,64 platelet-derived growth factor receptors (PDGFRs),65 and insulin-like growth factor receptors,66 and so on.
In the central nervous system, both GPCR and FGFR signaling are involved in the control of proliferation, migration, survival, and differentiation of neurons. More and more studies have showed that GPCRs form heterocomplexes with FGFRs and regulate the cell fate of neurons.67 Multiple methods have confirmed the interaction between FGFR1 and adenosine receptor A2AR. The function study revealed that this interaction is required for the enhanced activation of ERK1/2, which is important for the regulation of the synaptic plasticity.68 Another study showed that cannabinoid receptor 1 (CB1R)-FGFR1 complexes occur in the lipid rafts of the plasma membrane, leading to activation of ERK1/2, and play important roles in neuronal differentiation.69 CB1R activates Fyn and Src via PKC signaling, inducing the transactivation of FGFR1 by phosphorylating its kinase domain.69 The interactions between FGFR1 and muscarinic acetylocholine receptor (mAChR) subtype M1R and 5-hydroxytriptamine receptor 1A (5-HT1A) have been visualized.70 Stimulation of hippocampal neurons with M1R agonist oxotremorine-M activated FGFR1, and the crosstalk between mAChR and FGFR1 enhanced the neurite growth.71 Treatment of FGF2 and 5-HT1A agonist 7-(dipropylamino)-5,6,7,8-tetrahydronaphthalen-1-ol (8-OH-DPAT) can increase the FGFR1–5-HT1A complexes; activation of 5-HT1A by 8-OH-DPAT causes subsequent FGFR1 phosphorylation mediated by Src.70 Interestingly, the FGFR1-5-HT1A heterocomplexes display anti-depressive effects and thus may be the novel targets for the treatment of mood disorders.72
Other RTKs
FGF/FGFR signaling can also be modified by their interplay with other members of RTK family. The crosstalk among RTKs can occur at different levels, such as the ligand, receptor, and downstream cascades. Among them, different RTKs can form receptor heterocomplexes and subsequently cause tyrosine phosphorylation of one receptor by tyrosine kinase of the other one. Binding with other RTKs gives another way to modify FGF/FGFR activities more elegantly.
Eph receptors constitute the largest family of RTKs, including EphA (EphA1-EphA10) and EphB (EphB1-EphB6) receptors, and are activated by ephrin ligands.73 The Eph receptors contain structural features characteristic for RTKs. The Eph receptor-ephrin complexes regulate cell adhesion, organization of cytoskeleton, angiogenesis, neural development, and plasticity.74 EphA4 receptor interacts with FGFRs through the tyrosine kinase domain of Eph4 and the JM domain of FGFR1-4.75 More detailed analysis revealed that phosphorylation of the tyrosine residues within JM domain of Eph4 is required for the formation of EphA4–FGFR complexes. Kinase domains of EphA4 and FGFRs can trans-phosphorylate each other.75 Importantly, the ternary complex, involving FGFR1, EphA4, and FRS2α, was detected. FRS2α may act as a tethering molecule that integrates signals from both receptors and regulates the self-renewal, proliferation, and differentiation of neural stem/progenitor cells.76 Studies also showed that FGFR phosphorylate ephexin1, a targeting molecule of EphA receptors.77 Scaffolding protein Dlg-1, which directly interacts with EphA receptors, can also modulate FGFR signaling.78
PDGFRα and PDGFRβ are activated by multiple PDGFs: PDGF-AA, PDGF-BB, PDGF-AB, PDGF-CC, and PDGF-DD.79 PDGFR-mediated signaling can regulate cell motility, proliferation, angiogenesis, and are involved in a range of diseases.80 In vitro and in vivo experiments revealed that both PDGFRα and PDGFRβ interact with high affinity with FGFR1.81 The formation of PDGFRα-FGFR1 complexes is facilitated by the presence of ligands for both receptors. In receptor heterocomplex, PDGFRβ can directly phosphorylate FGFR1 on tyrosine residues.81 Interestingly, FRS2α functions as a bridging molecule between PDGFRβ and FGFR1, further supporting the speculation that FRS2α may act as a tethering molecule integrating signals from different RTKs.81
Nuclear FGFs and FGFRs
In addition to the FGF/FGFR complexes at plasma membrane, it has been recognized that canonical FGF ligands and FGFRs can enter the nucleus of multiple types of cells and tissues.82 Nuclear localization of FGFs/FGFRs lends an additional layer of regulatory complexity.83,84 Nuclear FGFs/FGFRs can exert their effects on proliferation, lineage commitment, and gene expressions. Dysregulation of nuclear FGFs/FGFRs has been found in congenital skeletal disorders and neoplastic transformation.85
Nuclear localization of FGFs and FGFRs has been demonstrated in multiple tissues in different pathophysiological conditions. During gonadal development, FGFR2 is firstly localized to the plasma membrane of proliferating sertoli progenitor cells, but in the early stage of specification and differentiation, FGFR2 is colocalized with SRY and SOX9 in the nucleus of sertoli cell.86 In the development of salivary gland, nuclear FGFR2 is specifically located in proliferating epithelial cells at the branch tips in response to FGF10.87 In human pancreatic cancer cells, FGFR1 and FGF2 are localized to the nucleus where they promote proliferation and invasion.88 In breast mucinous carcinoma, nuclear FGFR2 is commonly found colocalized with STAT5 and Runx2.89 The nuclear FGFR3 levels in breast, bladder, and pancreatic cancer cells are higher than those in corresponding non-tumor tissues.90
Several FGF ligands contain a nuclear localization signal to facilitate their nuclear import, and different mechanisms are involved in the receptor nuclear localization.91,92 In some cases, nuclear localization of full-length FGFRs occurs through a ligand-dependent mechanism. For example, FGF2, FGF1, and FGF10 localize to the nucleus with FGFR1.93,94 Structurally, all FGFRs contain a single-pass TMD, the major determinant of intracellular localization. Mutations in the TMD in FGFR1 and FGFR2 remarkably affect their subcellular distribution. FGFR2 mutations (FGFR2M391R and FGFR2Y381D) located in the TMD can reduce plasma membrane levels of FGFR2, and amplify its nuclear and nucleolar presence in growth plate chondrocytes derived from patients with skeletal disorder bent bone dysplasia syndrome (BBDS).95,96 Interestingly, posttranslational modifications, such as glycosylation, also contribute to the nuclear localization of FGFRs. In the skeletal disorder Crouzon syndrome, the FGFR2 mutation (FGFR2C278F) leads to incomplete FGFR2 glycosylation, blocks its membrane localization, and induces the perinuclear accumulation of receptor.97 It was found that FGFR1 and FGFR2 exert their nuclear import through a β-importin-dependent active nuclear pore-mediated mechanisms,93 and proteolytically cleaved FGFR1 and FGFR3 mediated by granzyme B and γ-secretase localize in the nucleus of invading cancer cells and multiple cell lines,94 but the detailed molecular events are still unclear.
Once in the nucleus, FGFs and FGFRs can promote gene expressions through multiple approaches, such as epigenetic mechanisms. In embryonic stem cells and neuronal cells, FGFR1 binds the proximal promoters and activates the transcription of pluripotency-related genes, Wnt/β-catenin signaling components, and P53.98 In preosteoblasts, FGFR2 and FGF2 localize to the nucleolus to recruit histone remodeling factors, such as the CBP homolog p300, to ribosomal DNA (rDNA) and activate RNA polymerase I-mediated transcription, increasing ribosome biogenesis and subsequently protein synthesis.95,96 Nuclear FGF/FGFR-mediated regulation of transcription suggests an alternative way through which FGFs/FGFRs can directly induce specific and rapid changes of gene expressions. In osteoprogenitor cells, nuclear FGFR2-mediated regulation of rDNA transcription promotes self-renewal over terminal osteoblast differentiation.95,96 In invading breast cancer cells, FGFR1 undergoes nuclear translocation and activates the transcription of genes critical for cell migration.94 The activating mutant FGFR2 Y376C in endometrial cancer has increased perinuclear localization and appears to be involved in disrupting cell polarity in metastatic cells.99 In pancreatic cancer, nuclear FGFR3 correlates with metastatic disease and poor overall prognosis.90
Compared with the well-established mechanisms in transmembrane signaling, the mechanisms for FGF/FGFR cascades in the nucleus are less studied. Nuclear localization of RTKs is not unique to the FGFs/FGFRs.100,101 It is very important to clarify the precise mechanisms for nuclear FGFR translocation, activation of downstream pathways, and target genes, as well as its functions in different pathophysiological conditions in the future study.
FGF signaling in skeleton development and repair/regeneration
Expressions of FGFs and FGFRs during skeleton development
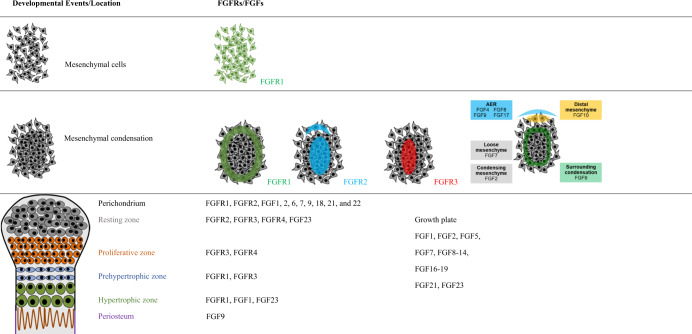
Both FGFs and FGFRs have characteristic spatiotemporal expression patterns throughout all stages of skeletal development (Table 1).102
Table 1.
FGF and FGFR expression in long bone development
During limb bud development, the active epithelial–mesenchymal interactions between ectoderm-expressed FGF (FGF8) and FGFR2b, and the mesenchyme-expressed FGF (FGF10) and FGFR1c, are indispensable for the outgrowth and patterning of limbs.103 FGFs 4, 8, 9, and 17 are specifically expressed in the mouse apical ectodermal ridge (AER), a major signaling center at the distal edge to ensure proper development of limb buds. FGF9 is located in regions corresponding to mesenchymal condensations in AER,104 and is only expressed in the mesenchyme surrounding the cartilaginous condensations at the later stage. FGF9 is then expressed in the perichondrium/periosteum and primary spongiosa.105 In rat, Lazarus et al.106 found that FGFs 1, 2, 6, 7, 9, 18, 21, and 22 are expressed in the perichondrium, while FGFs 2, 7, 18, and 22 are expressed in the growth plate. FGFs 1, 2, 17, and 19 are the predominant FGF ligands expressed in human fetal growth plate cartilage.107 FGF18 is expressed in the periosteum, the articular surface, synovial tissue, and in cells within the perichondrial groove of Ranvier.108 During intramembranous bone formation, FGF8 is expressed in developing calvarial osteoblasts, FGF9 is expressed in calvarial mesenchyme, and FGF18 is expressed in mesenchymal cells and differentiating osteoblasts, whereas FGF23 is mainly produced by differentiated osteoblasts and osteocytes.109
FGFR1 and FGFR2 are existed in mesenchymal cells prior to morphological indication of mesenchymal condensation. FGFR1 is evenly expressed in limb bud mesenchyme, while the expression of FGFR2 is increased in chondrogenic condensation area, as the first marker of chondrogenic condensation. Both FGFR1 and FGFR2 are expressed in the periphery of the condensation, where is the location of the origin cells of perichondrium and periosteum.109 In the established growth plates, FGFR3 is expressed mainly in the resting, proliferating, and prehypertrophic zone.110–112 As chondrocytes begin to hypertrophy, FGFR3 expression is shut down, while the expression of FGFR1 is elevated. It has also been found that FGFR2 is expressed in the resting zone, while FGFR4 is expressed in the resting and proliferative zones.106 FGFR3 is expressed more intensely in latent chondroprogenitor cells located in the groove of Ranvier and ring of LaCroix.113 The expressions of FGFR1 and FGFR2 in osteoblasts have been well characterized.112 FGFR3 is also found expressed in osteoblasts.114,115 In cranial sutures, FGFRs are expressed in a spatial-dependent manner. FGFR2 is predominantly expressed in osteoprogenitor cells, while FGFR1 is located in more differentiated osteoblasts.116 FGFR3 has lower expression in the periosteum and sutural osteogenic fronts at the late stage of suture development.117
FGF/FGFR-related genetic diseases with abnormal skeleton development in humans
The characteristic expression patterns of FGFs/FGFRs imply the critical roles of FGFs/FGFRs in skeletal development, and both gain-of-function (GOF) and loss-of-function (LOF) mutations in individual FGFRs or FGFs have been found to cause a variety of genetic skeletal diseases in humans.
Mutations and single-nucleotide polymorphisms (SNPs) of FGFs have been linked to multiple skeletal disorders. Constitutionally increased dosage of FGF3 and FGF4 genes is a risk factor of craniosynostosis.118 Heterozygous mutation in FGF3 gene causes deafness, congenital inner ear agenesis, microtia, and microdontia.119 Heterozygous mutation of FGF8 can lead to autosomal-dominant hypogonadotropic hypogonadism-6 with or without anosmia characterized by short stature, hyperlaxity of the digits, camptodactyly, and mild scoliosis.120 FGF8 mutation also accounts for a small percentage of Kallmann syndrome (KS).121 FGF9 heterozygous missense mutations S99N and R62G have been identified to be responsible for multiple synostoses syndrome 3, and some individuals showed sagittal suture synostosis and humeroradial synostoses in humans.122,123 LOF mutations in FGF10 cause an autosomal-dominant multiple congenital disorder characterized by lacrimal duct aplasia, malformed ears and deafness, and disturbed distal limb segments, named lacrimo-auriculo-dento-digital syndrome.124 FGF10 is identified as a genetic risk factor for nonsyndromic cleft lip with or without cleft palate.125 Truncated mutations of FGF16 are associated with X-linked recessive hand malformations with metacarpal 4/5 fusion.126 Congenital hypogonadotropic hypogonadism individuals caused by missense mutations of FGF17 displayed low bone mass.127 Missense mutations such as R176Q, R179W, and R179Q in FGF23 cause ADHR, frequently present with rickets, bone pain, and tooth abscesses.128 LOF mutations in FGF23 cause a rare autosomal recessive metabolic disorder, hyperphosphatemic familial tumoral calcinosis, characterized by the progressive ectopic calcifications and elevated serum phosphate levels.129
A GOF missense mutation in FGFR1 (P252A) leads to Pfeiffer syndrome (PS), a craniosynostosis syndrome with characteristic abnormalities, including broad thumbs and toes, brachydactyly or variable syndactyly, and elbow ankylosis.130,131 Several FGFR1 mutations, such as N330I and C379R, result in osteoglophonic dysplasia (OGD), characterized by craniofacial abnormalities, including craniosynostosis and depressed nasal bridge, rhizomelic dwarfism, and non-ossifying bone lesions.132 LOF mutations such as C277Y, R622X, and A167S in FGFR1 are responsible for autosomal-dominant KS, characterized by hypogonadotropic hypogonadism and anosmia. Some KS cases present skeletal abnormalities, such as scoliosis, limb anomalies, and loss of nasal cartilage.133 GOF mutations of FGFR2, mainly in the third Ig-like domain and adjacent linker regions (exons IIIa and IIIc), lead to multiple types of autosomal-dominant craniosynostoses, such as Apert syndrome (AS), Crouzon syndrome, and PS, as well as Beare-Stevenson cutis gyrata syndrome.134–138 Several de novo missense mutations of FGFR2 have been identified responsible for a perinatal lethal skeletal dysplasia entitled as BBDS-FGFR2 type characterized by deformities in multiple bone, including mineralization disorder of the calvarium, craniosynostosis, and dysmorphic facial features, as well as bent long bones and osteopenia.139 GOF mutations in FGFR3 affect predominantly bones developed through endochondral ossification causing hypochondroplasia, achondroplasia (ACH), and thanatophoric dysplasia (TD, type I/II).140,141 GOF mutations in FGFR3 have also been found to cause craniosynostoses. The A334T mutation of FGFR3 cuases mild craniosynostosis,142 while A391E mutation in FGFR3 TMD is responsible for Crouzon syndrome with acanthosis nigricans.143 FGFR3 P250R and P252R mutations cause Muenke syndrome, an autosomal-dominant disorder characterized by uni- or bi-coronal synostosis, macrocephaly, midfacial hypoplasia, and developmental delay.144 Some TD patients exhibit joint fusion and craniosynostoses.145 FGFR3 with R621H substitution in the tyrosine kinase domain and a homozygous missense mutation-T546K, leading to partial loss of FGFR3 function, cause camptodactyly, tall stature, and hearing loss syndrome.146,147 To date, no mutation of FGFR4 has been found responsible for genetic skeletal disorders in humans.
FGF/FGFR signaling in skeleton development and homeostasis
Accumulating studies dissecting the roles of FGFs/FGFRs in the development and homeostasis of skeleton have been carried out by using animal models and cell/tissue culture systems.
FGFs in skeleton development and homeostasis
FGF1 has been shown to play an important role in regulating the fate of bone marrow stromal cells (BMSCs) by inhibiting osteogenesis and promoting adipogenesis.148 FGF2 is expressed in osteoblasts and the stromal cells in the bone. Stored in the extracellular matrix, FGF2 promotes both osteoblastic and chondrogenic differentiation of cranial neural crest cells.149 Mice with non-targeted overexpression of FGF2 show shortened long bones caused by premature closure of the epiphyseal plate.150 Sobue et al.151 found that overexpression of FGF2 in mice leads to osteopenia and defective mineralization, proposing that FGF2 functions as a negative regulator of bone formation. The roles of the nuclear high molecular weight (HMW FGF2) and secreted low molecular weight (LMW FGF2) isoforms have been well clarified. The HMW FGF2 has an inhibitory effect on bone mineralization, while the LMW FGF2 promotes bone formation through the regulation of Wnt, BMP2, FGF23, and phosphate homeostasis.152,153 In the articular cartilage, FGF2 binds to perlecan in the pericellular matrix and acts as a mechanotransducer.154 Full-length FGF2 or LMW FGF2 ablation in mice leads to early onset of osteoarthritis (OA), whereas loss of HMW FGF2 isoform has a protective effect on the articular cartilage.155 FGF2 can upregulate the transcription of matrix metallopeptidases 1 and 13 (MMP1 and MMP13), stimulate ADAMTS 5 expression,156–158 and accelerate matrix degradation via a neuro-endocrine pathway in human adult articular chondrocytes.159 FGF3 together with BMP signaling regulates the specification of neural crest and the extension of anterior-posterior axis.160 FGF signaling (FGF3 and FGF8a) together with SHH hierarchically regulates the early specification of skull in zebrafish.161 FGF4 has been shown to be involved in the development and axial elongation of embryonic murine162,163 and Kratochwil et al.164 concluded that FGF4 is a direct target of Wnt signaling during tooth development in mice. FGF6 signaling transduction is mainly mediated by FGFR1 (osteoblasts and osteoclasts) and FGFR4 (osteogenic precursor cells and osteoblasts), which can activate RANKL (receptor activator of nuclear factor-κB (NF-κB)) to stimulate osteoclasts.165 FGF8 participates the regulation of osteogenic and chondrogenic fate in mesenchymal cells in the skull and hard palate.166,167 Hung et al.105 revealed that FGF9 can promote the hypertrophy of chondrocytes and regulate vascularization in growth plates. Transgenic overexpression of FGF9 in mouse chondrocytes led to decreased proliferation and terminal differentiation of chondrocytes, which mimics the phenotype of ACH.104 FGF9 is required for the normal expression of Gdf5 in the prospective joints through the regulation of Gdf5 promoter activity.168 FGF10 is present in the osteoprogenitors in condensation region of the frontal bone, and genetic knockdown (KD) of FGF10 can partially rescue the skeletal phenotype such as craniosynostosis and sternal abnormality in AS mouse model.169 FGF11 is involved in the simulation of osteoclast-mediated bone resorption induced by hypoxia.170 FGF17 can inhibit the proliferation of FGFR3-expressing rat chondrosarcoma chondrocytes.107 FGF18-deficient mice show delayed suture closure with decreased proliferation and delayed osteogenic differentiation of calvaria osteogenic mesenchymal cells, and increased proliferation and differentiation of chondrocytes, indicating that FGF18 positively regulates proliferation and differentiation during osteogenesis, while acts negatively in chondrogenesis.171,172 It has been reported that the deformities of the calvaria, ribs, hindlimb, forelimb, and axis in mice with mesenchyme-specific FGF18 inactivation are dependent on the expression of FGF18 originating from the mesenchymal compartment.108 Serum FGF21 concentration is positively correlated with lumbar BMD.173 FGF21 can lead to growth attenuation by antagonizing the stimulatory effects of growth hormone and even directly suppress the proliferation and differentiation of chondrocytes in the growth plate.174 FGF21 can enhance the osteogenic effect of BMP2.175 In addition, FGF21 is essential for lactation-induced skeletal changes.176 Transgenic mice with overexpression of FGF23 exhibit short stature, lower extremity deformities, and osteomalacia with low serum phosphate concentration.177 Conversely, FGF23-deficient mice exhibit hyperphosphatemia, ectopic mineralization, and poorly formed skeleton with an extremely low parathyroid hormone (PTH) level and elevated 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) level in the serum.178 FGF23 can suppress chondrocyte proliferation through suppression on IHH expression.179,180 FGF23 secreted from osteocytes may regulate mineralization through FGFR3 in a 1,25(OH)2 D3 and Klotho-independent manner via an autocrine/paracrine feedback loop.181
FGFRs in skeletal development and homeostasis
The roles of FGFRs in skeletal development and especially in genetic skeletal diseases have been further dissected by employing genetically modified animal models.
Zhou et al.182 found that mice carrying a P252R mutation in FGFR1 can mimic human PS with premature fusions of multiple sutures, accelerated osteoblast proliferation, and increased expressions of osteogenic genes, and further uncovered that CBFA1 may be a downstream target of FGF/FGFR1 signals in vitro. Trokovic et al.183 concluded that FGFR1 is expressed in pharyngeal region and create a permissive environment for neural crest cell migration in mice homozygous for a hypomorphic allele of FGFR1 with craniofacial defects. The hush puppy FGFR1 W691R mutation is unresponsive to FGF1 in calcium mobilization and downstream signaling through MAPK or PLCγ and can lead to ear defects and skull abnormalities in mice.184 By deletion of FGFR1 in osteochondro-progenitor cells and differentiated osteoblasts in mice, it is proposed that FGFR1 promotes the differentiation of mesenchymal progenitors into osteoblasts, but inhibits the maturation and mineralization of osteoblasts.112 Mice lacking FGFR1 in chondrocytes showed shortened stature and tibial length with expanded hypertrophic zone in growth plate, indicating the important role of FGFR1 during chondrocyte maturation.185 FGFR1 signaling in mature osteoblasts/osteocytes is required for the survival of osteocytes and bone mass maintaining in mice.186 In addition, our group revealed that FGFR1 can positively regulate the differentiation and resorption activity of osteoclasts.187
GOF mutation in FGFR2 (S252W) resulted in increased apoptosis of osteogenic cells,188 disturbed osteoblastic proliferation and differentiation, and the presence of ectopic cartilage at the midline sagittal suture.189 We observed that FGFR2-P253R mutation can directly affect both intramembranous and endochondral ossification in mice.190,191 Cells isolated from limbs of mice with FGFR2 S252W mutation can differentiate into chondrocytes in the osteogenic medium, suggesting that FGFR2 may affect the fate of mesenchymal cells.189 Further studies on BBDS resulting from FGFR2 mutations revealed that nuclear FGFR2 regulates the developing limb, musculoskeletal integration, and cell fate determination.96,192 Targeted disruption of FGFR2IIIc in mice leads to narrowed proliferative and hypertrophic zones in growth plate, and disturbed ossification with downregulation of IHH, PTHRP, and RUNX2.17 Yu et al.193 found that conditional deletion of FGFR2 in mesenchyme can lead to skeletal dwarfism and decreased bone mineral density with dramatically disturbed proliferation of osteoprogenitors and anabolic function of mature osteoblasts in mice. In zebrafish, FGFR2 is essential for the mesenchyme condensation, later chondrogenic differentiation, and survival of chondrocytes in late cranial cartilage development.194
Mice with FGFR3 mutation mimicking human ACH and TD II exhibit dome-shaped skulls and chondrodysplasia,195,196 while FGFR3 deficiency in mice causes increased bone length,197,198 indicating that FGFR3 is a negative regulator of endochondral bone formation. The expression levels of P16, P19, and P21 are upregulated in growth plates of ACH mice and FGF2 treatment can stimulate the expressions of P21 and P27 in RCS cells,195,199,200 suggesting that the upregulation of cell-cycle inhibitors may be involved in activated FGFR3-induced growth arrest of chondrocytes. FGFR3 downregulates PTH/PTHrP (PTH-related peptide) signaling partially through the Janus kinase/STAT pathway.201–203 Reduced telomerase activity may participate in the inhibitory effect of FGFR3 on the proliferation of chondrocytes.204 There are contradictories about the role of FGFR3 in the differentiation of chondrocytes. FGFR3 deficiency in mice causes enhanced chondrocyte hypertrophy;197,198 activated FGFR3 inhibits the hypertrophic differentiation of chondrocytes in cultured metatarsals. However, Minina et al.20 revealed that FGFR3 signaling can accelerate the hypertrophic differentiation of chondrocytes in cultured limbs.195 It has also been reported that FGFR3 promotes the terminal hypertrophic differentiation of chondrocytes partially through MAPK.20,205 Activation of endogenous FGFR3 by FGF2 stimulation leads to reversible premature senescence of RCS cells.206 FGFR3 inhibits the synthesis of chondrocyte ECM such as aggrecan and collagen 2199,207 and promotes the degradation of ECM via stimulation of several MMPs, including MMPs 3, 9, 10, and 13 in chondrocytes, as a negative regulator of ECM.208 FGFR3 signaling is involved in macroautophagy of growth plate chondrocytes, which is important for the postnatal skeleton development.209,210 Recently, it was found that activated mutations of FGFR3 result in long bone defects potentially due to the dysfunction of primary cilia, including shortened length, reduced IFT20 trafficking, and aberrant HH signaling,211,212 suggesting that FGFs/FGFR3 may be involved in the function of primary cilia. Furthermore, FGFR3 directly and indirectly regulates the osteogenesis process. Mice carrying FGFR3 P244R mutation display thinning cortical bone and decreased bone mineral density in long bones.213 Our group found that FGFR3 can stimulate the osteogenic differentiation of BMSCs.115 Mugniery et al.214 revealed that FGFR3 from disorganized growth plate has a direct effect and an indirect effect on osteoblasts. Activation of FGFR3 in chondrocytes leads to premature closure of synchondrosis with enhanced osteoblastic differentiation through upregulation of the BMPs messenger RNA (mRNA) expression and downregulation of BMP antagonist.215 Consistently, FGFR3 deficiency in chondrocytes promotes osteogenesis by stimulating differentiation and mineralization of osteoblasts through upregulation of IHH, BMP2, BMP4, BMP7, WNT4, and TGF-β1, and downregulation of NOGGIN expression.216 Both FGFR3 deficiency and constitutively activation lead to osteopenia and perturbed bone mineralization accompanied with changed osteoclastic activity,115,217 while FGFR3 has a direct positive effect on osteoclastic bone resorption.218
In general, FGFR1-3 all play critical roles in both chondrogenesis and osteogenesis, but FGFR3 is relatively more important in chondrogenesis.
The role of FGF signaling in skeleton repair
Accumulating evidences have supported the crucial roles of FGFs/FGFRs in the injury repair of skeleton, including both cartilage and bone.
Endogenous FGF signaling in skeleton injury repair
Injury and degeneration of cartilage
Cartilage is an essential part of the skeleton. Growth plate is critical for the growth of long bone, while the articular cartilage provides smooth and low-friction interaction between the bones of joints.
Growth plate is fragile in growing skeleton. Given the role of FGF signaling in growth plate, it may play potential role in growth plate injuries. However, the roles of FGF signaling in growth plate injuries and healing is largely unknown. In young rat growth plate injury model, FGF2 is expressed in fibrogenic response phase and osteogenic stage coinciding with mesenchymal cell infiltration and bony bridge formation, suggesting the possible involvement of FGF2 in the repair of injured growth plates.219 In addition, FGF2 is involved in the regulatory role of tumor necrosis factor-α (TNF-α) in injured growth plates220 and contributes to the pathogenesis of osteoradionecrosis, osteopenia, and growth arrest.221
OA is a degenerative disease affecting mainly the articular cartilage. Human adult articular chondrocytes express FGFR1-4 with evident higher levels of FGFR1 and FGFR3, while the expression levels of FGFs/FGFRs were altered in the articular cartilage of OA patients.222 In human osteoarthritic chondrocytes, FGFR1 expression is increased with a concomitant suppression of FGFR3 expression.223 In murine models, disruption of FGFR1 in adult articular cartilage can delay the cartilage degeneration progression with downregulation of MMP13.224 ACH individuals resulting from FGFR3 GOF mutation exhibit a lower incidence of OA.225 Consistent with this, we revealed that FGFR3 delays OA progression in the knee joints and temporomandibular joints partially through downregulation of IHH in both spontaneous and surgically induced OA models in mice.226,227 Recently, we revealed that FGFR3 deficiency enhances the chemotaxis of macrophages via upregulating CXCR7, exacerbating the destruction of synovial joints.228 Both FGFs 1 and 2 are associated with radiographic phenotypes of knee OA at early phase.229 FGF1 is considered as a catabolic factor through down-regulating of CCN2 by interaction and enhancing the degradation of cartilaginous ECM by MMP13.230 FGF2 has both beneficial and deleterious effects on articular cartilage. In human articular chondrocytes, FGF2 can accelerate matrix degradation via a neuro-endocrine pathway159 and stimulation of ADAMTS5 expression through upregulating the transcription of c-FOS/AP1 and CBFA1.231 On the contrary, FGF2 can promote the expression of TIMP1 (tissue inhibitor of metalloproteinases 1) and suppress interleukin-1 (IL-1)-induced aggrecanase activity.232,233 Ablation of full-length FGF2 in mice accelerates the development of spontaneous and surgically induced OA.155 Deletion of LMW FGF2 isoform can accelerate murine OA, while loss of HMW FGF2 isoforms plays a protective role.234 Elevated FGF23 is involved in the role of HMW FGF2 in OA development by modulating Wnt/β-catenin signaling.235 FGF8 promotes the degradation of cartilage, leading to exacerbation of OA through enhancing the production of protease MMP3 and prostaglandin E2 produced by the injured synovium.236 We revealed that the expression of FGF9 is decreased with aging.237 Ellsworth et al.238 showed that FGF18 can act as an anabolic factor in cultured articular chondrocytes through stimulating collagen 2, proteoglycan accumulation, and chondrocyte proliferation.
Bone regeneration
Multiple studies have demonstrated that FGFs and FGFRs recapitulate their expression pattern in skeleton development during fracture healing process. In rat closed femoral fracture model, FGFR1 and FGFR2 have similar expression pattern; they are expressed in inflammatory cells, periosteal cells, chondrocytes, osteoblasts, and osteoclasts in fracture callus during both endochondral and intramembranous bone formation processes.239,240 The expression of FGFR3 is existed in mesenchymal cells, prehypertrophic, and hypertrophic chondrocytes in the fracture callus at a relative later stage.240–242 In mouse long bone fracture model, FGFs 1, 2, 5, 6, 9, and 16-18 are expressed throughout the healing process:243 FGFs 1, 2, and 5 are mainly expressed at inflammatory stage; FGFs 16 and 18 peak at endochondral bone formation phase; FGF2, 9, 16, and 18 are highly expressed, while FGF1 and 17 show peak expression at the bony callus formation and remodeling stage. FGF1 expression is increased during the formation of a cartilaginous callus in fracture,244 especially in fibroblast-like mesenchymal cells.245 In rat femoral distraction osteogenesis model, the expression of FGF2 was detected in fibrous mesenchymal cells, immature osteoblastic-like cells, and the periosteum adjacent to the areas of chondroid tissues.246
Skeletal phenotypes in mice with genetically modifying FGFs/FGFRs and the expression patterns of FGFs/FGFRs during fracture healing indicate the indispensable function of FGF signaling in bone regeneration. The SNPs of FGFR1 are associated with fracture nonunion.247 We found that mice with FGFR2 GOF mutation (P253R) have enhanced bone formation induced by mechanical ablation of long bone marrow via upregulation of Wnt/β-catenin signaling.248 Our group using murine tibia fracture model reveal that FGFR3 plays a negative role in bone repairing through its regulation of both chondrogenesis and osteogenesis.242,249,250 In addition, FGFR3 inhibits the remodeling of injured tissue after cortical injury through downregulation of osteoclastic resorption.218 FGF1 may promote bone repair by inhibiting adipogenic differentiation and increasing the number of osteoblasts in the inflammatory environment.251,252 Using transgenic mice, Hurley’s group proved that LMW FGF2 accelerates the tibia fracture healing process through promoting chondrocyte and osteoblast differentiation and vascular invasion, and enhances the calvaria defect healing through canonical Wnt signaling.253,254 There is a strong positive association between plasma FGF21 levels and BMD in healthy women,255 although FGF21 promotes bone loss in mice.256 The serum FGF23 level may be a predictor of reduction of trabecular parameter and an indicator of nonunion.257–259
Application of FGF signaling modulators in skeleton repairment
Degeneration and injury of cartilage
FGFR1 promotes, while FGFR3 suppresses OA pathogenesis, suggesting that antagonists or neutralizing antibodies of FGFR1, and agonists or FGFs with high binding affinity for FGFR3, could be valuable therapeutics for OA. We revealed that pharmacologically antagonizing FGFR1 can alleviate OA progression in surgically induced mouse OA model and the osteoarthritic phenotype of cultured cartilage explants.260,261 As a high-affinity FGF ligand for FGFR3, exogenous FGF9 can attenuate cartilage degradation while aggravate osteophyte formation in murine post-traumatic OA model.237 In animal experiments, FGF18 has been repeatedly shown to have beneficial effects on OA and improve the healing of cartilage.262–264 To date, recombinant human FGF18 (rhFGF18) (trade name sprifermin) is the only FGF-based drug in clinical trials for OA. Clinic trial data show that intraarticular application of FGF18 can increase cartilage thickness and reduce cartilage loss without discernible local or systemic safety concerns.265–268 Exogenous FGF2 can enhance the repair of articular cartilage defect in vivo.153,269,270 FGF2 has also been used in combination with mesenchymal stem/progenitor cells to improve epiphyses repair.271,272 Due to the anabolic effect of FGF8 in the degradation of cartilage ECM, neutralizing antibody against FGF8 can partially alleviate the OA progression.236
Bone regeneration
Compared with the intervention of FGFRs, modulations of FGF signaling by ligands are closer to the clinical application. At present, more studies have been conducted on the application of exogenous FGFs in bone defect conditions.273,274
FGF1 in a sponge carrier has shown efficacy for bone regeneration as evidenced by more volume of new bone formation in rat critically sized cranial defect model.275 FGF1 with the fibrin carrier can promote bone regeneration of critically sized radial defect in rabbits.276 Kawaguchi et al.277 revealed that FGF2 in gelatin hydrogel could accelerate radiographic bone union of a surgical osteotomy in a dose-dependent manner, and promote tibial-shaft fracture repair with a safety profile in humans.278 FGF2 promotes the repair of bone injury mainly via inducing angiogenesis and enhancing the proliferation ability of osteoblastic lineage. However, the effect of FGF2 on bone formation in vivo is biphasic, with high-dose FGF2 having no stimulatory effect or inhibitory function. Sakano et al.279 found that injection of FGF2 (1 μg) markedly reduced the size of bone, and FGF2 completely inhibited ossification at a dose of 10 μg, during heterotopic bone formation induced by bone matrix powder implanted in murine hamstring muscles, indicating the inhibitory effect of FGF2 at a high dose on bone formation in vivo. Similar results have also acquired in a murine model putting collagen mini-pellet containing FGF2 into subperiosteal pouch, and in transosseous rat mandibular defects.280 Local delivery of FGF7 can enhance bone formation in rat mandible defects with enhanced osteogenesis and chemoattraction.281 Calvaria defects in either FGF9 or FGF18 haploinsufficiency mice showed impaired healing, which could be rescued by exogenous FGF ligands. FGF9-soaked collagen sponge causes sufficient bone regeneration in 2-mm diameter calvaria bone defects at postnatal day 7.282 Deletion of one FGF18 allele can markedly reduce long bone regeneration with dramatic impairment of neovascularization, osteoclast recruitment, and bone remodeling, and treatment with FGF18 protein rescued the disturbed healing capacity.283 FGF18 application together with BMP2 can stabilize BMP2-dependent bone regeneration of 3-mm diameter critical-sized bone defects in mouse calvarium.284 Kang et al.285 established FGF2-FGF18-loaded fiber scaffolds to release FGF2 and FGF18 in a sequential manner, and found that it is effective for bone regeneration in rat calvarium defect model.
Our knowledge of the complicated roles and mechanisms of FGF signaling in bone regeneration is limited. The precise role of individual FGFs and FGFRs in individual cell lineage at different stages during fracture healing and bone regeneration, the application dose, timing and duration of FGFs, and its combination with other bone-modulating signaling molecules, novel vectors and protein delivery systems, need to be further explored to effectively promote bone regeneration and achieve better clinical applications.
FGF signaling in lung development and diseases
The mammalian lung is derived through a series of epithelial branching events, leading to a complex branched airways and blood vessels, which eventually form a fully functioning air exchange organ. Lung development can be morphologically divided into several stages that correspond to key developmental transitions: the embryonic, pseudoglandular, canalicular, saccular, and alveolar stages.286 In chronological order, these stages involve endoderm induction, anterior-posterior and dorsal-ventral patterning, lung specification, lung budding, branching morphogenesis, and finally maturation.
Expressions of FGF ligands and receptors in the lung
The expressions of FGF ligands and receptors have been found during lung development. Using in situ hybridization and RNA-sequencing, Danopoulos et al.287 assessed the expressions and distribution of FGF ligands in the cultured human fetal lung. It is demonstrated that the expression of FGF7 is in both the epithelium and mesenchyme; FGF9 is mainly expressed in the distal epithelium, while FGF10 is diffusely expressed throughout the parenchyma, and some expression of FGF10 is found in the smooth muscle cells (SMCs). FGFR2 is highly expressed in proximal, distal epithelial cells, and SMCs. FGFR3 is mostly expressed in the epithelial cells, and expressed lower in the mesenchyme, while FGFR4 is highly expressed in the mesenchyme and distal epithelium. The expressions of FGF ligands and FGFRs (FGFR1-4) also have been reported in the developing rodent lung.288,289
Roles of FGF/FGFR signaling during lung development
FGF/FGFR signaling is essential for lung development. FGF1 stimulates lung epithelial cell proliferation and airway bud formation, and FGF7 causes cell proliferation in vitro inducing the formation of cysts from epithelia.290 Transgenic mice overexpressing FGF7 exhibit lung malformation.291 During the early phase of lung development, FGF9 controls epithelial branching and mesenchymal proliferation.292 Deletion or overexpression of FGF9 results in branching defects in mice with disturbance of the HH and Wnt/β-catenin pathway and the expressions of FGF10 and BMP4.293–295 FGF10 expression is drastically decreased in FGF9-deficient lungs from E14.5 onwards,296 and in FGF9-overexpressing lung, BMP4 expression is increasingly expressed in the proximal and distal airway epithelium, whereas FGF10 expression is upregulated locally in the distal mesenchyme.293 Deletion of FGF10 results in complete distal lung agenesis.297,298 In cultured human fetal lung both FGF7 and FGF10 can induce liquid secretion and enlargement in distal tips.299,300 Using in vitro organoid cultures from the distal tip epithelium of human embryonic lung at pseudoglandular stage, Nikolic et al.301 have revealed that FGF10 is not required for the initial establishment of SOX2+/SOX9+ progenitors and for human lung branching. A recent study shows that foregut spheroids treated with high levels of FGF10 and 1% fetal bovine serum can form human lung organoids containing airway-like structures, mesenchymal cells, and alveolar epithelial cell type I and type II markers.302 FGF18 plays a role in lung alveolar development during late embryonic lung development. FGF18 knockout mice show narrow alveolar space, thick interstitial mesenchymal compartments, and more embedded capillaries.303 Blocking the function of FGFR2 by a dominant-negative mutation results in blocked airway branching and epithelial differentiation.304 Mice deficient in both FGFR3 and FGFR4 show failure of alveogenesis, but deletion of either receptor alone does not disrupt lung development.305,306 A recent in vivo study demonstrated that FGFR3 and FGFR4 in mesenchymal cells have a function to control the organization of postnatal alveolar elastin, thereby driving the formation of alveolar septa for increasing the gas-exchange surface.289
Roles of FGF/FGFR signaling in lung diseases
SNPs and mutations of FGFs/FGFRs in human lung diseases
Genetic analysis has found that SNPs in FGFs are associated with various types of lung diseases. SNPs in FGF10 may be associated with susceptibility to chronic obstructive pulmonary disease (COPD).307 FGF10 SNPs are also associated with airway branch variants.308 SNPs in FGF3, FGF7, and FGFR4 are associated with respiratory distress syndrome (RDS). FGFR4 (rs1966265) is also associated with bronchopulmonary dysplasia (BPD), the common chronic lung disease of premature birth.309 Besides, mutations in FGFs and FGFRs also have been found in human lung diseases. Mutations in FGF10, FGFR2, or FGFR3 have been identified in LADD (lacrimo-auriculo-dento-digital) patients.124,310 Rare FGF10 mutations have been identified in lethal pulmonary hypoplasia.311 Defects in the formation of tracheal cartilaginous ring resulting in mortality, resulting from respiratory distress, have been reported in Crouzon, AS, and PS caused by activating mutations of FGFR2.312–314 Homozygous loss-of-function mutation (R255Q) of FGFR2 contributes to ectrodactyly and pulmonary acinar dysplasia.315 All these findings suggest the crucial roles of FGF signaling in lung diseases.
Abnormal expressions of FGFs/FGFRs in lung diseases
In human fetal congenital cystic adenomatoid malformation, the epithelial FGF9 expression is 4-fold higher than that of normal fetal lung, whereas FGF10 and FGFR2 gene expressions have no change in the lung mesenchyme.316 Reduced FGF10 expression has been shown in BPD.317 FGF18 expression is decreased in hypoplastic lungs from patients harboring congenital diaphragmatic hernia (CDH).318 Plasma FGF23 levels is significantly elevated in COPD patients.312 FGF1/FGFR signaling is aberrantly increased in idiopathic pulmonary fibrosis (IPF) and may lead to the pathogenesis of lung fibrosis by promoting fibroblast migration via increased MAPK signaling.319
Regulation of FGF/FGFR signaling in lung diseases using in vivo and in vitro models
Studies in rodent models and in vitro lung cells have further implicated the roles of FGF signaling pathway in lung diseases. In lung of CDH rat, FGF7 and FGF10 gene expressions are decreased significantly compared with controls.320 Studies using rat doxorubicin-induced EA-TEF (esophageal atresia-tracheoesophageal fistula) model have found that disturbed FGF10/CTSH signaling is associated with impaired airway branching and consequent impairment of epithelial cells in the lung.321 BPD model established by exposing newborn mice to sublethal hyperoxia shows decreased expressions of FGFR3 and FGFR4.322 Klotho knockout mice show COPD and airway inflammation with elevated FGFR4 in the lung, whereas airway inflammation was attenuated in mice with overexpression of klotho.312 FGF9 and FGF18 promote survival and migration of human lung fibroblasts from patients with IPF, and inhibit myofibroblast differentiation of human lung fibroblasts from patients with IPF.323 Recent studies have demonstrated that alveolar type 2 stem cells are maintained by FGF10-FGFR2B signaling. Loss of FGF10-FGFR2B signaling in bronchial epithelial cells leads to impaired generation of both neo-basal cells and alveolar epithelial cells after bleomycin injury, which can cause IPF.324 Deletion of FGFRs (FGFR1, 2, and 3) in lung mesenchyme decreases pulmonary fibrosis development in response to bleomycin.325 FGF7 and FGF10 can improve the lung repair and increase the epithelial survival after injury through FGFR2b signaling in rodents. FGF10 can also increase lung-resident mesenchymal stem cells and reduce the inflammatory response after acute lung injury (ALI).326 FGF10 has preventive roles in alveolar repair and resolution in ALI or acute RDS.327
FGF/FGFR signaling as a target for the therapies of lung diseases
FGF/FGFR signaling represents a privileged target for the therapeutic approach. Therapeutics targeting FGF signaling pathways are largely classified into “pro-FGF signaling” and “anti-FGF signaling” therapeutics. Recombinant FGFs or FGF analogs have been developed as pro-FGF signaling therapeutics to improve the beneficial effects of FGF signaling. On the other hand, tyrosine kinase inhibitors (TKIs), anti-FGFR antibodies or peptides, and FGF traps have been found as approaches aimed to block FGF signaling.328 A TKI, Nintedanib, which targets FGFRs 1-3, PDGF receptors α/β, and VEGF receptors 1-3, has been approved in the USA and the EU to treat IPF.329 Recent studies found that FGF1 may have preventative and therapeutic effects on transforming growth factor-β1 (TGF-β1)-induced pulmonary fibrosis through inducing AEC proliferation, inhibiting myofibroblast differentiation, regulating TGF-β1 signaling, and FGFR1 expression. Thus, modulating FGF1 signaling may be a potential therapeutic strategy for the treatment of pulmonary fibrosis.330 Considering that FGF2 acts as an angiogenic mediator involved in various lung disorders such as COPD, pulmonary fibrosis, pulmonary hypertension, asthma, and lung cancer, FGF2 could also be an crucial target for the treatment of these lung disorders.331 FGF7 stimulates proliferation of lung epithelial cells and has been considered as a potential therapy for lung injury.332 FGF9 is a strong candidate contributing to the progression of IPF, which makes it a potential target for the therapies of IPF.323 Because of its important roles in lung development and diseases, FGF10 becomes an intriguing target for preventing and treating lung diseases.
However, FGF family is comprised of various ligands and receptors with multiple effects on different cell types in the lung, limiting the potential therapeutic efficacy. For instance, in contrast to its anti-fibrotic effect in TGF-β1-induced lung fibrosis, FGF1 and FGFR1-4 are also expressed increasingly in IPF lungs, and FGF1 treatment led to decreased collagen production and increased apoptosis of IPF-derived lung fibroblasts, suggesting that FGF1 may lead to the pathogenesis of lung fibrosis.319 Recent studies reported that FGF9 and FGF18 decreased normal fibroblast apoptosis, but had no effect on fibroblasts from IPF patients. FGF9, but not FGF18, decreased basal and TGF-β1-mediated expression of collagen and myofibroblast differentiation of fibroblasts.323 All these studies suggest that individual members of FGF family may exert variable effects, depending on the responding cells and the involvement of other signalings. Thus, investigation of specific roles of distinct FGF ligands and receptors in different types of lung cells will help to target differential pathways with precision and optimize the efficacy of future therapies for patients with lung diseases.
FGF signaling in urinary system development and diseases
Expression pattern of FGFs /FGFRs in kidney development
The metanephric kidney develops from nephrogenic cord and Wolffian (nephric) duct, which then generate ureteric bud (UB) and the metanephric mesenchyme (MM), respectively.333 FGFR1-4 and FGFs are highly expressed in mammalian embryonic kidney and lower urinary tract and play critical roles in the development of kidney. Although all FGFRs were detected in embryonic kidneys, FGFR3 or FGFR4 global knockout mice does not show significant structural defects of the kidney or bladder,198,306 which indicates that FGFR1, FGFR2, and FGFRL1 play more necessary roles in kidney development. FGFR1 is mainly expressed in MM lineages (early MM, developing into nephrons starting with vesicles and cap mesenchyme), the ureteric lineage, and renal cortical stroma.334–337 FGFR2 is mainly present in the Wolffian duct, the tips and trunks of UB, and differentiating nephrons, but has fewer expressions in early MM and stromal mesenchyme adjacent to the Wolffian duct.338 FGFRL1 is located in renal vesicles.339 The expressions of FGF 1, 2, 7, 8, 9, 10, 12, and 20 during kidney development have been reported.338 FGF2 can be secreted by ureteric tips. FGF1, 7, and 10 are expressed in renal stroma. FGF8 is mainly observed in the renal vesicle. FGF9 mostly locates in the UB as well as in the cap mesenchyme. FGF12 only presents in the UB. FGF20 is detected in nephron progenitors.
FGFs/FGFRs in urinary system development
FGFs/FGFRs in nephron development
Early researches in rodents and Xenopus laevis explants have found that exogenous FGF2 can maintain the sustained mesenchymal tissue growth and in some conditions induce formation of epithelial nephrons.340–342 More definitive evidences indicate the essential roles of FGF signaling in nephron formation. Deletion of FGF8 with either Pax3Cre343 (in the MM) or brachyury (T) Cre (in mesodermal) line344 results in small kidneys with a complete block in nephron formation after the epithelial vesicle stage. Like the conditional FGF8 knockouts, global deletion of FGFRL1 also leads to blockade of nephron differentiation.339 These data indicate that FGFRL1 might be the candidate FGFR that binds to and mediates the effects of Fgf8 in the nephron lineages.
FGF signaling also has positive effects on the maintenance of nephron progenitors. Among the growth factors known to have expression in embryonic kidney, FGF1, 2, 9, and 20 were found to promote proliferation of nephron progenitors in vitro.345 Global knockout of FGF9 and FGF20 alone or together led to nephron progenitor apoptosis and subsequent renal agenesis.346 Exogenous FGF9 or FGF20 is sufficient to maintain the stemness of MM or sorted nephron progenitors in vitro.346 However, FGF1 knockout mice, alone and in combination with FGF2 knockout, have no nephron progenitor defects,347 and FGF2-null mice348 have no renal defects. Mice with double knockout of FGFR1 and FGFR2 in Pax3-positive cells display severe defect of MM, while mice with either FGFR1 or FGFR2 deficiency have well-developed kidneys.335 These results indicate that FGFR1 and FGFR2 may have a redundant role in establishing and sustaining early MM. Conditional deletion of FGFR1 and FGFR2 with Six2Cre (in nephron progenitors) reduces Six2-positive nephron progenitors leading to severe renal cystic dysplasia.349 FRS2α is the main driver of FGFR signaling through ectopically activating notch signaling in nephron progenitors.349 Double mutation mice, carrying the point mutation in the FRS2α binding site of FGFR2 and conditional deletion of FGFR1 with Pax3Cre, show nephron progenitor depletion at later stages of development.350 Considering the similarity of the phenotypes in knockout mice, FGF9 and FGF20 are the likely ligands for FGFR/FRS2α in nephron development.
FGFs/FGFRs in ureteric branching and induction
FGF7 and FGF10 bind to FGFR2 and regulate the growth and branching morphogenesis of the collecting duct system. FGF7-null mice show marked reduction in developing ureteric bud and mature collecting system with secondary loss of nephrons.351 Meanwhile, FGF7 administration could augment ureteric bud growth and increase the number of nephrons in vitro.351 FGF10-null mice also have smaller kidneys with fewer collecting ducts.352 FGF7 and FGF10 activate the b isoform of FGFR2. Consistently, mice deficient for FGFR2-IIIb have dysgenesis of the kidney similar to that observed in FGF7- and FGF10-null mice.353
Recent studies further investigated the role of FGFR1 and FGFR2 in renal development using conditional knockout mice, since global deficiency of FGFR1 or FGFR2 leads to embryonic lethality prior to kidney development. Conditional loss of FGFR2 in the Wolffian duct and its derivatives, including the ureteric bud using Hoxb7Cre, leads to renal hypoplasia, such as small ampullary, few ureteric branches, and thin trunks.336,354 Furthermore, neither knockout FGFR1 alone nor double knockout of FGFR1 and FGFR2 with Hoxb7cre led to additional abnormalities beyond single knockout of FGFR2.336 Global deletion of FGFR3 or FGFR4 in mice results in no obvious gross phenotype of kidney.198,306 These data together suggest that among four FGFRs, FGFR2 seems to be the most important one regulating ureteric bud branching morphogenesis and stromal mesenchyme patterning.
FGF signaling in kidney diseases
FGF and human genetic kidney diseases
Some mutations in FGFs or FGFRs in humans are associated with structural kidney and lower urinary tract diseases. Activating mutations of FGFR1, FGFR2, and FGFR3 lead to PS, AS, or TD. Some of these patients also have unilateral renal aplasia, hydroureter, vesicoureteral reflux, renal hypoplasia, and/or cystic dysplasia.355 Patients with Kallman syndrome due to LOF mutations in FGFR1 have unilateral renal aplasia. Inactivating mutations of FGF20 was found to cause bilateral renal aplasia.346
FGF signaling in CKD
Some endocrine FGFs (FGF21, FGF23) play important roles in CKD.
FGF21
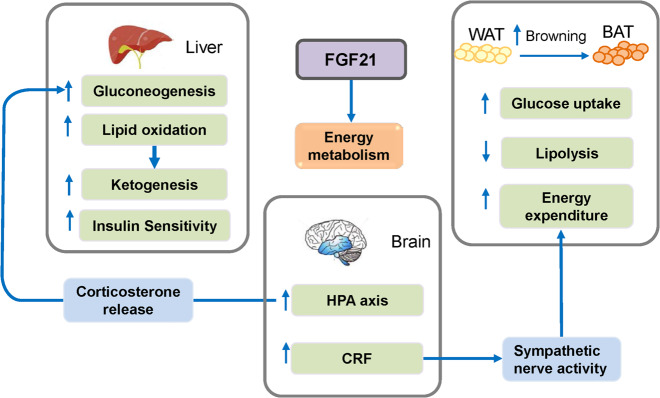
FGF21 binds to a complex of KLB and FGFR1c to induce catabolic metabolism. Increased serum FGF21 levels are detected in CKD patients as early as stage 2.356 Since FGF21 was reported to have anti-aging effects, increasing the levels of FGF21 might be useful for the longevity of CKD patients.36 However, increased FGF21 also has many side effects. High FGF21 level can induce growth retardation, which might be related to the growth hormone resistance in children with CKD.357 Overexpression of FGF21 leads to osteopenia and increased adipogenesis in bone marrow that may contribute to the progress of CKD-mineral and bone disorder (CKD-MBD).256 High FGF21 may also be involved in the neuropsychiatric symptoms in CKD patients. Overexpression of FGF21 in mice causes disturbed circadian rhythm that can be rescued by specific ablation of KLB in the suprachiasmatic nucleus.358 Some researchers speculate that the circadian rhythm disorder related with high FGF21 level may contribute to the blood pressure fluctuation in CKD patients.36 FGF21 also increases serum corticosterone concentration that has been found to cause depression.359,360 Both depression and high FGF21 are associated with high mortality in dialysis patients.361,362 In brief, FGF21-KLB axis could be a potential treatment target in CKD.
FGF23
FGF23 is secreted from bone tissue and binds to a complex of α-Klotho and FGFR1c, FGFR3c, or FGFR4 in kidney as a hormone to regulate systemic phosphate homeostasis and vitamin D metabolism.363 A secondary elevation of serum FGF23 levels is commonly detected in CKD patients that are partly due to decreased renal clearance.364 The increased FGF23 is beneficial for lowering serum phosphate level and reducing 1,25(OH)2D3, which further increases the PTH level. These disturbed hormones would lead to CKD-MBD, which causes abnormities of bone turnover, mineralization, bone volume, extraskeletal calcification, and increased mortality.365 Clinical studies indicate that elevated serum FGF23 concentrations can be used to predict kidney disease progression, especially in the early stages of diabetic nephropathy.366,367 However, neutralization of FGF23 with its antibody further enhances the increased serum phosphate level and vascular calcification that can cause increased risk of mortality.368 The direct role of elevated FGF23 in the cardiovascular events caused by CKD should be further studied. Furthermore, increased serum FGF23 level may be a beneficial compensatory response to maintain mineral homeostasis in early stage of CKD. FGF23 is not only a biomarker for the diagnosis and/or prognosis of CKD, but also a pathogenic factor for the progression of CKD and cardiovascular disease. Targeting the FGF23-Klotho endocrine axes might have therapeutic benefit for diseases of kidney in clinics.36 Whether blocking of FGF23 activities in patients with end-stage renal disease is an effective therapy to improve symptoms needs to be further studied.
Recently, FGF23 has been found to regulate immune system in CKD. Impaired immunological responses and susceptible to infections are common in CKD patients.369,370 Circulation FGF23 level is correlated with incidence of infections.371 Previous studies suggest that FGF23 might be intimately involved in inflammatory processes. FGF23 increases the number of macrophages and induces the expression of TNF-α in response to inoculation with Escherichia coli or lipopolysaccharide injection.372 The stimulation of TNF-α in M2 macrophages by FGF23 could be blocked by 1,25(OH)2D3.373 FGF23 inhibits arginase-1 expression in M2 macrophages.373 These studies suggest that FGF23 has pro-inflammatory functions. It was further reported that FGF23 prevented leukocyte recruitment and impaired host defense in CKD.374 FGF23-α-Klotho-FGFR2 axis plays a central role in this process by activating PKA and inhibiting Rap1 that will finally inactivate β2-integrin function.374 FGF23 could also facilitate the rolling of neutrophils.374
Fibrosis is a common feature of CKD, and involves leukocyte recruitment, angiogenesis, blood vessel leakage, and appearance of myofibroblasts. Secretion of FGF2375 and FGF23376 from podocytes, mesangial cells, interstitial mesenchymal cells, endothelia, or myofibroblasts was reported. FGF2 facilitates the trans-differentiation of tubular epithelial cells to mesenchymal cells, which accelerates the increase of matrix-producing cells.375 However, detailed mechanisms for the role FGF signaling in renal fibrosis remain to be explored.
FGF signaling in kidney injury and repair
Elevated FGF23 levels in the circulation and urine were reported in acute kidney injury (AKI) patients by numerous studies.377–381 Increased serum FGF23 level has been found to be an early marker of incident AKI. In three independent cardiac surgery cohorts, patients with AKI have higher levels of C-terminal FGF23 (cFGF23) than those who did not develop AKI as early as cardiopulmonary bypass ending.377,378,382 The predictive performance of cFGF23 was higher than other urinary injury biomarkers, including NAG (n-acetyl-b-(d)-glucosaminidase), KIM-1 (kidney injury molecule-1), and NGAL (neutrophil gelatinase-associated lipocalin) at the end of cardiopulmonary bypass.377 FGF23 is also thought to be a candidate prognostic marker for the adverse outcomes in AKI patients. Patients with the highest quartiles of cFGF23 and intact, biologically active protein (iFGF23) had a significantly increased risk of 60-day mortality than those having the lowest quartiles in two cohorts of critical illness involved AKI patients.383 Further study is required to clarify whether aberrant FGF23 contributes to the poor outcomes of AKI.
The mechanisms underlying the increased plasma FGF23 in AKI are not clear. Increased production of FGF23 in osteoblasts may be one of the major causes. Increased mRNA expressions of FGF23 in the bone, bone marrow, and renal tissues are found in several AKI mouse models.384–386 This could be reversed by pretreatment with PD173074, an FGFR inhibitor, or blocking the erythropoietin receptor.384,386 These results indicate that the increased circulating erythropoietin and erythropoietin receptor activation are involved in the mechanisms leading to increased plasma FGF23 in AKI. Resection of the obstructed kidney had no effect on the increased circulating iFGF23 levels,387 excluding the possibility that production of FGF23 by the kidneys contributes to plasma FGF23.
Considering the relevance of FGF signaling in kidney development and diseases, there may be potential therapeutic strategies to regulate the process of renal development and diseases by manipulating FGF signaling. For example, recombinant FGF10 may be useful in alleviating ureteric branching defects in Fraser syndrome (FRAS1 mutations).388 The requirement for FGFR2 signaling in lower urinary tract mesenchyme389 suggests that FGF-related therapies could be used to repair the smooth muscle defects in the ureter or bladder. FGF7 expression levels are increased after chemically induced kidney injury in rats.390 Intravenous administration of recombinant truncated human FGF7 largely prevented cyclophosphamide-induced urothelial injury in rats,391 indicating that FGF7 could be a potential therapy for patients with bladder urothelial injury.
FGF signaling in muscle and heart development and disease
FGF signaling in the skeletal muscle
Adult skeletal muscle possesses remarkable regeneration capacity; it can be rapidly repaired after the damage caused by exercise, trauma, toxins, or diseases.392 Satellite cells (SCs), which reside beneath the basal lamina of muscle fibers, are considered as the stem cells in the skeletal muscle. Normally SCs are mitotically quiescent, but upon regeneration they are activated, and give rise to myogenic precursors.392 After several rounds of proliferation and differentiation, most of these myogenic precursors form new muscle fibers, while a small population of these cells returns to quiescent SCs.392
FGFs in the skeletal muscle
FGFs are essential for the self-renewal of SCs and are needed for skeletal muscle maintenance and regeneration. FGF1, FGF2, FGF4, and FGF6 can be detected in SCs.393,394 FGF1 and FGF4 can be found in isolated myofiber cultures and in in vivo injured adult skeletal muscle tissue.394,395
FGF2
FGF2 is present in the extracellular matrix and basal lamina of skeletal muscles,396 and is produced by fibroblasts,397 myofibers,398 and SCs,399 while the relative contribution of FGF2 to these cells is difficult to distinguish. FGF2 has been used as a routine medium supplement in SC primary culture.400,401 Although SCs from young mice (3–6 months) do not need supplementation of FGF2 in the culture medium, SCs from geriatric mice (29–33 months) cannot proliferate without the addition of FGF2.402 FGF2 is considered as a mitogen for SCs; it triggers SC proliferation by repressing myogenesis.403,404 However, FGF2 is not able to stimulate cell division without serum.405,406 Recently, it is reported that excessive FGF2 removes age-associated proliferative inhibition of SCs.407 The upregulated expression of FGF2 in aged muscle fibers and downregulated expression of SPRY1 in aged SCs increase the FGF signaling under homeostatic conditions and break the quiescence of SCs, resulting in SC depletion and losing self-renewing capacity.408 SPRY1, an inhibitor of FGF signaling, is highly expressed in quiescent adult SCs in uninjured muscle,409 while muscle stem cell niche, the muscle fiber, expresses FGF2 under homeostatic conditions. Spry1 is needed for the maintenance of the endogenous adult Pax7-positive SCs in their native environment, but it is downregulated in proliferating myogenic progenitors in injured muscles.410 Overexpression of SPRY1 in SCs or inhibition of FGFR1 signaling can prevent SC depletion. Thus, blockade of FGF2/FGFR1 signaling might be a new therapeutic method to recover the regeneration capacity of skeletal muscles during aging.408 The expression of FGF2 is found to be increased during the muscle regeneration,398 and exogenous FGF2 could promote muscle regeneration in dystrophic mice.411 However, this effect is wiped out in FGF2-null mice,348 and injection of FGF-blocking antibodies also inhibits the regeneration process.412
FGF6
FGF6 can be detected in both embryonic and adult skeletal muscle tissues,413,414 and isolated myofibers.394 In adult mice, FGF6 is secreted by fast-twitch fibers, and its expression is increased after skeletal muscle injury.415 FGF6 mainly performs its function through binding to FGFR4.416 Presently, the role of FGF6 in the skeletal muscle is controversial. Interbreeding of FGF6-deficient mutants with dystrophic mdx mice (a model for Duchenne muscular dystrophy) results in tremendous dystrophic changes in skeletal muscles, including degeneration of myotube, emergence of many mononuclear cells, and collagen deposition. MyoD mRNA is normally upregulated in mdx; however, it is not observed in double mutant mice.415 It is also reported that FGF6-deficient mice show regeneration defects with myotube degeneration and severe fibrosis.415 The numbers of Myo+Myogenin+ activated SCs are severely reduced in mutant mice after injury, and which is not caused by the decreased quiescent SCs, probably by the lack of activated SCs.415 However, another team declared that no skeletal muscle phenotype is found in FGF6-deficient mice, and FGF6 might not play an essential role in muscle regeneration or its function is compensated by other FGFs.417 Using FGF6 global knockout mice and rescue experiments, Armand et al.418 found that FGF6 is participated in soleus regeneration of adult mice in a specific dose-dependent manner: FGF6 promotes the proliferation of the myogenic cells at high doses, while it regulates the differentiation of myogenic cells and muscle phenotype via a calcineurin signaling pathway at lower doses. Genetic deletion of FGF2 and FGF6 in mdx mice leads to much more severe dystrophic phenotypes in FGF2/FGF6/MDX triple-mutant mice than in mdx mice,419 which further supports that FGF6 plays an important role in muscle regeneration.
FGF15/19
Recently, FGF19 has been reported to have novel function in enlarging muscle fiber size, and in protecting the skeletal muscle from atrophy.420 Pharmacological dosage of FGF19 significantly increases human myotube size in vitro.420 Treatment of mice with FGF19 causes skeletal muscle hypertrophy, while genetic deletion of KLB eliminates the hypertrophic effect of FGF19 in mice.420 Both in vitro and in vivo, FGF19 stimulates the phosphorylation of ERK1/2 and the ribosomal protein S6 kinase (S6K1), which is an mTOR-dependent key regulator of muscle cell growth.420 Studies also found that FGF19 relieves the skeletal muscle wasting induced by glucocorticoid, obesity, or sarcopenia in mice. Therefore, FGF19 have the therapeutic potential for promotion of the skeletal muscle mass and treatment of muscle wasting.420
FGFRs in the skeletal muscle
Among the four FGFRs, SCs express high levels of FGFR1 and FGFR4, low levels of FGFR3, and little or no detectable FGFR2.404,421 However, studying the relative contributions of the FGFRs to SCs is rather difficult, because they usually activate multiple intracellular signaling pathways and their functions are often compensated by each other when inhibited by one of the FGFR.
FGFR1
FGFR1 is highly expressed in freshly isolated SCs and myogenic cultures, and it has been considered in the context of adult myogenesis.394,422 FGFR1-null mice cannot gastrulate.423,424 Myogenic-specific (MyoDCre-driven) ablation of FGFR1 in mice seems to have no overt effect on the histology characteristics of muscle and the progress of muscle regeneration following cardiotoxin-induced injury.404 In contrast, SCs could not respond to the stimulation of FGF2 in isolated myofibers from FGFR1-ablated mice,404 which suggests that other FGFRs may compensate the function of FGFR1 during SC differentiation. FGFR1 downstream signals include both ERK regulating SC proliferation425 and p38α and p38β (p38α/β) MAPK pathways that is involved in the exit of SCs from quiescence,426,427 asymmetric division of SCs,427 and differentiation of SCs in vivo.427 Recently, it is reported that SCs from aged mice autonomously lose their self-renewal ability due to alterations in FGFR1, p38α, and p38β MAPK signaling.428 Ectopic activation of phospho-FGFR1 partially rescues their age-associated self-renew ability with asymmetric localization of phospho-p38α/β MAPK in dividing SCs.428 These results highlight an age-associated deregulation of homeostatic network of SCs and hints a therapeutic potential for the treatment of muscle wasting.
FGFR4
FGFR4 is expressed in intact myofibers, muscle connective tissue, isolated proliferating and differentiating SCs in culture.394 FGFR4 plays a role in cell fate determination during embryonic muscle development.429 However, FGFR4-null mice are healthy and fertile with no evident muscle defects, which hints that FGFR4 is dispensable during embryonic development.306
FGF signaling in the heart
Unlike other tissues and organs such as muscle, blood, and liver, the mammalian heart possesses very limited regenerative capacity. Mammalian cardiomyocytes could robustly proliferate in the second heart field during early organogenesis.430 However, recent lineage tracing studies dubbed c-Kit-positive cardiac stem cells (CSCs), which had no cardiogenic activity and could not support heart repair in adulthood.431–434 Instead, the injured myocardium develops scar and fibrosis.435 Thus, researchers have been tempted to uncover the mechanisms of the cardiogenesis and regeneration, which may make it possible to stimulate and manipulate the regenerative potential of heart. FGF signaling pathways, especially FGFs, have been shown to be highly involved in the cardiac development, diseases, and repair.
FGF1
FGF1 together with TNF-related weak inducer of apoptosis (TWEAK), by binding to FGFR1, could induce cardiomyocyte cycle re-entry.436 This effect can be blocked by inhibiting the TNF receptor superfamily member FGF-inducible molecule 14. TWEAK induces the activation of cardiomyocyte cycle, which can be inhibited by blocking FGFR1 signaling.436 Co-stimulation experiments showed that FGF1 and TWEAK could regulate the cardiomyocyte cycle induction via PI3K/AKT signaling.436 It is also reported that the treatment of FGF1 stimulation and p38 inhibition have protective effect on ischemic heart disease by inhibiting cardiomyocyte apoptosis.437,438 In vitro postnatal mammalian cardiomyocytes can proliferate under the FGF1 stimulation and p38 MAPK (p38) inhibition,439 and the combination treatment also increases cardiomyocyte mitosis after acute myocardial injury in 8–10-week-old rats. Four weeks after injury, the treatment reduces heart scarring, wall thinning, and markedly rescues cardiac function.439 However, cardiac-specific overexpression of FGF1 only delays the formation of myocardial infarct, but has no significant effect on maximal infarct size.440 In contrast, inhibition of p38 fails to rescue heart function despite increased cardiomyocyte mitosis. These results imply that FGF1 might promote the survival of newly generated cardiomyocytes through the enhancement of angiogenesis.439 Even so, the combination of FGF1 stimulation and p38 inhibition may have therapeutic effect by improving human cardiac regeneration.435
FGF2
FGF2 is widely expressed in murine heart. In FGF2 transgenic mice, the hearts exhibit exacerbated cardiac hypertrophy assessed by myocyte cross-sectional area and heart weight-to-body weight ratios, which is eliminated in the presence of ERK inhibitor, but not p38 pathway inhibitor.441 In contrast, the chronic elevation of blood pressure, fibrosis, and hypertrophy induced by two-kidney one-clip can be attenuated in FGF2 knockout mice.442 Isoproterenol-induced and myocardial infarct-induced cardiac fibrosis and hypertrophy can also be attenuated in FGF2 knockout mice.441,443 Besides, FGF2 is a cardio-protector in myocardial infarction models and ischemia/reperfusion (I/R) injury.444 The expression of FGF2 is shown to be upregulated after a cardiac injury.445 FGF2 inhibits the autophagy and increased the clearance of ubiquitinated protein through PI3K/AKT/mTOR signaling in mouse myocardial I/R injury model.446 FGF2 also suppresses endoplasmic stress and mitochondrial dysfunction through PI3K/AKT and RAS/MAPK signaling pathways.446 Therefore, FGF2 is being tried for treating ischemic conditions in several clinically relevant trials.447–449
FGF9
FGF9, expressed in the endocardium and epicardium, regulates cardiomyocyte proliferation during embryogenesis,450 and newborn FGF9 knockout mice develop a dilated cardiomyopathy due to premature differentiation of cardiomyocytes.450 FGF9 is also shown to improve systolic function and heart failure mortality by stimulating the hypertrophy of non-infarcted left ventricular after myocardial infarction with increased microvessel density (MVD), reduced fetal gene expression, and interstitial fibrosis in myocardium-specific transgenic FGF9 mice.451 However, FGF9 only stimulates the network formation and the proliferation of endothelial cells (ECs) without induction effects on myocardial hypertrophy in culture.451 It is reported that FGF9 can mediate the differentiation of monocytes to M2 macrophages; FGF9 treatment of an infarcted myocardium in diabetic mice increased anti-inflammatory cytokines and M2 macrophage differentiation, which resulted in reduced adverse remodeling and improved cardiac function.452 Therefore, FGF9 may have novel therapeutic potential for this type of myocardial infarction.
FGF10
FGF10 is found in the second heart field during early heart development,430 and also expressed in progenitors for the right ventricle and outflow tract.453 Neonatal mouse hearts possess the regenerative ability, but gradually lose this ability after postnatal day 7.454 FGF10 is reported to promote regional fetal cardiomyocyte proliferation and cell-cycle re-entry of adult cardiomyocytes, but has no effect on fibroblasts that is mediated by FOXO3/P27.454 In addition, FGF10 deficiency mice display misplacement of the heart in the thoracic cavity with right ventricular hypoplasia due to reduced cardiomyocyte proliferation.455 In contrast, overexpression of FGF10 in the myocardium of mice promotes cardiomyocyte proliferation after heart injury without the increase of epithelial-to-mesenchymal transition and fibrosis;456 thus, FGF10 may be a potential drug for cardiac repair.
FGF signaling in angiogenesis, lymphangiogenesis, and related diseases
Angiogenesis or lymphangiogenesis is the process of vascular or lymphatic formation during physiological and pathological conditions, such as embryogenesis, trauma, inflammation, and tumor development. Since lymphatics can be derived from the sprouting of veins, lymphangiogenesis is considered to be associated with angiogenesis.457 FGF/FGFR signaling has been demonstrated to play important roles in angiogenesis and lymphangiogenesis.
Expressions of FGFs/FGFRs during angiogenesis and lymphangiogenesis
FGFR1 is expressed in vascular ECs and FGFR1 knockdown leads to upregulated FGFR3 expression in the endothelium.458 FGFR2 was found expressed in murine aortic endothelium.459 ECs express the FGFR1IIIc, FGFR2IIIc, and FGFR3IIIc isoforms of FGFRs, but not the IIIb isoforms nor FGFR4, and vascular SMCs (VSMCs) express the similar isoforms of FGFRs; several FGFs are expressed in ECs (FGFs 1, 2, 5, 7, 8, 16, and 18) and VSMCs (FGF1, 2, 5, 8, 16, and 18). FGFR1 and FGFR3 are expressed in lymphatic ECs (LECs) during lymphangiogenesis as demonstrated by several studies,458,460 and they were reported to be critical for the lymphatic formation.
FGF signaling in vascular and lymphatic formation
FGF signaling can influence the whole process of angiogenesis. Activation of FGFR1 or FGFR2 has been demonstrated to have a positive effect on vascular endothelial proliferation.461 One important step of angiogenesis is extracellular matrix degradation. Some FGFs, including FGF1, FGF2, and FGF4, promote the expressions of MMPs in ECs.462 FGF2 can stimulate shedding of MMP2 and MMP9 in cell surface membrane vesicles from ECs, which is able to stimulate the angiogenesis of ECs seeded in Matrigel.463 Another essential step of angiogenesis is endothelium migration. FGF1, FGF2, FGF8, and FGF10 were demonstrated to stimulate endothelium chemotaxis.464 The pro-chemotactic effect of FGF2 depends on activation of MAPK.465
The role of FGFR3 in lymphangiogenesis is controversial. It is revealed that FGFR3 is a novel target gene of PROX1, which is essential for lymphatic development. Knockdown FGFR3 by small interfering RNA (siRNA) inhibited LEC proliferation.460 Meanwhile, 9-cis retinoic acid (9-cisRA) was reported to activate FGF signaling and enhance lymphatic formation and regeneration by promoting the proliferation, migration, and tube formation of LECs.466 FGFR3 expression in LECs was upregulated after 9-cisRA treatment. 9-cisRA-induced LEC proliferation and migration were significantly inhibited by soluble FGFR3 recombinant protein as well as FGFR inhibitor PD173074.466 However, Yu et al.458 showed that FGFR3 alone is not enough to influence lymphangiogenesis. Vascular and lymphatic vessel defects were observed in FGFR1/FGFR3 double mutant mice, but single knockout of FGFR1 or FGFR3 led to no abnormality in lymphatic front migration in embryonic mouse skin examined by whole-mount staining for VEGFR3 (vascular endothelial growth factor receptor 3) and PECAM1 (platelet and endothelial cell adhesion molecule-1). The controversial effects of FGFR3 on lymphatics may be due to its differential influence on LECs during embryonic phase or adulthood.
FGF/FGFR-related diseases with abnormal angiogenesis and lymphangiogenesis
There are few clinical reports about the relationships between FGFs/FGFRs and diseases with abnormal angiogenesis and lymphangiogenesis. Some experimental results demonstrate that FGFs/FGFRs may play an essential role in diseases with abnormal vascular formation. Many tumor cell lines produce FGF2.467 Inhibition of FGFR1 by FGF2 antisense complementary DNAs (cDNAs) suppressed vascularization and growth of human melanomas in nude mice.468 Furthermore, FGF levels were correlated with intratumoral MVD, an important parameter for tumor progression.469 In some tumors like melanoma, FGF2 level has a strong correlation with MVD and clinical outcome of the patients.469 However, whether FGFs/FGFRs also influence tumor parenchyma needs to be further clarified. Inflammation is an important trigger for angiogenesis. It is revealed that monocytes, mononuclear phagocytes, and mast cells express FGF2.470 Inflammatory cytokines including IL-1β can stimulate FGF2 production in ECs.471 Inflammatory mediators might stimulate angiogenesis through increasing FGF signaling in endothelium. EC death can lead to increased FGF2 release. Hypoxia upregulates VEGF and FGF2 production and increases endothelial responsiveness to FGF2.472 The activity of FGF/FGFR signaling may be strongly associated with inflammation and influence angiogenesis at multiple levels.
FGF signaling and EndMT
Endothelial-to-mesenchymal transition (EndMT) is the process through which ECs transform into mesenchymal cells. EndMT plays important roles in the pathogenesis of various human diseases, including cardiac fibrosis, atherosclerosis, and heterotopic ossification (HO).473 EndMT was first confirmed in animal models in which Tie1+ endothelials adopted cardiac fibroblast fate during cardiac fibrosis development.474 Further investigations found that Tie2+ vascular ECs contributed to HO formation in fibrodysplasia ossificans progressive and BMP4-induced HO mouse models.475 Currently, TGF-β1 signaling is regarded as the main inducer of EndMT.473 FGF signaling has recently been demonstrated to downregulate TGF-β signaling and inhibit EndMT. Basal FGF signaling maintains endothelial homeostasis through inhibiting the expressions of TGF-β, TGF-βR1, and SMAD2 via controlling the let-7 microRNA (miRNA) levels.476 Meanwhile, in vitro and in vivo experiments showed that inflammatory cytokines, including interferon-γ, TNF-α, and IL-1β, decreased FGFR1 expression, leading to reduced FGF signaling activation in ECs.476 Another study reported that FGF2 can induce miRNA-20a expression, which represses TGF-β signaling in endothelium and inhibits EndMT.477 Therefore, it is plausible that FGF signaling downregulation by inflammatory cytokines contributes to vascular neointima formation and fibrosis driven by TGF-β-induced EndMT.
Therapeutic modulation of angiogenesis and lymphangiogenesis
Therapeutic angiogenesis is a promising approach to the recovery of ischemic diseases. It was shown that intracoronary FGF2 administration preserved myocardial function by increasing vascularization.478 Some clinical trials demonstrated that FGF2 administration can improve the symptoms of patients with coronary artery disease or peripheral artery disease.448,479 In addition, inhibition of FGF2/FGFR1 by antisense cDNAs blocked intratumoral angiogenesis and arrested the growth of human melanomas grown subcutaneously in nude mice.468
FGF-based angiogenic therapy has been shown to be a potential treatment for patients with ischemic diseases. However, many details including timing, dosage, application alone, or in combinations with other drugs and effective delivery approach need to be further clarified. There are few reports about the therapeutic modulation of lymphangiogenesis based on FGF signaling. 9-cisRA was reported to have a therapeutic effect on lymphatic regeneration and secondary lymphedema in experimental mouse models, which could be dependent on FGF signaling in LECs.466
FGF signaling in inflammatory response
Inflammation is a complex adaptive response that can be induced by endogenous and exogenous substances/stimuli.480 Besides the recognition of inducers, inflammatory response includes multiple process such as the production of multiple inflammatory mediators, including inflammatory factors, chemokines, and vasoactive amines, which are released by immune cells like macrophages and mast cells.480 There are lots of studies reported that FGFs/FGFRs play important roles in the regulation of inflammationory response.
FGFs in inflammation
FGF1 in inflammation
FGF1 can accentuate inflammatory response.481 Generally, FGF1 is highly expressed in the inflammatory cells and tissues. High levels of FGF1 can be found in multiple tissues of inflammatory arthritic joints, including bone, cartilage, synovium, ligament, and tendon.482 Besides, most T cells in synovial tissue in rheumatoid arthritis express FGFR1 for FGF1.483 FGF1 can enhance IL-2 production and activation of NF-κB in T cells.483 Rossini et al.484 found that both FGF1 and FGFR1 are expressed in filtrating lymphocytes and macrophages during the renal inflammation, and FGFR1 is highly expressed in tubules, suggesting that FGF1 might have both autocrine and paracrine functions. Hackshaw and Shi485 reported that FGF1 affects the calcium mobilization and increases the level of cytosolic calcium in macrophages. FGF1 causes ATP release from spinal astrocytes and opens gap junction channels after spinal cord injury, which may aggravate the inflammation in neurological disease and injury.486,487 Recently, Huang et al.488 engineered the FGF1 mutants (termed FGF1ΔHBS) with reduced ability to activate FGFR, and found that FGF1ΔHBS inhibited inflammation and oxidative stress in CKD via activating PI3K/AKT and GSK-3β/Nrf2 signaling pathways, which inhibited the ASK1/JNK.489 The results suggest that FGF1 bears the responsibility of anti-inflammation, especially in certain chronic inflammatory diseases. Besides, FGF1 has the ability of anti-inflammation in diabetic nephropathy via inhibition of JNK (c-Jun N-terminal kinase) and NF-κB pathways.490 Thus, the effects of FGF1 on inflammation may vary from different diseases and conditions.
FGF2 in inflammation
FGF2 is also involved in several inflammation-related diseases such as multiple sclerosis and rheumatoid arthritis.491 Ectopic expression of FGF2 exacerbates inflammatory response and symptom of colitis and collagen-induced arthritis models.492,493 FGF2 contributes to the inflammation in articular cartilage during the process of OA.494 Besides, the level of FGF2 is increased during the whole blood inflammatory reaction induced by the artificial surface.495 During the infection of HIV, FGF2 shows a positive correlation with the number of CD4+ T cell.496 FGF2 induces the expression of RANKL via ERK1/2 activation in human bone marrow mesenchymal stromal cells, which suggests that FGF2 may play the osteoimmunological role during bone regeneration.497 Pawlowski et al.498 found that FGF2 is highly expressed in fibroblasts and adipocytes, and FGF2 may contribute to perpetuation of inflammation in the orbital tissue of Graves’ orbitopathy. FGF2 has close relationship with inflammatory response during angiogenesis such as activation of pro-inflammatory chemokines in ECs and engagement of monocyte/macrophage.499 FGF2 increases the concentrations of cellular IL-1β in human VSMCs.500 The above studies indicate that FGF2 has the function of pro-inflammation. However, exogenic FGF2 could attenuate inflammatory response such as the decreased expression of IL-1β in epileptogenesis-associated neuroinflammation.501 In addition, inhaling recombinant FGF2 decreases lung inflammation in asthma and COPD.502,503 Thus, targeting FGF2 is a potential method to alleviate certain inflammatory diseases such as neuroinflammation, asthma, and COPD.
FGF3/FGF21/FGF23 in inflammation
Unlike FGF1 and FGF2, there are few studies that reported the relationship between FGF3 and inflammation. The level of FGF3 in sinonasal tissues is significantly upregulated in acute allergic rhinitis and chronic sinonasal inflammation mouse models.504,505 However, FGF3 level in middle ears is significantly downregulated in mouse model for acute otitis media.506 Combining these results, we speculate that the role of FGF3 in inflammation may be distinct in the different tissues/organs.
FGF21 can be induced by inflammatory stimuli.507–509 FGF21 is associated with the suppression of cardiac, renal, and hepatic inflammation.510–512 FGF21 is thought to be one of the potential immunotherapy targets for cardiovascular inflammation and pancreatic fibrogenesis as it can alter the macrophage polarization states.513,514 Exogenous FGF21 was found to alleviate soakage of inflammatory cells in the lung potentially via elevation of IL-10.515 FGF21 inhibits macrophage migration and significantly reduces inflammatory factor expression in oxidized low-density lipoprotein-induced THP-1 macrophages.516 In addition, FGF21 can also repress inflammatory factors induced by insulin resistance.517 FGF21 has anti-inflammatory effect on preadipocytes via FRS2/ERK1/2 signaling pathway.518 Besides, FGF21 can suppress the production of IL-1β mediated by NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3) inflammasome.519 In general, inflammation increases the expression of FGF21, which is an anti-inflammatory factor in many diseases.
The relationship between inflammation and FGF23 may be bidirectional.520 Lang et al.521 suggested that the increase of FGF23 induced by inflammatory signaling may amplify inflammation by suppressing the synthesis of the anti-inflammatory 1,25(OH)2D3 in inflammatory diseases. Besides, FGF23 can induce multiple inflammatory signaling pathways like TNF-α signaling. In addition, FGF23 activates calcineurin signaling by activating FGFR4 in hepatocytes, which causes the increased level of inflammatory cytokines in CKD.522 In summary, inflammatory response can induce the expression of FGF23 and FGF23 can act as a pro-inflammatory factor.
FGFRs in inflammation
In addition to the FGFs, the receptors of FGFs also play important roles in inflammatory response. FGFR1 promotes inflammation via activating NF-κB signaling pathway in prostate cancer cells.523 However, FGF2/FGFR1 pathway has inhibitive effects on astrocyte-mediated neuroinflammation after infrasound exposure.524 In turn, there is a profound reduction in FGFR1 in human umbilical vein ECs treated by TNF-α and IL-1β, while other inflammatory cytokines such as IL-6 could not inhibit the expression of FGFR1.476 Besides, our group recently identified that FGFR3 deficiency promoted chemotaxis of macrophages via activation of NF-κB/CXCR7 signaling pathway, which reveals the negative role of FGFR3 in synovial inflammatory response.228 More studies about the roles of FGFRs in inflammation are needed in the future.
Inflammatory response is regulated by multiple factors in a variety of cellular behaviors.525 Targeting pro-inflammatory factors such as IL-6 and TNF-α has been shown to be an effective therapy for some inflammatory diseases, and therapeutic antibodies are also promising strategy to treat inflammatory diseases.526,527 From the above studies, we can conclude that FGF signaling has close relationships with inflammatory response, and whether it exerts a pro-inflammatory or an anti-inflammatory role mainly depends on the types of FGFs and inflammation of diseases. Application of specific modulatory molecules such as antibodies against pro-inflammatory FGFs/FGFRs like FGF23 will benefit for certain inflammation-related diseases.
FGF signaling in metabolism
Among the 22 members of the FGF family, FGF15/19, FGF21, and FGF23 comprise the FGF19 subfamily that functions as endocrine hormones to regulate bile acid (BA), fatty acid, glucose, and mineral metabolism.
FGF15/FGF19 in energy homeostasis
FGF15 and its human ortholog FGF19 (FGF15/19) are gut-derived circulating hormone that represses hepatic BA synthesis through FGFR4 and the coreceptor KLB complex.528 Furthermore, FGF15/19 also regulates global body energy and glucose homeostasis (Fig. 3).529–531
Fig. 3.
The regulation of FGF15/19 on energy metabolism. FGF15/19 regulates energy metabolism both peripherally and centrally. In the liver, FGF15/19 inhibits BA production and promotes gallbladder filling. As for lipid and glucose metabolism, FGF15/19 improves glycogen synthesis, but suppresses lipogenesis and gluconeogenesis. In the adipose tissue, FGF15/19 promotes energy expenditure and fatty acid oxidation. In the brain, FGF15/19 promotes the expression of CRF in the hypothalamus and stimulates sympathetic nerve activity, and then increases energy expenditure in the adipose tissue. Furthermore, FGF15/19 promotes peripheral insulin sensitivity and glucose metabolism by repressing HPA axis and AGRP/NPY neuron activity. AGRP agouti-related protein, BA bile acid, HPA hypothalamic-pituitary-adrenal, NPY neuropeptide Y
FGF15 is highly expressed in the ileum, jejunum, and duodenum of adult mice.532,533 FGF19 is expressed in human ileum and gallbladder epithelial cells,534,535 and is not detected normally in human liver.536 The expression and production of FGF15/19 are regulated by many factors, such as BAs, nutrition, and so on.537,538
Effect on liver and gallbladder
BA homeostasis
FGF15/19 negatively regulates BA synthesis. FGF19 treatment inhibits the expression of cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting and major regulatory enzyme of BAs, by an autocrine/paracrine mechanism in hepatocytes.529,539 Deletion of FGF15 in mice results in enhanced BA production by upregulating CYP7A1 expression in the liver, while FGF15 administration inhibits BA production by decreasing CYP7A1 mRNA levels.533
The alternation of gallbladder filling and emptying regulates the bile flowing into the intestine. FGF15/FGF19 is required for gallbladder filling as evidenced by the absence of bile in the gallbladder of FGF15 knockout mice, and FGF15 or FGF19 treatment leads to significant increase in gallbladder volume, which is partially caused by a cAMP-dependent relaxation of gallbladder smooth muscle.540
Hepatic glucose and lipid metabolism
Fed FGF15 knockout mice showed decreased hepatic glycogen stores in the liver, and administration of FGF19 significantly promotes glycogen accumulation and protein synthesis in the liver of fasted mice, which is independent of insulin action.541 FGF15/19 also suppresses hepatic metabolic, such as the tricarboxylic acid cycle flux and gluconeogenesis, through inhibiting CREB-PGC-1α (cyclic AMP response element binding protein-peroxisome proliferator-activated receptor-γ coactivator-1α) signaling.542
FGF15/19 represses liver fat storage. FGF19 transgenic mice show decreased expression of lipogenic enzymes and liver triglyceride levels.530 FGF19 inhibits the expression of lipogenic enzymes and the insulin lipogenic action in rat primary hepatocytes through activating STAT3 signaling and repressing PGC-1β expression,543 and also enhances the expression of fatty acid oxidation-related proteins.544 Long-term treatment by FGF19 reduces liver lipid accumulation in vivo and protects liver from diet-induced steatosis.545
Effect on body energy and glucose homeostasis
FGF15/19 is beneficial for global energy balance. FGF19 transgenic mice have a significantly reduced fat mass resulted from increased metabolic rate that leads to enhanced energy expenditure, and do not become diabetic or obese when fed a high-fat diet (HFD).530 In HFD fed mice, FGF19 increases the metabolic rate simultaneously with an increased fatty acid oxidation, and alleviates the obesity in ob/ob mice.529 Adeno-associated virus (AAV) delivery of FGF15 and FGF19 reduces fat mass and increases energy expenditure in diet-induced obesity (DIO) mice, and FGF19 can also overt diabetes in db/db mice.531
In addition to the direct effects of FGF15/19 on body energy metabolism, FGF15/19 also regulates the energy and glucose metabolism by affecting brain after binding to FGFR4 and KLB in the brain.358,546 FGF19 activates ERK signaling in the hypothalamus.547 Intracerebroventricular (ICV) injection of FGF19 induces the sympathetic nerve activity to BAT and increases energy expenditure,548 and also improves peripheral insulin sensitivity and glucose metabolism by reducing hypothalamic agouti-related protein/neuropeptide Y neuron activity and activating of ERK1/2 signaling in obese and insulin-resistant states.547 Furthermore, FGF15/19 signaling in the central nervous system has an insulin-independent glucose-lowering effect. Acute ICV FGF19 injection reduces food intake and body weight, and improves glucose tolerance without changing plasma insulin levels.546,549 The suppressed hypothalamic-pituitary-adrenal (HPA) axis and subsequent decreased hepatic acetyl CoA level are responsible for mediating the insulin-independent, glucose-lowering effects of FGF19.549
Metabolic role of FGF21
FGF21 is mainly expressed in the liver, adipose tissue and pancreas,550,551 and also expressed in the muscle.552 Under physiologic conditions, FGF21 in the blood is mostly derived from the liver.551 FGF21 activates FGF signaling by binding to FGFR1c and its coreceptor protein KLB in the liver, adipose tissue, and brain.532
FGF21 is a hormone regulating glucose and lipid homeostasis, and insulin sensitivity. FGF21 can cause weight loss, decrease plasma glucose and triglycerides level, and improve insulin sensitivity in obese and diabetic animal models without affecting total caloric intake.553,554 Mice with overexpressed FGF21 resist to DIO. In both ob/ob and db/db mice,553,554 treatment of FGF21 decreased serum glucose and triglycerides to near normal levels. FGF21 regulates glucose and lipid metabolism mainly by affecting liver, adipose tissue, and brain (Fig. 4).
Fig. 4.
The regulation of FGF21 on energy metabolism. FGF21 regulates energy metabolism in peripheral and central manners. In the liver, FGF21 promotes gluconeogenesis and lipid oxidation, and thus improves ketogenesis and insulin sensitivity. In the adipose tissue, FGF21 simulates glucose uptake in both WAT and BAT and induces WAT browning, as well as promotes energy expenditure, while FGF21 inhibits lipolysis. In the brain, FGF21 stimulates the HPA axis, thus contributing to corticosterone release and ultimately promoting gluconeogenesis in the liver. Furthermore, FGF21 improves CRF expression in the hypothalamus and stimulates sympathetic nerve activity, and then promotes energy expenditure in BAT. BAT brown adipose tissue, CRF corticotropin-releasing factor, HPA hypothalamic-pituitary-adrenal, WAT white adipose tissue
The effect on liver
Nutritional stresses, such as starvation, amino acid restriction, ketogenic, and HFD, can strongly induce the expression and release of FGF21 in liver.555
FGF21 decreases insulin resistance, enhances fat oxidation, and suppresses hepatic steatosis in the liver of DIO and ob/ob mice,553,554 which is related to the increased level of adiponectin in vivo.556 FGF21 participates in high-fat, low-carbohydrate ketogenic diet-induced triglyceride clearance, hepatic lipid oxidation, and ketogenesis. Downregulated hepatic FGF21 in ketogenic diet-fed mice altered the expressions of lipid and ketone metabolism-related genes in the liver, and leads to fatty liver, lipemia, and decreased serum ketone.557 FGF21 stimulates hepatic gluconeogenesis and ketogenesis in the liver during fasting and starvation558,559 by inducing the expression of PGC-1α.559 FGF21 knockout mice fail to induce PGC-1α expression and have impaired gluconeogenesis and ketogenesis in response to a prolonged fast.559 However, the mechanisms for the regulation of FGF21 on liver metabolism need to be further explored.
The effect on adipose tissue
In addition to liver, adipose tissue is another source of systemic FGF21. White adipose tissue (WAT) stores energy, and brown adipose tissue (BAT) expends energy to generate heat through a process known as adaptive thermogenesis.560 FGF21 in WAT is induced by fasting/refeeding regimens and the thiazolidinedione drugs.558 FGF21 in BAT is induced by cold exposure.561
FGF21 stimulates glucose uptake in adipocytes in an insulin-independent manner through induction of GLUT1 expression,562 and inhibits lipolysis of adipocytes.563 However, FGF21 stimulates lipolysis in WAT during starvation.558
The thermogenic activity of BAT and browning of WAT are important components of energy expenditure, which can be induced by FGF21.554,564 Cold exposure induces expression of mitochondrial uncoupling protein 1 (UCP1) in BAT. UCP1 uncouples oxidative phosphorylation, releasing chemical energy as heat.565 FGF21 improves the expression of UCP1 in WAT by upregulating PGC-1α protein level and promoting browning of WAT in adaptive thermogenesis.559,566 FGF21 knockout mice show diminished browning of WAT and a decreased adaption to chronic cold exposure.566 In addition, FGF21 also upregulates UCP1 mRNA expression through CREB567 signaling, and induces phosphorylation of STAT3 to activate the oxidative metabolism in adipose tissues.567
FGF21 also promotes adipocyte differentiation and insulin sensitivity by stimulating peroxisome proliferator-activated receptor-γ (PPAR-γ) transcriptional activity through inhibiting its SUMOylation in WAT568,569 in DIO mice. FGF21 knockout mice show decreased WAT mass with reduced PPAR-γ activity, adipocyte size, and insulin sensitivity in DIO mice.569
The effect on brain
In addition to regulating liver and adipose tissue, FGF21 also involves in energy metabolism through regulating brain. FGF21 is not expressed in the central nervous system,532 but can cross the blood–brain barrier to enter into the brain.570 ICV injection of FGF21 in obese rats increases hepatic insulin sensitivity and energy expenditure.571 FGF21 improves the expression of neuropeptide corticotropin-releasing factor in the hypothalamus and stimulates sympathetic nerve activity, and then promotes energy expenditure in BAT.572 Furthermore, FGF21 activates the HPA axis for the release of corticosterone that stimulates hepatic gluconeogenesis.359
The effect of FGF23 on mineral metabolism
FGF23 is mainly secreted by osteoblasts and osteocytes in bone tissue,573 and regulates systemic phosphate homeostasis and vitamin D metabolism through binding FGFR and the coreceptor α-Klotho complex in cell membranes of target tissues574 (Fig. 5).
Fig. 5.
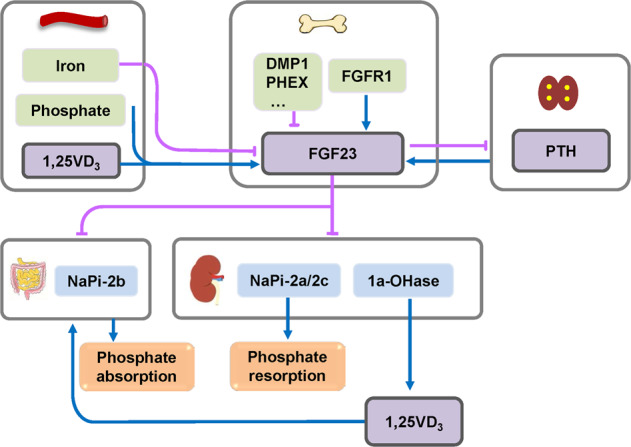
The regulation of FGF23 and its effect on phosphate homeostasis. The bone-derived FGF23 is regulated by several factors such as iron, phosphate, and 1,25(OH)2D3 and PTH in blood, as well as DMP1, PHEX, and FGFR1 in the bone. FGF23 downregulates serum phosphate. In the kidney, FGF23 reduces 1,25(OH)2D3 level and inhibits renal phosphate resorption by inhibiting the expression of NaPi-2a/2c. In the intestine, FGF23 inhibits phosphate absorption by reducing NaPi-2b expression or indirectly suppressing 1,25(OH)2D3. In the parathyroid, FGF23 inhibits PTH synthesis and secretion, and then contributes to its own negative feedback regulation. 1,25VD3 1,25(OH)2D3, NaPi-2 type IIa sodium-phosphate co-transporter, PTH parathyroid hormone
The effect on metabolism of phosphate, sodium, and calcium
Clinical studies identified the important role of FGF23 in regulating phosphate metabolism. Mutations in an RXXR site in FGF23 lead to ADHR characterized by low serum phosphorus level, osteomalacia, and rickets, as well as short stature and bone pain.128 FGF23 is also the cause of tumor-induced osteomalacia and fibrous dysplasia because of its overexpression in tumors and osteogenic cells in fibrous dysplastic lesions.575 Furthermore, multiple FGF23 gene mutations lead to reduced FGF23 level in patients, which is responsible for hyperphosphatemic familial tumoral calcinosis, a genetic disease characterized by hyperphosphatemia and tumor-like soft tissue calcifications.576 In mouse models, overexpression of FGF23 in the liver, osteoblasts, or ubiquitously in mice lead to decreased serum phosphate concentration and rachitic bone.177,577,578
Phosphate homeostasis is simultaneously regulated by several organs, including the kidney, intestine, and bone.579 Type II sodium-dependent phosphate co-transporters (NPT2) are responsible for the absorption of extracellular phosphate.580 Type IIa sodium-phosphate co-transporter (NPT2a, NaPi-2a) is mainly expressed in the brush-border membrane of proximal tubules of the kidney.581 FGF23 inhibits renal phosphate reabsorption and leads to phosphate loss by inhibiting the expression of NaPi-2a/2c through binding to a FGFR1-α-Klotho coreceptor complex and activating ERK signaling.582 NaPi-2b is expressed in the luminal membrane of the ileum and regulates phosphate absorption in the intestine.583 FGF23 can reduce NaPi-2b level to inhibit phosphate absorption in the intestine.584 1,25(OH)2D3 also promotes phosphate absorption in the intestine.585 FGF23 can reduce 1,25(OH)2D3 level by inhibiting 25-hydroxyvitamin D-1a-hydroxylase and increasing 25-hydroxyvitamin D-24-hydroxylase expression in the kidney,586 and then may indirectly suppress 1,25(OH)2D3-mediated intestinal phosphate absorption.
In addition, FGF23 also regulates the metabolism of sodium and calcium. FGF23 promotes sodium reabsorption by increasing the sodium chloride co-transporter expression in the distal renal tubules resulting in volume expansion and hypertension.587 FGF23 directly promotes calcium reabsorption in the kidney by regulating transient receptor potential vanilloid-5 channels in the distal renal tubules.588 Furthermore, 1,25(OH)2D3 promotes calcium absorption in the small intestine.589 PTH promotes calcium absorption in the kidney via increasing the 1,25(OH)2D3 level and accelerates calcium release from the bone by stimulating bone resorption. FGF23 can systematically regulate serum calcium by decreasing serum levels of 1,25(OH)2D3 and PTH.
The regulation of FGF23
FGF signaling participates in the regulation of FGF23. Several OGD patients caused by activating mutations of FGFR1 present hypophosphatemia and increased serum level of FGF23.132 Inhibition of FGFR1 decreased FGF23 mRNA expression in the bone.590 Integrative nuclear FGFR1 promotes FGF23 transcription by activating the transcription factor CREB.591 HMW isoform of FGF2 (HMWFGF2), the ligand for nuclear FGFR1, stimulates FGF23 expression.592 Transgenic mice with overexpression of HMW FGF2 in immature and mature osteoblasts display increased FGF23 level, hypophosphatemia, and rickets.592
Some proteins regulating phosphate homeostasis are also expressed in osteoblasts and osteocytes, such as DMP1 and PHEX,593,594 and regulate FGF23 expression. Inactivating mutations in DMP1 and PHEX lead to XLH (X-linked hypophosphatemic rickets) and ARHR (autosomal recessive hypophosphatemic rickets), respectively, accompanying with increased serum FGF23 level.594,595 Both DMP1 and PHEX knockout mice exhibit hypophosphatemic rickets and increased FGF23 expression.594,596 Although PHEX is a peptidase expressed in the bone, it can inhibit the expression FGF23 without regulating FGF23 degradation.597
Some circulating proteins also regulate FGF23 level. FGF23 is regulated by feedback loops, including the phosphate level, 1,25(OH)2D3, and PTH. Either dietary phosphate or administration of 1,25(OH)2D3 can increase the serum FGF23 level in humans and rodents,598,599 which depends on both translational and posttranslational regulation of FGF23.600 FGF23 inhibits PTH synthesis and secretion,601,602 and then contributes to its own negative feedback regulation. Patients with hyperparathyroidism have high FGF23 level,603 and some studies including cell culture experiments showed that PTH induces FGF23 expression in human and rodent cells through activating the orphan nuclear receptor Nurr1.604,605
Furthermore, iron can regulate FGF23 expression. Iron deficiency not only increases FGF23 transcription,606 but also its cleavage.607 However, the detailed mechanism is still unclear.
FGF signaling in tumors
A typical regulation of the FGF/FGFR system occurs in multiple human tumors, leading to the deregulated activation of ligand-dependent or -independent FGFR signaling.
The expressions and mutations of FGF signaling molecules in tumors
FGF signal is highly related to the initiation and progression of several tumors including urothelial carcinoma, multiple myeloma, prostate cancer, and hepatocellular carcinoma (Table 2).
Table 2.
Somatic GOF mutations of FGFRs in cancers
| Gene | Type | Site | Cancers |
|---|---|---|---|
| FGFR1 | Amplification | Breast cancer (ER+) | |
| Gastric cancer | |||
| Lung cancer (SCC, SC) | |||
| Ovarian cancer | |||
| Urothelial cancer | |||
| Fusion | FGFR1-TACC1 | Glioblastoma | |
| BCR-FGFR1, CNTRL-FGFR1, ZMYM2-FGFR1, etc. | MPN | ||
| Mutation | N546K | Ewing sarcoma | |
| N546K, K656E | Glioblastoma | ||
| FGFR2 | Amplification | Breast cancer (TNBC) | |
| Gastric cancer | |||
| Fusion | FGFR2-AFF3, FGFR2-CASP7 | Breast cancer | |
| FGFR2-BICC1, FGFR2-PPHLN1, etc. | Cholangiocarcinoma | ||
| FGFR2-CIT | Lung cancer | ||
| Mutation | R203C, N549K, K659N | Breast cancer | |
| S252W, P253R, N549K, K659E | Endometrial cancer | ||
| S252W, P253R, K659E | Lung cancer | ||
| FGFR3 | Amplification | Ovarian and urothelial cancers | |
| Fusion | FGFR3-TACC3 | Glioblastoma and lung cancer | |
| ETV6-FGFR3 | Lymphoma | ||
| t(4;14) (p16;q32) | Multiple myeloma | ||
| FGFR3-BAIAP2L1, FGFR3-JAKMIP1, FGFR3-TACC3 | Urothelial cancer | ||
| Mutation | R248C, S249C, G370C, Y373C, G380R, K650M | Gallbladder cancer | |
| R248C, S249C, G370C, K650E | Lung cancer | ||
| R248C, Y373C, K650E/M | Multiple myeloma | ||
| R248C, S249C, G370C, S371C, Y373C, N540S, K650E/M | Urothelial cancer | ||
| FGFR4 | Mutation | N535K, V550E | Rhabdomyosarcoma |
Expressions of FGFs
FGF5 is overexpressed in breast cancer tissue.608 Guo et al.609 reported that FGF6 was significantly decreased in non-metastatic liver cancer lesion tissues and increased in metastatic liver carcinoma tissue. FGF7 is expressed in normal mucosal gland epithelium and in stromal fibroblasts, and FGF7 protein levels were elevated in gastric inflammation and gastric adenocarcinoma.610 Overexpression of FGF8 in prostate cancer is highly related to the decreased patient survival and persists in androgen-independent disease.611 FGF8, as cell growth regulator, can mediate the tumor suppression effect of Annexin-A7 in prostate tumorigenesis.612 FGF9 is expressed in many non-small cell lung carcinoma (NSCLC) primary tumors and derived cell lines. The NSCLC patients with high FGF9 expression had shorter overall survival.613 Aberrant signaling of FGF10 through FGFR2b, and in some instances FGFR1b, contributes to the progression of a number of human cancers, including breast cancer, prostate cancer, and pancreatic adenocarcinoma, as well as gastric cancer (GC), skin cancers, and lung squamous cell carcinomas.614 FGF12 gene was overexpressed in esophageal squamous cells.615 FGF13 was highly upregulated in aggressively metastatic breast tumors and pancreatic endocrine tumors.616 FGF14 was preferentially methylated in colorectal cancer.616 The expression of FGF16 is markedly increased in ovarian tumors.617 FGF17 is overexpressed as a potential mediator of FGF8 function in human prostate cancer.618 In genomically stable and chromosomal instable subtypes of GC, FGF18 was overexpressed with relevance to poor survival.619 FGF18/FGFR3IIIc was upregulated and could drive growth of tumor cell in CD44+ subpopulation of colon adenoma cells.620 Aberrant signaling through FGF19 and its receptor FGFR4 seems to be the oncogenic driver for a subset of human hepatocellular carcinoma (HCCs) and is associated with poor prognosis.621 Ectopic expression of FGF20 in NIH 3T3 cells rendered the cells transformed in vitro and tumorigenic in nude mice.622 The mRNA level of FGF20 was upregulated in adenomas in mice and FGF20 is found to be a critical element in Wnt signaling-induced oncogenesis.623 Huang et al.624 found that overexpression of FGF21 delayed the appearance of diethylnitrosamine-induced liver tumors and proposed that FGF21 might delay development of adenomas through activation of resident hepatocyte FGFR4 at early time. Liu et al.625 demonstrated that FGF22 expression was tightly associated with the poor overall survival. FGF23 is present at an increased level and promotes the progression of prostate cancer.626
Mutations of FGFs and FGFRs in tumors
The risk of relapse in the subgroup of progesterone-receptor-negative patients of breast tumors was five times greater for those with int-2/FGF3 amplification than for those without this alteration.627 High-throughput tissue microarray analysis showed that gene amplifications of FGF3 and FGF4 were observed in urinary bladder cancer.628 Kim and his colleagues629 revealed that three SNPs in the FGF23 gene (rs11063118, rs13312789, and rs7955866) were associated with an increased risk of prostate cancer. Mutations of FGFRs are commonly observed in many tumors, including the breast cancer, lung cancer, liver cancer, GC, uterine cancer, and bladder cancer.630 FGFR1 amplification is one of the most common focal amplifications in breast cancer.631 FGFR1 amplification was observed in 32% of small cell lung cancer samples.632 A single somatic FGFR1 mutation (c.C754A p.P252T) was also detected in a bronchoalveolar cancer.633 Constitutional and somatic FGFR1 alterations were frequently observed in dysembryoplastic neuroepithelial tumor (DNET) and played a key role in the pathogenesis of DNET.634 FGFR2 amplifications have been observed in nearly 10% of GCs, playing a critical role in the proliferation and survival of GC cell.635 GC cell lines with FGFR2 amplifications were highly sensitive to FGFR inhibitors.636 Dutt et al.637 reported that somatic mutations of FGFR2 were present in 12% of endometrial carcinomas, and inhibition of FGFR2 kinase activity in endometrial carcinoma cell line bearing such FGFR2 mutations could inhibit its transformation and survival, implicating FGFR2 as a novel therapeutic target in endometrial carcinoma. FGFR2 fusions were reported to be present in up to 13% of liver cancers such as intrahepatic cholangiocarcinoma.638,639 FGFR2 amplifications occur in triple-negative breast cancer, and are associated with high sensitivity to FGFR inhibitors.640 FGFR2 is shown to be associated with a higher risk of sporadic post-menopausal breast cancer.641 Amplifications of FGFR3 have been described rarely in cancer, while activation of FGFR3 by mutation was quite common.642 FGFR3 alterations (mutations or translocation) are among the most frequent genetic events in bladder carcinoma. Single-nucleotide substitution mutations of FGFR3 were present in 35% of bladder carcinomas.643 The mutations of FGFR3 could lead to an aberrant activation of FGFR3 signaling, conferring an oncogenic dependence, while inhibition of FGFR3 signaling decreased cell viability in vitro and tumor growth in vivo.644 FGFR3 mutations were also identified in cervical cancers,645 multiple myeloma,38 prostate cancer,646 testicular tumors,647 and lung adenocarcinoma.648 FGFR1-3 gene fusions have been observed in breast cancer to occur with multiple gene partners (i.e., TACC1-3, BAIAP2L1, AFF3, SLC45A3, and AHCYL1).640 A very low level of amplifications of FGFR3 and FGFR4 were detected in breast cancer.649 Mutations in FGFR4 in human rhabdomyosarcoma (RMS) could lead to its activation and contribute to RMS progression as an oncogene.650 The mutation of FGFR4 gene transcript in MDA-MB-453 mammary carcinoma cells lead to the substitution of glycine by arginine at position 388, which increased cell motility. The FGFR4 Arg388 allele was related to the metastasis of colon cancer in patients.651
FGFs and FGFRs in tumorigenesis
FGF/FGFR signaling is involved in the major steps of tumor progression, including cancer cell survival and proliferation, angiogenesis, invasion, and metastatic dissemination and response to therapy.
FGFs and FGFRs in tumor growth
The expression of FGF4 was increased in germ cell tumors, especially in non-seminomas, which could promote malignant growth of cultured embryonal carcinomas by targeting all-trans-retinoic acid.652 FGF2 can induce breast cancer growth through ligand-independent activation and stimulate the MYC gene expression through recruitment of ERα and PRB4 isoform to MYC regulatory sequences.653 The results from Betsuyaku, T.’s group showed that the FGF2 aptamer that can block FGF2 activity could inhibit the growth of FGF2-FGFR pathway-dependent lung cancer cells.654 Increased expression of FGF4 in ovarian cancer stem-like cells/cancer-initiating cells is involved in the upregulating tumor initiation capacity of fibroblasts.655 Fang et al.656 demonstrated that miR-188-5p suppressed the tumor cell proliferation and metastasis by directly targeting FGF5 in HCC. The neutralizing antibody to FGF8b could significantly inhibit cell growth of prostate cancer.611 In mouse Leydig tumor cells, FGF9/FGFR2 signaling can increase its proliferation by activating ERK1/2, Rb/E2F1, and cell-cycle pathways.657 Downregulation of FGF18 suppressed the tumor formation abilities, induced G1-phase cell-cycle arrest and enhanced anticancer drug sensitivity.619 The antibody of FGF19 could inhibit the growth of colon tumor xenografts in vivo and effectively prevent HCCs in FGF19 transgenic mice, suggesting that the inactivation of FGF19 could be beneficial treatment for cancers and other malignancies involving interaction of FGF19 and FGFR4.658 Low concentration exogenous FGF19 promoted the growth of prostate cancer cells, while inhibition of FGF19 in prostate cancer cells could decrease proliferation in vitro and tumor growth in vivo.629 FGF9 greatly contributes to Pregnane X receptor-mediated tumor aggressiveness in humans and mice.659 In endoplasmic reticulum stress-induced HCC cells, FGF19 overexpression promoted cell survival and increased resistance to apoptosis, whereas FGF19 silencing counteracted these effects.660 FGF19 gene amplification has been found to be corresponding with an increased dependency upon FGF19/FGFR4 autocrine signaling mediated by ERK/AKT-p70S6K-S6 activation in head and neck squamous cell carcinomas.661 FGF23 enhances the proliferation, invasion, and anchorage-independent growth of prostate cancer cell lines in vitro, while FGF23 KD also decreases tumor growth in vivo.626 Activation of FGFR1 leads to rapid tumor growth as a result of increased proliferation in prostate cancer cells.662 FGFR2 promotes breast cancer tumorigenicity by maintaining tumor-initiating cells.663 FGFR3 is overexpressed in the early stages of bladder cancer, and targeting the extracellular domain of FGFR3 with human single-chain Fv antibodies could suppress the proliferation of bladder carcinoma cell line.664
FGFs and FGFRs in the invasion and migration tumors
Henriksson et al.665 reported that colorectal cancer cells activate adjacent fibroblasts, which results in enhanced FGF1/FGFR3 signaling and subsequent increased invasion of tumor cells. Abrogation of the nuclear translocation of FGFR1 and FGF2 in pancreatic cancer cells significantly inhibit cancer cell invasion.88 FGF7/KGF could trigger cell transformation and invasion of immortalized human prostatic epithelial PNT1A cells.666 FGF7/FGFR2/THBS1 promotes the invasion and migration in human GC.667 FGF9 secreted by cancer-associated fibroblasts is considered as a possible mediator by promoting the anti-apoptosis and invasive capability of GC cells.668 FGF10/FGFR2 signal can significantly promote the cell migration and invasion in pancreatic cancer.669 FGF16 enhanced the invasion of SKOV-3 ovarian cancer cells through activation of MAPK signaling pathway.617 The members of FGF8 subfamily including FGF8, FGF17, and FGF18 are involved in autocrine and paracrine signaling in HCC and enhance the survival of tumor cells, tube formation, and neoangiogenesis.670 FGF18 has been reported to control the migration, invasion, and tumorigenicity of ovarian cancer cells through NF-κB activation, which increased the production of oncogenic cytokines and chemokines.671 FGF9 greatly contributes to Pregnane X receptor-mediated tumor aggressiveness in humans and mice.659
FGFs and FGFRs in tumor angiogenesis
The onset of angiogenesis is a discrete step that occurs at any stage of tumor progression. FGF ligands and receptors promote angiogenesis in a variety of tumors.672 Wang and Becker673 showed that delivery of an episomal vector containing antisense FGF2 or FGFR1 cDNA could completely prevent the growth of tumors partially through the blockage of angiogenesis in the human melanoma grown as a subcutaneous tumor model in nude mice. FGF2 can induce tumor growth and neovascularization in vivo.674 FGF2 and MMP2 may cause increased angiogenesis and invasion of bone marrow plasma cells in several unidentified monoclonal gamma globulin disease and multiple myeloma cases.675 FGF binding protein can be used as an angiogenesis conversion molecule in human tumors via promoting the release of biologically active FGF2 and leading to tumor growth.676 The type 1 repeats of thrombospondin-1 (TSP1) can block angiogenesis driven by FGF2 or vascular VEGF and inhibit tumor growth.677 IL-10 blocks the proliferation of microvascular ECs induced by VEGF and FGF2 in vitro and has a direct effect on preventing angiogenesis in human lymphomas.678 It has been reported that the average serum FGF2 level was significantly increased (~7 times) in testicular cancer patients, and the expression level of FGF2 was also significantly increased in tumor biopsies.679 Targeting the mRNA of early growth response (EGR1) an upstream of FGF2, can inhibit the expression of EGR1 protein and block tumor angiogenesis.680 Human melanoma cell survival and growth depend on autocrine action of FGF2.681 In addition, neutralized FGF2 with antibodies could block the angiogenesis in melanoma cell lines transplanted nude mice models.682 In addition, FGF2 is shown to be involved in angiogenesis in the formation of pituitary tumors.683 FGF1 can cause increased angiogenesis that contributes to the poor survival rate of patients with advanced serous ovarian cancer.684 Two angiogenic factors PDGF-BB and FGF2 in tumors can synergistically promote the neovascularization and metastasis in murine tumor model.685 Targeted inhibition of PDGF receptors can downregulate the expression of FGF2 and epithelial growth factor FGF7, thereby reducing angiogenesis.686
Therapeutics and strategies for targeting FGF signaling
FGF signaling plays critical roles in tissue/organ development and homeostasis, and dysregulated FGF signaling has been found in a variety of diseases and injuries (see above). It is a promising therapeutic strategy for these diseases/injuries by modifying or correcting the aberrant FGF signaling. So far, FGF-based therapeutics are largely classified into three classes, including enhancing FGF signaling therapeutics, blocking FGF signaling therapeutics, and gene therapy.
Enhancing FGF signaling therapeutics
FGFs are involved in numerous pathophysiological processes;1 recombinant FGF or FGF analogs have been developed as first-generation strategies to augment the beneficial effects of FGFs/FGFRs (shown in Table 3).
Table 3.
Therapeutics targeting FGF signaling
| Class | Drug | Targets | Diseases | Drug development |
|---|---|---|---|---|
| Recombinant FGFs or FGF analogs | rmFGF1 | FGF1 receptor | T2DM | Preclinical |
| rhFGF1 | FGF1 receptor | T2DM | Preclinical | |
| rhFGF2 (trafermin) | FGF2 receptor |
Skin ulcers Periodontitis |
Approved Japan P3 (NCT01015404) |
|
| FGF7 (palifermin) | FGF7 receptor | Oral mucositis | Approved USA | |
| FGF10 (repifermin) | FGF10 receptor | Mucositis | Clinical trials was terminated in 2004 | |
| rhFGF18 (sprifermin) | FGF18 receptor | Osteoarthritis | P2 (NCT01919164) | |
| FGF19-4/5/6 | FGF19 receptor | Tumorigenicity | Preclinical | |
| FGF19 variant (FGF19v) | FGF19 receptor | Mitogenic | Preclinical | |
| NGM282 (M70) | FGF19 receptor |
T2DM PSC |
P2 (NCT01943045) P2 (NCT02704364) |
|
| Obeticholic acid and Px-104 | FGF19 receptor |
Primary/secondary bile acid malabsorption Obesity NAFLD |
P2 (NCT01585025) P2 (NCT01625026) P2 (NCT01265498) P2 (NCT01999101) |
|
| LY2405319 | FGF21 receptor | T2DM | P1 (NCT01869959) | |
| FGF21variant (PEG-FGF21G71C, Fc-FGF21(RG)) | FGF21 receptor | T2DM | Preclinical | |
| PF-05231023 (CVX-343) | FGF21 receptor | T2DM | P1 (NCT01285518) | |
| Non-selective TKIs | Lucitanib (E3810) | FGFR1/2, VEGFR1/2/3, and PDGFRα/β | Cancer with FGFR alteration |
P2 (NCT02747797) P3 (NCT00165672) |
| Nintedanib (BIBF1120) | FGFR1/2/3, VEGFR1/2/3, and PDGFRα/β | Cancer with FGFR alteration | Submitted P3 | |
| Dovitinib (CHIR258 or TKI258) | VEGFR1/2/3, FGFR1/2/3, PDGFRβ, c-Kit, RET, TrkA, CSF-1R, and FLT3 | Cancer with FGFR alteration |
P2 (NCT01719549) P2 (NCT01732107) |
|
| Regorafenib | P2 (NCT01929616) | |||
| Brivanib | P2 (NCT03516071) | |||
| Ponatinib | Approved for market | |||
| Lenvatinib | FGFR1/2/3 | Cancer with FGFR | P2 (NCT03609359) | |
| Pazopanib | alteration | P2 (NCT01253369) | ||
| Orantinib | P3 (NCT01465464) | |||
| Sunitinib | P2 (NCT00768144) | |||
| Cediranib | P3 (NCT00399035) | |||
| Selective TKIs | AZD4547 | FGFR1/2/3 | Cancer with FGFR alteration |
P2 (NCT01824901) P2 (NCT01791985) P2 (NCT02824133) P2 (NCT01213160) |
| BGJ398 (NVP-BGJ398) | FGFR1/2/3 | Cancer with FGFR alteration |
P2 (NCT01975701) P2 (NCT02150967) P2 (NCT02160041) |
|
| JNJ-42756493 (erdafitinib) | FGFR1/2/3/4 | Cancer with FGFR alteration |
P2 (NCT02365597) P2 (NCT02699606) |
|
| LY287445, Debio-1347, TAS-120, and BAY-1163877 | FGFR1/2/3/4 | Cancer with FGFR alteration | Preclinical | |
| Neutralizing monoclonal antibodies (mAbs) | KRN23 | FGF23 | XLH | P3 (NCT02537431) |
| Bemarituzumab (FPA144) | FGFR2b | Neoplasms | P1 (NCT02318329) | |
| BAY1179470 | FGFR2 | Neoplasms | P1 (NCT01881217) | |
| MFGR1877S | FGFR3 | Neoplasms | P1 (NCT01122875) | |
| hIgG1-1A2 | FGF2 | |||
| GAL-F2 | FGF2 | |||
| 3F12E7 | FGF2 | |||
| KM1334 | FGF8b | Neoplasms | Preclinical | |
| FGF10 mAb | FGF10 | |||
| FN1 and FC1 | FGF23 | |||
| R1MAb1 | FGFR1 | |||
| FGF traps | FP-1039 (GSK3052230) | FGF1/2/4 | Neoplasms | P1 (NCT01868022) |
| SM27 | FGF2 | Angiogenesis | Preclinical | |
| NSC12 | FGF2 | Lung tumors | Preclinical | |
| sFGFR2IIIc (S252W) | FGFR2 | AS | Preclinical | |
| sFGFR3 | FGF2/9/18 | Chondrodysplasia | Preclinical | |
| Peptide P3 | FGFR3 | Chondrodysplasia | Preclinical | |
| Gene therapy | XRP0038 (NV1FGF) | FGF1 receptor | Peripheral vascular diseases | P2 (NCT00566657) |
| Expression of FGF18 cDNA | FGF18 receptor | Murine models | Preclinical | |
| AAV9-Fgfr2-shRNA | Fgfr2-P253R allele | AS | Preclinical | |
| CRISPR/Cas9 | Fgfr3-G374R | Achondroplasia | Preclinical |
T2DM type 2 diabetes mellitus, PSC primary sclerosing cholangitis, NAFLD non-alcoholic fatty liver disease, XLH X-linked hypophosphatemia, AS Apert syndrome, P1 phase I clinical trial, P2 phase II clinical trial, P3 phase III clinical trial
Canonical FGFs, encoded by FGF1, FGF4, FGF7, FGF8, and FGF9 subfamily gene, by binding to heparan sulfate proteoglycans largely exert their effects locally.1 A single injection of mouse recombinant FGF1 causes potent, dose- and insulin-dependent glucose lowering in diabetic mice without hypoglycemia.687 Recombinant human FGF1 (rhFGF1) is also able to normalize blood glucose in diabetic mice.687 In addition, trafermin (rhFGF2) has been supported for their use in the patients with skin ulcers,688 and in phase III clinical trial, trafermin was further approved for its application in patients with periodontal surgery. Palifermin, a truncated form of FGF7, has been approved for the treatment of patients with oral mucositis.688 In pediatric patients, palifermin may provide advantage to prevent chemotherapy-induced mucositis.689 Repifermin, a truncated form of FGF10, with the pharmacological effects similar to that of FGF7, promotes the healing of ulcerated oral and intestinal mucosal tissue, and reduces the complications in preclinical tests.690 However, the clinical trials about the effect of repifermin on mucositis were terminated in 2004 as no effective evidence for reducing the incidence or severity.691 In addition, rhFGF18 have been approved for treating OA and cartilage injury of the knee in phase II clinical trial.266
Endocrine FGFs, encoded by FGF19 subfamily gene, which bind and activate FGFRs with the Klotho family protein, regulate a wide range of metabolic processes.692 Based on the structure–function principle, separating mitogenic and metabolic activities of FGF19 through mutagenesis of five N-terminal and heparin-binding regions of FGF19 yielded a series of FGF19 variants, which retain the beneficial metabolic effects, while reduce the side effects of FGF19 on tumorigenicity.693 In addition, a new constructed FGF19 variant (25-194 of FGF19 and 1-20 of FGF21), impaired in activating FGFR4 and still had beneficial effects on glucose and lipid metabolism.694 These studies provide a strategy for engineering FGF19 as a potential therapy for related diseases/injuries. These variants were found to be devoid of BA regulatory activity. However, another FGF19 variant NGM282 (M70) retains the beneficial BA metabolism effects, while is devoid of murine mitogenic activity by inactivating the STAT3 pathway.695 To date, M70 as one FGF19 variant was studied through phase II clinical trials for their use in patients with primary sclerosing cholangitis and diabetes mellitus. In addition, several FGF19-inducing strategies (farnesoid X receptor agonists) such as obeticholic acid and Px-104 were tested through phase II clinical trials and provided with further support for their use in the patients with primary/secondary BA malabsorption and nonalcoholic fatty liver disease, respectively.
Several strategies have been used to optimize the “druggability” of FGF21. LY2405319, a novel FGF21 variant, was reconstructed by introducing an additional disulfide bond firstly in the C terminal of FGF21 by mutations (L118C, A134C), and then further optimized by deleting His-Pro-Ile-Pro in the N terminal of FGF21 along with a mutation to replace the major site of O-linked glycosylation (Ser167Ala). Subcutaneous administration of LY2405319 in DIO mice exhibited a potency similar to FGF21, resulting in decreased plasma glucose along with a reduction in body weight.696 To date, LY2405319 has been tested through phase I clinical trial to reduce body weight and fasting insulin, and is noteworthy in improving dyslipidemia in patients with type 2 diabetes mellitus.697 Another FGF21 variant is reconstructed through the introduction of p-acetyl phenylalanine into the N-terminal residue of rhFGF21 for the attachment of PEG (PEGylated rhFGF21).698 PEGylated rhFGF21 has the ability to normalize insulin-mediated glucose utilization in diabetic murine models,699 but exhibits remarkably lower bioactivity than FGF21, along with induction of renal vacuole formation.700 Song et al.701 further optimized FGF21 by introducing G71C mutation to generate the mimetic PEG-FGF21G71C, which exhibits increased half-life. Subsequently, an alternative strategy was adopted to yield Fc-FGF21 by fusing Fc fragment of human IgG1 to the N-terminal end of FGF21 to improve the pharmacokinetic properties of FGF21, which exhibited a prominently increased half-life compared to the native FGF21.702 Since, the C-terminal region of Fc-FGF21, especially between Pro171 and Ser172, was rapidly degraded, Pro171Gly mutation was introduced to retain biological activity, while eliminate the proteolytic degradation.703 Moreover, FGF21 has the additional concern of forming aggregates during protein production. By combining Pro171Gly and Leu98Arg mutations into one molecule, a novel variant named Fc-FGF21 (RG) was generated with resistance to aggregation and proteolysis.703 Another approach to improve plasma half-life is to fuse FGF21 to a scaffold monoclonal antibody (mAb).704,705
Blocking FGFs signaling therapeutics
Given that a variety of human diseases and injuries caused by excessive FGF signaling. So far, the measures blocking FGF signaling can be generally classified to TKIs, neutralizing mAbs, and FGF traps.
TKIs
Nonselective TKIs
Nonselective TKIs have been developed as first-generation strategies to blocking FGFs signaling. These TKIs have the benefit of concurrently targeting tumor proliferation and angiogenesis, while also displaying a remarkable effect against FGFR signaling pathways, together with a multiplicity of adverse effects that limit their use in clinic.
Lucitanib (E3810) is a triple TKI, which targeting FGFRs, VEGFRs, and PDGFRs. E3810 showed a promising efficacy and a manageable side effect in patients with both FGF-aberrant or angiogenesis-sensitive tumor types.706 Until 2018, E3810 were completed phase II clinical trials, which inhibits the growth of tumor by antiangiogenesis.
Nintedanib (BIBF1120) is another novel triple angiokinase inhibitor, with less activity against SRC, RET, and FLT3.707 BIBF1120 competitively binds to the ATP-binding pocket of these receptors, and blocks the intracellular signaling critical for the proliferation and survival of angiogenesis-related cells.707 Up-to-date, BIBF1120 has been approved for the treatment of pulmonary fibrosis and as a second-line therapy for NSCLC in combination with docetaxel. Phase III clinical trials are still ongoing to study the response of patients selected for specific FGFR alterations.708
Dovitinib (CHIR258 or TKI258) is an oral ATP-competitive multikinase inhibitor that targets FGFRs, VEGFRs, and PDGFRβ.709 TKI258 has a promising inhibitory activity in cell lines with FGFR translocations or amplification.710 In phase II trials, TKI258 can stabilize disease in multiple myeloma bearing t (4;14) translocation by blocking FGFR3 activity.711
Beyond that, several other nonselective TKIs are shown in Table 3, which have been developed and are in preclinical and clinical evaluation.712 However, these nonselective TKIs induce a series of side effects: cardiotoxicity or proteinuria on account of the concurrent VEGFR inhibition, as well as cutaneous reactions, digestive disorders, and gastrointestinal disease, for example.153
Selective TKIs
To overcome the off-target effects, second-generation selective FGFR TKIs have been developed.
AZD4547 is a potent reversible TKI specific for FGFRs.713 Of note, AZD4547 is able to sharply diminish cancer stem-like cells by inducing MET via MEK/ERK pathway downstream of FGFR signaling.714 In addition, administered AZD4547 prominently impaired ductal branching and stem cell-like characteristics in mammary epithelial cell and spontaneous tumor cells.715 In phase I/II trials, AZD4547 further showed promising inhibitory activity in models of cancer with FGFR alteration.
BGJ398 (NVP-BGJ398) is a selective reversible ATP-competitive inhibitor targeting FGFRs, which showed superior potency to ponatinib and dovitinib, and exerted a more potent therapeutic effect against chemotherapy-refractory cholangiocarcinoma containing FGFR2 fusions.638 Of note, in phase I/II trials, BGJ398 promoted tumor reduction in patients with FGFR-related advanced solid tumors.716
JNJ42756493 (erdafitinib) with potent TKI activity can target all FGFRs, which suppresses phospho-FGFR and phospho-ERK resulting in dose-dependent antitumor activity.717 Further in phase I/II trials, the administered erdafitinib has an inhibitory activity in patients with advanced solid tumors characterized by FGFR translocations or FGFR3–TACC3 fusions.717–719
Other selective TKIs are shown in Table 3, and showed promising results in preclinical and clinical evaluation on different oncotypes.712
Unfortunately, drug resistance limits the success of TKIs with mutations at the “gatekeeper” residue, leading to tumor progression. Structural analyses showed that the FGFR1 “gatekeeper” mutation (V561M) can induce a potently increased autophosphorylation, in part, by a network of interacting residues forming a hydrophobic spine to stabilize the active conformation. Further kinetic assays established that V561M confers significant resistance to E3810, while it retains affinity for AZD4547 due to a flexible linker that allows multiple inhibitor binding modes.720 In addition, JNJ42756493 binds to the ATP pocket of the FGFR1 KD with unique structural conformations, and its inhibitory efficacy is reduced by 200-fold in the FGFR3 “gatekeeper” mutation (V555M), while an increase in efficacy for TKI258.721 In contrast, some FGFR2 “gatekeeper” mutations drive acquired resistance to TKI258 by causing steric hindrance to the binding of the TKI to the receptor (such as N550K, E566G, and K660E) or by stabilizing the active conformation of the kinase (V651I).722 Moreover, multiple recurrent patients have point mutations in the FGFR2 KD at progression, and each mutation drives acquired resistance to BGJ398, and was surmountable by structurally distinct FGFR inhibitors.723 Thus, designing inhibitor with flexibility to overcome drug resistance may be an vital way for exploiting effective inhibitor against mutation.
Neutralizing mAbs
When compared to TKIs, neutralizing mAbs have unique advantages of low toxicity due to the absence of off-target effects.
Burosumab (formerly KRN23) is a fully human IgG1 mAb that binds to and blocks the biologic activity of FGF23. Injection of Burosumab normalized both phosphate and vitamin D concentrations in hypophosphatemia mouse models.724 In 2019, phase II clinical trials for Burosumab was completed and provided support for its use for XLH.
Bemarituzumab (FPA144) is a rhIgG1 mAb that specially binds to the IgG III region of the FGFR2b receptor isoform to prevent ligand binding and downstream signaling activation. In phase I clinical trial, a single dose of FPA144 was conducted in gastroesophageal adenocarcinoma (GEA) patients with FGFR2b overexpression, which remarkably inhibited GEA growth.
MGFR1877S is an mAb targeting FGFR3 by hampering its dimerization, which is well tolerated with low toxicities in patients with multiple myeloma and solid tumors in phase I clinical trials.708
Beyond that, there are other mAbs awaiting further confirmation in preclinical and clinical testing, such as BAY1179470, hIgG1-1A2, GAL-F2, 3F12E7, KM1334, FGF10 mAb, FN1, FC1, and R1MAb1, as detailed in Table 3.
FGF traps
An alternative strategy to modulate the activity of the FGF/FGFR signaling is to use the molecules able to bind and neutralize multiple FGF ligands. This strategy represents a novel path for the development of FGF traps.
FP-1039 (GSK3052230) is an FGF ligand trap that binds and neutralizes multiple FGFs and thus inhibits the activation of FGFR1. In preclinical trials, FP-1039 blocked FGF2-stimulated tumor cell proliferation and inhibited tumor growth in xenograft models.672 In phase I clinical trials, associated with paclitaxel and carboplatin, or docetaxel, intraperitoneal injection of FP-1039 was well tolerated in patients with solid malignancies.725 However, FP-1039 does not effectively inhibit endocrine FGFs (FGF19, FGF21, and FGF23).726 Therefore, FP-1039 has the potential to effectively block the neoplasms or advanced cancer-promoting FGFs, with less toxicity compared to small molecules such as FGFR kinase inhibitor.
The development of FGF trap agents has also relied on the structural characterization of the interactions of FGFs with their natural “interactome,” including thrombospondin-1 (TSP1), HSPGs, and pentraxin-3 (PTX3).727 Structural analysis of the complex between FGF2 and TSP1 identified a new small-molecule SM27 that inhibits FGF2-induced angiogenesis through binding to FGF2.728 Similar to the integrative TSP1, SM27 perturbs FGF2 dynamics in distant regions, including the FGFR1 binding site, by binding the heparin affinity site of FGF2, thus preventing FGF2 binding to HSPG and FGFR1.729 Therefore, SM27 acts as a dual direct and allosteric inhibitor of the binding between FGF2 and its receptors, which has unique benefits for the development of novel cancer drug. In addition, structural analysis of the complex between FGF2 and the N terminal of PTX3730 identified an acetylated pentapeptide ARPCA as the minimal FGF2 binding peptide that inhibits FGF8b-induced angiogenesis.731 Besides, based on pharmacophore modeling of the ARPCA/FGF2 interaction, NSC12 was identified as multi-FGF trap that can participate in the formation of the HSPG/FGF2/FGFR1 ternary complex. In tumor models, administration of NSC12 can block the growth, angiogenesis, and metastasis of FGF-dependent lung tumors.732
In addition, a soluble FGFR2 mutant with S252W (sFGFR2IIIc (S252W)) was found to partially alleviate the AS in mice by alleviating the premature closure of coronal suture in cultured calvarias and transgenic mice.733,734 Moreover, sFGFR3, a recombinant protein, acts as a FGFR trap to prevent FGF ligand binding to FGFR3. In ACH mice, subcutaneous injection of sFGFR3, to compete with endogenous FGFR3 ligands, showed a dose-dependent rescue of chondrodysplasia phenotypes.735 Besides, in TD II model, administration of peptide P3 with the ability to downregulate the activity of FGFR3 rescues the lethal phenotype and partially restores the structural distortion of growth plates.736
Gene therapy
At present, gene therapy is inevitable, especially in the era of precision medicine. Expression of FGF18 by AAV-mediated gene transfer in the pinnae of nude mice resulted in a noteworthy increased thickness due to an FGF18-mediated increase in chondrocyte proliferation and ECM production.238 Conditional expression of FGF18 in stromal cells surrounding proximal airway cartilage in normal mouse lung is capable of enhancing proximal programs during lung morphogenesis.737 Up-to-date, only few FGF signaling-related gene therapies have entered clinical trials. NV1FGF is a plasmid-based angiogenic gene delivery system for local expression of FGF1. Intramuscular administration of NV1FGF resulted in a noteworthy reduced risk of major amputation in patients with critical limb ischemia.738 In 2017, phase II clinical trials for NV1FGF was completed and provided further support for its use in patients with severe peripheral artery occlusive disease.
The above-described molecules such as sFGFR2IIIc (S252W)733,734 or MEK inhibitor739 or glycosaminoglycans740 can partially alleviate the AS, but may bring undesired effects as they do not specifically antagonize the mutant FGFR2 itself. In contrast, RNA interference (RNAi) could inhibit the expression of mutant alleles at the transcriptional level. A short hairpin RNA (shRNA) targeting the dominant mutant form of FGFR2 (FGFR2 (S252W)) prevents the phenotypes of AS in mice.739 Safety and efficiency are the two major concerns for the application of RNAi-related therapeutics. AAV has unique advantages of gene transfer for therapeutic treatment of a number of diseases, including congenital blindness, hemophilia, and spinal muscular atrophy.741,742 Our group screened a siRNA specifically targeting the FGFR2-P253R allele, when this siRNA was delivered to the skulls in AS mouse model using AAV9 (AAV9-FGFR2-shRNA), it attenuated the premature closure of coronal suture and the decreased calvaria bone volume.743 Such biological strategy, in combination with other therapies including surgeries, provides experimental clues for the biological therapies of other genetic skeletal diseases.
In recent years, CRISPR/Cas9-based method has been is developed for gene therapy. Some studies have verified the advantage of CRISPR/Cas9 technology for the correction of human hereditary genetic diseases, such as liver diseases,744 cataract disorder,745 Duchenne muscular dystrophy,746 tyrosinemia,747 thalassemia,748 and so forth. Miao et al.749 found that Cas9 protein can achieve higher frequency of precise correction of the FGFR3-G374R mutation than Cas9 mRNA. These strategies completely suppressed phenotypes of ACH without off-target effects checked by whole-genome sequencing. CRISPR/Cas9 technology can precisely correct individual mutations with high fidelity and is potentially translatable for clinical therapies of human diseases, especially genetic diseases in the future.
Conclusion and perspective
Knowledge of the role of FGF/FGFR signaling in pathological and physiological conditions has advanced considerably in the past decades. In this review, we summarized the structure and function of FGF signaling molecules and the detailed regulatory mechanisms. FGF/FGFR system contributes to the pathophysiology of multiple disorders in humans, including genetic diseases, dysplastic diseases, various types of cancer, metabolic disorders, and degenerative diseases, as well as injuries and regeneration. Much remains to be learned. The spatiotemporal expression patterns, accurate roles, and underlying mechanisms of individual FGFs/FGFRs in the development and diseases/injuries are largely unknown.
Activation of FGF signaling is tightly controlled with diverse transduction specificity, which mainly depends on the molecular structures of FGFs/FGFRs. With the advance of multiple disciplines including structure biology, we have acquired more information about FGFs/FGFRs, such as their structures, binding partners, key amino acids mediating the specific binding and signaling pathways. We need to know from the viewpoint of structure why individual FGFs have variable binding affinities of respective FGFRs; why the same FGF ligand bind distinct group of FGFRs at different concentration; the downstream signaling pathways activated by individual FGF through respective FGFR at different concentrations and in physiological and pathologic circumstance; can we switch the binding affinity of individual FGFs, based on their structure, to HS and FGFRs to have novel therapeutic effects on aberrant FGF signaling-related disease? With this information, we will have the possibility to fine tune FGF-related signaling to achieve better therapeutic outcome in the future.
There are complex interactions among individual FGFs and FGFRs. Most FGF can bind multiple FGFRs with differential binding affinities. So far, there are few studies about the differential signaling pathways activated by individual FGF through corresponding FGFRs. Considering the differential even opposite effects of each FGFR in the homeostasis maintenance and occurrence of diseases, for example, FGFR1 promotes while FGFR3 suppresses OA pathogenesis, the effects of individual FGF on OA and cartilage injuries are the summed effects of all signaling pathways of FGFRs activated by the applied FGF. More studies are needed to know the individual FGFRs activated by the applied FGFs at specific concentrations.
To obtain these knowledges, we need new strategies such as omics technology, single-cell analysis, and in vivo imaging, as well as utilization of more species of model animals and more spatiotemporally tunable genetic approaches. For example, our commonly used strategy to study the role of individual FGFs or FGFRs in the disease pathogenesis has limitation. We need to use conditional approach to spatiotemporally delete or overexpress individual FGFs or FGFRs in a certain type of cells, for example, chondrocytes, aimed to dissect the role of individual FGFs or FGFRs in the development and maintenance of the targeted cells. In addition, it is appreciated that mutations of individual FGFs or FGFRs can have detrimental effects, but a systematic understanding of intracellular pathway activation and dynamics is still lacking.750
To mimic the effects obtained from omics and conditional knockout study, we need to use targeted therapy approaches, which means to precisely modulate individual FGFs, FGFRs, and downstream signaling in specific types of cells at specific disease stages. The good news is that we are having more and more approach to exert these targeted treatments. For example, aptamer-based cell lineage or tissue targeting approaches are increasingly utilized. Several aptamers have been discovered to specifically target bone-forming site, osteoblasts, osteoclasts, and osteocytes in the skeletal tissue. We can similarly find aptamers specifically targeting for distinct cells at different growth phases, or inflammatory cells, paracancerous, and non-tumorous tissues, and so on.
FGF pathway interacts extensively with other signaling pathways during a variety of development and disease processes. Clarifying the interactions among FGF signaling, and these signaling pathways, such as BMP/TGF-β, PTH, hedgehog, and retinoid pathways, will provide us with the molecular bases for searching for combined therapies.751
Interventions targeting FGFs/FGFRs represent new approaches for the treatment of a wide range of diseases including genetic disorders, cancer, metabolic disease, degenerative disease, and injury repair. Developments in this field will likely be facilitated by structure-based drug design of agonists and antagonists for FGF signaling.
Funding
The National Key Research and Development Program of China (2018YFA0800802), National Natural Science Foundation of China (81530071, 81772306, 81721001, 81991513), Innovative Research Team in University (IRT1216) and Key research and development projects of science and technology innovation of social undertakings and people’s livelihood security in Chongqing (cstc2017shms-zdyfX0027).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yangli Xie, Nan Su
Contributor Information
Yangli Xie, Email: xieyangli841015@163.com.
Huabing Qi, Email: hbqi3@163.com.
Lin Chen, Email: linchen70@163.com.
References
- 1.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 3.Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell. Biol. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell B, Breeze AL. Structure, activation and dysregulation of fibroblast growth factor receptor kinases: perspectives for clinical targeting. Biochem. Soc. Trans. 2018;46:1753–1770. doi: 10.1042/BST20180004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Z, et al. Two FGF receptor kinase molecules act in concert to recruit and transphosphorylate phospholipase Cgamma. Mol. Cell. 2016;61:98–110. doi: 10.1016/j.molcel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 8.Furthauer M, et al. Sprouty4 acts in vivo as a feedback-induced antagonist of FGF signaling in zebrafish. Development. 2001;128:2175–2186. doi: 10.1242/dev.128.12.2175. [DOI] [PubMed] [Google Scholar]
- 9.Mailleux AA, et al. Evidence that SPROUTY2 functions as an inhibitor of mouse embryonic lung growth and morphogenesis. Mech. Dev. 2001;102:81–94. doi: 10.1016/s0925-4773(01)00286-6. [DOI] [PubMed] [Google Scholar]
- 10.Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat. Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- 11.Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- 12.Torii S, et al. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Zhang ZY. The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J. Biol. Chem. 2001;276:32382–32391. doi: 10.1074/jbc.M103369200. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami Y, et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- 15.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb. Perspect. Biol. 2013;5:a015958. doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eswarakumar VP, et al. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 18.Miraoui H, Marie PJ. Fibroblast growth factor receptor signaling crosstalk in skeletogenesis. Sci. Signal. 2010;3:re9. doi: 10.1126/scisignal.3146re9. [DOI] [PubMed] [Google Scholar]
- 19.Qi H, et al. FGFR3 induces degradation of BMP type I receptor to regulate skeletal development. Biochim. Biophys. Acta. 2014;1843:1237–1247. doi: 10.1016/j.bbamcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minina E, et al. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev. Cell. 2002;3:439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 21.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol. Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 22.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 23.Gong SG. Isoforms of receptors of fibroblast growth factors. J. Cell. Physiol. 2014;229:1887–1895. doi: 10.1002/jcp.24649. [DOI] [PubMed] [Google Scholar]
- 24.Yeh BK, et al. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl Acad. Sci. USA. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X, Lee K, Asa SL, Ezzat S. Epigenetic silencing through DNA and histone methylation of fibroblast growth factor receptor 2 in neoplastic pituitary cells. Am. J. Pathol. 2007;170:1618–1628. doi: 10.2353/ajpath.2007.061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarabipour S, Hristova K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016;7:10262. doi: 10.1038/ncomms10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Triantis V, et al. Glycosylation of fibroblast growth factor receptor 4 is a key regulator of fibroblast growth factor 19-mediated down-regulation of cytochrome P450 7A1. Hepatology. 2010;52:656–666. doi: 10.1002/hep.23708. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler JA, Clinkenbeard EL. Regulation of fibroblast growth factor 23 by iron, EPO, and HIF. Curr. Mol. Biol. Rep. 2019;5:8–17. doi: 10.1007/s40610-019-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucinska M, et al. Differential regulation of fibroblast growth factor receptor 1 trafficking and function by extracellular galectins. Cell Commun. Signal. 2019;17:65. doi: 10.1186/s12964-019-0371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porebska N, et al. Targeting cellular trafficking of fibroblast growth factor receptors as a strategy for selective cancer treatment. J. Clin. Med. 2018;8:7. doi: 10.3390/jcm8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JP, Kusche-Gullberg M. Heparan sulfate: biosynthesis, structure, and function. Int. Rev. Cell. Mol. Biol. 2016;325:215–273. doi: 10.1016/bs.ircmb.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Pellegrini L. Role of heparan sulfate in fibroblast growth factor signalling: a structural view. Curr. Opin. Struct. Biol. 2001;11:629–634. doi: 10.1016/s0959-440x(00)00258-x. [DOI] [PubMed] [Google Scholar]
- 33.Goetz R, et al. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlessinger J, et al. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, et al. Alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature. 2018;553:461–466. doi: 10.1038/nature25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuro-o M. The Klotho proteins in health and disease. Nat. Rev. Nephrol. 2019;15:27–44. doi: 10.1038/s41581-018-0078-3. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, et al. C-terminal tail of FGF19 determines its specificity toward Klotho co-receptors. J. Biol. Chem. 2008;283:33304–33309. doi: 10.1074/jbc.M803319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goetz R, et al. Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Mol. Cell. Biol. 2012;32:1944–1954. doi: 10.1128/MCB.06603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goetz R, et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-Klotho complex formation. Proc. Natl Acad. Sci. USA. 2010;107:407–412. doi: 10.1073/pnas.0902006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White KE, et al. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60:2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 41.Shimada T, et al. Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology. 2002;143:3179–3182. doi: 10.1210/endo.143.8.8795. [DOI] [PubMed] [Google Scholar]
- 42.Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat. Rev. Mol. Cell. Biol. 2011;12:189–197. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- 43.Latko M, et al. Cross-talk between fibroblast growth factor receptors and other cell surface proteins. Cells. 2019;8:455. doi: 10.3390/cells8050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Heras E, Howell FV, Williams G, Doherty P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J. Biol. Chem. 2006;281:35208–35216. doi: 10.1074/jbc.M608655200. [DOI] [PubMed] [Google Scholar]
- 46.Qian X, et al. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene. 2014;33:3411–3421. doi: 10.1038/onc.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boscher C, Mege RM. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal. 2008;20:1061–1072. doi: 10.1016/j.cellsig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Williams EJ, Furness J, Walsh FS, Doherty P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron. 1994;13:583–594. doi: 10.1016/0896-6273(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 49.Carafoli F, Saffell JL, Hohenester E. Structure of the tandem fibronectin type 3 domains of neural cell adhesion molecule. J. Mol. Biol. 2008;377:524–534. doi: 10.1016/j.jmb.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kon E, et al. N-cadherin-regulated FGFR ubiquitination and degradation control mammalian neocortical projection neuron migration. Elife. 2019;8:e47673. doi: 10.7554/eLife.47673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francavilla C, et al. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J. Cell Biol. 2009;187:1101–1116. doi: 10.1083/jcb.200903030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachmann M, Kukkurainen S, Hytonen VP, Wehrle-Haller B. Cell adhesion by integrins. Physiol. Rev. 2019;99:1655–1699. doi: 10.1152/physrev.00036.2018. [DOI] [PubMed] [Google Scholar]
- 53.Mori S, Takada Y. Crosstalk between fibroblast growth factor (FGF) receptor and integrin through direct integrin binding to FGF and resulting integrin-FGF-FGFR ternary complex formation. Med. Sci. 2013;1:20–36. [Google Scholar]
- 54.Mori S, et al. The integrin-binding defective FGF2 mutants potently suppress FGF2 signalling and angiogenesis. Biosci. Rep. 2017;37:BSR20170173. doi: 10.1042/BSR20170173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rusnati M, et al. alphavbeta3 integrin mediates the cell-adhesive capacity and biological activity of basic fibroblast growth factor (FGF-2) in cultured endothelial cells. Mol. Biol. Cell. 1997;8:2449–2461. doi: 10.1091/mbc.8.12.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mori S, et al. Direct binding of integrin alphavbeta3 to FGF1 plays a role in FGF1 signaling. J. Biol. Chem. 2008;283:18066–18075. doi: 10.1074/jbc.M801213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ronn LC, et al. Neurite outgrowth induced by a synthetic peptide ligand of neural cell adhesion molecule requires fibroblast growth factor receptor activation. J. Neurochem. 2000;75:665–671. doi: 10.1046/j.1471-4159.2000.0750665.x. [DOI] [PubMed] [Google Scholar]
- 58.Meiri KF, Saffell JL, Walsh FS, Doherty P. Neurite outgrowth stimulated by neural cell adhesion molecules requires growth-associated protein-43 (GAP-43) function and is associated with GAP-43 phosphorylation in growth cones. J. Neurosci. 1998;18:10429–10437. doi: 10.1523/JNEUROSCI.18-24-10429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiryushko D, Korshunova I, Berezin V, Bock E. Neural cell adhesion molecule induces intracellular signaling via multiple mechanisms of Ca2+ homeostasis. Mol. Biol. Cell. 2006;17:2278–2286. doi: 10.1091/mbc.E05-10-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weis WI, Kobilka BK. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liebmann C, Bohmer FD. Signal transduction pathways of G protein-coupled receptors and their cross-talk with receptor tyrosine kinases: lessons from bradykinin signaling. Curr. Med. Chem. 2000;7:911–943. doi: 10.2174/0929867003374589. [DOI] [PubMed] [Google Scholar]
- 62.Natarajan K, Berk BC. Crosstalk coregulation mechanisms of G protein-coupled receptors and receptor tyrosine kinases. Methods Mol. Biol. 2006;332:51–77. doi: 10.1385/1-59745-048-0:51. [DOI] [PubMed] [Google Scholar]
- 63.Cattaneo F, et al. Cell-surface receptors transactivation mediated by G protein-coupled receptors. Int J. Mol. Sci. 2014;15:19700–19728. doi: 10.3390/ijms151119700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z. Transactivation of epidermal growth factor receptor by g protein-coupled receptors: recent progress, challenges and future research. Int. J. Mol. Sci. 2016;17:95. doi: 10.3390/ijms17010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alderton F, et al. Tethering of the platelet-derived growth factor beta receptor to G-protein-coupled receptors. A novel platform for integrative signaling by these receptor classes in mammalian cells. J. Biol. Chem. 2001;276:28578–28585. doi: 10.1074/jbc.M102771200. [DOI] [PubMed] [Google Scholar]
- 66.Rozengurt E, Sinnett-Smith J, Kisfalvi K. Crosstalk between insulin/insulin-like growth factor-1 receptors and G protein-coupled receptor signaling systems: a novel target for the antidiabetic drug metformin in pancreatic cancer. Clin. Cancer Res. 2010;16:2505–2511. doi: 10.1158/1078-0432.CCR-09-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Liberto V, Mudo G, Belluardo N. Crosstalk between receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCR) in the brain: Focus on heteroreceptor complexes and related functional neurotrophic effects. Neuropharmacology. 2019;152:67–77. doi: 10.1016/j.neuropharm.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 68.Flajolet M, et al. FGF acts as a co-transmitter through adenosine A(2A) receptor to regulate synaptic plasticity. Nat. Neurosci. 2008;11:1402–1409. doi: 10.1038/nn.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asimaki O, et al. Cannabinoid 1 receptor-dependent transactivation of fibroblast growth factor receptor 1 emanates from lipid rafts and amplifies extracellular signal-regulated kinase 1/2 activation in embryonic cortical neurons. J. Neurochem. 2011;116:866–873. doi: 10.1111/j.1471-4159.2010.07030.x. [DOI] [PubMed] [Google Scholar]
- 70.Borroto-Escuela DO, et al. Fibroblast growth factor receptor 1- 5-hydroxytryptamine 1A heteroreceptor complexes and their enhancement of hippocampal plasticity. Biol. Psychiatry. 2012;71:84–91. doi: 10.1016/j.biopsych.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 71.Di Liberto V, et al. Existence of muscarinic acetylcholine receptor (mAChR) and fibroblast growth factor receptor (FGFR) heteroreceptor complexes and their enhancement of neurite outgrowth in neural hippocampal cultures. Biochim. Biophys. Acta Gen. ubj. 2017;1861:235–245. doi: 10.1016/j.bbagen.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 72.Borroto-Escuela DO, Tarakanov AO, Fuxe K. FGFR1-5-HT1A heteroreceptor complexes: implications for understanding and treating major depression. Trends Neurosci. 2016;39:5–15. doi: 10.1016/j.tins.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 73.Barquilla A, Pasquale EB. Eph receptors and ephrins: therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2015;55:465–487. doi: 10.1146/annurev-pharmtox-011112-140226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lisabeth, E. M., Falivelli, G. & Pasquale, E. B. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 5, a009159 (2013). [DOI] [PMC free article] [PubMed]
- 75.Yokote H, et al. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc. Natl Acad. Sci. USA. 2005;102:18866–18871. doi: 10.1073/pnas.0509741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawada T, et al. Ternary complex formation of EphA4, FGFR and FRS2alpha plays an important role in the proliferation of embryonic neural stem/progenitor cells. Genes Cells. 2010;15:297–311. doi: 10.1111/j.1365-2443.2010.01391.x. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. Regulation of ephexin1, a guanine nucleotide exchange factor of Rho family GTPases, by fibroblast growth factor receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 2007;282:31103–31112. doi: 10.1074/jbc.M704430200. [DOI] [PubMed] [Google Scholar]
- 78.Lee S, Shatadal S, Griep AE. Dlg-1 interacts with and regulates the activities of fibroblast growth factor receptors and EphA2 in the mouse lens. Invest. Ophthalmol. Vis. Sci. 2016;57:707–718. doi: 10.1167/iovs.15-17727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen PH, Chen X, He X. Platelet-derived growth factors and their receptors: structural and functional perspectives. Biochim. Biophys. Acta. 2013;1834:2176–2186. doi: 10.1016/j.bbapap.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen PY, Simons M, Friesel R. FRS2 via fibroblast growth factor receptor 1 is required for platelet-derived growth factor receptor beta-mediated regulation of vascular smooth muscle marker gene expression. J. Biol. Chem. 2009;284:15980–15992. doi: 10.1074/jbc.M809399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bryant DM, Stow JL. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic. 2005;6:947–954. doi: 10.1111/j.1600-0854.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 83.Arese M, et al. Nuclear activities of basic fibroblast growth factor: potentiation of low-serum growth mediated by natural or chimeric nuclear localization signals. Mol. Biol. Cell. 1999;10:1429–1444. doi: 10.1091/mbc.10.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piasecka D, et al. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors - implications for therapy of luminal breast cancer. J. Exp. Clin. Cancer Res. 2019;38:230. doi: 10.1186/s13046-019-1236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuzon CT, Rigueur D, Merrill AE. Nuclear fibroblast growth factor receptor signaling in skeletal development and disease. Curr. Osteoporos. Rep. 2019;17:138–146. doi: 10.1007/s11914-019-00512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmahl J, et al. Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development. 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- 87.Steinberg Z, et al. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 2005;132:1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- 88.Coleman SJ, et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol. Med. 2014;6:467–481. doi: 10.1002/emmm.201302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cerliani JP, et al. Interaction between FGFR-2, STAT5, and progesterone receptors in breast cancer. Cancer Res. 2011;71:3720–3731. doi: 10.1158/0008-5472.CAN-10-3074. [DOI] [PubMed] [Google Scholar]
- 90.Zhou L, et al. Nuclear translocation of fibroblast growth factor receptor 3 and its significance in pancreatic cancer. Int. J. Clin. Exp. Pathol. 2015;8:14640–14648. [PMC free article] [PubMed] [Google Scholar]
- 91.Claus P, et al. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J. Biol. Chem. 2003;278:479–485. doi: 10.1074/jbc.M206056200. [DOI] [PubMed] [Google Scholar]
- 92.Lin YZ, Yao SY, Hawiger J. Role of the nuclear localization sequence in fibroblast growth factor-1-stimulated mitogenic pathways. J. Biol. Chem. 1996;271:5305–5308. doi: 10.1074/jbc.271.10.5305. [DOI] [PubMed] [Google Scholar]
- 93.Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J. Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J. Cell Biol. 2012;197:801–817. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Neben CL, et al. Bent bone dysplasia syndrome reveals nucleolar activity for FGFR2 in ribosomal DNA transcription. Hum. Mol. Genet. 2014;23:5659–5671. doi: 10.1093/hmg/ddu282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neben CL, et al. FGFR2 mutations in bent bone dysplasia syndrome activate nucleolar stress and perturb cell fate determination. Hum. Mol. Genet. 2017;26:3253–3270. doi: 10.1093/hmg/ddx209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hatch NE, et al. Intracellular retention, degradation, and signaling of glycosylation-deficient FGFR2 and craniosynostosis syndrome-associated FGFR2C278F. J. Biol. Chem. 2006;281:27292–27305. doi: 10.1074/jbc.M600448200. [DOI] [PubMed] [Google Scholar]
- 98.Terranova C, et al. Global developmental gene programing involves a nuclear form of fibroblast growth factor receptor-1 (FGFR1) PLoS ONE. 2015;10:e0123380. doi: 10.1371/journal.pone.0123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stehbens SJ, et al. FGFR2-activating mutations disrupt cell polarity to potentiate migration and invasion in endometrial cancer cell models. J. Cell Sci. 2018;131:jcs213678. doi: 10.1242/jcs.213678. [DOI] [PubMed] [Google Scholar]
- 100.Carpenter G, Liao HJ. Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 2013;5:a008979. doi: 10.1101/cshperspect.a008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr. Opin. Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 102.Chen L, Deng CX. Roles of FGF signaling in skeletal development and human genetic diseases. Front. Biosci. 2005;10:1961–1976. doi: 10.2741/1671. [DOI] [PubMed] [Google Scholar]
- 103.Xu X, Weinstein M, Li C, Deng C. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell Tissue Res. 1999;296:33–43. doi: 10.1007/s004410051264. [DOI] [PubMed] [Google Scholar]
- 104.Garofalo S, et al. Skeletal dysplasia and defective chondrocyte differentiation by targeted overexpression of fibroblast growth factor 9 in transgenic mice. J. Bone Miner. Res. 1999;14:1909–1915. doi: 10.1359/jbmr.1999.14.11.1909. [DOI] [PubMed] [Google Scholar]
- 105.Hung IH, Yu K, Lavine KJ, Ornitz DM. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev. Biol. 2007;307:300–313. doi: 10.1016/j.ydbio.2007.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lazarus JE, et al. Fibroblast growth factor expression in the postnatal growth plate. Bone. 2007;40:577–586. doi: 10.1016/j.bone.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 107.Krejci P, Krakow D, Mekikian PB, Wilcox WR. Fibroblast growth factors 1, 2, 17, and 19 are the predominant FGF ligands expressed in human fetal growth plate cartilage. Pediatr. Res. 2007;61:267–272. doi: 10.1203/pdr.0b013e318030d157. [DOI] [PubMed] [Google Scholar]
- 108.Hagan AS, et al. Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18) Dev. Dyn. 2019;248:882–893. doi: 10.1002/dvdy.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ornitz DM, Marie PJ. Fibroblast growth factors in skeletal development. Curr. Top. Dev. Biol. 2019;133:195–234. doi: 10.1016/bs.ctdb.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 110.Delezoide AL, et al. Spatio-temporal expression of FGFR 1, 2 and 3 genes during human embryo-fetal ossification. Mech. Dev. 1998;77:19–30. doi: 10.1016/s0925-4773(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 111.Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16:205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev. Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinson, D. et al. Fibroblast growth factor receptor-3 as a marker for precartilaginous stem cells. Clin. Orthop. Relat. Res. S163–S175, (1999). [DOI] [PubMed]
- 114.Xiao L, et al. Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J. Biol. Chem. 2004;279:27743–27752. doi: 10.1074/jbc.M314323200. [DOI] [PubMed] [Google Scholar]
- 115.Su N, et al. Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum. Mol. Genet. 2010;19:1199–1210. doi: 10.1093/hmg/ddp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iseki S, Wilkie AO, Morriss-Kay GM. Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development. 1999;126:5611–5620. doi: 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- 117.Rice DP, et al. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 118.Grillo L, et al. Increased FGF3 and FGF4 gene dosage is a risk factor for craniosynostosis. Gene. 2014;534:435–439. doi: 10.1016/j.gene.2013.09.120. [DOI] [PubMed] [Google Scholar]
- 119.Tekin M, et al. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am. J. Hum. Genet. 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J. Clin. Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hardelin JP, Dode C, et al. The complex genetics of Kallmann syndrome: KAL1, FGFR1, FGF8, PROKR2, PROK2. Sex. Dev. 2008;2:181–193. doi: 10.1159/000152034. [DOI] [PubMed] [Google Scholar]
- 122.Wu XL, et al. Multiple synostoses syndrome is due to a missense mutation in exon 2 of FGF9 gene. Am. J. Hum. Genet. 2009;85:53–63. doi: 10.1016/j.ajhg.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rodriguez-Zabala M, et al. FGF9 mutation causes craniosynostosis along with multiple synostoses. Hum. Mutat. 2017;38:1471–1476. doi: 10.1002/humu.23292. [DOI] [PubMed] [Google Scholar]
- 124.Rohmann E, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 125.Li W, et al. Exploring the interaction between FGF Genes and T-box genes among chinese nonsyndromic cleft lip with or without cleft palate case-parent trios. Environ. Mol. Mutagen. 2019;60:602–606. doi: 10.1002/em.22286. [DOI] [PubMed] [Google Scholar]
- 126.Jamsheer A, et al. Whole exome sequencing identifies FGF16 nonsense mutations as the cause of X-linked recessive metacarpal 4/5 fusion. J. Med. Genet. 2013;50:579–584. doi: 10.1136/jmedgenet-2013-101659. [DOI] [PubMed] [Google Scholar]
- 127.Miraoui H, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am. J. Hum. Genet. 2013;92:725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Consortium A. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- 129.Larsson T, et al. A novel recessive mutation in fibroblast growth factor-23 causes familial tumoral calcinosis. J. Clin. Endocrinol. Metab. 2005;90:2424–2427. doi: 10.1210/jc.2004-2238. [DOI] [PubMed] [Google Scholar]
- 130.Muenke M, et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat. Genet. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- 131.Vogels A, Fryns JP. Pfeiffer syndrome. Orphanet J. Rare Dis. 2006;1:19. doi: 10.1186/1750-1172-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.White KE, et al. Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am. J. Hum. Genet. 2005;76:361–367. doi: 10.1086/427956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jarzabek K, et al. Evidence that FGFR1 loss-of-function mutations may cause variable skeletal malformations in patients with Kallmann syndrome. Adv. Med. Sci. 2012;57:314–321. doi: 10.2478/v10039-012-0036-4. [DOI] [PubMed] [Google Scholar]
- 134.Cunningham ML, et al. Syndromic craniosynostosis: from history to hydrogen bonds. Orthod. Craniofac. Res. 2007;10:67–81. doi: 10.1111/j.1601-6343.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- 135.Wilkie AO. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 136.Sharma VP, et al. Atypical Crouzon syndrome with a novel Cys62Arg mutation in FGFR2 presenting with sagittal synostosis. Cleft Palate Craniofac. J. 2012;49:373–377. doi: 10.1597/11-185. [DOI] [PubMed] [Google Scholar]
- 137.Wilkinson CC, et al. Syndromic craniosynostosis, fibroblast growth factor receptor 2 (FGFR2) mutations, and sacrococcygeal eversion presenting as human tails. Childs Nerv. Syst. 2012;28:1221–1226. doi: 10.1007/s00381-012-1813-x. [DOI] [PubMed] [Google Scholar]
- 138.Park J, et al. Functional characterization of a novel FGFR2 mutation, E731K, in craniosynostosis. J. Cell. Biochem. 2012;113:457–464. doi: 10.1002/jcb.23368. [DOI] [PubMed] [Google Scholar]
- 139.Merrill AE, et al. Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am. J. Hum. Genet. 2012;90:550–557. doi: 10.1016/j.ajhg.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He X, Xie F, Ren ZR. Rapid detection of G1138A and G1138C mutations of the FGFR3 gene in patients with achondroplasia using high-resolution melting analysis. Genet. Test. Mol. Biomark. 2012;16:297–301. doi: 10.1089/gtmb.2011.0113. [DOI] [PubMed] [Google Scholar]
- 141.Krejci P. The paradox of FGFR3 signaling in skeletal dysplasia: why chondrocytes growth arrest while other cells over proliferate. Mutat. Res. Rev. Mutat. Res. 2014;759:40–48. doi: 10.1016/j.mrrev.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 142.Barroso E, et al. Mild isolated craniosynostosis due to a novel FGFR3 mutation, p.Ala334Thr. Am. J. Med. Genet. A. 2011;155A:3050–3053. doi: 10.1002/ajmg.a.34199. [DOI] [PubMed] [Google Scholar]
- 143.Wilkes D, et al. A recurrent mutation, ala391glu, in the transmembrane region of FGFR3 causes Crouzon syndrome and acanthosis nigricans. J. Med. Genet. 1996;33:744–748. doi: 10.1136/jmg.33.9.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Agochukwu NB, Solomon BD, Gropman AL, Muenke M. Epilepsy in Muenke syndrome: FGFR3-related craniosynostosis. Pediatr. Neurol. 2012;47:355–361. doi: 10.1016/j.pediatrneurol.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tavormina PL, et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat. Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 146.Toydemir RM, et al. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am. J. Hum. Genet. 2006;79:935–941. doi: 10.1086/508433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Makrythanasis P, et al. A novel homozygous mutation in FGFR3 causes tall stature, severe lateral tibial deviation, scoliosis, hearing impairment, camptodactyly, and arachnodactyly. Hum. Mutat. 2014;35:959–963. doi: 10.1002/humu.22597. [DOI] [PubMed] [Google Scholar]
- 148.Simann M, et al. Canonical FGFs prevent osteogenic lineage commitment and differentiation of human bone marrow stromal cells via ERK1/2 signaling. J. Cell. Biochem. 2017;118:263–275. doi: 10.1002/jcb.25631. [DOI] [PubMed] [Google Scholar]
- 149.Sarkar S, et al. FGF2 promotes skeletogenic differentiation of cranial neural crest cells. Development. 2001;128:2143–2152. doi: 10.1242/dev.128.11.2143. [DOI] [PubMed] [Google Scholar]
- 150.Coffin JD, et al. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol. Biol. Cell. 1995;6:1861–1873. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sobue T, et al. Over-expression of fibroblast growth factor-2 causes defective bone mineralization and osteopenia in transgenic mice. J. Cell. Biochem. 2005;95:83–94. doi: 10.1002/jcb.20389. [DOI] [PubMed] [Google Scholar]
- 152.Coffin JD, Homer-Bouthiette C, Hurley MM. Fibroblast growth factor 2 and its receptors in bone biology and disease. J. Endocr. Soc. 2018;2:657–671. doi: 10.1210/js.2018-00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Meo Burt P, et al. FGF2 high molecular weight isoforms contribute to osteoarthropathy in male mice. Endocrinology. 2016;157:4602–4614. doi: 10.1210/en.2016-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vincent TL, et al. FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthr. Cartil. 2007;15:752–763. doi: 10.1016/j.joca.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 155.Chia SL, et al. Fibroblast growth factor 2 is an intrinsic chondroprotective agent that suppresses ADAMTS-5 and delays cartilage degradation in murineosteoarthritis. Arthritis Rheum. 2009;60:2019–2027. doi: 10.1002/art.24654. [DOI] [PubMed] [Google Scholar]
- 156.Muddasani P, et al. Basic fibroblast growth factor activates the MAPK and NFkappaB pathways that converge on Elk-1 to control production of matrix metalloproteinase-13 by human adult articular chondrocytes. J. Biol. Chem. 2007;282:31409–31421. doi: 10.1074/jbc.M706508200. [DOI] [PubMed] [Google Scholar]
- 157.Nummenmaa E, et al. Effects of FGF-2 and FGF receptor antagonists on MMP enzymes, aggrecan, and type II collagen in primary human OA chondrocytes. Scand. J. Rheumatol. 2015;44:321–330. doi: 10.3109/03009742.2014.1000372. [DOI] [PubMed] [Google Scholar]
- 158.Nixon AJ, et al. Gene therapy in musculoskeletal repair. Ann. NY Acad. Sci. 2007;1117:310–327. doi: 10.1196/annals.1402.065. [DOI] [PubMed] [Google Scholar]
- 159.Im HJ, et al. Basic fibroblast growth factor accelerates matrix degradation via a neuro-endocrine pathway in human adult articular chondrocytes. J. Cell. Physiol. 2008;215:452–463. doi: 10.1002/jcp.21317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 160.Anderson MJ, Schimmang T, Lewandoski M. An FGF3-BMP signaling axis regulates caudal neural tube closure, neural crest specification and anterior–posterior axis extension. PLoS Genet. 2016;12:e1006018. doi: 10.1371/journal.pgen.1006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McCarthy N, Sidik A, Bertrand JY, Eberhart JK. An Fgf-Shh signaling hierarchy regulates early specification of the zebrafish skull. Dev. Biol. 2016;415:261–277. doi: 10.1016/j.ydbio.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murohashi M, et al. An FGF4–FRS2alpha–Cdx2 axis in trophoblast stem cells induces Bmp4 to regulate proper growth of early mouse embryos. Stem Cells. 2010;28:113–121. doi: 10.1002/stem.247. [DOI] [PubMed] [Google Scholar]
- 163.Boulet AM, Capecchi MR. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012;371:235–245. doi: 10.1016/j.ydbio.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kratochwil K, et al. FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(−/−) mice. Genes Dev. 2002;16:3173–3185. doi: 10.1101/gad.1035602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bosetti M, et al. Regulation of osteoblast and osteoclast functions by FGF-6. J. Cell. Physiol. 2010;225:466–471. doi: 10.1002/jcp.22225. [DOI] [PubMed] [Google Scholar]
- 166.Schmidt L, et al. Increased FGF8 signaling promotes chondrogenic rather than osteogenic development in the embryonic skull. Dis. Model. Mech. 2018;11:dmm031526. doi: 10.1242/dmm.031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xu J, et al. FGF8 signaling alters the osteogenic cell fate in the hard palate. J. Dent. Res. 2018;97:589–596. doi: 10.1177/0022034517750141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tang L, et al. A point mutation in Fgf9 impedes joint interzone formation leading to multiple synostoses syndrome. Hum. Mol. Genet. 2017;26:1280–1293. doi: 10.1093/hmg/ddx029. [DOI] [PubMed] [Google Scholar]
- 169.Hajihosseini MK, et al. Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev. Dyn. 2009;238:376–385. doi: 10.1002/dvdy.21648. [DOI] [PubMed] [Google Scholar]
- 170.Knowles HJ. Hypoxia-induced fibroblast growth factor 11 stimulates osteoclast-mediated resorption of bone. Calcif. Tissue Int. 2017;100:382–391. doi: 10.1007/s00223-016-0228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ohbayashi N, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hu W, et al. Fibroblast growth factor 21 is associated with bone mineral density, but not with bone turnover markers and fractures in chinese postmenopausal women. J. Clin. Densitom. 2019;22:179–184. doi: 10.1016/j.jocd.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 174.Wu S, Levenson A, Kharitonenkov A, De Luca F. Fibroblast growth factor 21 (FGF21) inhibits chondrocyte function and growth hormone action directly at the growth plate. J. Biol. Chem. 2012;287:26060–26067. doi: 10.1074/jbc.M112.343707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ishida K, Haudenschild DR. Interactions between FGF21 and BMP-2 in osteogenesis. Biochem. Biophys. Res. Commun. 2013;432:677–682. doi: 10.1016/j.bbrc.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 176.Bornstein S, et al. FGF-21 and skeletal remodeling during and after lactation in C57BL/6J mice. Endocrinology. 2014;155:3516–3526. doi: 10.1210/en.2014-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shimada T, et al. FGF-23 transgenic mice demonstrate hypophosphatemic rickets with reduced expression of sodium phosphate cotransporter type IIa. Biochem. Biophys. Res. Commun. 2004;314:409–414. doi: 10.1016/j.bbrc.2003.12.102. [DOI] [PubMed] [Google Scholar]
- 178.Liu S, et al. Pathogenic role of Fgf23 in Hyp mice. Am. J. Physiol. Endocrinol. Metab. 2006;291:E38–E49. doi: 10.1152/ajpendo.00008.2006. [DOI] [PubMed] [Google Scholar]
- 179.Shalhoub V, et al. Fibroblast growth factor 23 (FGF23) and alpha-klotho stimulate osteoblastic MC3T3.E1 cell proliferation and inhibit mineralization. Calcif. Tissue Int. 2011;89:140–150. doi: 10.1007/s00223-011-9501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kawai M, et al. FGF23 suppresses chondrocyte proliferation in the presence of soluble alpha-Klotho both in vitro and in vivo. J. Biol. Chem. 2013;288:2414–2427. doi: 10.1074/jbc.M112.410043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Murali SK, et al. FGF23 regulates bone mineralization in a 1,25(OH)2 D3 and Klotho-independent manner. J. Bone Miner. Res. 2016;31:129–142. doi: 10.1002/jbmr.2606. [DOI] [PubMed] [Google Scholar]
- 182.Zhou YX, et al. A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum. Mol. Genet. 2000;9:2001–2008. doi: 10.1093/hmg/9.13.2001. [DOI] [PubMed] [Google Scholar]
- 183.Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Calvert JA, et al. A missense mutation in Fgfr1 causes ear and skull defects in hush puppy mice. Mamm. Genome. 2011;22:290–305. doi: 10.1007/s00335-011-9324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Karolak MR, Yang X, Elefteriou F. FGFR1 signaling in hypertrophic chondrocytes is attenuated by the Ras-GAP neurofibromin during endochondral bone formation. Hum. Mol. Genet. 2015;24:2552–2564. doi: 10.1093/hmg/ddv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McKenzie J, et al. Osteocyte death and bone overgrowth in mice lacking fibroblast growth factor receptors 1 and 2 in mature osteoblasts and osteocytes. J. Bone Miner. Res. 2019;34:1660–1675. doi: 10.1002/jbmr.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lu X, et al. Fibroblast growth factor receptor 1 regulates the differentiation and activation of osteoclasts through Erk1/2 pathway. Biochem. Biophys. Res. Commun. 2009;390:494–499. doi: 10.1016/j.bbrc.2009.09.123. [DOI] [PubMed] [Google Scholar]
- 188.Chen L, et al. A Ser252Trp [corrected] substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone. 2003;33:169–178. doi: 10.1016/s8756-3282(03)00222-9. [DOI] [PubMed] [Google Scholar]
- 189.Wang Y, et al. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development. 2005;132:3537–3548. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- 190.Yin L, et al. A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone. 2008;42:631–643. doi: 10.1016/j.bone.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 191.Luo F, et al. Deformed skull morphology is caused by the combined effects of the maldevelopment of calvarias, cranial base and brain in FGFR2-P253R mice mimicking human Apert syndrome. Int. J. Biol. Sci. 2017;13:32–45. doi: 10.7150/ijbs.16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Salva JE, Roberts RR, Stucky TS, Merrill AE. Nuclear FGFR2 regulates musculoskeletal integration within the developing limb. Dev. Dyn. 2019;248:233–246. doi: 10.1002/dvdy.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yu K, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 194.Larbuisson A, Dalcq J, Martial JA, Muller M. Fgf receptors Fgfr1a and Fgfr2 control the function of pharyngeal endoderm in late cranial cartilage development. Differentiation. 2013;86:192–206. doi: 10.1016/j.diff.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 195.Chen L, et al. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J. Clin. Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum. Mol. Genet. 2001;10:1255–1264. doi: 10.1093/hmg/10.12.1255. [DOI] [PubMed] [Google Scholar]
- 197.Deng C, et al. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 198.Colvin JS, et al. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 199.Krejci P, et al. FGF2 inhibits proliferation and alters the cartilage-like phenotype of RCS cells. Exp. Cell Res. 2004;297:152–164. doi: 10.1016/j.yexcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 200.Parafioriti A, et al. Increased p21 expression in chondrocytes of achondroplasic children independently from the presence of the G380R FGFR3 mutation. J. Orthop. Sci. 2009;14:623–630. doi: 10.1007/s00776-009-1355-6. [DOI] [PubMed] [Google Scholar]
- 201.Iwata T, et al. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum. Mol. Genet. 2000;9:1603–1613. doi: 10.1093/hmg/9.11.1603. [DOI] [PubMed] [Google Scholar]
- 202.Zhou S, et al. FGFR3 deficiency causes multiple chondroma-like lesions by upregulating hedgehog signaling. PLoS Genet. 2015;11:e1005214. doi: 10.1371/journal.pgen.1005214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chen L, et al. A Ser(365)->Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum. Mol. Genet. 2001;10:457–465. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- 204.Smith LB, Belanger JM, Oberbauer AM. Fibroblast growth factor receptor 3 effects on proliferation and telomerase activity in sheep growth plate chondrocytes. J. Anim. Sci. Biotechnol. 2012;3:39. doi: 10.1186/2049-1891-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dailey L, Laplantine E, Priore R, Basilico C. A network of transcriptional and signaling events is activated by FGF to induce chondrocyte growth arrest and differentiation. J. Cell Biol. 2003;161:1053–1066. doi: 10.1083/jcb.200302075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Krejci P, et al. FGFR3 signaling induces a reversible senescence phenotype in chondrocytes similar to oncogene-induced premature senescence. Bone. 2010;47:102–110. doi: 10.1016/j.bone.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Foldynova-Trantirkova S, Wilcox WR, Krejci P. Sixteen years and counting: the current understanding of fibroblast growth factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum. Mutat. 2012;33:29–41. doi: 10.1002/humu.21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Krejci P, et al. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J. Cell Sci. 2005;118:5089–5100. doi: 10.1242/jcs.02618. [DOI] [PubMed] [Google Scholar]
- 209.Cinque L, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272–275. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 210.Wang X, et al. FGFR3/fibroblast growth factor receptor 3 inhibits autophagy through decreasing the ATG12–ATG5 conjugate, leading to the delay of cartilage development in achondroplasia. Autophagy. 2015;11:1998–2013. doi: 10.1080/15548627.2015.1091551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Martin L, et al. Constitutively-active FGFR3 disrupts primary cilium length and IFT20 trafficking in various chondrocyte models of achondroplasia. Hum. Mol. Genet. 2018;27:1–13. doi: 10.1093/hmg/ddx374. [DOI] [PubMed] [Google Scholar]
- 212.Kunova Bosakova M, et al. Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum. Mol. Genet. 2018;27:1093–1105. doi: 10.1093/hmg/ddy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Twigg SR, et al. Skeletal analysis of the Fgfr3(P244R) mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev. Dyn. 2009;238:331–342. doi: 10.1002/dvdy.21790. [DOI] [PubMed] [Google Scholar]
- 214.Mugniery E, et al. An activating Fgfr3 mutation affects trabecular bone formation via a paracrine mechanism during growth. Hum. Mol. Genet. 2012;21:2503–2513. doi: 10.1093/hmg/dds065. [DOI] [PubMed] [Google Scholar]
- 215.Matsushita T, et al. FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Hum. Mol. Genet. 2009;18:227–240. doi: 10.1093/hmg/ddn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wen X, et al. Chondrocyte FGFR3 regulates bone mass by inhibiting osteogenesis. J. Biol. Chem. 2016;291:24912–24921. doi: 10.1074/jbc.M116.730093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Valverde-Franco G, et al. Defective bone mineralization and osteopenia in young adult FGFR3-/- mice. Hum. Mol. Genet. 2004;13:271–284. doi: 10.1093/hmg/ddh034. [DOI] [PubMed] [Google Scholar]
- 218.Su N, et al. Deletion of FGFR3 in osteoclast lineage cells results in increased bone mass in mice by inhibiting osteoclastic bone resorption. J. Bone Miner. Res. 2016;31:1676–1687. doi: 10.1002/jbmr.2839. [DOI] [PubMed] [Google Scholar]
- 219.Zhou FH, Foster BK, Sander G, Xian CJ. Expression of proinflammatory cytokines and growth factors at the injured growth plate cartilage in young rats. Bone. 2004;35:1307–1315. doi: 10.1016/j.bone.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 220.Zhou FH, et al. TNF-alpha mediates p38 MAP kinase activation and negatively regulates bone formation at the injured growth plate in rats. J. Bone Miner. Res. 2006;21:1075–1088. doi: 10.1359/jbmr.060410. [DOI] [PubMed] [Google Scholar]
- 221.Damron TA, et al. Temporal changes in PTHrP, Bcl-2, Bax, caspase, TGF-beta, and FGF-2 expression following growth plate irradiation with or without radioprotectant. J. Histochem. Cytochem. 2004;52:157–167. doi: 10.1177/002215540405200203. [DOI] [PubMed] [Google Scholar]
- 222.Daouti S, et al. Development of comprehensive functional genomic screens to identify novel mediators of osteoarthritis. Osteoarthr. Cartil. 2005;13:508–518. doi: 10.1016/j.joca.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 223.Yan D, et al. Fibroblast growth factor receptor 1 is principally responsible for fibroblast growth factor 2-induced catabolic activities in human articular chondrocytes. Arthritis Res. Ther. 2011;13:R130. doi: 10.1186/ar3441. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 224.Weng T, et al. Genetic inhibition of fibroblast growth factor receptor 1 in knee cartilage attenuates the degeneration of articular cartilage in adult mice. Arthritis Rheum. 2012;64:3982–3992. doi: 10.1002/art.34645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Klag KA, Horton WA. Advances in treatment of achondroplasia and osteoarthritis. Hum. Mol. Genet. 2016;25:R2–R8. doi: 10.1093/hmg/ddv419. [DOI] [PubMed] [Google Scholar]
- 226.Tang J, et al. Fibroblast growth factor receptor 3 inhibits osteoarthritis progression in the knee joints of adult mice. Arthritis Rheumatol. 2016;68:2432–2443. doi: 10.1002/art.39739. [DOI] [PubMed] [Google Scholar]
- 227.Zhou S, et al. Conditional deletion of Fgfr3 in chondrocytes leads to osteoarthritis-like defects in temporomandibular joint of adult mice. Sci. Rep. 2016;6:24039. doi: 10.1038/srep24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kuang L, et al. FGFR3 deficiency enhances CXCL12-dependent chemotaxis of macrophages via upregulating CXCR7 and aggravates joint destruction in mice. Ann. Rheum. Dis. 2020;79:112–122. doi: 10.1136/annrheumdis-2019-215696. [DOI] [PubMed] [Google Scholar]
- 229.Kisand K, Tamm AE, Lintrop M, Tamm AO. New insights into the natural course of knee osteoarthritis: early regulation of cytokines and growth factors, with emphasis on sex-dependent angiogenesis and tissue remodeling. A pilot study. Osteoarthr. Cartil. 2018;26:1045–1054. doi: 10.1016/j.joca.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 230.El-Seoudi A, et al. Catabolic effects of FGF-1 on chondrocytes and its possible role in osteoarthritis. J. Cell Commun. Signal. 2017;11:255–263. doi: 10.1007/s12079-017-0384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Im HJ, et al. Basic fibroblast growth factor stimulates matrix metalloproteinase-13 via the molecular cross-talk between the mitogen-activated protein kinases and protein kinase Cdelta pathways in human adult articular chondrocytes. J. Biol. Chem. 2007;282:11110–11121. doi: 10.1074/jbc.M609040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chong KW, et al. Fibroblast growth factor 2 drives changes in gene expression following injury to murine cartilage in vitro and in vivo. Arthritis Rheum. 2013;65:2346–2355. doi: 10.1002/art.38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sawaji Y, Hynes J, Vincent T, Saklatvala J. Fibroblast growth factor 2 inhibits induction of aggrecanase activity in human articular cartilage. Arthritis Rheum. 2008;58:3498–3509. doi: 10.1002/art.24025. [DOI] [PubMed] [Google Scholar]
- 234.Burt PM, Xiao L, Doetschman T, Hurley MM. Ablation of low-molecular-weight FGF2 isoform accelerates murine osteoarthritis while loss of high-molecular-weight FGF2 isoforms offers protection. J. Cell. Physiol. 2019;234:4418–4431. doi: 10.1002/jcp.27230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Meo Burt P, Xiao L, Hurley MM. FGF23 regulates Wnt/beta-catenin signaling-mediated osteoarthritis in mice overexpressing high-molecular-weight FGF2. Endocrinology. 2018;159:2386–2396. doi: 10.1210/en.2018-00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Uchii M, et al. Role of fibroblast growth factor 8 (FGF8) in animal models of osteoarthritis. Arthritis Res. Ther. 2008;10:R90. doi: 10.1186/ar2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhou S, et al. Exogenous fibroblast growth factor 9 attenuates cartilage degradation and aggravates osteophyte formation in post-traumatic osteoarthritis. Osteoarthr. Cartil. 2016;24:2181–2192. doi: 10.1016/j.joca.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 238.Ellsworth JL, et al. Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthr. Cartil. 2002;10:308–320. doi: 10.1053/joca.2002.0514. [DOI] [PubMed] [Google Scholar]
- 239.Nakajima A, et al. Spatial and temporal gene expression for fibroblast growth factor type I receptor (FGFR1) during fracture healing in the rat. Bone. 2001;29:458–466. doi: 10.1016/s8756-3282(01)00604-4. [DOI] [PubMed] [Google Scholar]
- 240.Rundle, C. H. et al. Expression of the fibroblast growth factor receptor genes in fracture repair. Clin. Orthop. Relat. Res. 253–263 (2002). [DOI] [PubMed]
- 241.Nakajima A, Shimizu S, Moriya H, Yamazaki M. Expression of fibroblast growth factor receptor-3 (FGFR3), signal transducer and activator of transcription-1, and cyclin-dependent kinase inhibitor p21 during endochondral ossification: differential role of FGFR3 in skeletal development and fracture repair. Endocrinology. 2003;144:4659–4668. doi: 10.1210/en.2003-0158. [DOI] [PubMed] [Google Scholar]
- 242.Su N, et al. Gain-of-function mutation of FGFR3 results in impaired fracture healing due to inhibition of chondrocyte differentiation. Biochem. Biophys. Res. Commun. 2008;376:454–459. doi: 10.1016/j.bbrc.2008.08.165. [DOI] [PubMed] [Google Scholar]
- 243.Schmid GJ, Kobayashi C, Sandell LJ, Ornitz DM. Fibroblast growth factor expression during skeletal fracture healing in mice. Dev. Dyn. 2009;238:766–774. doi: 10.1002/dvdy.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bolander ME. Regulation of fracture repair by growth factors. Proc. Soc. Exp. Biol. Med. 1992;200:165–170. doi: 10.3181/00379727-200-43410a. [DOI] [PubMed] [Google Scholar]
- 245.Bourque WT, Gross M, Hall BK. Expression of four growth factors during fracture repair. Int. J. Dev. Biol. 1993;37:573–579. [PubMed] [Google Scholar]
- 246.Pacicca DM, et al. Expression of angiogenic factors during distraction osteogenesis. Bone. 2003;33:889–898. doi: 10.1016/j.bone.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 247.Guimaraes JM, et al. Polymorphisms in BMP4 and FGFR1 genes are associated with fracture non-union. J. Orthop. Res. 2013;31:1971–1979. doi: 10.1002/jor.22455. [DOI] [PubMed] [Google Scholar]
- 248.Xu W, et al. Inducible activation of FGFR2 in adult mice promotes bone formation after bone marrow ablation. J. Bone Miner. Res. 2017;32:2194–2206. doi: 10.1002/jbmr.3204. [DOI] [PubMed] [Google Scholar]
- 249.Xie Y, et al. FGFR3 deficient mice have accelerated fracture repair. Int. J. Biol. Sci. 2017;13:1029–1037. doi: 10.7150/ijbs.19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chen H, et al. PTH 1–34 ameliorates the osteopenia and delayed healing of stabilized tibia fracture in mice with achondroplasia resulting from gain-of-function mutation of FGFR3. Int. J. Biol. Sci. 2017;13:1254–1265. doi: 10.7150/ijbs.21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Le Blanc S, et al. Fibroblast growth factors 1 and 2 inhibit adipogenesis of human bone marrow stromal cells in 3D collagen gels. Exp. Cell Res. 2015;338:136–148. doi: 10.1016/j.yexcr.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 252.Wang J, Liu S, Li J, Yi Z. The role of the fibroblast growth factor family in bone-related diseases. Chem. Biol. Drug Des. 2019;94:1740–1749. doi: 10.1111/cbdd.13588. [DOI] [PubMed] [Google Scholar]
- 253.Hurley MM, et al. Accelerated fracture healing in transgenic mice overexpressing an anabolic isoform of fibroblast growth factor 2. J. Cell. Biochem. 2016;117:599–611. doi: 10.1002/jcb.25308. [DOI] [PubMed] [Google Scholar]
- 254.Xiao L, et al. Fibroblast growth factor-2 isoform (low molecular weight/18 kDa) overexpression in preosteoblast cells promotes bone regeneration in critical size calvarial defects in male mice. Endocrinology. 2014;155:965–974. doi: 10.1210/en.2013-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lee P, et al. Fibroblast growth factor 21 (FGF21) and bone: is there a relationship in humans? Osteoporos. Int. 2013;24:3053–3057. doi: 10.1007/s00198-013-2464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wei W, et al. Fibroblast growth factor 21 promotes bone loss by potentiating the effects of peroxisome proliferator-activated receptor gamma. Proc. Natl Acad. Sci. USA. 2012;109:3143–3148. doi: 10.1073/pnas.1200797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rupp T, et al. High FGF23 levels are associated with impaired trabecular bone microarchitecture in patients with osteoporosis. Osteoporos. Int. 2019;30:1655–1662. doi: 10.1007/s00198-019-04996-7. [DOI] [PubMed] [Google Scholar]
- 258.Goebel S, et al. FGF23 is a putative marker for bone healing and regeneration. J. Orthop. Res. 2009;27:1141–1146. doi: 10.1002/jor.20857. [DOI] [PubMed] [Google Scholar]
- 259.Clinkenbeard EL, White KE. Systemic control of bone homeostasis by FGF23 signaling. Curr. Mol. Biol. Rep. 2016;2:62–71. doi: 10.1007/s40610-016-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Xu W, et al. A novel fibroblast growth factor receptor 1 inhibitor protects against cartilage degradation in a murine model of osteoarthritis. Sci. Rep. 2016;6:24042. doi: 10.1038/srep24042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Tan Q, et al. A novel FGFR1-binding peptide attenuates the degeneration of articular cartilage in adult mice. Osteoarthr. Cartil. 2018;26:1733–1743. doi: 10.1016/j.joca.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 262.Yao X, et al. Fibroblast growth factor 18 exerts anti-osteoarthritic effects through PI3K-AKT signaling and mitochondrial fusion and fission. Pharm. Res. 2019;139:314–324. doi: 10.1016/j.phrs.2018.09.026. [DOI] [PubMed] [Google Scholar]
- 263.Howard D, Wardale J, Guehring H, Henson F. Delivering rhFGF-18 via a bilayer collagen membrane to enhance microfracture treatment of chondral defects in a large animal model. J. Orthop. Res. 2015;33:1120–1127. doi: 10.1002/jor.22882. [DOI] [PubMed] [Google Scholar]
- 264.Power J, et al. Intra-articular injection of rhFGF-18 improves the healing in microfracture treated chondral defects in an ovine model. J. Orthop. Res. 2014;32:669–676. doi: 10.1002/jor.22580. [DOI] [PubMed] [Google Scholar]
- 265.Eckstein F, et al. Brief report: intraarticular sprifermin not only increases cartilage thickness, but also reduces cartilage loss: location-independent post hoc analysis using magnetic resonance imaging. Arthritis Rheumatol. 2015;67:2916–2922. doi: 10.1002/art.39265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lohmander LS, et al. Intraarticular sprifermin (recombinant human fibroblast growth factor 18) in knee osteoarthritis: a randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol. 2014;66:1820–1831. doi: 10.1002/art.38614. [DOI] [PubMed] [Google Scholar]
- 267.Onuora S. Osteoarthritis: Sprifermin shows cartilage-protective effects in knee OA. Nat. Rev. Rheumatol. 2014;10:322. doi: 10.1038/nrrheum.2014.68. [DOI] [PubMed] [Google Scholar]
- 268.Hochberg MC, et al. Effect of intra-articular sprifermin vs placebo on femorotibial joint cartilage thickness in patients with osteoarthritis: The FORWARD Randomized Clinical Trial. JAMA. 2019;322:1360–1370. doi: 10.1001/jama.2019.14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tang ZF, Li HY. Effects of fibroblast growth factors 2 and low intensity pulsed ultrasound on the repair of knee articular cartilage in rabbits. Eur. Rev. Med. Pharm. Sci. 2018;22:2447–2453. doi: 10.26355/eurrev_201804_14838. [DOI] [PubMed] [Google Scholar]
- 270.Cuevas P, Burgos J, Baird A. Basic fibroblast growth factor (FGF) promotes cartilage repair in vivo. Biochem. Biophys. Res. Commun. 1988;156:611–618. doi: 10.1016/s0006-291x(88)80887-8. [DOI] [PubMed] [Google Scholar]
- 271.Sanghani A, Chimutengwende-Gordon M, Adesida A, Khan W. Applications of stem cell therapy for physeal injuries. Curr. Stem Cell Res. Ther. 2013;8:451–455. doi: 10.2174/1574888x1130800063. [DOI] [PubMed] [Google Scholar]
- 272.Chung R, Xian CJ. Recent research on the growth plate: mechanisms for growth plate injury repair and potential cell-based therapies for regeneration. J. Mol. Endocrinol. 2014;53:T45–T61. doi: 10.1530/JME-14-0062. [DOI] [PubMed] [Google Scholar]
- 273.Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. J. Cell. Physiol. 2012;227:3731–3743. doi: 10.1002/jcp.24083. [DOI] [PubMed] [Google Scholar]
- 274.Gothard D, et al. Tissue engineered bone using select growth factors: a comprehensive review of animal studies and clinical translation studies in man. Eur. Cell Mater. 2014;28:166–207. doi: 10.22203/ecm.v028a13. [DOI] [PubMed] [Google Scholar]
- 275.Arias-Gallo J, Chamorro-Pons M, Avendano C, Gimenez-Gallego G. Influence of acidic fibroblast growth factor on bone regeneration in experimental cranial defects using spongostan and Bio-Oss as protein carriers. J. Craniofac. Surg. 2013;24:1507–1514. doi: 10.1097/SCS.0b013e31828f2469. [DOI] [PubMed] [Google Scholar]
- 276.Mackenzie DJ, et al. Recombinant human acidic fibroblast growth factor and fibrin carrier regenerates bone. Plast. Reconstr. Surg. 2001;107:989–996. doi: 10.1097/00006534-200104010-00013. [DOI] [PubMed] [Google Scholar]
- 277.Kawaguchi H, et al. Local application of recombinant human fibroblast growth factor-2 on bone repair: a dose-escalation prospective trial on patients with osteotomy. J. Orthop. Res. 2007;25:480–487. doi: 10.1002/jor.20315. [DOI] [PubMed] [Google Scholar]
- 278.Kawaguchi H, et al. A local application of recombinant human fibroblast growth factor 2 for tibial shaft fractures: a randomized, placebo-controlled trial. J. Bone Miner. Res. 2010;25:2735–2743. doi: 10.1002/jbmr.146. [DOI] [PubMed] [Google Scholar]
- 279.Sakano S, et al. Inhibitory effect of bFGF on endochondral heterotopic ossification. Biochem. Biophys. Res. Commun. 2002;293:680–685. doi: 10.1016/S0006-291X(02)00273-5. [DOI] [PubMed] [Google Scholar]
- 280.Kimoto T, et al. Continuous administration of basic fibroblast growth factor (FGF-2) accelerates bone induction on rat calvaria-an application of a new drug delivery system. J. Dent. Res. 1998;77:1965–1969. doi: 10.1177/00220345980770120301. [DOI] [PubMed] [Google Scholar]
- 281.Poudel SB, et al. Local delivery of recombinant human FGF7 enhances bone formation in rat mandible defects. J. Bone Miner. Metab. 2017;35:485–496. doi: 10.1007/s00774-016-0784-5. [DOI] [PubMed] [Google Scholar]
- 282.Behr B, Panetta NJ, Longaker MT, Quarto N. Different endogenous threshold levels of fibroblast growth factor-ligands determine the healing potential of frontal and parietal bones. Bone. 2010;47:281–294. doi: 10.1016/j.bone.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 283.Behr B, et al. Fgf-18 is required for osteogenesis but not angiogenesis during long bone repair. Tissue Eng. Part A. 2011;17:2061–2069. doi: 10.1089/ten.tea.2010.0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Fujioka-Kobayashi M, et al. Cholesteryl group- and acryloyl group-bearing pullulan nanogel to deliver BMP2 and FGF18 for bone tissue engineering. Biomaterials. 2012;33:7613–7620. doi: 10.1016/j.biomaterials.2012.06.075. [DOI] [PubMed] [Google Scholar]
- 285.Kang MS, et al. Therapeutic-designed electrospun bone scaffolds: mesoporous bioactive nanocarriers in hollow fiber composites to sequentially deliver dual growth factors. Acta Biomater. 2015;16:103–116. doi: 10.1016/j.actbio.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 286.deMello DE, Reid LM. Embryonic and early fetal development of human lung vasculature and its functional implications. Pediatr. Dev. Pathol. 2000;3:439–449. doi: 10.1007/s100240010090. [DOI] [PubMed] [Google Scholar]
- 287.Danopoulos S, et al. Discordant roles for FGF ligands in lung branching morphogenesis between human and mouse. J. Pathol. 2019;247:254–265. doi: 10.1002/path.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Powell PP, et al. Differential expression of fibroblast growth factor receptors 1 to 4 and ligand genes in late fetal and early postnatal rat lung. Am. J. Respir. Cell. Mol. Biol. 1998;19:563–572. doi: 10.1165/ajrcmb.19.4.2994. [DOI] [PubMed] [Google Scholar]
- 289.Ma DL, et al. Luminescent chemosensors by using cyclometalated iridium(iii) complexes and their applications. Chem. Sci. 2017;8:878–889. doi: 10.1039/c6sc04175b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Cardoso WV, et al. FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev. Dyn. 1997;208:398–405. doi: 10.1002/(SICI)1097-0177(199703)208:3<398::AID-AJA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 291.Simonet WS, et al. Pulmonary malformation in transgenic mice expressing human keratinocyte growth factor in the lung. Proc. Natl Acad. Sci. USA. 1995;92:12461–12465. doi: 10.1073/pnas.92.26.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Yin Y, Wang F, Ornitz DM. Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development. 2011;138:3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.White AC, et al. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- 294.Yin Y, et al. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev. Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.del Moral PM, et al. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev. Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 296.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 297.Sekine K, et al. Fgf10 is essential for limb and lung formation. Nat. Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 298.De Moerlooze L, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal–epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. [Google Scholar]
- 299.Graeff RW, Wang G, McCray PB., Jr KGF and FGF-10 stimulate liquid secretion in human fetal lung. Pediatr. Res. 1999;46:523–529. doi: 10.1203/00006450-199911000-00006. [DOI] [PubMed] [Google Scholar]
- 300.Danopoulos S, et al. Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018;314:L144–L149. doi: 10.1152/ajplung.00379.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Nikolic MZ, et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife. 2017;6:e26575. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Miller AJ, et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019;14:518–540. doi: 10.1038/s41596-018-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Usui H, et al. Fgf18 is required for embryonic lung alveolar development. Biochem. Biophys. Res Commun. 2004;322:887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- 304.Peters K, et al. Targeted expression of a dominant negative FGF receptor blocks branching morphogenesis and epithelial differentiation of the mouse lung. EMBO J. 1994;13:3296–3301. doi: 10.1002/j.1460-2075.1994.tb06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Srisuma S, et al. Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. Am. J. Respir. Crit. Care Med. 2010;181:838–850. doi: 10.1164/rccm.200904-0544OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- 307.Ren JT, et al. Relationship between the gene polymorphism in fibroblast growth factor-10 and susceptibility to chronic obstructive pulmonary disease 220 cases. ZhonghuaJie He He Hu Xi Za Zhi. 2013;36:935–939. [PubMed] [Google Scholar]
- 308.Smith BM, et al. Human airway branch variation and chronic obstructive pulmonary disease. Proc. Natl Acad. Sci. USA. 2018;115:E974–E981. doi: 10.1073/pnas.1715564115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rezvani M, et al. Association of a FGFR-4 gene polymorphism with bronchopulmonary dysplasia and neonatal respiratory distress. Dis. Markers. 2013;35:633–640. doi: 10.1155/2013/932356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Milunsky JM, et al. LADD syndrome is caused by FGF10 mutations. Clin. Genet. 2006;69:349–354. doi: 10.1111/j.1399-0004.2006.00597.x. [DOI] [PubMed] [Google Scholar]
- 311.Karolak JA, et al. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am. J. Hum. Genet. 2019;104:213–228. doi: 10.1016/j.ajhg.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Devine P, et al. Completely cartilaginous trachea in a child with Crouzon syndrome. Am. J. Dis. Child. 1984;138:40–43. doi: 10.1001/archpedi.1984.02140390032010. [DOI] [PubMed] [Google Scholar]
- 313.Cinalli G, et al. Chronic tonsillar herniation in Crouzon’s and Apert’s syndromes: the role of premature synostosis of the lambdoid suture. J. Neurosurg. 1995;83:575–582. doi: 10.3171/jns.1995.83.4.0575. [DOI] [PubMed] [Google Scholar]
- 314.Gonzales M, et al. Vertebral anomalies and cartilaginous tracheal sleeve in three patients with Pfeiffer syndrome carrying the S351C FGFR2 mutation. Clin. Genet. 2005;68:179–181. doi: 10.1111/j.1399-0004.2005.00477.x. [DOI] [PubMed] [Google Scholar]
- 315.Barnett CP, et al. Ectrodactyly and lethal pulmonary acinar dysplasia associated with homozygous FGFR2 mutations identified by exome sequencing. Hum. Mutat. 2016;37:955–963. doi: 10.1002/humu.23032. [DOI] [PubMed] [Google Scholar]
- 316.Jancelewicz T, Nobuhara K, Hawgood S. Laser microdissection allows detection of abnormal gene expression in cystic adenomatoid malformation of the lung. J. Pediatr. Surg. 2008;43:1044–1051. doi: 10.1016/j.jpedsurg.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 317.Benjamin JT, et al. FGF-10 is decreased in bronchopulmonary dysplasia and suppressed by Toll-like receptor activation. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;292:L550–L558. doi: 10.1152/ajplung.00329.2006. [DOI] [PubMed] [Google Scholar]
- 318.Boucherat O, et al. Decreased lung fibroblast growth factor 18 and elastin in human congenital diaphragmatic hernia and animal models. Am. J. Respir. Crit. Care Med. 2007;175:1066–1077. doi: 10.1164/rccm.200601-050OC. [DOI] [PubMed] [Google Scholar]
- 319.MacKenzie B, et al. Increased FGF1-FGFRc expression in idiopathic pulmonary fibrosis. Respir. Res. 2015;16:83. doi: 10.1186/s12931-015-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Teramoto H, Yoneda A, Puri P. Gene expression of fibroblast growth factors 10 and 7 is downregulated in the lung of nitrofen-induced diaphragmatic hernia in rats. J. Pediatr. Surg. 2003;38:1021–1024. doi: 10.1016/s0022-3468(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 321.Wang J, Liu H, Gao L, Liu X. Impaired FGF10 signaling and epithelial development in experimental lung hypoplasia with esophageal atresia. Front. Pediatr. 2018;6:109. doi: 10.3389/fped.2018.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Park MS, et al. Altered expressions of fibroblast growth factor receptors and alveolarization in neonatal mice exposed to 85% oxygen. Pediatr. Res. 2007;62:652–657. doi: 10.1203/PDR.0b013e318159af61. [DOI] [PubMed] [Google Scholar]
- 323.Joannes A, et al. FGF9 and FGF18 in idiopathic pulmonary fibrosis promote survival and migration and inhibit myofibroblast differentiation of human lung fibroblasts in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016;310:L615–L629. doi: 10.1152/ajplung.00185.2015. [DOI] [PubMed] [Google Scholar]
- 324.Yuan T, et al. FGF10-FGFR2B signaling generates basal cells and drives alveolar epithelial regeneration by bronchial epithelial stem cells after lung injury. Stem Cell Rep. 2019;12:1041–1055. doi: 10.1016/j.stemcr.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Guzy RD, et al. Pulmonary fibrosis requires cell-autonomous mesenchymal fibroblast growth factor (FGF) signaling. J. Biol. Chem. 2017;292:10364–10378. doi: 10.1074/jbc.M117.791764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Tong L, et al. Fibroblast growth factor-10 (FGF-10) mobilizes lung-resident mesenchymal stem cells and protects against acute lung injury. Sci. Rep. 2016;6:21642. doi: 10.1038/srep21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Fang X, Bai C, Wang X. Potential clinical application of KGF-2 (FGF-10) for acute lung injury/acute respiratory distress syndrome. Expert Rev. Clin. Pharm. 2010;3:797–805. doi: 10.1586/ecp.10.59. [DOI] [PubMed] [Google Scholar]
- 328.Katoh M. Therapeutics targeting FGF signaling network in human diseases. Trends Pharm. Sci. 2016;37:1081–1096. doi: 10.1016/j.tips.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 329.Keating GM. Nintedanib: a review of its use in patients with idiopathic pulmonary fibrosis. Drugs. 2015;75:1131–1140. doi: 10.1007/s40265-015-0418-6. [DOI] [PubMed] [Google Scholar]
- 330.Shimbori C, et al. Fibroblast growth factor-1 attenuates TGF-beta1-induced lung fibrosis. J. Pathol. 2016;240:197–210. doi: 10.1002/path.4768. [DOI] [PubMed] [Google Scholar]
- 331.Laddha AP, Kulkarni YA. VEGF and FGF-2: promising targets for the treatment of respiratory disorders. Respir. Med. 2019;156:33–46. doi: 10.1016/j.rmed.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 332.Ware LB. Keratinocyte growth factor as an epithelial protective agent: where do we stand? Int. J. Radiat. Oncol. Biol. Phys. 2004;60:1345–1346. doi: 10.1016/j.ijrobp.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 333.Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Am. J. Physiol. Ren. Physiol. 2011;301:F245–F251. doi: 10.1152/ajprenal.00186.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Orr-Urtreger A, et al. Developmental expression of two murine fibroblast growth factor receptors, flg and bek. Development. 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- 335.Poladia DP, et al. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev. Biol. 2006;291:325–339. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 336.Zhao H, et al. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev. Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Walker KA, Sims-Lucas S, Bates CM. Fibroblast growth factor receptor signaling in kidney and lower urinary tract development. Pediatr. Nephrol. 2016;31:885–895. doi: 10.1007/s00467-015-3151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Gerber SD, et al. The murine Fgfrl1 receptor is essential for the development of the metanephric kidney. Dev. Biol. 2009;335:106–119. doi: 10.1016/j.ydbio.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 340.Barasch J, et al. Ureteric bud cells secrete multiple factors, including bFGF, which rescue renal progenitors from apoptosis. Am. J. Physiol. 1997;273:F757–F767. doi: 10.1152/ajprenal.1997.273.5.F757. [DOI] [PubMed] [Google Scholar]
- 341.Brennan HC, Nijjar S, Jones EA. The specification and growth factor inducibility of the pronephric glomus in Xenopus laevis. Development. 1999;126:5847–5856. doi: 10.1242/dev.126.24.5847. [DOI] [PubMed] [Google Scholar]
- 342.Plisov SY, et al. TGF beta 2, LIF and FGF2 cooperate to induce nephrogenesis. Development. 2001;128:1045–1057. doi: 10.1242/dev.128.7.1045. [DOI] [PubMed] [Google Scholar]
- 343.Grieshammer U, et al. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 344.Perantoni AO, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 345.Brown AC, et al. FGF/EGF signaling regulates the renewal of early nephron progenitors during embryonic development. Development. 2011;138:5099–5112. doi: 10.1242/dev.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Barak H, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Miller DL, et al. Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol. Cell. Biol. 2000;20:2260–2268. doi: 10.1128/mcb.20.6.2260-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Zhou M, et al. Fibroblast growth factor 2 control of vascular tone. Nat. Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Di Giovanni V, et al. Fibroblast growth factor receptor-Frs2alpha signaling is critical for nephron progenitors. Dev. Biol. 2015;400:82–93. doi: 10.1016/j.ydbio.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Sims-Lucas S, et al. Fgfr1 and the IIIc isoform of Fgfr2 play critical roles in the metanephric mesenchyme mediating early inductive events in kidney development. Dev. Dyn. 2011;240:240–249. doi: 10.1002/dvdy.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Qiao J, et al. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- 352.Ohuchi H, et al. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 353.Revest JM, et al. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev. Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 354.Sims-Lucas S, et al. Three-dimensional imaging reveals ureteric and mesenchymal defects in Fgfr2-mutant kidneys. J. Am. Soc. Nephrol. 2009;20:2525–2533. doi: 10.1681/ASN.2009050532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Su N, Jin M, Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2014;2:14003. doi: 10.1038/boneres.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Anuwatmatee S, et al. Fibroblast growth factor 21 in chronic kidney disease. Clin. Chim. Acta. 2019;489:196–202. doi: 10.1016/j.cca.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 357.Inagaki T, et al. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab. 2008;8:77–83. doi: 10.1016/j.cmet.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Bookout AL, et al. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat. Med. 2013;19:1147–1152. doi: 10.1038/nm.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Liang Q, et al. FGF21 maintains glucose homeostasis by mediating the cross talk between liver and brain during prolonged fasting. Diabetes. 2014;63:4064–4075. doi: 10.2337/db14-0541. [DOI] [PubMed] [Google Scholar]
- 360.Ding H, et al. Depression-like behaviors induced by chronic corticosterone exposure via drinking water: time-course analysis. Neurosci. Lett. 2018;687:202–206. doi: 10.1016/j.neulet.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 361.Farrokhi F, et al. Association between depression and mortality in patients receiving long-term dialysis: a systematic review and meta-analysis. Am. J. Kidney Dis. 2014;63:623–635. doi: 10.1053/j.ajkd.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 362.Kohara M, et al. Association between circulating fibroblast growth factor 21 and mortality in end-stage renal disease. PLoS ONE. 2017;12:e0178971. doi: 10.1371/journal.pone.0178971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Fukumoto S. Actions and mode of actions of FGF19 subfamily members. Endocr. J. 2008;55:23–31. doi: 10.1507/endocrj.kr07e-002. [DOI] [PubMed] [Google Scholar]
- 364.Isakova T, et al. fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease. Kidney Int. 2011;79:1370–1378. doi: 10.1038/ki.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Moe SM, et al. The pathophysiology of early-stage chronic kidney disease-mineral bone disorder (CKD-MBD) and response to phosphate binders in the rat. J. Bone Miner. Res. 2011;26:2672–2681. doi: 10.1002/jbmr.485. [DOI] [PubMed] [Google Scholar]
- 366.Lee CH, et al. Circulating fibroblast growth factor 21 levels predict progressive kidney disease in subjects with type 2 diabetes and normoalbuminuria. J. Clin. Endocrinol. Metab. 2015;100:1368–1375. doi: 10.1210/jc.2014-3465. [DOI] [PubMed] [Google Scholar]
- 367.El-Saeed AM, El-Mohasseb GF. Circulating fibroblast growth factors 21 and 23 as biomarkers of progression in diabetic nephropathy in type 2 diabetes with normoalbuminuria. Egypt J. Immunol. 2017;24:93–99. [PubMed] [Google Scholar]
- 368.Shalhoub V, et al. FGF23 neutralization improves chronic kidney disease-associated hyperparathyroidism yet increases mortality. J. Clin. Invest. 2012;122:2543–2553. doi: 10.1172/JCI61405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Dalrymple LS, Go AS. Epidemiology of acute infections among patients with chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2008;3:1487–1493. doi: 10.2215/CJN.01290308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Thompson S, et al. Cause of death in patients with reduced kidney function. J. Am. Soc. Nephrol. 2015;26:2504–2511. doi: 10.1681/ASN.2014070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Chonchol M, et al. Low vitamin D and high fibroblast growth factor 23 serum levels associate with infectious and cardiac deaths in the HEMO Study. J. Am. Soc. Nephrol. 2016;27:227–237. doi: 10.1681/ASN.2014101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Masuda Y, et al. Expression of Fgf23 in activated dendritic cells and macrophages in response to immunological stimuli in mice. Biol. Pharm. Bull. 2015;38:687–693. doi: 10.1248/bpb.b14-00276. [DOI] [PubMed] [Google Scholar]
- 373.Han X, et al. Counter-regulatory paracrine actions of FGF-23 and 1,25(OH)2 D in macrophages. FEBS Lett. 2016;590:53–67. doi: 10.1002/1873-3468.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Rossaint J, et al. FGF23 signaling impairs neutrophil recruitment and host defense during CKD. J. Clin. Invest. 2016;126:962–974. doi: 10.1172/JCI83470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Strutz F. The role of FGF-2 in renal fibrogenesis. Front. Biosci. (Sch. Ed.) 2009;1:125–131. doi: 10.2741/S12. [DOI] [PubMed] [Google Scholar]
- 376.Duffield JS. Cellular and molecular mechanisms in kidney fibrosis. J. Clin. Invest. 2014;124:2299–2306. doi: 10.1172/JCI72267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Leaf DE, et al. Fibroblast growth factor 23 levels are elevated and associated with severe acute kidney injury and death following cardiac surgery. Kidney Int. 2016;89:939–948. doi: 10.1016/j.kint.2015.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Hanudel MR, et al. Effects of acute kidney injury and chronic hypoxemia on fibroblast growth factor 23 levels in pediatric cardiac surgery patients. Pediatr. Nephrol. 2016;31:661–669. doi: 10.1007/s00467-015-3257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Leaf DE, et al. Dysregulated mineral metabolism in patients with acute kidney injury and risk of adverse outcomes. Clin. Endocrinol. (Oxf.) 2013;79:491–498. doi: 10.1111/cen.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Brown JR, et al. Fibroblast growth factor-23 and the long-term risk of hospital-associated AKI among community-dwelling older individuals. Clin. J. Am. Soc. Nephrol. 2014;9:239–246. doi: 10.2215/CJN.05830513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Ali FN, Hassinger A, Price H, Langman CB. Preoperative plasma FGF23 levels predict acute kidney injury in children: results of a pilot study. Pediatr. Nephrol. 2013;28:959–962. doi: 10.1007/s00467-012-2395-2. [DOI] [PubMed] [Google Scholar]
- 382.Volovelsky O, et al. Early postoperative measurement of fibroblast growth factor 23 predicts severe acute kidney injury in infants after cardiac surgery. Clin. Nephrol. 2018;90:165–171. doi: 10.5414/CN109359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Leaf DE, et al. Fibroblast growth factor 23 associates with death in critically ill patients. Clin. J. Am. Soc. Nephrol. 2018;13:531–541. doi: 10.2215/CJN.10810917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Hassan A, et al. The fibroblast growth factor receptor mediates the increased FGF23 expression in acute and chronic uremia. Am. J. Physiol. Ren. Physiol. 2016;310:F217–F221. doi: 10.1152/ajprenal.00332.2015. [DOI] [PubMed] [Google Scholar]
- 385.Christov M, et al. Plasma FGF23 levels increase rapidly after acute kidney injury. Kidney Int. 2013;84:776–785. doi: 10.1038/ki.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Smith ER, Tan SJ, Holt SG, Hewitson TD. FGF23 is synthesised locally by renal tubules and activates injury-primed fibroblasts. Sci. Rep. 2017;7:3345. doi: 10.1038/s41598-017-02709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Mace ML, et al. Kidney fibroblast growth factor 23 does not contribute to elevation of its circulating levels in uremia. Kidney Int. 2017;92:165–178. doi: 10.1016/j.kint.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 388.Michos O, et al. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Ikeda Y, et al. Fgfr2 is integral for bladder mesenchyme patterning and function. Am. J. Physiol. Ren. Physiol. 2017;312:F607–F618. doi: 10.1152/ajprenal.00463.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ichimura T, et al. Induction of FGF-7 after kidney damage: a possible paracrine mechanism for tubule repair. Am. J. Physiol. 1996;271:F967–F976. doi: 10.1152/ajprenal.1996.271.5.F967. [DOI] [PubMed] [Google Scholar]
- 391.Yamzon J, L. KH, Kuremoto KI. FGF-10/FGFR2B signaling during acute cyclophosphamide-induced bladder urothelial injury in mice. J. Urol. 2011;185:e547–e548. [Google Scholar]
- 392.Motohashi N, Asakura A. Muscle satellite cell heterogeneity and self-renewal. Front. Cell Dev. Biol. 2014;2:1. doi: 10.3389/fcell.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Sheehan SM, Allen RE. Skeletal muscle satellite cell proliferation in response to members of the fibroblast growth factor family and hepatocyte growth factor. J. Cell Physiol. 1999;181:499–506. doi: 10.1002/(SICI)1097-4652(199912)181:3<499::AID-JCP14>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 394.Kastner S, Elias MC, Rivera AJ, Yablonka-Reuveni Z. Gene expression patterns of the fibroblast growth factors and their receptors during myogenesis of rat satellite cells. J. Histochem. Cytochem. 2000;48:1079–1096. doi: 10.1177/002215540004800805. [DOI] [PubMed] [Google Scholar]
- 395.Conte C, et al. Fibroblast growth factor 1 induced during myogenesis by a transcription-translation coupling mechanism. Nucleic Acids Res. 2009;37:5267–5278. doi: 10.1093/nar/gkp550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.DiMario J, Buffinger N, Yamada S, Strohman RC. Fibroblast growth factor in the extracellular matrix of dystrophic (mdx) mouse muscle. Science. 1989;244:688–690. doi: 10.1126/science.2717945. [DOI] [PubMed] [Google Scholar]
- 397.Rao N, et al. Fibroblasts influence muscle progenitor differentiation and alignment in contact independent and dependent manners in organized co-culture devices. Biomed. Microdev. 2013;15:161–169. doi: 10.1007/s10544-012-9709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Anderson JE, Mitchell CM, McGeachie JK, Grounds MD. The time course of basic fibroblast growth factor expression in crush-injured skeletal muscles of SJL/J and BALB/c mice. Exp. Cell Res. 1995;216:325–334. doi: 10.1006/excr.1995.1041. [DOI] [PubMed] [Google Scholar]
- 399.Hannon K, et al. Differentially expressed fibroblast growth factors regulate skeletal muscle development through autocrine and paracrine mechanisms. J. Cell Biol. 1996;132:1151–1159. doi: 10.1083/jcb.132.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Motohashi, N., Asakura, Y. & Asakura, A. Isolation, culture, and transplantation of muscle satellite cells. J. Vis. Exp. 50846 (2014). [DOI] [PMC free article] [PubMed]
- 402.Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev. Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Olwin BB, Hauschka SD. Identification of the fibroblast growth factor receptor of Swiss 3T3 cells and mouse skeletal muscle myoblasts. Biochemistry. 1986;25:3487–3492. doi: 10.1021/bi00360a001. [DOI] [PubMed] [Google Scholar]
- 404.Yablonka-Reuveni Z, Danoviz ME, Phelps M, Stuelsatz P. Myogenic-specific ablation of Fgfr1 impairs FGF2-mediated proliferation of satellite cells at the myofiber niche but does not abolish the capacity for muscle regeneration. Front. Aging Neurosci. 2015;7:85. doi: 10.3389/fnagi.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Clegg CH, Linkhart TA, Olwin BB, Hauschka SD. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 1987;105:949–956. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Kudla AJ, et al. The FGF receptor-1 tyrosine kinase domain regulates myogenesis but is not sufficient to stimulate proliferation. J. Cell Biol. 1998;142:241–250. doi: 10.1083/jcb.142.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Li J, Han S, Cousin W, Conboy IM. Age-specific functional epigenetic changes in p21 and p16 in injury-activated satellite cells. Stem Cells. 2015;33:951–961. doi: 10.1002/stem.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Fukada S, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 410.Shea KL, et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Lefaucheur JP, Sebille A. Basic fibroblast growth factor promotes in vivo muscle regeneration in murine muscular dystrophy. Neurosci. Lett. 1995;202:121–124. doi: 10.1016/0304-3940(95)12223-0. [DOI] [PubMed] [Google Scholar]
- 412.Lefaucheur JP, Sebille A. Muscle regeneration following injury can be modified in vivo by immune neutralization of basic fibroblast growth factor, transforming growth factor beta 1 or insulin-like growth factor I. J. Neuroimmunol. 1995;57:85–91. doi: 10.1016/0165-5728(94)00166-l. [DOI] [PubMed] [Google Scholar]
- 413.Han JK, Martin GR. Embryonic expression of Fgf-6 is restricted to the skeletal muscle lineage. Dev. Biol. 1993;158:549–554. doi: 10.1006/dbio.1993.1212. [DOI] [PubMed] [Google Scholar]
- 414.deLapeyriere O, et al. Expression of the Fgf6 gene is restricted to developing skeletal muscle in the mouse embryo. Development. 1993;118:601–611. doi: 10.1242/dev.118.2.601. [DOI] [PubMed] [Google Scholar]
- 415.Floss T, Arnold HH, Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Cool SM, et al. Temporal and spatial expression of fibroblast growth factor receptor 4 isoforms in murine tissues. Histochem. J. 2002;34:291–297. doi: 10.1023/a:1023326524562. [DOI] [PubMed] [Google Scholar]
- 417.Fiore F, Sebille A, Birnbaum D. Skeletal muscle regeneration is not impaired in Fgf6 −/− mutant mice. Biochem. Biophys. Res. Commun. 2000;272:138–143. doi: 10.1006/bbrc.2000.2703. [DOI] [PubMed] [Google Scholar]
- 418.Armand AS, et al. FGF6 regulates muscle differentiation through a calcineurin-dependent pathway in regenerating soleus of adult mice. J. Cell. Physiol. 2005;204:297–308. doi: 10.1002/jcp.20302. [DOI] [PubMed] [Google Scholar]
- 419.Neuhaus P, et al. Reduced mobility of fibroblast growth factor (FGF)-deficient myoblasts might contribute to dystrophic changes in the musculature of FGF2/FGF6/mdx triple-mutant mice. Mol. Cell. Biol. 2003;23:6037–6048. doi: 10.1128/MCB.23.17.6037-6048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Benoit B, et al. Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nat. Med. 2017;23:990–996. doi: 10.1038/nm.4363. [DOI] [PubMed] [Google Scholar]
- 421.Cornelison DD, et al. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev. Biol. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 422.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- 423.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 424.Deng CX, et al. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 425.Jones NC, Fedorov YV, Rosenthal RS, Olwin BB. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J. Cell. Physiol. 2001;186:104–115. doi: 10.1002/1097-4652(200101)186:1<104::AID-JCP1015>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 426.Jones NC, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Troy A, et al. Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38alpha/beta MAPK. Cell Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Bernet JD, et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Lagha M, et al. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 2008;22:1828–1837. doi: 10.1101/gad.477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.El Agha E, Kosanovic D, Schermuly RT, Bellusci S. Role of fibroblast growth factors in organ regeneration and repair. Semin. Cell Dev. Biol. 2016;53:76–84. doi: 10.1016/j.semcdb.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 431.Beltrami AP, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 432.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Sultana N, et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat. Commun. 2015;6:8701. doi: 10.1038/ncomms9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Liu Q, et al. Genetic lineage tracing identifies in situ Kit-expressing cardiomyocytes. Cell Res. 2016;26:119–130. doi: 10.1038/cr.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Engel FB, Hsieh PC, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl Acad. Sci. USA. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Novoyatleva T, et al. FGF1-mediated cardiomyocyte cell cycle reentry depends on the interaction of FGFR-1 and Fn14. FASEB J. 2014;28:2492–2503. doi: 10.1096/fj.13-243576. [DOI] [PubMed] [Google Scholar]
- 437.Cuevas P, et al. Fibroblast growth factor-1 prevents myocardial apoptosis triggered by ischemia reperfusion injury. Eur. J. Med. Res. 1997;2:465–468. [PubMed] [Google Scholar]
- 438.Baines CP, Molkentin JD. STRESS signaling pathways that modulate cardiac myocyte apoptosis. J. Mol. Cell Cardiol. 2005;38:47–62. doi: 10.1016/j.yjmcc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 439.Engel FB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Buehler A, et al. Angiogenesis-independent cardioprotection in FGF-1 transgenic mice. Cardiovasc. Res. 2002;55:768–777. doi: 10.1016/s0008-6363(02)00494-7. [DOI] [PubMed] [Google Scholar]
- 441.House SL, et al. Fibroblast growth factor 2 mediates isoproterenol-induced cardiac hypertrophy through activation of the extracellular regulated kinase. Mol. Cell Pharmacol. 2010;2:143–154. doi: 10.4255/mcpharmacol.10.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Pellieux C, et al. Dilated cardiomyopathy and impaired cardiac hypertrophic response to angiotensin II in mice lacking FGF-2. J. Clin. Invest. 2001;108:1843–1851. doi: 10.1172/JCI13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Virag JA, et al. Fibroblast growth factor-2 regulates myocardial infarct repair: effects on cell proliferation, scar contraction, and ventricular function. Am. J. Pathol. 2007;171:1431–1440. doi: 10.2353/ajpath.2007.070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.House SL, et al. Fibroblast growth factor 2 is an essential cardioprotective factor in a closed-chest model of cardiac ischemia-reperfusion injury. Physiol Rep. 2015;3:e12278. doi: 10.14814/phy2.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Detillieux KA, Sheikh F, Kardami E, Cattini PA. Biological activities of fibroblast growth factor-2 in the adult myocardium. Cardiovasc. Res. 2003;57:8–19. doi: 10.1016/s0008-6363(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 446.Wang ZG, et al. bFGF regulates autophagy and ubiquitinated protein accumulation induced by myocardial ischemia/reperfusion via the activation of the PI3K/Akt/mTOR pathway. Sci. Rep. 2015;5:9287. doi: 10.1038/srep09287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Ruel M, et al. Long-term effects of surgical angiogenic therapy with fibroblast growth factor 2 protein. J. Thorac. Cardiovasc. Surg. 2002;124:28–34. doi: 10.1067/mtc.2002.121974. [DOI] [PubMed] [Google Scholar]
- 448.Simons M, et al. Pharmacological treatment of coronary artery disease with recombinant fibroblast growth factor-2: double-blind, randomized, controlled clinical trial. Circulation. 2002;105:788–793. doi: 10.1161/hc0802.104407. [DOI] [PubMed] [Google Scholar]
- 449.Simons M, Ware JA. Therapeutic angiogenesis in cardiovascular disease. Nat. Rev. Drug Discov. 2003;2:863–871. doi: 10.1038/nrd1226. [DOI] [PubMed] [Google Scholar]
- 450.Lavine KJ, et al. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 451.Korf-Klingebiel M, et al. Conditional transgenic expression of fibroblast growth factor 9 in the adult mouse heart reduces heart failure mortality after myocardial infarction. Circulation. 2011;123:504–514. doi: 10.1161/CIRCULATIONAHA.110.989665. [DOI] [PubMed] [Google Scholar]
- 452.Singla DK, Singla RD, Abdelli LS, Glass C. Fibroblast growth factor-9 enhances M2 macrophage differentiation and attenuates adverse cardiac remodeling in the infarcted diabetic heart. PLoS ONE. 2015;10:e0120739. doi: 10.1371/journal.pone.0120739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 454.Rochais F, et al. FGF10 promotes regional foetal cardiomyocyte proliferation and adult cardiomyocyte cell-cycle re-entry. Cardiovasc. Res. 2014;104:432–442. doi: 10.1093/cvr/cvu232. [DOI] [PubMed] [Google Scholar]
- 455.Marguerie A, et al. Congenital heart defects in Fgfr2-IIIb and Fgf10 mutant mice. Cardiovasc Res. 2006;71:50–60. doi: 10.1016/j.cardiores.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 456.Rubin N, et al. FGF10 signaling enhances epicardial cell expansion during neonatal mouse heart repair. J. Cardiovasc. Dis. Diagn. 2013;1:101. doi: 10.4172/2329-9517.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Nicenboim J, et al. Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature. 2015;522:56–61. doi: 10.1038/nature14425. [DOI] [PubMed] [Google Scholar]
- 458.Yu P, et al. FGF-dependent metabolic control of vascular development. Nature. 2017;545:224–228. doi: 10.1038/nature22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Dell'Era P, et al. Paracrine and autocrine effects of fibroblast growth factor-4 in endothelial cells. Oncogene. 2001;20:2655–2663. doi: 10.1038/sj.onc.1204368. [DOI] [PubMed] [Google Scholar]
- 460.Shin JW, et al. Prox1 promotes lineage-specific expression of fibroblast growth factor (FGF) receptor-3 in lymphatic endothelium: a role for FGF signaling in lymphangiogenesis. Mol. Biol. Cell. 2006;17:576–584. doi: 10.1091/mbc.E05-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Cross MJ, Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharm. Sci. 2001;22:201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- 462.Mignatti P, Rifkin DB. Nonenzymatic interactions between proteinases and the cell surface: novel roles in normal and malignant cell physiology. Adv. Cancer Res. 1999;78:103–157. doi: 10.1016/s0065-230x(08)61024-6. [DOI] [PubMed] [Google Scholar]
- 463.Giulia T, et al. Shedding of the matrix metalloproteinases MMP-2, MMP-9, and MT1-MMP as membrane vesicle-associated components by endothelial cells. Am. J. Pathol. 2002;160:673–680. doi: 10.1016/S0002-9440(10)64887-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Gillis P, et al. Keratinocyte growth factor induces angiogenesis and protects endothelial barrier function. J. Cell Sci. 1999;112(Part 12):2049–2057. doi: 10.1242/jcs.112.12.2049. [DOI] [PubMed] [Google Scholar]
- 465.Shono T, Kanetake H, Kanda S. The role of mitogen-activated protein kinase activation within focal adhesions in chemotaxis toward FGF-2 by murine brain capillary endothelial cells. Exp. Cell Res. 2001;264:275–283. doi: 10.1006/excr.2001.5154. [DOI] [PubMed] [Google Scholar]
- 466.Choi I, et al. 9-Cis retinoic acid promotes lymphangiogenesis and enhances lymphatic vessel regeneration: therapeutic implications of 9-cis retinoic acid for secondary lymphedema. Circulation. 2012;125:872–882. doi: 10.1161/CIRCULATIONAHA.111.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Moscatelli D, Presta M, Joseph-Silverstein J, Rifkin DB. Both normal and tumor cells produce basic fibroblast growth factor. J. Cell. Physiol. 1986;129:273–276. doi: 10.1002/jcp.1041290220. [DOI] [PubMed] [Google Scholar]
- 468.Wang Y, Becker D. Antisense targeting of basic fibroblast growth factor and dibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat. Med. 1997;3:887–893. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 469.Presta M, et al. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 470.Domenico R, et al. Angiogenic activity of rat mast cells in the chick embryo chorioallantoic membrane is down-regulated by treatment with recombinant human alpha-2a interferon and partly mediated by fibroblast growth factor-2. Haematologica. 2002;87:465–471. [PubMed] [Google Scholar]
- 471.Hyung Taek L, Jeong Goo L, Moonseok N, Kay EDP. FGF-2 induced by interleukin-1 beta through the action of phosphatidylinositol 3-kinase mediates endothelial mesenchymal transformation in corneal endothelial cells. J. Biol. Chem. 2004;279:32325. doi: 10.1074/jbc.M405208200. [DOI] [PubMed] [Google Scholar]
- 472.Jian L, Shworak NW, Michael S. Increased responsiveness of hypoxic endothelial cells to FGF2 is mediated by HIF-1alpha-dependent regulation of enzymes involved in synthesis of heparan sulfate FGF2-binding sites. J. Cell Sci. 2002;115:1951–1959. doi: 10.1242/jcs.115.9.1951. [DOI] [PubMed] [Google Scholar]
- 473.Piera-Velazquez S, Jimenez SA. Endothelial to mesenchymal transition: role in physiology and in the pathogenesis of human diseases. Physiol. Rev. 2019;99:1281–1324. doi: 10.1152/physrev.00021.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 475.Medici D, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat. Med. 2010;16:1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Chen PY, et al. FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2012;2:1684–1696. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Ana, C. P. C., Jan-Renier, A. J. M., Marja, G. L. B. & Guido, K. FGF-2 inhibits endothelial–mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-β signaling. J. Cell Sci.129, 569–579 (2016). [DOI] [PubMed]
- 478.Post MJ, Laham R, Sellke FW, Simons M. Therapeutic angiogenesis in cardiology using protein formulations. Cardiovasc. Res. 2001;49:522–531. doi: 10.1016/s0008-6363(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 479.Lederman RJ, et al. Design of the therapeutic angiogenesis with recombinant fibroblast growth factor-2 for intermittent claudication (TRAFFIC) trial. Am. J. Cardiol. 2001;88:192–195. doi: 10.1016/s0002-9149(01)01622-8. [DOI] [PubMed] [Google Scholar]
- 480.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 481.Jonker JW, et al. A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485:391–394. doi: 10.1038/nature10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Sano H, et al. Detection of high levels of heparin binding growth factor-1 (acidic fibroblast growth factor) in inflammatory arthritic joints. J. Cell Biol. 1990;110:1417–1426. doi: 10.1083/jcb.110.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Byrd VM, Ballard DW, Miller GG, Thomas JW. Fibroblast growth factor-1 (FGF-1) enhances IL-2 production and nuclear translocation of NF-kappaB in FGF receptor-bearing Jurkat T cells. J. Immunol. 1999;162:5853–5859. [PubMed] [Google Scholar]
- 484.Rossini M, et al. Immunolocalization of fibroblast growth factor-1 (FGF-1), its receptor (FGFR-1), and fibroblast-specific protein-1 (FSP-1) in inflammatory renal disease. Kidney Int. 2005;68:2621–2628. doi: 10.1111/j.1523-1755.2005.00734.x. [DOI] [PubMed] [Google Scholar]
- 485.Hackshaw KV, Shi Y. Fibroblast growth factors mobilize peritoneal macrophage intracellular calcium. Life Sci. 1994;54:661–670. doi: 10.1016/0024-3205(94)00549-4. [DOI] [PubMed] [Google Scholar]
- 486.Garre JM, et al. FGF-1 induces ATP release from spinal astrocytes in culture and opens pannexin and connexin hemichannels. Proc. Natl Acad. Sci. USA. 2010;107:22659–22664. doi: 10.1073/pnas.1013793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Garre JM, Yang G, Bukauskas FF, Bennett MV. FGF-1 triggers Pannexin-1 hemichannel opening in spinal astrocytes of rodents and promotes inflammatory responses in acute spinal cord slices. J. Neurosci. 2016;36:4785–4801. doi: 10.1523/JNEUROSCI.4195-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Huang Z, et al. Uncoupling the mitogenic and metabolic functions of FGF1 by tuning FGF1-FGF receptor dimer stability. Cell Rep. 2017;20:1717–1728. doi: 10.1016/j.celrep.2017.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Wang D, et al. FGF1(DeltaHBS) ameliorates chronic kidney disease via PI3K/AKT mediated suppression of oxidative stress and inflammation. Cell Death Dis. 2019;10:464. doi: 10.1038/s41419-019-1696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Liang G, et al. Fibroblast growth factor 1 ameliorates diabetic nephropathy by an anti-inflammatory mechanism. Kidney Int. 2018;93:95–109. doi: 10.1016/j.kint.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Harada M, et al. Temporal expression of growth factors triggered by epiregulin regulates inflammation development. J. Immunol. 2015;194:1039–1046. doi: 10.4049/jimmunol.1400562. [DOI] [PubMed] [Google Scholar]
- 492.Shao X, et al. FGF2 cooperates with IL-17 to promote autoimmune inflammation. Sci. Rep. 2017;7:7024. doi: 10.1038/s41598-017-07597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Song X, et al. Growth factor FGF2 cooperates with interleukin-17 to repair intestinal epithelial damage. Immunity. 2015;43:488–501. doi: 10.1016/j.immuni.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 494.Boehme, K. A. & Rolauffs, B. Onset and progression of human osteoarthritis—can growth factors, inflammatory cytokines, or differential miRNA expression concomitantly induce proliferation, ECM degradation, and inflammation in articular cartilage? Int. J. Mol. Sci. 19, 2282 (2018). [DOI] [PMC free article] [PubMed]
- 495.Lappegard KT, et al. The artificial surface-induced whole blood inflammatory reaction revealed by increases in a series of chemokines and growth factors is largely complement dependent. J. Biomed. Mater. Res. A. 2008;87:129–135. doi: 10.1002/jbm.a.31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Keating SM, et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of women. AIDS. 2011;25:1823–1832. doi: 10.1097/QAD.0b013e3283489d1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Bocelli-Tyndall C, et al. FGF2 induces RANKL gene expression as well as IL1beta regulated MHC class II in human bone marrow-derived mesenchymal progenitor stromal cells. Ann. Rheum. Dis. 2015;74:260–266. doi: 10.1136/annrheumdis-2013-204235. [DOI] [PubMed] [Google Scholar]
- 498.Pawlowski P, et al. Markers of inflammation and fibrosis in the orbital fat/connective tissue of patients with Graves' orbitopathy: clinical implications. Mediat. Inflamm. 2014;2014:412158. doi: 10.1155/2014/412158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Presta M, et al. Inflammatory cells and chemokines sustain FGF2-induced angiogenesis. Eur. Cytokine Netw. 2009;20:39–50. doi: 10.1684/ecn.2009.0155. [DOI] [PubMed] [Google Scholar]
- 500.Schultz K, Murthy V, Tatro JB, Beasley D. Endogenous interleukin-1 alpha promotes a proliferative and proinflammatory phenotype in human vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H2927–H2934. doi: 10.1152/ajpheart.00700.2006. [DOI] [PubMed] [Google Scholar]
- 501.Bovolenta R, et al. Hippocampal FGF-2 and BDNF overexpression attenuates epileptogenesis-associated neuroinflammation and reduces spontaneous recurrent seizures. J. Neuroinflamm. 2010;7:81. doi: 10.1186/1742-2094-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Kim YS, et al. The role of FGF-2 in smoke-induced emphysema and the therapeutic potential of recombinant FGF-2 in patients with COPD. Exp. Mol. Med. 2018;50:1–10. doi: 10.1038/s12276-018-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Jeon SG, et al. Recombinant basic fibroblast growth factor inhibits the airway hyperresponsiveness, mucus production, and lung inflammation induced by an allergen challenge. J. Allergy Clin. Immunol. 2007;119:831–837. doi: 10.1016/j.jaci.2006.12.653. [DOI] [PubMed] [Google Scholar]
- 504.Sautter NB, Delaney KL, Hausman FA, Trune DR. Tissue remodeling gene expression in a murine model of chronic rhinosinusitis. Laryngoscope. 2012;122:711–717. doi: 10.1002/lary.22148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Sautter NB, Delaney KL, Trune DR. Altered expression of tissue remodeling genes in a mouse model of acute allergic rhinitis. Int. Forum Allergy Rhinol. 2011;1:262–267. doi: 10.1002/alr.20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Sautter NB, Delaney KL, Hausman FA, Trune DR. Tissue remodeling in the acute otitis media mouse model. Int. J. Pediatr. Otorhinolaryngol. 2011;75:1368–1371. doi: 10.1016/j.ijporl.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Feingold KR, et al. FGF21 is increased by inflammatory stimuli and protects leptin-deficient ob/ob mice from the toxicity of sepsis. Endocrinology. 2012;153:2689–2700. doi: 10.1210/en.2011-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Gariani K, et al. Increased FGF21 plasma levels in humans with sepsis and SIRS. Endocr. Connect. 2013;2:146–153. doi: 10.1530/EC-13-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Refsgaard Holm M, et al. Fibroblast growth factor 21 in patients with cardiac cachexia: a possible role of chronic inflammation. ESC Heart Fail. 2019;6:983–991. doi: 10.1002/ehf2.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Planavila A, et al. Fibroblast growth factor 21 protects the heart from oxidative stress. Cardiovasc Res. 2015;106:19–31. doi: 10.1093/cvr/cvu263. [DOI] [PubMed] [Google Scholar]
- 511.Zhang C, et al. Attenuation of hyperlipidemia- and diabetes-induced early-stage apoptosis and late-stage renal dysfunction via administration of fibroblast growth factor-21 is associated with suppression of renal inflammation. PLoS ONE. 2013;8:e82275. doi: 10.1371/journal.pone.0082275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Lee KJ, et al. Expression of fibroblast growth factor 21 and beta-Klotho regulates hepatic fibrosis through the nuclear factor-kappaB and c-Jun N-terminal kinase pathways. Gut Liver. 2018;12:449–456. doi: 10.5009/gnl17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Mindur JE, Swirski FK. Growth factors as immunotherapeutic targets in cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2019;39:1275–1287. doi: 10.1161/ATVBAHA.119.311994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Wang N, et al. Fibroblast growth factor 21 ameliorates pancreatic fibrogenesis via regulating polarization of macrophages. Exp. Cell Res. 2019;382:111457. doi: 10.1016/j.yexcr.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 515.Li JY, et al. FGF-21 elevated IL-10 production to correct LPS-induced inflammation. Inflammation. 2018;41:751–759. doi: 10.1007/s10753-018-0729-3. [DOI] [PubMed] [Google Scholar]
- 516.Wang N, et al. Fibroblast growth factor 21 regulates foam cells formation and inflammatory response in Ox-LDL-induced THP-1 macrophages. Biomed. Pharmacother. 2018;108:1825–1834. doi: 10.1016/j.biopha.2018.09.143. [DOI] [PubMed] [Google Scholar]
- 517.Wang N, et al. Improving hyperglycemic effect of FGF-21 is associated with alleviating inflammatory state in diabetes. Int. Immunopharmacol. 2018;56:301–309. doi: 10.1016/j.intimp.2018.01.048. [DOI] [PubMed] [Google Scholar]
- 518.Wang N, et al. Fibroblast frowth factor 21 exerts its anti-inflammatory effects on multiple cell types of adipose tissue in obesity. Obesity (Silver Spring) 2019;27:399–408. doi: 10.1002/oby.22376. [DOI] [PubMed] [Google Scholar]
- 519.Liu MH. FGF-21 alleviates diabetes-associated vascular complications: Inhibiting NF-kappaB/NLRP3 inflammasome-mediated inflammation? Int. J. Cardiol. 2015;185:320–321. doi: 10.1016/j.ijcard.2015.03.165. [DOI] [PubMed] [Google Scholar]
- 520.Holecki M, et al. Inflammation but not obesity or insulin resistance is associated with increased plasma fibroblast growth factor 23 concentration in the elderly. Clin. Endocrinol. 2015;82:900. doi: 10.1111/cen.12759. [DOI] [PubMed] [Google Scholar]
- 521.Lang F, et al. Phosphate homeostasis, inflammation and the regulation of FGF-23. Kidney Blood Press. Res. 2018;43:1742–1748. doi: 10.1159/000495393. [DOI] [PubMed] [Google Scholar]
- 522.Singh S, et al. Fibroblast growth factor 23 directly targets hepatocytes to promote inflammation in chronic kidney disease. Kidney Int. 2016;90:985–996. doi: 10.1016/j.kint.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Wang C, et al. Ectopic fibroblast growth factor receptor 1 promotes inflammation by promoting nuclear factor-kappaB signaling in prostate cancer cells. J. Biol. Chem. 2018;293:14839–14849. doi: 10.1074/jbc.RA118.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Shi YJ, et al. Inhibitive effects of FGF2/FGFR1 pathway on astrocyte-mediated inflammation in vivo and in vitro after infrasound exposure. Front. Neurosci. 2018;12:582. doi: 10.3389/fnins.2018.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Ruslan M, Tiffany H. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 526.Georg S, et al. How cytokine networks fuel inflammation: toward a cytokine-based disease taxonomy. Nat. Med. 2013;19:822–824. doi: 10.1038/nm.3260. [DOI] [PubMed] [Google Scholar]
- 527.Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 528.Somm E, Jornayvaz FR. Fibroblast growth factor 15/19: from basic functions to therapeutic perspectives. Endocr. Rev. 2018;39:960–989. doi: 10.1210/er.2018-00134. [DOI] [PubMed] [Google Scholar]
- 529.Fu L, et al. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 530.Tomlinson E, et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–1747. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- 531.Zhou M, et al. Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J. Hepatol. 2017;66:1182–1192. doi: 10.1016/j.jhep.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 532.Fon Tacer K, et al. Research resource: comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 534.Jung D, et al. FXR-induced secretion of FGF15/19 inhibits CYP27 expression in cholangiocytes through p38 kinase pathway. Pflug. Arch. 2014;466:1011–1019. doi: 10.1007/s00424-013-1364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Zhang JH, et al. Potent stimulation of fibroblast growth factor 19 expression in the human ileum by bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G940–G948. doi: 10.1152/ajpgi.00398.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Naugler WE, et al. Fibroblast growth factor signaling controls liver size in mice with humanized livers. Gastroenterology. 2015;149:728–740. doi: 10.1053/j.gastro.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Stroeve JH, et al. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab. Invest. 2010;90:1457–1467. doi: 10.1038/labinvest.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.de Wit NJ, et al. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med. Genomics. 2008;1:14. doi: 10.1186/1755-8794-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Song KH, et al. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Choi M, et al. Identification of a hormonal basis for gallbladder filling. Nat. Med. 2006;12:1253–1255. doi: 10.1038/nm1501. [DOI] [PubMed] [Google Scholar]
- 541.Kir S, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Potthoff MJ, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Bhatnagar S, Damron HA, Hillgartner FB. Fibroblast growth factor-19, a novel factor that inhibits hepatic fatty acid synthesis. J. Biol. Chem. 2009;284:10023–10033. doi: 10.1074/jbc.M808818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Massafra V, et al. Quantitative liver proteomics identifies FGF19 targets that couple metabolism and proliferation. PLoS ONE. 2017;12:e0171185. doi: 10.1371/journal.pone.0171185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Alvarez-Sola G, et al. Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut. 2017;66:1818–1828. doi: 10.1136/gutjnl-2016-312975. [DOI] [PubMed] [Google Scholar]
- 546.Ryan KK, et al. Fibroblast growth factor-19 action in the brain reduces food intake and body weight and improves glucose tolerance in male rats. Endocrinology. 2013;154:9–15. doi: 10.1210/en.2012-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Marcelin G, et al. Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism. Mol. Metab. 2014;3:19–28. doi: 10.1016/j.molmet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Lan T, et al. FGF19, FGF21, and an FGFR1/beta-Klotho-activating antibody act on the nervous system to regulate body weight and glycemia. Cell Metab. 2017;26:709–718. doi: 10.1016/j.cmet.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Perry RJ, et al. FGF1 and FGF19 reverse diabetes by suppression of the hypothalamic-pituitary-adrenal axis. Nat. Commun. 2015;6:6980. doi: 10.1038/ncomms7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Zhang X, et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes. 2008;57:1246–1253. doi: 10.2337/db07-1476. [DOI] [PubMed] [Google Scholar]
- 551.Markan KR, et al. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes. 2014;63:4057–4063. doi: 10.2337/db14-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Tezze C, Romanello V, Sandri M. FGF21 as modulator of metabolism in health and disease. Front. Physiol. 2019;10:419. doi: 10.3389/fphys.2019.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Xu J, et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58:250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Coskun T, et al. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149:6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 555.Potthoff MJ. FGF21 and metabolic disease in 2016: a new frontier in FGF21 biology. Nat. Rev. Endocrinol. 2017;13:74–76. doi: 10.1038/nrendo.2016.206. [DOI] [PubMed] [Google Scholar]
- 556.Lin Z, et al. Adiponectin mediates the metabolic effects of FGF21 on glucose homeostasis and insulin sensitivity in mice. Cell Metab. 2013;17:779–789. doi: 10.1016/j.cmet.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 557.Badman MK, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 558.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 559.Potthoff MJ, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl Acad. Sci. USA. 2009;106:10853–10858. doi: 10.1073/pnas.0904187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Hondares E, et al. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. J. Biol. Chem. 2011;286:12983–12990. doi: 10.1074/jbc.M110.215889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Kharitonenkov A, et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Li X, et al. Inhibition of lipolysis may contribute to the acute regulation of plasma FFA and glucose by FGF21 in ob/ob mice. FEBS Lett. 2009;583:3230–3234. doi: 10.1016/j.febslet.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 564.Cuevas-Ramos D, Mehta R, Aguilar-Salinas CA. Fibroblast growth factor 21 and browning of white adipose tissue. Front. Physiol. 2019;10:37. doi: 10.3389/fphys.2019.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Villarroya F, Peyrou M, Giralt M. Transcriptional regulation of the uncoupling protein-1 gene. Biochimie. 2017;134:86–92. doi: 10.1016/j.biochi.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 566.Fisher FM, et al. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Wu AL, et al. Amelioration of type 2 diabetes by antibody-mediated activation of fibroblast growth factor receptor 1. Sci. Transl. Med. 2011;3:113ra126. doi: 10.1126/scitranslmed.3002669. [DOI] [PubMed] [Google Scholar]
- 568.Katafuchi T, et al. PPARgamma-K107 SUMOylation regulates insulin sensitivity but not adiposity in mice. Proc. Natl Acad. Sci. USA. 2018;115:12102–12111. doi: 10.1073/pnas.1814522115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Dutchak PA, et al. Fibroblast growth factor-21 regulates PPARgamma activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148:556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Tan BK, et al. Fibroblast growth factor 21 (FGF21) in human cerebrospinal fluid: relationship with plasma FGF21 and body adiposity. Diabetes. 2011;60:2758–2762. doi: 10.2337/db11-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Sarruf DA, et al. Fibroblast growth factor 21 action in the brain increases energy expenditure and insulin sensitivity in obese rats. Diabetes. 2010;59:1817–1824. doi: 10.2337/db09-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Owen BM, et al. FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab. 2014;20:670–677. doi: 10.1016/j.cmet.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Yoshiko Y, et al. Mineralized tissue cells are a principal source of FGF23. Bone. 2007;40:1565–1573. doi: 10.1016/j.bone.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 574.Quarles LD. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat. Rev. Endocrinol. 2012;8:276–286. doi: 10.1038/nrendo.2011.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Shimada T, et al. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl Acad. Sci. USA. 2001;98:6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Benet-Pages A, Orlik P, Strom TM, Lorenz-Depiereux B. An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum. Mol. Genet. 2005;14:385–390. doi: 10.1093/hmg/ddi034. [DOI] [PubMed] [Google Scholar]
- 577.Larsson T, et al. Transgenic mice expressing fibroblast growth factor 23 under the control of the alpha1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology. 2004;145:3087–3094. doi: 10.1210/en.2003-1768. [DOI] [PubMed] [Google Scholar]
- 578.Bai X, et al. Transgenic mice overexpressing human fibroblast growth factor 23 (R176Q) delineate a putative role for parathyroid hormone in renal phosphate wasting disorders. Endocrinology. 2004;145:5269–5279. doi: 10.1210/en.2004-0233. [DOI] [PubMed] [Google Scholar]
- 579.Hu MC, Shi M, Moe OW. Role of alphaKlotho and FGF23 in regulation of type II Na-dependent phosphate co-transporters. Pflug. Arch. 2019;471:99–108. doi: 10.1007/s00424-018-2238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Beck L, et al. Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc. Natl Acad. Sci. USA. 1998;95:5372–5377. doi: 10.1073/pnas.95.9.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Madjdpour C, et al. Segment-specific expression of sodium-phosphate cotransporters NaPi-IIa and -IIc and interacting proteins in mouse renal proximal tubules. Pflug. Arch. 2004;448:402–410. doi: 10.1007/s00424-004-1253-x. [DOI] [PubMed] [Google Scholar]
- 582.Gattineni J, et al. FGF23 decreases renal NaPi-2a and NaPi-2c expression and induces hypophosphatemia in vivo predominantly via FGF receptor 1. Am. J. Physiol. Ren. Physiol. 2009;297:F282–F291. doi: 10.1152/ajprenal.90742.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Sabbagh Y, et al. Intestinal phosphate transport. Adv. Chronic Kidney Dis. 2011;18:85–90. doi: 10.1053/j.ackd.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Miyamoto K, et al. Inhibition of intestinal sodium-dependent inorganic phosphate transport by fibroblast growth factor 23. Ther. Apher. Dial. 2005;9:331–335. doi: 10.1111/j.1744-9987.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 585.Fukumoto S. Phosphate metabolism and vitamin D. Bonekey Rep. 2014;3:497. doi: 10.1038/bonekey.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Shimada T, et al. FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J. Bone Miner. Res. 2004;19:429–435. doi: 10.1359/JBMR.0301264. [DOI] [PubMed] [Google Scholar]
- 587.Andrukhova O, et al. FGF23 regulates renal sodium handling and blood pressure. EMBO Mol. Med. 2014;6:744–759. doi: 10.1002/emmm.201303716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Andrukhova O, et al. FGF23 promotes renal calcium reabsorption through the TRPV5 channel. EMBO J. 2014;33:229–246. doi: 10.1002/embj.201284188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Jacquillet G, Unwin RJ. Physiological regulation of phosphate by vitamin D, parathyroid hormone (PTH) and phosphate (Pi) Pflug. Arch. 2019;471:83–98. doi: 10.1007/s00424-018-2231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Wohrle S, et al. FGF receptors control vitamin D and phosphate homeostasis by mediating renal FGF-23 signaling and regulating FGF-23 expression in bone. J. Bone Miner. Res. 2011;26:2486–2497. doi: 10.1002/jbmr.478. [DOI] [PubMed] [Google Scholar]
- 591.Peng H, et al. Integrative nuclear FGFR1 signaling (INFS) pathway mediates activation of the tyrosine hydroxylase gene by angiotensin II, depolarization and protein kinase C. J. Neurochem. 2002;81:506–524. doi: 10.1046/j.1471-4159.2002.00833.x. [DOI] [PubMed] [Google Scholar]
- 592.Xiao L, et al. Nuclear isoforms of fibroblast growth factor 2 are novel inducers of hypophosphatemia via modulation of FGF23 and KLOTHO. J. Biol. Chem. 2010;285:2834–2846. doi: 10.1074/jbc.M109.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.The HYP Consortium. A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. Nat. Genet. 1995;11:130–136. doi: 10.1038/ng1095-130. [DOI] [PubMed] [Google Scholar]
- 594.Feng JQ, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat. Genet. 2006;38:1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Fukumoto S, Yamashita T. Fibroblast growth factor 23 in oncogenic osteomalacia and X-linked hypophosphatemia. N. Engl. J. Med. 2003;349:505–506. [PubMed] [Google Scholar]
- 596.Strom TM, et al. Pex gene deletions in Gy and Hyp mice provide mouse models for X-linked hypophosphatemia. Hum. Mol. Genet. 1997;6:165–171. doi: 10.1093/hmg/6.2.165. [DOI] [PubMed] [Google Scholar]
- 597.Liu S, et al. Regulation of fibroblastic growth factor 23 expression but not degradation by PHEX. J. Biol. Chem. 2003;278:37419–37426. doi: 10.1074/jbc.M304544200. [DOI] [PubMed] [Google Scholar]
- 598.Saito H, et al. Circulating FGF-23 is regulated by 1alpha,25-dihydroxyvitamin D3 and phosphorus in vivo. J. Biol. Chem. 2005;280:2543–2549. doi: 10.1074/jbc.M408903200. [DOI] [PubMed] [Google Scholar]
- 599.Collins MT, et al. Fibroblast growth factor-23 is regulated by 1alpha,25-dihydroxyvitamin D. J. Bone Miner. Res. 2005;20:1944–1950. doi: 10.1359/JBMR.050718. [DOI] [PubMed] [Google Scholar]
- 600.Ito N, et al. Extracellular phosphate modulates the effect of 1alpha,25-dihydroxy vitamin D3 (1,25D) on osteocyte like cells. J. Steroid Biochem. Mol. Biol. 2013;136:183–186. doi: 10.1016/j.jsbmb.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 601.Olauson H, et al. Parathyroid-specific deletion of Klotho unravels a novel calcineurin-dependent FGF23 signaling pathway that regulates PTH secretion. PLoS Genet. 2013;9:e1003975. doi: 10.1371/journal.pgen.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Ben-Dov IZ, et al. The parathyroid is a target organ for FGF23 in rats. J. Clin. Invest. 2007;117:4003–4008. doi: 10.1172/JCI32409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Kobayashi K, et al. Regulation of plasma fibroblast growth factor 23 by calcium in primary hyperparathyroidism. Eur. J. Endocrinol. 2006;154:93–99. doi: 10.1530/eje.1.02053. [DOI] [PubMed] [Google Scholar]
- 604.Meir T, et al. Parathyroid hormone activates the orphan nuclear receptor Nurr1 to induce FGF23 transcription. Kidney Int. 2014;86:1106–1115. doi: 10.1038/ki.2014.215. [DOI] [PubMed] [Google Scholar]
- 605.Rhee Y, et al. Parathyroid hormone receptor signaling in osteocytes increases the expression of fibroblast growth factor-23 in vitro and in vivo. Bone. 2011;49:636–643. doi: 10.1016/j.bone.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Lewerin C, et al. Low serum iron is associated with high serum intact FGF23 in elderly men: the Swedish MrOS study. Bone. 2017;98:1–8. doi: 10.1016/j.bone.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 607.Farrow EG, et al. Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice. Proc. Natl Acad. Sci. USA. 2011;108:E1146–E1155. doi: 10.1073/pnas.1110905108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Huang Y, Wang H, Yang Y. Expression of fibroblast growth factor 5 (FGF5) and its influence on survival of breast cancer patients. Med. Sci. Monit. 2018;24:3524–3530. doi: 10.12659/MSM.907798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Guo S, et al. A gene-based recessive diplotype exome scan discovers FGF6, a novel hepcidin-regulating iron-metabolism gene. Blood. 2019;133:1888–1898. doi: 10.1182/blood-2018-10-879585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Shaoul R, et al. Elevated expression of FGF7 protein is common in human gastric diseases. Biochem. Biophys. Res. Commun. 2006;350:825–833. doi: 10.1016/j.bbrc.2006.08.198. [DOI] [PubMed] [Google Scholar]
- 611.Dorkin TJ, et al. FGF8 over-expression in prostate cancer is associated with decreased patient survival and persists in androgen independent disease. Oncogene. 1999;18:2755–2761. doi: 10.1038/sj.onc.1202624. [DOI] [PubMed] [Google Scholar]
- 612.Bera A, Leighton XM, Pollard H, Srivastava M, Cyclin E. and FGF8 are downstream cell growth regulators in distinct tumor suppressor effects of ANXA7 in hormone-resistant cancer cells of breast versus prostate origin. Trends Cancer Res. 2018;13:55–62. [PMC free article] [PubMed] [Google Scholar]
- 613.Hegab AE, et al. Tumor associated macrophages support the growth of FGF9-induced lung adenocarcinoma by multiple mechanisms. Lung Cancer. 2018;119:25–35. doi: 10.1016/j.lungcan.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 614.Clayton NS, Grose RP. Emerging roles of fibroblast growth factor 10 in cancer. Front. Genet. 2018;9:499. doi: 10.3389/fgene.2018.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Bhushan A, et al. Identification and validation of fibroblast growth factor 12 gene as a novel potential biomarker in esophageal cancer using Cancer Genomic Datasets. OMICS. 2017;21:616–631. doi: 10.1089/omi.2017.0116. [DOI] [PubMed] [Google Scholar]
- 616.Bublik DR, et al. Regulatory module involving FGF13, miR-504, and p53 regulates ribosomal biogenesis and supports cancer cell survival. Proc. Natl Acad. Sci. USA. 2017;114:E496–E505. doi: 10.1073/pnas.1614876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Basu M, Mukhopadhyay S, Chatterjee U, Roy SS. FGF16 promotes invasive behavior of SKOV-3 ovarian cancer cells through activation of mitogen-activated protein kinase (MAPK) signaling pathway. J. Biol. Chem. 2014;289:1415–1428. doi: 10.1074/jbc.M113.535427. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 618.Heer R, et al. Fibroblast growth factor 17 is over-expressed in human prostate cancer. J. Pathol. 2004;204:578–586. doi: 10.1002/path.1668. [DOI] [PubMed] [Google Scholar]
- 619.Zhang J, et al. FGF18, a prominent player in FGF signaling, promotes gastric tumorigenesis through autocrine manner and is negatively regulated by miR-590-5p. Oncogene. 2019;38:33–46. doi: 10.1038/s41388-018-0430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Koneczny I, et al. Autocrine fibroblast growth factor 18 signaling mediates Wnt‐dependent stimulation of CD44‐positive human colorectal adenoma cells. Mol. Carcinogen. 2015;54:789–799. doi: 10.1002/mc.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Jeffers M, et al. Identification of a novel human fibroblast growth factor and characterization of its role in oncogenesis. Cancer Res. 2001;61:3131–3138. [PubMed] [Google Scholar]
- 623.Chamorro MN, et al. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Huang X, et al. Forced expression of hepatocyte-specific fibroblast growth factor 21 delays initiation of chemically induced hepatocarcinogenesis. Mol. Carcinogen. 2006;45:934–942. doi: 10.1002/mc.20241. [DOI] [PubMed] [Google Scholar]
- 625.Liu HY, Zhao H, Li WX. Integrated analysis of transcriptome and prognosis data identifies FGF22 as a prognostic marker of lung adenocarcinoma. Technol. Cancer Res. Treat. 2019;18:1533033819827317. doi: 10.1177/1533033819827317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Feng S, et al. FGF23 promotes prostate cancer progression. Oncotarget. 2015;6:17291–17301. doi: 10.18632/oncotarget.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Champeme MH, Bieche I, Hacene K, Lidereau R. Int-2/FGF3 amplification is a better independent predictor of relapse than c-myc and c-erbB-2/neu amplifications in primary human breast cancer. Mod. Pathol. 1994;7:900–905. [PubMed] [Google Scholar]
- 628.Zaharieva BM, et al. High‐throughput tissue microarray analysis of 11q13 gene amplification (CCND1, FGF3, FGF4, EMS1) in urinary bladder cancer. J. Pathol. 2003;201:603–608. doi: 10.1002/path.1481. [DOI] [PubMed] [Google Scholar]
- 629.Kim HJ, et al. Single nucleotide polymorphisms in fibroblast growth factor 23 gene, FGF23, are associated with prostate cancer risk. BJU. Int. 2014;114:303–310. doi: 10.1111/bju.12396. [DOI] [PubMed] [Google Scholar]
- 630.Katoh M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 2019;16:105–122. doi: 10.1038/s41571-018-0115-y. [DOI] [PubMed] [Google Scholar]
- 631.Reis-Filho JS, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin. Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 632.Thomas A, et al. Characterization of fibroblast growth factor receptor 1 in small-cell lung cancer. J. Thorac. Oncol. 2014;9:567–571. doi: 10.1097/JTO.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Davies H, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 634.Rivera B, et al. Germline and somatic FGFR1 abnormalities in dysembryoplastic neuroepithelial tumors. Acta Neuropathol. 2016;131:847–863. doi: 10.1007/s00401-016-1549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Kunii K, et al. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- 636.Takeda M, et al. AZD2171 shows potent antitumor activity against gastric cancer over-expressing fibroblast growth factor receptor 2/keratinocyte growth factor receptor. Clin. Cancer Res. 2007;13:3051–3057. doi: 10.1158/1078-0432.CCR-06-2743. [DOI] [PubMed] [Google Scholar]
- 637.Dutt A, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl Acad. Sci. USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Wang Y, et al. Antitumor effect of FGFR inhibitors on a novel cholangiocarcinoma patient derived xenograft mouse model endogenously expressing an FGFR2-CCDC6 fusion protein. Cancer Lett. 2016;380:163–173. doi: 10.1016/j.canlet.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Rizvi S, et al. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018;15:95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Sobhani, N. et al. Current status of fibroblast growth factor receptor-targeted therapies in breast cancer. Cells7, 76 (2018). [DOI] [PMC free article] [PubMed]
- 641.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Nord H, et al. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. Int. J. Cancer. 2010;126:1390–1402. doi: 10.1002/ijc.24954. [DOI] [PubMed] [Google Scholar]
- 643.Cappellen D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 644.Mahe, M. et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol. Med. 10, e8163 (2018). [DOI] [PMC free article] [PubMed]
- 645.Rosty C, et al. Clinical and biological characteristics of cervical neoplasias with FGFR3 mutation. Mol. Cancer. 2005;4:15. doi: 10.1186/1476-4598-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Hernández, S. et al. FGFR3 mutations in prostate cancer: association with low-grade tumors. Mod. Pathol.22, 848–856 (2009). [DOI] [PubMed]
- 647.Goriely A, et al. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat. Genet. 2009;41:1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Chandrani P, et al. Drug-sensitive FGFR3 mutations in lung adenocarcinoma. Ann. Oncol. 2017;28:597–603. doi: 10.1093/annonc/mdw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Navid, S. et al. The fibroblast growth factor receptors in breast cancer: from oncogenesis to better treatments. Int. J. Mol. Sci. 21, 2011 (2020). [DOI] [PMC free article] [PubMed]
- 650.Taylor JG, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Bange J, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–847. [PubMed] [Google Scholar]
- 652.Maerz WJ, et al. FGF4 dissociates anti-tumorigenic from differentiation signals of retinoic acid in human embryonal carcinomas. Oncogene. 1998;17:761–767. doi: 10.1038/sj.onc.1201992. [DOI] [PubMed] [Google Scholar]
- 653.Giulianelli S, et al. FGF2 induces breast cancer growth through ligand-independent activation and recruitment of ERalpha and PRBDelta4 isoform to MYC regulatory sequences. Int. J. Cancer. 2019;145:1874–1888. doi: 10.1002/ijc.32252. [DOI] [PubMed] [Google Scholar]
- 654.Hamamoto J, et al. The FGF2 aptamer inhibits the growth of FGF2-FGFR pathway driven lung cancer cells. Biochem. Biophys. Res. Commun. 2018;503:1330–1334. doi: 10.1016/j.bbrc.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 655.Yasuda K, et al. Fibroblasts induce expression of FGF4 in ovarian cancer stem-like cells/cancer-initiating cells and upregulate their tumor initiation capacity. Lab. Invest. 2014;94:1355–1369. doi: 10.1038/labinvest.2014.122. [DOI] [PubMed] [Google Scholar]
- 656.Fang F, et al. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J. Hepatol. 2015;63:874–885. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 657.Chang, M. M. et al. FGF9/FGFR2 increase cell proliferation by activating ERK 1/2, Rb/E2F1, and cell cycle pathways in mouse Leydig tumor cells. Cancer Sci.109, 3503–3518 (2018). [DOI] [PMC free article] [PubMed]
- 658.Desnoyers LR, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- 659.Wang H, et al. Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J. Clin. Invest. 2011;121:3220–3232. doi: 10.1172/JCI41514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Teng Y, et al. FGF19 protects hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4-GSK3beta-Nrf2 signaling. Cancer Res. 2017;77:6215–6225. doi: 10.1158/0008-5472.CAN-17-2039. [DOI] [PubMed] [Google Scholar]
- 661.Gao, L. et al. FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene38, 2394–2404 (2019). [DOI] [PubMed]
- 662.Freeman KW, et al. Conditional activation of fibroblast growth factor receptor (FGFR) 1, but not FGFR2, in prostate cancer cells leads to increased osteopontin induction, extracellular signal-regulated kinase activation, and in vivo proliferation. Cancer Res. 2003;63:6237–6243. [PubMed] [Google Scholar]
- 663.Kim S, et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PLoS ONE. 2013;8:e51671. doi: 10.1371/journal.pone.0051671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Martinez-Torrecuadrada J, et al. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin. Cancer Res. 2005;11:6280–6290. doi: 10.1158/1078-0432.CCR-05-0282. [DOI] [PubMed] [Google Scholar]
- 665.Henriksson ML, et al. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am. J. Pathol. 2011;178:1387–1394. doi: 10.1016/j.ajpath.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Ropiquet F, et al. FGF7/KGF triggers cell transformation and invasion on immortalised human prostatic epithelial PNT1A cells. Int. J. Cancer. 1999;82:237–243. doi: 10.1002/(sici)1097-0215(19990719)82:2<237::aid-ijc14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 667.Huang T, et al. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int. J. Oncol. 2017;50:1501–1512. doi: 10.3892/ijo.2017.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Sun C, et al. FGF9 from cancer-associated fibroblasts is a possible mediator of invasion and anti-apoptosis of gastric cancer cells. BMC Cancer. 2015;15:333. doi: 10.1186/s12885-015-1353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Nomura S, et al. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br. J. Cancer. 2008;99:305–313. doi: 10.1038/sj.bjc.6604473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Gauglhofer C, et al. Up-regulation of the fibroblast growth factor 8 subfamily in human hepatocellular carcinoma for cell survival and neoangiogenesis. Hepatology. 2011;53:854–864. doi: 10.1002/hep.24099. [DOI] [PubMed] [Google Scholar]
- 671.Wei W, et al. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J. Clin. Invest. 2013;123:4435–4448. doi: 10.1172/JCI70625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672.Harding TC, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci. Transl. Med. 2013;5:178ra139. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 673.Wang Y, Becker D. Antisense targeting of basic fibroblast growth factor and fibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat. Med. 1997;3:887–893. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 674.Sharma B, et al. Antisense targeting of perlecan blocks tumor growth and angiogenesis in vivo. J. Clin. Invest. 1998;102:1599–1608. doi: 10.1172/JCI3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Vacca A, et al. Bone marrow neovascularization, plasma cell angiogenic potential, and matrix metalloproteinase-2 secretion parallel progression of human multiple myeloma. Blood. 1999;93:3064–3073. [PubMed] [Google Scholar]
- 676.Czubayko F, et al. A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nat. Med. 1997;3:1137–1140. doi: 10.1038/nm1097-1137. [DOI] [PubMed] [Google Scholar]
- 677.Iruela-Arispe ML, et al. Inhibition of angiogenesis by thrombospondin-1 is mediated by 2 independent regions within the type 1 repeats. Circulation. 1999;100:1423–1431. doi: 10.1161/01.cir.100.13.1423. [DOI] [PubMed] [Google Scholar]
- 678.Cervenak L, et al. Abolished angiogenicity and tumorigenicity of Burkitt lymphoma by interleukin-10. Blood. 2000;96:2568–2573. [PubMed] [Google Scholar]
- 679.Aigner A, et al. Marked increase of the growth factors pleiotrophin and fibroblast growth factor-2 in serum of testicular cancer patients. Ann. Oncol. 2003;14:1525–1529. doi: 10.1093/annonc/mdg416. [DOI] [PubMed] [Google Scholar]
- 680.Fahmy RG, et al. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat. Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- 681.Graeven U, et al. Modulation of angiogenesis and tumorigenicity of human melanocytic cells by vascular endothelial growth factor and basic fibroblast growth factor. Cancer Res. 2001;61:7282–7290. [PubMed] [Google Scholar]
- 682.Rofstad EK, Halsor EF. Vascular endothelial growth factor, interleukin 8, platelet-derived endothelial cell growth factor, and basic fibroblast growth factor promote angiogenesis and metastasis in human melanoma xenografts. Cancer Res. 2000;60:4932–4938. [PubMed] [Google Scholar]
- 683.Mukdsi JH, et al. Pattern of FGF-2 isoform expression correlated with its biological action in experimental prolactinomas. Acta Neuropathol. 2006;112:491–501. doi: 10.1007/s00401-006-0101-9. [DOI] [PubMed] [Google Scholar]
- 684.Birrer MJ, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J. Clin. Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 685.Nissen LJ, et al. Angiogenic factors FGF2 and PDGF-BB synergistically promote murine tumor neovascularization and metastasis. J. Clin. Invest. 2007;117:2766–2777. doi: 10.1172/JCI32479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Pietras K, Pahler J, Bergers G, Hanahan D. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med. 2008;5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Suh JM, et al. Endocrinization of FGF1 produces a neomorphic and potent insulin sensitizer. Nature. 2014;513:436–439. doi: 10.1038/nature13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Zhang J, Li Y. Therapeutic uses of FGFs. Semin. Cell Dev. Biol. 2016;53:144–154. doi: 10.1016/j.semcdb.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 689.Nixon, J. R. The multiple synostoses syndrome. A plea for simplicity. Clin.Orthop. Relat. Res.135, 48–51 (1978). [PubMed]
- 690.Han DS, et al. Keratinocyte growth factor-2 (FGF-10) promotes healing of experimental small intestinal ulceration in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;279:G1011–G1022. doi: 10.1152/ajpgi.2000.279.5.G1011. [DOI] [PubMed] [Google Scholar]
- 691.Freytes CO, et al. Phase I/II randomized trial evaluating the safety and clinical effects of repifermin administered to reduce mucositis in patients undergoing autologous hematopoietic stem cell transplantation. Clin. Cancer Res. 2004;10:8318–8324. doi: 10.1158/1078-0432.CCR-04-1118. [DOI] [PubMed] [Google Scholar]
- 692.Degirolamo C, Sabba C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat. Rev. Drug Discov. 2016;15:51–69. doi: 10.1038/nrd.2015.9. [DOI] [PubMed] [Google Scholar]
- 693.Wu X, et al. Separating mitogenic and metabolic activities of fibroblast growth factor 19 (FGF19) Proc. Natl Acad. Sci. USA. 2010;107:14158–14163. doi: 10.1073/pnas.1009427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Wu AL, et al. FGF19 regulates cell proliferation, glucose and bile acid metabolism via FGFR4-dependent and independent pathways. PLoS ONE. 2011;6:e17868. doi: 10.1371/journal.pone.0017868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Zhou M, et al. Separating tumorigenicity from bile acid regulatory activity for endocrine hormone FGF19. Cancer Res. 2014;74:3306–3316. doi: 10.1158/0008-5472.CAN-14-0208. [DOI] [PubMed] [Google Scholar]
- 696.Kharitonenkov A, et al. Rational design of a fibroblast growth factor 21-based clinical candidate, LY2405319. PLoS ONE. 2013;8:e58575. doi: 10.1371/journal.pone.0058575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Gaich G, et al. The effects of LY2405319, an FGF21 analog, in obese human subjects with type 2 diabetes. Cell Metab. 2013;18:333–340. doi: 10.1016/j.cmet.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 698.Huang Z, et al. A better anti-diabetic recombinant human fibroblast growth factor 21 (rhFGF21) modified with polyethylene glycol. PLoS ONE. 2011;6:e20669. doi: 10.1371/journal.pone.0020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Camacho RC, et al. Pegylated Fgf21 rapidly normalizes insulin-stimulated glucose utilization in diet-induced insulin resistant mice. Eur. J. Pharm. 2013;715:41–45. doi: 10.1016/j.ejphar.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 700.Xu J, et al. Polyethylene glycol modified FGF21 engineered to maximize potency and minimize vacuole formation. Bioconjug. Chem. 2013;24:915–925. doi: 10.1021/bc300603k. [DOI] [PubMed] [Google Scholar]
- 701.Song L, et al. A solid-phase PEGylation strategy for protein therapeutics using a potent FGF21 analog. Biomaterials. 2014;35:5206–5215. doi: 10.1016/j.biomaterials.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 702.Veniant MM, et al. Long-acting FGF21 has enhanced efficacy in diet-induced obese mice and in obese rhesus monkeys. Endocrinology. 2012;153:4192–4203. doi: 10.1210/en.2012-1211. [DOI] [PubMed] [Google Scholar]
- 703.Hecht R, et al. Rationale-based engineering of a potent long-acting FGF21 analog for the treatment of type 2 diabetes. PLoS ONE. 2012;7:e49345. doi: 10.1371/journal.pone.0049345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Huang J, et al. Development of a novel long-acting antidiabetic FGF21 mimetic by targeted conjugation to a scaffold antibody. J. Pharm. Exp. Ther. 2013;346:270–280. doi: 10.1124/jpet.113.204420. [DOI] [PubMed] [Google Scholar]
- 705.Weng Y, et al. Pharmacokinetics (PK), pharmacodynamics (PD) and integrated PK/PD modeling of a novel long acting FGF21 clinical candidate PF-05231023 in diet-induced obese and leptin-deficient obese mice. PLoS ONE. 2015;10:e0119104. doi: 10.1371/journal.pone.0119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.Soria JC, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann. Oncol. 2014;25:2244–2251. doi: 10.1093/annonc/mdu390. [DOI] [PubMed] [Google Scholar]
- 707.Awasthi N, Schwarz RE. Profile of nintedanib in the treatment of solid tumors: the evidence to date. Onco Targets Ther. 2015;8:3691–3701. doi: 10.2147/OTT.S78805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Carter EP, Fearon AE, Grose RP. Careless talk costs lives: fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol. 2015;25:221–233. doi: 10.1016/j.tcb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 709.Porta C, Giglione P, Liguigli W, Paglino C. Dovitinib (CHIR258, TKI258): structure, development and preclinical and clinical activity. Fut. Oncol. 2015;11:39–50. doi: 10.2217/fon.14.208. [DOI] [PubMed] [Google Scholar]
- 710.Andre F, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin. Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 711.Trudel S, et al. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105:2941–2948. doi: 10.1182/blood-2004-10-3913. [DOI] [PubMed] [Google Scholar]
- 712.Porta R, et al. FGFR a promising druggable target in cancer: molecular biology and new drugs. Crit. Rev. Oncol. Hematol. 2017;113:256–267. doi: 10.1016/j.critrevonc.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 713.Gavine PR, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 714.Maehara O, et al. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis. 2017;38:1073–1083. doi: 10.1093/carcin/bgx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Zhao Q, et al. FGFR inhibitor, AZD4547, impedes the stemness of mammary epithelial cells in the premalignant tissues of MMTV-ErbB2 transgenic mice. Sci. Rep. 2017;7:11306. doi: 10.1038/s41598-017-11751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Nogova L, et al. Evaluation of BGJ398, a fibroblast growth factor receptor 1–3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase I, dose-escalation and dose-expansion study. J. Clin. Oncol. 2017;35:157–165. doi: 10.1200/JCO.2016.67.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Perera TPS, et al. Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol. Cancer Ther. 2017;16:1010–1020. doi: 10.1158/1535-7163.MCT-16-0589. [DOI] [PubMed] [Google Scholar]
- 718.Tabernero J, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2015;33:3401–3408. doi: 10.1200/JCO.2014.60.7341. [DOI] [PubMed] [Google Scholar]
- 719.Di Stefano AL, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin. Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Sohl CD, et al. Illuminating the molecular mechanisms of tyrosine kinase inhibitor resistance for the FGFR1 gatekeeper mutation: the Achilles' heel of targeted therapy. ACS Chem. Biol. 2015;10:1319–1329. doi: 10.1021/acschembio.5b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Lau KH, et al. Opposing effects of Sca-1(+) cell-based systemic FGF2 gene transfer strategy on lumbar versus caudal vertebrae in the mouse. Gene Ther. 2016;23:500–509. doi: 10.1038/gt.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Byron SA, et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia. 2013;15:975–988. doi: 10.1593/neo.121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Goyal L, et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 2017;7:252–263. doi: 10.1158/2159-8290.CD-16-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Aono Y, et al. Therapeutic effects of anti-FGF23 antibodies in hypophosphatemic rickets/osteomalacia. J. Bone Miner. Res. 2009;24:1879–1888. doi: 10.1359/jbmr.090509. [DOI] [PubMed] [Google Scholar]
- 725.Tolcher AW, et al. A phase I, first in human study of FP-1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Ann. Oncol. 2016;27:526–532. doi: 10.1093/annonc/mdv591. [DOI] [PubMed] [Google Scholar]
- 726.Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 727.Giorgio C, et al. Pharmacological evaluation of new bioavailable small molecules targeting Eph/ephrin interaction. Biochem. Pharmacol. 2018;147:21–29. doi: 10.1016/j.bcp.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 728.Colombo G, et al. Non-peptidic thrombospondin-1 mimics as fibroblast growth factor-2 inhibitors: an integrated strategy for the development of new antiangiogenic compounds. J. Biol. Chem. 2010;285:8733–8742. doi: 10.1074/jbc.M109.085605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Pagano K, et al. Direct and allosteric inhibition of the FGF2/HSPGs/FGFR1 ternary complex formation by an antiangiogenic, thrombospondin-1-mimic small molecule. PLoS ONE. 2012;7:e36990. doi: 10.1371/journal.pone.0036990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Camozzi M, et al. Identification of an antiangiogenic FGF2-binding site in the N terminus of the soluble pattern recognition receptor PTX3. J. Biol. Chem. 2006;281:22605–22613. doi: 10.1074/jbc.M601023200. [DOI] [PubMed] [Google Scholar]
- 731.Leali D, et al. Fibroblast growth factor 2-antagonist activity of a long-pentraxin 3-derived anti-angiogenic pentapeptide. J. Cell Mol. Med. 2010;14:2109–2121. doi: 10.1111/j.1582-4934.2009.00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Castelli R, et al. Synthesis, structural elucidation, and biological evaluation of NSC12, an orally available fibroblast growth factor (FGF) ligand Trap for the treatment of FGF-dependent lung tumors. J. Med. Chem. 2016;59:4651–4663. doi: 10.1021/acs.jmedchem.5b02021. [DOI] [PubMed] [Google Scholar]
- 733.Yokota M, et al. Therapeutic effect of nanogel-based delivery of soluble FGFR2 with S252W mutation on craniosynostosis. PLoS ONE. 2014;9:e101693. doi: 10.1371/journal.pone.0101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Morita J, et al. Soluble form of FGFR2 with S252W partially prevents craniosynostosis of the apert mouse model. Dev. Dyn. 2014;243:560–567. doi: 10.1002/dvdy.24099. [DOI] [PubMed] [Google Scholar]
- 735.Garcia S, et al. Postnatal soluble FGFR3 therapy rescues achondroplasia symptoms and restores bone growth in mice. Sci. Transl. Med. 2013;5:203ra124. doi: 10.1126/scitranslmed.3006247. [DOI] [PubMed] [Google Scholar]
- 736.Jin M, et al. A novel FGFR3-binding peptide inhibits FGFR3 signaling and reverses the lethal phenotype of mice mimicking human thanatophoric dysplasia. Hum. Mol. Genet. 2012;21:5443–5455. doi: 10.1093/hmg/dds390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Whitsett JA, et al. Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. J. Biol. Chem. 2002;277:22743–22749. doi: 10.1074/jbc.M202253200. [DOI] [PubMed] [Google Scholar]
- 738.Nikol S, et al. Therapeutic angiogenesis with intramuscular NV1FGF improves amputation-free survival in patients with critical limb ischemia. Mol. Ther. 2008;16:972–978. doi: 10.1038/mt.2008.33. [DOI] [PubMed] [Google Scholar]
- 739.Shukla V, et al. RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nat. Genet. 2007;39:1145–1150. doi: 10.1038/ng2096. [DOI] [PubMed] [Google Scholar]
- 740.McDowell LM, et al. Inhibition or activation of Apert syndrome FGFR2 (S252W) signaling by specific glycosaminoglycans. J. Biol. Chem. 2006;281:6924–6930. doi: 10.1074/jbc.M512932200. [DOI] [PubMed] [Google Scholar]
- 741.Valdmanis PN, Kay MA. Future of rAAV gene therapy: platform for RNAi, gene editing, and beyond. Hum. Gene Ther. 2017;28:361–372. doi: 10.1089/hum.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 743.Luo F, et al. Adeno-associated virus-mediated RNAi against mutant alleles attenuates abnormal calvarial phenotypes in an Apert Syndrome Mouse Model. Mol. Ther. Nucleic Acids. 2018;13:291–302. doi: 10.1016/j.omtn.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Yang Y, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Wu Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 746.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Ou Z, et al. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human beta-thalassemia in mice. Sci. Rep. 2016;6:32463. doi: 10.1038/srep32463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Miao K, et al. Optimizing CRISPR/Cas9 technology for precise correction of the Fgfr3-G374R mutation in achondroplasia in mice. J. Biol. Chem. 2019;294:1142–1151. doi: 10.1074/jbc.RA118.006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Vasudevan HN, Soriano P. A Thousand and One Receptor Tyrosine Kinases: Wherein the Specificity? Curr.Top. Dev. Biol. 2016;117:393–404. doi: 10.1016/bs.ctdb.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Xie Y, et al. Intermittent PTH (1-34) injection rescues the retarded skeletal development and postnatal lethality of mice mimicking human achondroplasia and thanatophoric dysplasia. Hum. Mol. Genet. 2012;21:3941–3955. doi: 10.1093/hmg/dds181. [DOI] [PubMed] [Google Scholar]