Abstract
Aims
Licorice (Glycyrrhiza uralensis Fisch.) is a crucial medicinal herb as it accumulates glycyrrhizin and liquiritin in roots. Licorice root-associated bacterial communities shaped by soil characteristics are supposed to regulate the accumulation of root secondary metabolites.
Methods
The soil characteristics, root secondary metabolites, and root-associated bacterial communities were analyzed in licorice plants of different ages to explore their temporal dynamics and interaction mechanisms.
Results
Temporal variation in soil characteristics and root secondary metabolites was distinct. The alpha-diversity of root-associated bacterial communities decreased with root proximity, and the community composition was clustered in the rhizosphere. Different taxa that were core-enriched from the dominant taxa in the bulk soil, rhizosphere soil, and root endosphere displayed varied time–decay relationships. Soil total potassium (TK) as a key factor regulated the temporal variation in some individual taxa in the bulk and rhizosphere soils; these taxa were associated with the adjustment of root secondary metabolites across different TK levels.
Conclusions
Licorice specifically selects root-associated core bacteria over the course of plant development, and TK is correlated with root secondary metabolites and individual core-enriched taxa in the bulk and rhizosphere soils, which may have implications for practical licorice cultivation.
Electronic supplementary material
The online version of this article (10.1007/s11104-020-04692-0) contains supplementary material, which is available to authorized users.
Keywords: Soil nutrients, Glycyrrhizin, Liquiritin, Bacterial community, Temporal dynamics, Glycyrrhiza uralensis Fisch
Introduction
Licorice (Glycyrrhiza) is a genus of herbaceous perennial plants in the family Leguminosae, with approximately 20 species known as popular medicinal plants in arid and semi-arid regions worldwide (Lewis et al., 2005; Xie et al. 2018). Glycyrrhiza uralensis Fisch. is the most common species of licorice and its roots contain two major secondary metabolites, glycyrrhizin and liquiritin, which belong to triterpene saponins and flavonoids, respectively (Hayashi and Sudo 2009). Licorice roots are widely used in the food, beverage, and cosmetics industries (Zhang and Ye 2009). The roots are also used in the pharmaceutical industry for antiviral (human immunodeficiency virus [HIV] and severe acute respiratory syndrome [SARS]) and antiulcer medication, and for regulating blood coagulation and thrombosis (Cinatl et al. 2003; Hosseinzadeh and Nassiri-Asl 2016). Wild licorice have been exhausted and are gradually being replaced by cultivated licorice, which decreases the quality of licorice (Chen et al. 2017).
The growth of plants and production of secondary metabolites are intimately related to microbial communities. Therefore, the dynamics of the microbiome associated with medicinal plants have been explored extensively (Huang et al. 2018; Koberl et al. 2013). There are several studies that have reported the beneficial effects of arbuscular mycorrhizal symbiosis on plant growth and secondary metabolite accumulations in the roots of licorice (Chen et al. 2017; Xie et al. 2018). Other studies have shown that the growth and efficacy of the medicinal herb Danshen (Salvia miltiorrhiza) can be promoted by beneficial plant-associated microbes, such as those of the Pantoea, Pseudomonas, and Sphingomonas genera (Huang et al. 2018). However, to our knowledge, there is no information available on licorice root-associated bacterial communities and their links with secondary metabolites.
The temporal dynamics of soil microbial communities are usually investigated in chronosequences of land restoration in natural conditions (Lauber et al. 2013; Liu et al. 2019) and different developmental stages of crop plants root microbiota (Zhang et al. 2018). A vital indicator for the dynamics of microbial succession is the temporal turnover of the time–decay relationships, which have been used to describe decreasing community similarities over time (Nekola and White 1999). So far, the time–decay turnover patterns have not yet been investigated in root-associated bacterial communities. Additionally, the structural and functional variability in root-associated microbiomes have been studied in some perennial plants, such as ryegrass (Lolium perenne), Drummond’s rockcress (Boechera stricta), alpine rockcress (Arabis alpina), and fruit (Citrus) (Chen et al. 2016; Dombrowski et al. 2017; Wagner et al. 2016; Xu et al. 2018). However, there is currently a lack of research on the temporal dynamics of root-associated bacterial communities in perennial licorice.
Soil represents the most diverse ecosystem on earth with an exceptionally high microbial diversity (Fierer and Jackson 2006). During the growth process, plants select and enrich microorganisms from bulk soil to root-associated compartments (such as the rhizosphere and endosphere), each of which harbors a distinct microbiome (Edwards et al. 2015). The rhizosphere soil is profoundly influenced by the secretion of plant root exudates, which accordingly results in a ‘hotspot’ environment (Peiffer et al. 2013; Schlaeppi et al. 2014). In contrast, the root endosphere features a highly specific microbiome that is mainly affected by plant genotype (Vandenkoornhuyse et al. 2015). Several studies have reported on the root-associated dominant or core bacteria using Arabidopsis thaliana as the model plant (Bulgarelli et al. 2012; Lundberg et al. 2012). Information on the core microbiota is critical for understanding the assembly and stability of root-associated microbial communities. Licorice plants of different ages may be accompanied by consecutively and specifically enriched bacterial communities among distinct soil–root compartments. However, the selective core enrichment process of root-associated bacterial communities in licorice has rarely been investigated.
Although root-associated microbiota have been investigated for a long time, there is still little agreement on how these communities are shaped and what factors regulate their composition (Bulgarelli et al. 2012; Fan et al. 2018). The temporal dynamics of microbial communities in natural environments are influenced by multiple factors simultaneously. Therefore, it is difficult to determine the relative contributions made by individual factors to the overall microbial succession process (Jiao et al. 2017). The assembly and composition of the root-associated microbiome could be influenced by soil conditions such as the pH, nitrogen and phosphorus contents (Edwards et al. 2015; Xiao et al. 2017). Therefore, it is desirable to obtain a predictive understanding of how root-associated bacterial communities in licorice plants of different ages respond to soil characteristics. In this context, variation in soil characteristics including primary nutrients, soil texture, and micronutrients, is essential for normal growth and secondary metabolite production in plants (Fageria et al. 2002). Therefore, understanding how soil variables regulate the individual taxa of the microbial communities associated with the accumulation of root secondary metabolites in licorice is crucial for the optimization of plant–soil interactions for practical licorice cultivation.
Many studies have analyzed soil microbial communities using Illumina paired-end sequencing of 16S rRNA and internal transcribed spacer (ITS) amplicons (Jiao et al. 2017; Xiao et al. 2017). In this study, single-end sequencing was adopted using the IonS5™XL platform. This provided longer sequences to read without splicing (Mehrotra et al. 2017), and thus could help to elucidate the root-associated bacterial communities more comprehensively and accurately. The aims of this study were to (1) elucidate the temporal dynamics of root-associated bacterial communities together with variation in soil characteristics and secondary metabolite concentrations in roots; (2) investigate the core-enriched taxa and their time–decay relationships; and (3) provide a comprehensive understanding of the key factor(s) regulating the temporal dynamics of individual taxa related to root secondary metabolites in licorice.
Materials and methods
Study site
The study area is located in the Yuzhong North Mountains (104°18′–104°19′ E, 36°8′–36°9’ N), in Lanzhou, Gansu province (~150 km from downtown Lanzhou) in northeast China. This area has an altitude of 2400–2500 m in the mountains and it is characterized by a temperate semi-arid continental climate. It is known as the cradle and cultivation base of licorice (Glycyrrhiza uralensis Fisch.) with wide distributions of licorice plants. Its average annual and accumulated temperatures are 6.7 °C and 2754–3285 °C, respectively. The average annual sunshine duration is 2500–2600 h and average annual precipitation is 400 mm. The soil is classified as loessial (Calcaric Cambisol according to FAO classification). The licorice species in the study area were all identified as G. uralensis Fisch. The general fertilization regime was conducted with nitrogen (urea: 85 kg ha−1), phosphorus (P2O5: 40 kg ha−1), and potassium (KCl: 35 kg ha−1). The cultivated sites were subjected to similar fertilization and management practices, with close distance (< 500 m) from the wild site and each other.
Experimental design and sampling strategy
One-year-old uniform licorice seedlings were transplanted from a nursery into cultivated sites for one to four years until sampling in July 2018 (the transplanting was staggered so that all samples were collected in the same year). Licorice seedlings were properly cultivated after transplantation, but the plants that had been cultivated for three years had been excavated ahead of our sampling. Therefore, the samples selected from the cultivated sites were referred to as 1-y, 2-y, and 4-y (Supplementary Table S1). According to the descriptions of local residents, the wild licorice had been growing for over 10 years.
The sampling comprised five plots as biological replicates following a Z-shaped pattern in each site, where licorice grew evenly. For each plot, five random soil cores from 0 to 20 cm depth and with a 10 cm diameter were collected and mixed to form a single sample as bulk soil. A group of three to five licorice plants were excavated by digging around the group to keep the roots as intact as possible in each plot. All samples were placed into sterile plastic bags, kept on dry ice, and taken to the laboratory immediately. Bulk soils were homogenized by passing the samples through a 2 mm sieve. The soils were then divided into two subsamples. Licorice roots were processed to obtain the rhizosphere soil according to a previously described method (Xiao et al. 2017). The scrubbed roots (the root endosphere) were also divided into two subsamples. Subsample of the bulk soils and the licorice roots were used for the analysis of soil characteristics and root secondary metabolites, respectively. The remaining (sub)samples were stored at −80 °C in preparation for microbial analysis. In total, 60 samples (four sites × three soil–root compartments × five plots) were obtained for further processing.
Analysis of soil characteristics and root secondary metabolites
Bulk soil characteristics were divided into three groups including primary nutrients, micronutrients, and texture. Soil pH was determined using a routine method with a fresh soil to water ratio of 1:5. The soil water content (SWC) was measured gravimetrically by drying 5 g fresh soil until the soil reached a constant weight. For the analysis of soil organic matter (SOM), total carbon (TC), total nitrogen (TN), total phosphorus (TP), and total potassium (TK), the samples were air-dried and sieved (1 mm mesh), and the contents were determined by combustion (Fan et al. 2018). The soil texture was tested using a laser particle sizer (LS13320, Beckman Coulter, USA) with air-dried soil (Fan et al. 2018). For the soil available nitrogen (AN), phosphorus (AP), and potassium (AK) analyses, the samples were dispersed or extracted, and the contents were determined using an atomic absorption spectrometer (Fan et al. 2018). Specifically, the TN was quantified following the Kjeldahl method and the AN was quantified by extraction with 1 mol L−1 KCl, steam distillation, and titration following the Alkaline diffusion method. The TP was extracted using 1 N HCl after ignition at 550 °C, the AP were extracted using 0.5 mol L−1 NaHCO3 (pH = 8.5), and then they were measured following the Mo-Sb colorimetric method. The TK was digested in a nickel crucible with sodium hydroxide at 750 °C, the AK was extracted using 1 mol L−1 ammonium acetate, and then they were measured by using flame photometry.
Soil micronutrients including copper (Cu), iron (Fe), and zinc (Zn) were analyzed using atomic absorption spectroscopy (Fageria et al. 2002). Moreover, three representative root secondary metabolites in licorice were analyzed, namely glycyrrhizin, liquiritin, and isoliquiritin (liquiritin isomer). Chromatography–mass spectrometry was performed according to standard procedure (Zhang and Ye 2009) with slight modifications. The detailed method is available in Supplementary S1.
DNA extraction, high-throughput sequencing, and bioinformatic analysis
The total DNA was extracted from bulk and rhizosphere soil samples (0.5 g each) using the Fast DNA®SPIN Kit (MP Biochemicals, Solon, USA). The total DNA was extracted from the root samples (0.5 g each) using a DNA secure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s procedures. The DNA concentration and purity were estimated using a Nanodrop 1000 spectrophotometer (Thermofisher Inc., Wilmington, Germany) and electrophoresis in 1% (w/v) agarose gel (Xiao et al. 2017). The hypervariable V4-V5 region of the 16S rRNA gene was amplified using the primers pair 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′) (Edwards et al. 2015). The polymerase chain reaction (PCR) amplifications were performed in triplicate for each sample, and PCR products were mixed in equidensity ratios. Then, the mixed PCR products were purified using a GeneJET™ Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using Ion Plus Fragment Library Kit (48 rxns; Thermo Scientific). The library quality was assessed using the Qubit@ 2.0 Fluorometer (Thermo Scientific). Finally, the library was sequenced on an IonS5™XL platform (Thermofisher Inc., Massachusetts, USA) and 400 bp single-end reads were generated by Novogene (Beijing, China).
Low-quality sequences were sheared using Cutadapt (Martin 2011) and quality-filtered using the QIIME pipeline (v1.7.0) (Caporaso et al. 2010). After the removal of chimeric sequences using the USEARCH tool in the UCHIME algorithm (Edgar et al. 2011), the remaining sequences were assigned to operational taxonomic units (OTUs) at similarities of 97% using the UPARSE pipeline (Edgar et al. 2011). OTUs lacking more than two sequences were removed. Taxonomic information was annotated for a representative sequence of each OTU using the RDP classifier at a confidence level of 80% (Wang et al. 2007) using the SILVA database release 128.
Statistical analysis
All statistical analyses were conducted in the R environment (v3.6.1; http://www.r-project.org/). Most of the results were visualized using the ‘ggplot2’ package (Gómez Rubio 2017), unless otherwise indicated. The soil characteristics and root secondary metabolite concentrations were log-transformed before the significance of factors (plant sites) was tested by analysis of variance (ANOVA), followed by comparisons of means using Tukey HSD parametric tests (‘multcomp’ package) (Hothorn et al. 2008). Principal component analysis (PCA) was used to investigate the distribution of these variables on a scaled parameter matrix and visualized using the ‘FactoMineR’ package (Lê et al. 2008). The Spearman’s correlation of these variables was analyzed and visualized using the ‘corrplot’ package (Wei and Simko 2013).
Alpha-diversity indices were calculated using QIIME (v1.7.0) and significant differences were found using Kruskal-Wallis non-parametric tests along with ANOVA (‘agricolae’ package) (De Mendiburu 2014). Nonmetric multidimensional scaling (NMDS) was implemented, with significant differences in bacterial community composition tested by three different but complementary non-parametric multivariate statistical analysis methods (‘vegan’ package) (Oksanen et al. 2007): permutational multivariate analysis of variance using distance matrices (Adonis), analysis of similarities (Anosim), and multiple response permutation procedure (MRPP). Heatmaps were illustrated based on Z-score-normalized relative abundance of taxa using the ‘pheatmap’ package (Kolde 2015). A circos plot was created based on the relative abundance of taxa using the ‘circlize’ package (Gu et al. 2014). Boxplots were used to illustrate the variation in alpha- and beta-diversity values.
The most common and ubiquitous bacterial taxa across all sites were identified as dominant taxa using the criteria of a recent study (Delgado-Baquerizo et al. 2018) with slight modifications. Analysis of the differential taxa was performed using a negative binomial generalized linear model in the ‘edgeR’ package (Nikolayeva and Robinson 2014). The OTU counts from bulk soil were taken as a control, and corresponding P-values were corrected for multiple tests using a false discovery rate (FDR) set at 0.05 (Benjamini and Hochberg 1995). Core-enriched taxa were selected from overlapping OTUs in Venn diagrams using the ‘Venndiagram’ package (Chen and Boutros 2011). Ternary plots were visualized to show the distribution of these taxa using the ‘ggtern’ package (Hamilton 2015). The functions of these taxa were annotated using FAPROTAX (Louca et al. 2016). Time–decay relationships were estimated by fitting a linear model between the changes in community similarity (based on 1 – [Bray-Curtis dissimilarity]) and the age of licorice plants; the slopes were used to measure the rate of community turnover (Nekola and White 1999).
The correlations among the matrices of the three compartment communities, soil characteristics, and secondary metabolites in the roots were examined using the Mantel test (‘vegan’ package). Multiple regression on distance matrices (MRM) analysis and the random forest (RF) regression algorithm were used to investigate the effect level of significantly correlated variables on bacterial communities (assessed by Bray-Curtis distance and Beta-NMDS1) using the ‘ecodist’ (Goslee and Urban 2007) and ‘rfPermute’ (Jiao et al. 2019) packages, respectively. Linear regression was used to investigate the correlations between log-transformed TK content and the average relative abundance of core-enriched taxa; significant correlations were calculated for each taxon (‘corrplot’ package). Individual taxa were then selected from these significantly correlated taxa again, using the ‘maSigPro’ package (Conesa et al. 2006). These taxa were visualized using curve plots to illustrate their temporal variability; their correlations with secondary metabolites were calculated using the ‘corrplot’ package.
Results
Temporal variation in soil characteristics and root secondary metabolites
The ANOVA results showed that the age of licorice plants had significant effects on most variables (P < 0.05). Most soil characteristics such as SOM, TC, TN, and Cu, significantly increased with increasing age of cultivated plants; the C/N ratio significantly decreased correspondingly (Table 1). Other soil characteristics such as SWC, TP, AP, TK, AK, Zn, silt, and clay also increased, whereas AN, Fe, and sand decreased. In addition, the isoliquiritin concentrations in licorice roots steadily decreased with increasing age of cultivated plants. However, the liquiritin and glycyrrhizin concentrations first decreased and then recovered slightly (Table 2). This trend was similar to the temporal variation in soil AN, TP, and TK levels. Many soil characteristics (SWC, TP, AP, TK, AK, Fe, and silt) occurred at their lowest levels in the wild site that lacked fertilization and agricultural management. However, all three secondary metabolites had higher concentrations in wild than cultivated licorice.
Table 1.
Soil characteristics in sites with different licorice plant ages (n = 5); values are mean ± standard deviation
| Soil primary nutrients | Soil micronutrients | Soil texture | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | pH | SWC (%) | SOM (g/kg) | TC (g/kg) | TN (g/kg) | AN (mg/kg) | C/N | TP (g/kg) | AP (mg/kg) | TK (g/kg) | AK (mg/kg) | Cu (mg/kg) | Fe (g/kg) | Zn (mg/kg) | Sand (%) | Silt (%) | Clay (%) |
| 1-y | 8.66 ± 0.15 a | 0.08 ± 0.03 a | 6.46 ± 0.77 a | 20.54 ± 1.48 a | 0.27 ± 0.09 a | 36.16 ± 24.66 c | 76.07 ± 16.44 c | 0.67 ± 0.03 a | 5.75 ± 4.15 b | 15.57 ± 0.21 b | 95.46 ± 40.97 a | 18.46 ± 0.23 a | 25.2 ± 0.79 b | 57.14 ± 2.28 a | 26.96 ± 3.24 b | 71.05 ± 2.93 a | 1.99 ± 0.34 a |
| 2-y | 9.05 ± 0.02 c | 0.08 ± 0.02 a | 10.15 ± 0.61 b | 24.95 ± 0.4 b | 0.44 ± 0.04 b | 12.12 ± 1.76 b | 56.70 ± 10b | 0.66 ± 0.02 a | 7.14 ± 1.55 bc | 15.42 ± 0.21 b | 144 ± 7.8 a | 19.14 ± 0.55 b | 24.39 ± 1.12 ab | 57.57 ± 2.05 a | 24.74 ± 2.13 b | 73.18 ± 2.27 a | 2.08 ± 0.18 ab |
| 4-y | 8.77 ± 0.01 ab | 0.13 ± 0.01 b | 15.67 ± 3.51 c | 27.71 ± 0.42 c | 0.77 ± 0.07 c | 17.54 ± 1.32 bc | 35.99 ± 6 a | 0.8 ± 0.05 b | 14.12 ± 2.75 c | 16 ± 0.2 c | 302.08 ± 52.33 b | 20.12 ± 0.3 c | 24.36 ± 0.75 ab | 62.25 ± 0.81 b | 19.31 ± 1.63 a | 78.18 ± 1.49 b | 2.51 ± 0.26 b |
| Wild | 8.88 ± 0.02 b | 0.07 ± 0 a | 9.81 ± 0.23 b | 23.51 ± 0.04 b | 0.45 ± 0.01 b | 2.87 ± 0.18 a | 52.24 ± 4 b | 0.64 ± 0.01 a | 2.09 ± 0.1 a | 14.81 ± 0.01 a | 88.52 ± 1.01 a | 19.13 ± 0.11 b | 22.93 ± 1.38 a | 58.45 ± 1.59 a | 26.23 ± 3.32 b | 71.31 ± 3.07 a | 2.46 ± 0.29 ab |
| F | 22.74 | 14.64 | 21.84 | 70.38 | 58.43 | 6.43 | 9.05 | 34.87 | 18.66 | 38.03 | 44.19 | 20.23 | 4.06 | 8.63 | 8.32 | 8.62 | 4.54 |
| P | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.01 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.05 | < 0.01 | < 0.01 | < 0.01 | < 0.05 |
Note: pH, soil pH; SWC, soil water content; SOM, soil organic matter; TC, total carbon; TN, total nitrogen; AN, available nitrogen; C/N, total carbon/nitrogen ratio; TP, total phosphorus; AP, available phosphorus; TK, total potassium; AK, available potassium; Cu, copper; Fe, iron; and Zn, zinc. Values within the same column followed by different letters indicate significant differences (P < 0.05, ANOVA, Tukey HSD)
Table 2.
Concentrations of secondary metabolites in the roots of licorice plants of different ages (n = 5); values are mean ± standard deviation
| Site | Liq (%) | Isoliq (%) | Acid (%) |
|---|---|---|---|
| 1-y | 1.68 ± 0.42 b | 0.34 ± 0.07 ab | 2.41 ± 0.41 ab |
| 2-y | 0.7 ± 0.53 a | 0.26 ± 0.16 a | 1.58 ± 1.19 a |
| 4-y | 0.99 ± 0.14 a | 0.18 ± 0.04 a | 2.11 ± 0.42 ab |
| Wild | 1.82 ± 0.1 b | 0.46 ± 0.05 b | 3.19 ± 0.1 b |
| F | 12.01 | 8.47 | 5.09 |
| P | < 0.001 | < 0.01 | < 0.05 |
Note: Liq, liquiritin; Isoliq, isoliquiritin; and Acid, glycyrrhizin. Values within the same column followed by different letters indicate significant differences (P < 0.05, ANOVA, Tukey HSD)
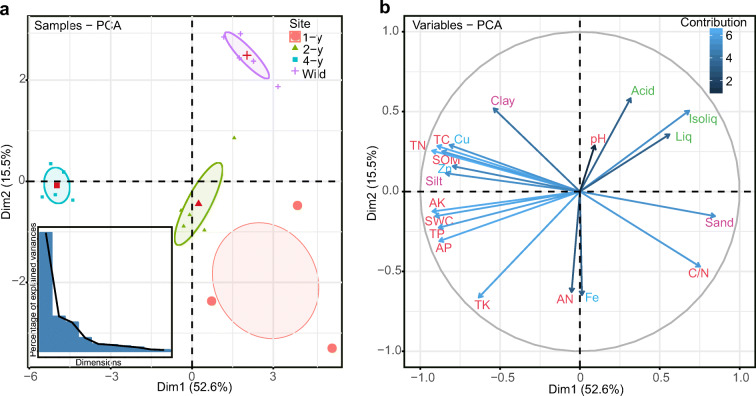
The PCA based on scaled soil characteristics and secondary metabolite concentrations showed that the samples were clearly separated according to the age of licorice plants (Fig. 1A). The contributions of variables to principle components were different and their correlations were determined according to the angles among the variables (Fig. 1B), which were further confirmed by the correlation analysis. The concentrations of the three secondary metabolites were strongly positively correlated with each other, but were significantly negatively correlated with most of the soil primary nutrients (TK, TP, AK, SWC, AP, SOM, TC, and TN). Among these, only TK was related to all three secondary metabolites. Significant correlations were also illustrated between soil characteristics (Supplementary Fig. S1). In general, soil characteristics and root secondary metabolites had varying degrees of temporal variation and exhibited intimate connections.
Fig. 1.
Principal component analysis (PCA) of soil characteristics and secondary metabolites in the roots of licorice plants of different ages. (a) Ordering distribution and interpretation of the first ten principal components. (b) The contribution of the variables to the principal components. The depth of the color of the arrow line represents the degree of contribution. The angle of the arrow line represents correlations. pH, soil pH; SWC, soil water content; SOM, soil organic matter; TC, total carbon; TN, total nitrogen; AN, available nitrogen; C/N, total carbon/nitrogen ratio; TP, total phosphorus; AP, available phosphorus; TK, total potassium; AK, available potassium; Cu, copper; Fe, iron; Zn, zinc; Liq, liquiritin; Isoliq, isoliquiritin; and Acid, glycyrrhizin
Temporal succession of root-associated bacterial communities
Based on the sequencing of 16S rRNA amplicons, 4,464,796 quality reads were obtained (after filtering) from the 60 samples (20 samples per soil–root compartment). The number of reads per sample ranged from 51,305 to 87,560 with an average of 74,413 ± 9175 reads. In total 3,223,764 reads could be classified as bacteria, and most (92.7%) could be classified at the phylum level. A total of 11,430 OTUs were clustered and the number of OTUs per sample ranged from 72 to 3233, with an average of 1626 ± 1087 OTUs. After homogenization, subsequent analysis was carried out based on a minimum of 26,262 reads per sample.
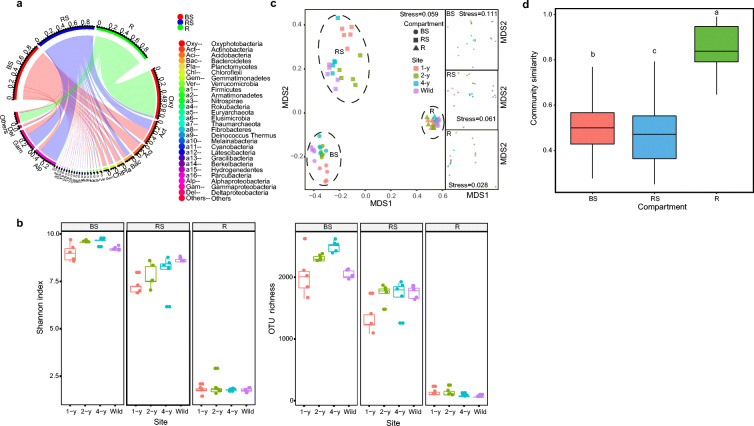
The most of abundant phyla were Oxyphotobacteria (BS: 0.1%; RS: 3.0%; R: 82.6%), Proteobacteria (BS: 21.6%; RS: 60.3%; R: 1.2%), Actinobacteria (BS: 21.7%; RS: 18.9%; R: 0.1%), Acidobacteria (BS: 19.3%; RS: 3.1%; R: 0.01%), Bacteroidetes (BS: 6.9%; RS: 8.5%; R: 0.1%), Planctomycetes (BS: 8.7%; RS: 1.7%; R: 0.004%), Chloroflexi (BS: 6.2%; RS: 1.8%; R: 0.003%), and Gemmatimonadetes (BS: 6.9%; RS: 0.7%; R: 0.002%). Different soil–root compartments had distinct relative abundance of taxa, which also varied across the licorice sites (Supplementary Fig. S2). Overall, the bulk soil communities were dominated by Actinobacteria, Acidobacteria, Planctomycetes, Chloroflexi, Gemmatimonadetes, and Deltaproteobacteria. The rhizosphere communities were dominated by Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Firmicutes. In particular, Oxyphotobacteria accounted for the majority (82.6%) of the root endosphere communities (Fig. 2A).
Fig. 2.
(a) Circos plot showing the phylum distribution of licorice root-associated bacterial communities in the three soil–root compartments. Because the relative abundance of Proteobacteria was greater than 25%, bacteria in this phylum are shown at the class level. (b) Boxplots of the alpha-diversity (Shannon index and operational taxonomic unit [OTU] richness) of root-associated bacteria across different sites in the three soil–root compartments. (c) Nonmetric multidimensional scaling (NMDS) of root-associated bacterial community composition across different sites in three soil–root compartments based on Bray-Curtis distances. The three insets represent the three soil–root compartments. (d) Boxplots of root-associated bacterial community similarity among the three soil–root compartments. Different letters indicate significant differences (P < 0.05, Kruskal-wallis test). BS, bulk soil; RS, rhizosphere soil; R, root endosphere
The alpha diversity of the root-associated bacterial communities was significantly influenced by soil–root compartment and licorice age (Supplementary Table S2). The Shannon index and OTU richness decreased with root proximity, while they increased in the rhizosphere soil communities with increasing age of licorice plants. Similarly, both indices of the bulk soil communities increased from the 1-y to 4-y cultivated licorice (this was not observed for wild licorice). For the root endosphere communities, the indices were not markedly different, apart from the OTU richness in the wild site compared with that of the 1-y and 2-y sites, which significantly differed (Fig. 2B).
In the NMDS plot, different soil–root compartments were clearly separated. The distinct sampling sites were well clustered in the bulk and rhizosphere soils. Specifically, rhizosphere soil communities were completely divided into four clusters corresponding to the three ages of the cultivated licorice and the wild licorice (Fig. 2C). The community similarity was ranked as rhizosphere soil < bulk soil < root endosphere (Fig. 2D). The stress values of NMDS analysis ranged from 0.028 to 0.111 in the whole bacterial communities and the three compartments, which reflected the accuracy of the method. The NMDS results were also confirmed by three multiple statistical approaches (Table 3). For example, compartment and age both had significant effects on the composition of whole communities (P < 0.05). When the whole communities were split into three compartments, the age of licorice plants had greater effects (P < 0.05) on the rhizosphere soil communities (Adonis: R2 = 0.601, Anosim: R = 0.842, MRPP: δ = 0.355) than on the bulk soil communities (R2 = 0.485, R = 0.660, δ = 0.379). However, age had no significant effects on the composition of the root endosphere communities (P > 0.05) with respect to community similarity. Taken together, these results revealed distinct temporal successions of the diversity and composition of the root-associated bacterial communities in the three soil–root compartments with increasing age of licorice plants.
Table 3.
Three statistical analyses of the whole root-associated bacterial community composition among different factors contributing to the variation in sequencing data based on Bray-Curtis distances
| Factors | Adonis | Anosim | MRPP | |||
|---|---|---|---|---|---|---|
| R2 | P | R | P | δ | P | |
| Compartment | 0.709 | 0.001 | 0.996 | 0.001 | 0.387 | 0.001 |
| Age | 0.063 | 0.001 | 0.003 | 0.385 | 0.731 | 0.13 |
| Age | ||||||
| BS | 0.485 | 0.001 | 0.660 | 0.001 | 0.379 | 0.001 |
| RS | 0.601 | 0.001 | 0.842 | 0.001 | 0.355 | 0.001 |
| R | 0.277 | 0.12 | 0.098 | 0.125 | 0.127 | 0.103 |
Note: Adonis, permutational multivariate analysis of variance using distance matrices; Anosim, analysis of similarities; MRPP, multiple response permutation procedure. Significance analysis was based on 999 permutation tests. Bold P-values indicate significant differences (P < 0.05). BS, bulk soil; RS, rhizosphere soil; and R, root endosphere
Time–decay relationships of core-enriched taxa
First, the dominant taxa that were highly abundant and ubiquitous in the licorice plants of different ages were screened. The taxa in the top 20.0% of relative abundance (highly abundant taxa) and occurring in more than half of all soil samples (ubiquitous taxa) were collected. This yielded 360 OTUs, accounting for 3.2% of all observed taxa. However, on average, these dominant taxa accounted for 65.3% of the sequences. Additionally, the dominant communities had close relationships (r = 0.98; P < 0.001; Supplementary Fig. S3A) with the remaining communities and exhibited similar distribution patterns with the whole communities (Supplementary Fig. S3B and Table S3).
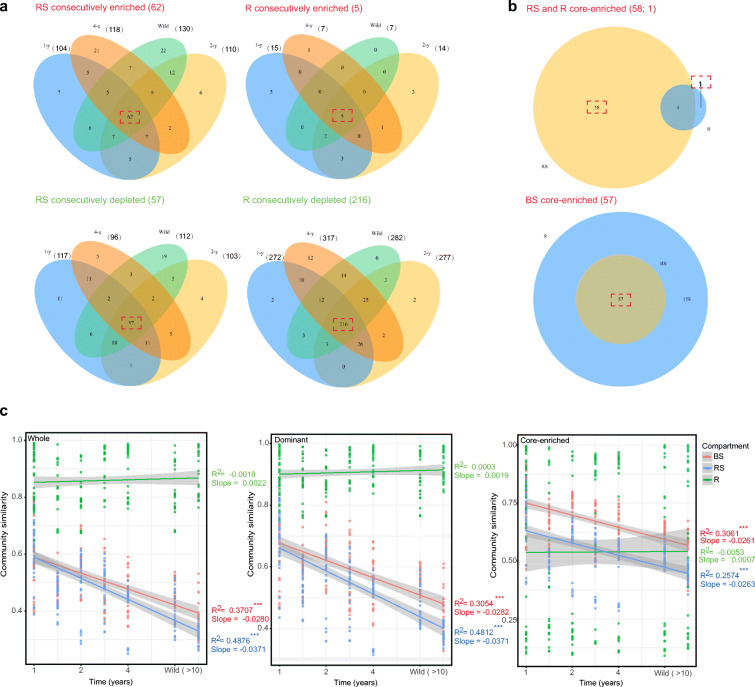
Second, the number of enriched taxa was analyzed in the rhizosphere soil and root endosphere compared with the bulk soil based on the dominant taxa. In total, 104 (1-y), 110 (2-y), 118 (4-y), and 130 (wild) OTUs were enriched in rhizosphere soil across the different licorice sites. The growing number of enriched taxa revealed an increasing trend with temporal succession. On the contrary, the number of enriched OTUs displayed a decreasing trend in the root endosphere, with 15, 14, 7, and 7 OTUs in the 1-y, 2-y, 4-y, and wild sites, separately. Interestingly, the number of root endosphere-enriched OTUs was reduced by half in the 4-y site and then became stable. Despite these differences, 62 OTUs were found to be overlapped and consecutively enriched in the rhizosphere soil across the four ages of licorice. Similarly, there were 5 OTUs enriched consecutively in the root endosphere (Fig. 3A).
Fig. 3.
Venn diagrams of the enriched taxa across different licorice sites in the rhizosphere soil (RS), root endosphere (R), and bulk soil (BS). (a) The rhizosphere soil and root endosphere individually and consecutively enriched and depleted OTUs compared with the bulk soil in the corresponding sites. The bulk soil enriched OTUs are depleted OTUs in the rhizosphere soil and root endosphere. (b) Overlapping between the rhizosphere soil and root endosphere consecutively enriched and depleted OTUs. Red font, enriched OTUs; green font, depleted OTUs. The number of individually, consecutively, and core-enriched discriminating OTUs is indicated in brackets. (c) Time–decay relationships of the whole, dominant, and core-enriched taxa. The rate of community turnover across time (slopes) and the fitting degree (R2) of the line are provided. Significance of the linear fitting model is marked with *** (P < 0.001)
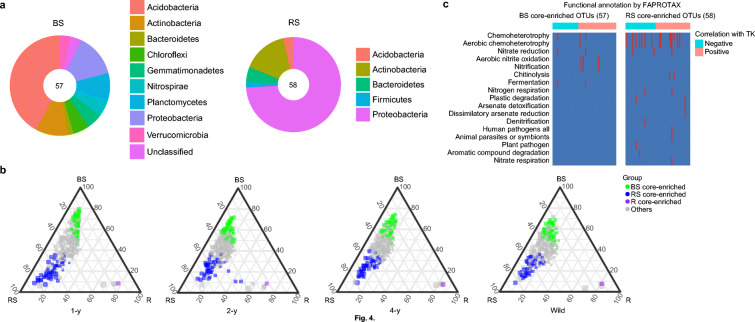
Third, core-enriched OTUs were selected in the rhizosphere soil (58 OTUs) and root endosphere (1 OTUs) from the overlapping of consecutively enriched OTUs between these two compartments. It was found that more OTUs were consecutively enriched in the bulk soil when compared with the root endosphere (216 OTUs) than when compared with the rhizosphere soil (57 OTUs) across the four licorice ages. Thus, the bulk soil had 57 core-enriched OTUs (Fig. 3B). A substantial number of these OTUs in the bulk and rhizosphere soils belonged to the phyla Acidobacteria and Proteobacteria, respectively (Fig. 4A). The core-enriched OTUs also had distinct distributions and relative abundances in the three compartments across the four ages of licorice (Fig. 4B). The results from FAPROTAX showed that these core-enriched OTUs were related to 17 functional pathways (Fig. 4C). In general, most of the OTUs had chemoheterotrophic features. Some core-enriched OTUs of the bulk soil were related to nitrification and fermentation. In contrast, some core-enriched OTUs of the rhizosphere soil were related to denitrification, plant pathogen, and aromatic compound degradation.
Fig. 4.
Distribution and functions of root-associated core-enriched taxa in licorice plants of different ages. (a) Taxonomic composition of the core-enriched OTUs in the bulk and rhizosphere soils at the phylum level. (b) The ternary diagrams of core-enriched OTUs across different licorice sites (1-y, 2-y, 4-y and wild) in the three soil–root compartments. (c) Metabolic and ecological functions of core-enriched OTUs in the bulk and rhizosphere soils based on FAPROTAX. Each column represents an OTU. The presence of functions is shown in red color. BS, bulk soil; RS, rhizosphere soil; and R, root endosphere
Finally, the time–decay relationships of the whole, dominant, and core-enriched communities were elucidated among the three soil–root compartments (Fig. 3C). The whole and dominant bacterial communities had similar time–decay relationships (P < 0.001) in the bulk and rhizosphere soils. The rhizosphere community displayed a stronger rate of decay (slopes = −0.037) than the bulk soil community (slopes = −0.028). However, the core-enriched community of both the bulk and rhizosphere soils displayed decreased turnover slopes at almost the same rate of decay (slopes = −0.026). In the root endosphere, the whole, dominant, and core-enriched communities had no significant time–decay relationships due to the relatively stable community composition.
Effect of soil potassium on individual taxa
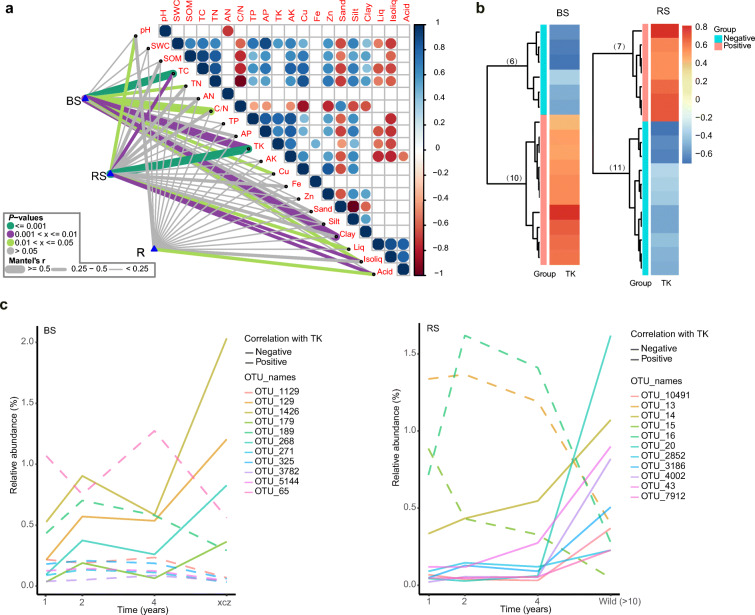
Through visualizing of the Mantel test, it was found that soil TK, TC, TN, C/N, Cu, and clay as well as root isoliquiritin and glycyrrhizin levels, had significant correlations with the core-enriched bacterial communities of the bulk soil. These variables were deemed latent factors affecting these communities. Distinct patterns occurred in the rhizosphere soil and root endosphere (Fig. 5A). Among the latent factors, soil TK had higher correlations with the core-enriched bacterial communities of the bulk and rhizosphere soils than the other factors. Additionally, soil TK consistently displayed a significantly higher explained variance for core-enriched bacterial communities in the two compartments according to the results of MRM (Table 4) and RF (Supplementary Table S4) analyses, which confirmed the results of the Mantel test. Therefore, soil TK could be a key factor regulating the temporal dynamics of core-enriched bacterial communities in the bulk and rhizosphere soils.
Fig. 5.
(a) Correlation analysis among soil characteristics, root secondary metabolites, and root-associated core-enriched bacterial communities (Bray-Curtis distances) in the three soil–root compartments based on the Mantel test. The color of the line represents the significance of the differences (P-values). The size of the line represents correlation coefficients (Mantel’s r). (b) Correlation heatmaps between TK and the core-enriched OTUs in the bulk and rhizosphere soils. The number of significant correlations is indicated in brackets. (c) Temporal variation curves of individual taxa with significant correlations and temporal variability in the bulk and rhizosphere soils. pH, soil pH; SWC, soil water content; SOM, soil organic matter; TC, total carbon; TN, total nitrogen; AN, available nitrogen; C/N, total carbon/nitrogen ratio; TP, total phosphorus; AP, available phosphorus; TK, total potassium; AK, available potassium; Cu, copper; Fe, iron; Zn, zinc; Liq, liquiritin; Isoliq, isoliquiritin; and Acid, glycyrrhizin. BS, bulk soil; RS, rhizosphere soil; and R, root endosphere
Table 4.
Results of the multiple regression on distance matrices (MRM) analysis of the core-enriched bacterial communities (Bray–Curtis distances) in the bulk soil (BS) and rhizosphere soil (RS) for each latent factor
| Compartment | Variables | Coefficients | P-values |
|---|---|---|---|
| BS (R2 = 34.11%, P < 0.001) | TK | 0.096 | 0.005 |
| Clay | 0.059 | 0.195 | |
| Isoliq | 0.033 | 0.773 | |
| Cu | 0.032 | 0.180 | |
| TC | 0.021 | 0.022 | |
| C/N | 0.002 | 0.129 | |
| TN | −0.346 | 0.034 | |
| RS (R2 = 36.13%, P < 0.001) | TK | 0.168 | 0.001 |
| pH | 0.139 | 0.024 | |
| Acid | 0.033 | 0.151 | |
| TC | 0.012 | 0.030 | |
| Liq | 0.008 | 0.821 | |
| Isoliq | −0.144 | 0.420 |
Note: pH, pH; TC, total nitrogen; TN, total nitrogen; C/N, total carbon/nitrogen ratio; TK, total potassium; Cu, copper; Liq, liquiritin; Isoliq, isoliquiritin; and Acid, glycyrrhizin. R2 represents the variation in community dissimilarity that can be explained by the latent factors (significantly correlated variables). The partial regression coefficients and associated P-values of the final model are reported from permutation test (nperm = 999)
Two divergent clusters of individuals from the core-enriched taxa were then characterized: negatively (TK-Neg) and positively (TK-Pos) related to TK in the bulk and rhizosphere soils (Fig. 5B). Their characterization was confirmed by the linear regression model (Supplementary Fig. S4). Based on the ‘maSigPro’ analysis, the taxa that were significantly related to soil TK were further selected along with time series. In particular, 11 individual taxa showed significant temporal variation in the bulk and rhizosphere soils with various annotations (Supplementary Table S5). Next, the overall temporal variation in these taxa and their significant correlations with secondary metabolites were illustrated (Fig. 5C; Supplementary Fig. S5). In detail, all TK-Neg taxa showed consistent variation in bulk soil and displayed positive correlation with some of the secondary metabolites. These taxa all belonged to the phyla Acidobacteria and Actinobacteria. However, some taxa (including OTU 20, 43, and 10,491) showed the same trends with temporal variation in soil TK in the rhizosphere soil regardless of their negative correlations. These taxa were affiliated to the phyla Proteobacteria and Actinobacteria, which were further annotated as Inquilinus, Nocardioides, and Promicromonospora at the genus level, respectively. These taxa were all positively correlated with root glycyrrhizin and isoliquiritin, and their highest relative abundances were found in the wild site, which exhibited the lowest soil TK content. Therefore, all TK-Pos taxa displayed their lowest relative abundances in the wild site.
In addition, the temporal variation of some TK-Pos taxa in the bulk soil (including OTU 189, 325, 5144, and 271) opposed the TK variation. Some of these taxa showed negative correlations with the three secondary metabolites; they belonged to the phyla Acidobacteria and Bacteroidetes (in which they were annotated to the class of Ignavibacteria). This result was also found in the rhizosphere soil. OTU 16 and 13 were negatively correlated with the secondary metabolites. They were annotated to the Xanthomonadaceae family and the Variovorax genus, respectively, both of which belong to the phylum Proteobacteria. Collectively, these individual taxa displayed significant temporal variation and habitat adaptation related to soil TK. Additionally, they may play different roles in adjusting the accumulation of root secondary metabolites in licorice, which provides clues for later isolation of these individual taxa.
Discussion
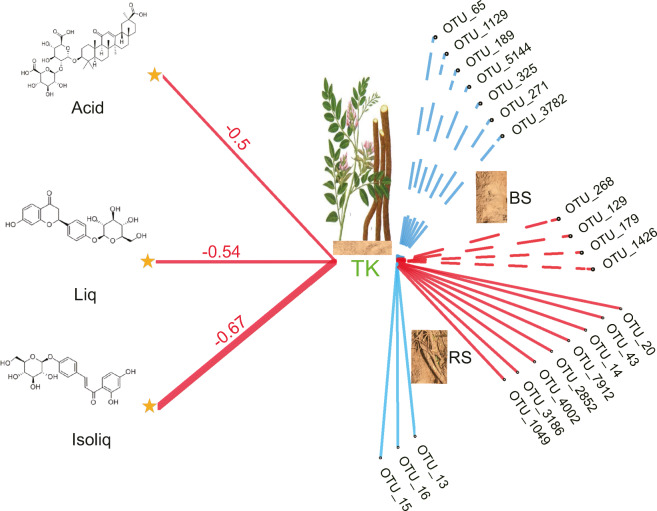
Specific root exudates have been shown to exhibit high selectivity for the soil microbiome in rhizosphere soil (Edwards et al. 2015; Huang et al. 2018). In the current study, the rhizosphere soil communities were clearly discriminated from the bulk soil communities. This was due to the greater effects of licorice age and habitat filtering on the rhizosphere soil. The research focus was narrowed down to a few hundred dominant taxa in the root-associated bacterial communities, and the core-enriched taxa in different soil–root compartments were investigated. Soil TK was defined as a key factor affecting the core-enriched bacterial communities according to multiple but complementary methods. On this basis, we further explored the effects of the key factor on individual taxa associated with the roots of licorice. This approach differed from other studies that have always focused on the community level when investigating the relationships between soil characteristics and microbial communities (Chen et al. 2016; Fan et al. 2018). This analysis facilitated rapid and accurate forecasting of soil characteristics that mediated the links between root secondary metabolites and the individual root-associated taxa (Fig. 6). Consequently, we proposed a focused and novel framework for revealing the regulatory effects of soil characteristics on core-enriched and individual bacterial taxa associated with the synthesis of root secondary metabolites in licorice.
Fig. 6.
Conceptual diagram of total soil potassium (TK) mediating the links between root secondary metabolites in licorice and individual taxa (OTUs) in the bulk soil (BS) and rhizosphere soil (RS). The color of the line represents significantly positive (blue) and negative (red) correlations. The size of line represents correlation coefficients (r). Liq, liquiritin; Isoliq, isoliquiritin; and Acid, glycyrrhizin. Etabolites in licorice
Effects of soil characteristics on root secondary metabolites
Normally, the content of nutrients in the soil decreases during perennial growth due to plant nutrient uptake (Chen et al. 2016; Shakya et al. 2013) and removal in the harvested portion of the crop. In contrast, most of the soil characteristics tested in this study significantly increased as the age of licorice plants increased. For example, the increase in SOM, TN, and TC may be mainly due to the effects of normal irrigation and fertilization every year, but also on account of the accumulation of litters from plant leaf and stem. The SWC displayed an increasing trend in the cultivated sites and was negatively correlated with the concentrations of secondary metabolites across all sites. This may be explained by the fact that licorice is resistant to water deficient environment and widely used for ecological restoration due to its drought-tolerant features (Li et al. 2011; Xie et al. 2018). In addition, there is more accumulation of licorice root secondary metabolites under environmental stress conditions (Xie et al. 2018). The increasing trend in silt and clay contents in the cultivated sites compared with the wild site not only confirmed licorice’s role in soil water conservation and improvement, but identified the impacts of the better field management in the cultivated sites.
The three secondary metabolites had higher values in the wild site, which was not well managed or fertilized. Therefore, licorice plants may be inherently excellent at maintaining higher secondary metabolite concentrations in roots, despite lower levels of SWC and other soil primary nutrients. In the present study, soil TK was negatively related to the glycyrrhizin, liquiritin, and isoliquiritin concentrations in the licorice roots, implying its potential roles in the accumulation of secondary metabolites. Potassium is also involved in many enzyme activation systems in plants, which promotes the stem strength and improves the stress resistance of plants (Wang and Wu 2017). Soil potassium is directly related to the production of phytonutrients in oregano (Origanum vulgare L.) (Jan et al. 2018).
The distributions of soil characteristics and secondary metabolites were divided into four groups corresponding to the composition of bulk soil bacterial communities. This division confirmed that the age of licorice plants had significant effects on some of the soil characteristics and secondary metabolites (as was found from the results of the ANOVA and significance tests). Root secondary metabolites can be massively accumulated in licorice under drought or nutritional stress conditions (Chen et al. 2017; Xie et al. 2018). This is consistent with the results that the root secondary metabolites were all significantly negatively correlated with some soil primary nutrients in our study. Thus, excessive soil nutrients are not necessarily beneficial to the accumulation of root secondary metabolites in licorice. The three secondary metabolites (glycyrrhizin, liquiritin, and isoliquiritin) tested in this study had mutually reinforcing relationships because of their similar metabolic pathways (Xie et al. 2018).
Distribution of different root-associated bacterial communities
The phylum-level composition of the bacterial communities associated with the licorice roots was similar to that described in previous studies, regardless of plant species (Chen et al. 2016; Peiffer et al. 2013). Interestingly, the phylum Oxyphotobacteria accounted for 82.6% of the root endosphere communities in the present study. Oxyphotobacteria is the product of the evolution of oxygenic photosynthesis in the ancestors of Cyanobacteria and it is the most abundant phylum in oceanic picoplankton (Bibby et al. 2001). This result has expanded the distribution range of this phylum and needs to be further clarified. Additionally, the fact that some Cyanobacteria have photosynthetic nitrogen-fixing function is interesting and inner diazotrophic bacteria have been well documented for rice (Zhang et al. 2019), maize (Roesch et al. 2008), sugarcane (Thaweenut et al. 2011) and some gramineous energy plants (Kirchhof et al. 1997), but the function of Oxyphotobacteria in licorice root should be further identified. However, the root nodules on the licorice in different age groups were not enough to investigate the rhizobia communities, resulting from the effects of licorice growing environment and artificial management. Thus, the root nodules and rhizobia were not included in our study. The symbionts were not recovered in the root endophytic population, which might be as consequence of our data processing and rare root nodules during our sampling. However, we would pay further attention to the symbionts in root endophytic communities according to the recent study (Zhang et al. 2019) in the future.
The alpha-diversity of the root-associated bacterial communities significantly decreased with increasing root proximity. This result was consistent with other study that has explained ecological niche selection by plants (Edwards et al. 2015). The Shannon index and OTU richness both increased in the bulk and rhizosphere soils with the increasing age of licorice plants. This trend may be related to the increased soil primary nutrients. In the root endosphere, the Shannon index was stable and the OTU richness displayed little change among the four licorice sites. This confirmed the stability of root endosphere communities.
The composition of the root-associated bacterial communities was well separated according to the three soil–root compartments. Additionally, the rhizosphere soil communities were well divided into four clusters compared with those of the bulk soil due to a greater effect of age. This is because licorice root exudates have greater effects on rhizosphere bacteria than bulk soil bacteria; the higher input of rhizodeposits into the rhizosphere may result in substantial changes in the bacterial community (Shakya et al. 2013; Wagner et al. 2016). Similarly, the composition of the root endosphere communities was stable and no significant differences were observed in their alpha-diversity between the licorice sites. A plausible reason is that these communities, which inhabited the roots of the same licorice species, were less affected by bulk soil characteristics and mainly determined by plant genotype. Similar results have also been obtained in the model plant A. thaliana (Bulgarelli et al. 2012; Hardoim et al. 2015). The genotypic variation amongst licorice plants needs to be further studied to confirm our inference.
Enrichment process and time–decay relationships of core-enriched taxa
Dominant microbial communities play important roles in nutrient cycling in agricultural soils and in microbial colonization of plants (Jiao et al. 2019; Lundberg et al. 2012). Accordingly, the dominant bacterial communities associated with the roots of licorice were identified representatively in this study. Licorice root compartments selectively enriched bacterial communities from the bulk soil and harbored different numbers of enriched taxa across various ages. This enrichment process could be due to the development stages of licorice plants, which was supported by a previous study (Zhang et al. 2018). With increasing licorice age, the rhizosphere soil enriched more taxa, whereas the root endosphere enriched fewer taxa. The amount and chemical composition of root exudates have been found to change during distinct rice growth stages (Zhang et al. 2018). Thus, it is inferred that the licorice plants selected specific communities over time, enriched a variety of taxa in the rhizosphere soil to prevent environmental interference or pathogenic infection. On the contrary, the root endosphere communities were relatively stable. It is possible that more taxa were enriched at first to adapt to the new environment, whereas some unstable taxa were eliminated during voluntary selection by the plants to recover stable original conditions similar to those in wild licorice.
The core-enriched taxa were then defined. This method has been used to select commonly enriched taxa of the same rice species growing in different farmlands (Zhang et al. 2019). However, only one taxon without annotations was obtained from the root endosphere and the root endosphere communities were always stable as mentioned above. Thus, the conserved core-enriched taxa in the bulk and rhizosphere soils were worthy of attention because of precise selections for them. The functional pathways of these core-enriched taxa also had habitat preferences. For example, rhizosphere soil is a relatively anaerobic microenvironment suitable for the metabolism of denitrifying bacteria. Additionally, rhizosphere communities are more affected by plant root exudates that can recruit pathogenic microorganisms (Dombrowski et al. 2017; Huang et al. 2018). These habitat characteristics corresponded to the functions of the taxa present. These functions include chemoheterotrophy, which was found to be the basic lifestyle of most of the bacterial communities in the bulk and rhizosphere soils of the licorice in this study.
Time–decay relationships in microbial communities are consistent among similar environments (Shade et al. 2013). Herein, it was found that the significant turnover slopes of root-associated bacterial communities were much smaller (−0.028 to −0.037) compared with previous studies, which reported the turnover slopes to be between 0 and − 0.3 in air, soil, and freshwater habitats (Jiao et al. 2017). A possible reason for this wide difference is that licorice selected relatively conserved root-associated bacterial communities. There were no decay patterns in root endosphere communities, which was consistent with their stability. The whole and dominant bacterial communities had steeper turnover slopes in the rhizosphere soil, which indicates a faster rate of community temporal turnover (Nekola and White 1999). In addition, this result indicated that the rhizosphere soil bacterial communities were more vulnerable and underwent rapid elimination and replacement of species due to interactions with different root exudates and soil characteristics (Huang et al. 2018; Koberl et al. 2013). However, the turnover slopes of the core-enriched communities both decreased, because these communities were relatively conserved during licorice development.
The links between secondary metabolites and individual taxa mediated by soil potassium
The core-enriched bacterial communities in the three soil–root compartments were affected by different latent factors resulting from niche differentiation. Many studies have reported that the dynamics of the rhizomicrobiome are affected by different soil characteristics, such as soil pH, nitrogen, and water conditions, to varying degrees (Edwards et al. 2015; Xiao et al. 2017; Xie et al. 2018). Similar relationships are elucidated in another study, indicating that soil characteristics can influence the metabolite content in herbs through different assembly of bacterial communities (Huang et al. 2018).
In this study, soil TK was defined as a key factor driving the temporal dynamics of core-enriched bacterial communities in the bulk and rhizosphere soils. Some studies have reported that soil potassium is significantly correlated with rhizosphere bacterial communities in wild oat (Nuccio et al. 2016) and with the relative abundance of Proteobacteria in Japanese barberry (Coats et al. 2014). Some individual taxa were then selected from the core-enriched taxa based on standards of significant correlation with TK and temporal variation in these taxa for better targeting. These individual taxa were sensitive to temporal variation in soil TK and were significantly correlated with the concentrations of different secondary metabolites. This finding demonstrated that soil TK mediates the links between root secondary metabolites and the individual taxa in this study.
Many studies have reported that Acidobacteria is a phylum of oligotrophic bacteria (Chen et al. 2016; Xu et al. 2018). Therefore, it was likely that these taxa that were negatively correlated with TK had a higher relative abundance due to the lower TK content in the wild site. The positively correlated taxa displayed lower relative abundances in the wild site, which was similar to the result of the rhizosphere soil. These taxa may regulate the accumulation of root secondary metabolites in licorice at lower TK levels as indicated by the distinct correlations between their relative abundances and secondary metabolites. Moreover, the class Ignavibacteria, which belongs to the phylum Bacteroidetes, showed opposing temporal variation to TK. Bacteria in this class were negatively related to secondary metabolites in the bulk soil. This is because soil TK is mainly consisted of inorganic potassium, which cannot be used by Ignavibacteria. Ignavibacteria prefers organic matter, including secondary metabolites, available in the bulk soil. This is supported by a study reporting that Ignavibacteria are chemoorganotrophic bacteria (Podosokorskaya et al. 2013).
Proteobacteria and Actinobacteria species are always copiotrophic in bulk soil (Edwards et al. 2015). However, the genus Inquilinus of the phylum Proteobacteria preferred lower TK in the rhizosphere soil in this study. Inquilinus has been isolated from ginseng fields and is able to promote the growth of ginseng (Chun Hwi et al. 2012; Hae-Min et al. 2011). This may also be true for licorice as Inquilinus was positively correlated with the secondary metabolites in the licorice roots. Some studies have found that Nocardioides and Promicromonospora of the Actinobacteria phylum have a growth-promoting effect on ginseng and pine tree roots, respectively (Chun Hwi et al. 2012; Kaewkla and Franco 2017). In addition, the inoculation with the plant growth-promoting rhizobacteria improve the tolerance of licorice to salt stress (Egamberdieva et al., 2016). These taxa may therefore have the potential ability to stimulate the licorice resistance to environmental disturbance and the accumulation of root secondary metabolites in licorice, given their positive correlation with glycyrrhizin and isoliquiritin.
In the rhizosphere soil, the family Xanthomonadaceae and the genus Variovorax displayed opposing temporal variation to soil TK and were negatively correlated with secondary metabolites in the licorice roots. It has been reported that Xanthomonadaceae is enriched during the consecutive monoculture of Rehmannia glutinosa, which is detrimental to plant growth (Wu et al. 2018). Additionally, Variovorax is one of the bacterial genera that displays increased abundance in response to apple replant disease (Lucas et al. 2018). These two taxa may also be harmful to licorice growth and inhibit the production of secondary metabolites despite of the Variovorax paradoxus is commonly described as plant growth promoting rhizobacteria (Kudoyarova et al., 2019). That may be due to the different environmental conditions and plant species. Collectively, these individual taxa exhibited habitat-specific features and were associated with the adjustment of secondary metabolite accumulation in the licorice roots under regulation by TK. The interactions of these taxa are very complex and may not be isolated from each other. Further research is needed to investigate the vital role of TK in the regulation of root secondary metabolites in licorice under controlled conditions.
Conclusions
This study investigated the temporal succession of root-associated bacterial communities and simultaneous variation in soil characteristics and root secondary metabolites in licorice plants of different ages. The core-enriched communities differed among the root-associated bacterial communities, and their time–decay relationships also varied. Soil TK was defined as the key factor regulating individual core-enriched taxa. These individual taxa with distinct habitat adaptation to soil TK played different roles in adjusting the accumulation of root secondary metabolites in licorice. Overall, the results provide valuable information for better understanding the soil potassium-regulated temporal dynamics of root-associated core bacterial communities in licorice. Not only do the results shed light on the core community level, but also on the level of individual taxa. Future research should focus on the isolation of these individual taxa and on confirming their interactions with soil potassium and root secondary metabolites in licorice in controlled experiments. This research should be applied to manage the fertilization and soil bacterial communities efficiently in practical licorice cultivation.
Electronic supplementary material
(DOCX 2201 kb)
(CSV 4 kb)
Acknowledgments
We thank Xiaowu Yan and Ming Li for assistance in soil sampling. We also thank the anonymous reviewers for comments on the manuscript. This work was funded by the National Natural Science Foundation of China (No. 41830755 and 41807030).
Compliance with ethical standards
All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shuo Jiao, Email: shuojiao@nwsuaf.edu.cn.
Gehong Wei, Email: weigehong@nwsuaf.edu.cn.
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Bibby TS, Nield J, Partensky F, Barber J. Antenna ring around photosystem I. Nature. 2001;413:590. doi: 10.1038/35098153. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Brookes PC, Xu J, Zhang J, Zhang C, Zhou X, Luo Y. Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biol Biochem. 2016;98:1–10. [Google Scholar]
- Chen M, Yang G, Sheng Y, Li P, Qiu H, Zhou X, Huang L, Chao Z. Glomus mosseae inoculation improves the root system architecture, photosynthetic efficiency and flavonoids accumulation of liquorice under nutrient stress. Front Plant Sci. 2017;8:931. doi: 10.3389/fpls.2017.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun Hwi C, Jung-Sook L, Dong-Shan A, Tae Woong W, Song-Gun K. Nocardioides panacisoli sp. nov., isolated from the soil of a ginseng field. Int J Syst Evol Microbiol. 2012;60:591–595. doi: 10.1099/ijs.0.012690-0. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Morgenstern B, Bauer G, Chandra PP, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats VC, Pelletreau KN, Rumpho ME. Amplicon pyrosequencing reveals the soil microbial diversity associated with invasive Japanese barberry (Berberis thunbergii DC.) Mol Ecol. 2014;23:1318–1332. doi: 10.1111/mec.12544. [DOI] [PubMed] [Google Scholar]
- Conesa A, Nueda MJ, Ferrer A, Talon M. MaSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- De Mendiburu F. Agricolae: statistical procedures for agricultural research. R package version. 2014;1:1–3. [Google Scholar]
- Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- Dombrowski N, Schlaeppi K, Agler MT, Hacquard S, Kemen E, Garrido-Oter R, Wunder J, Coupland G, Schulze-Lefert P. Root microbiota dynamics of perennial Arabis alpina are dependent on soil residence time but independent of flowering time. ISME J. 2017;11:43–55. doi: 10.1038/ismej.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A. 2015;112:911–920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva D, Li L, Lindström K, Räsänen LA. A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis fish.) under salt stress. Appl Microbiol Biotechnol. 2016;100:2829–2841. doi: 10.1007/s00253-015-7147-3. [DOI] [PubMed] [Google Scholar]
- Fageria NK, Baligar VC, Clark RB. Micronutrients in crop production. Adv Agron. 2002;77:185–268. [Google Scholar]
- Fan K, Weisenhorn P, Gilbert JA, Shi Y, Bai Y, Chu H. Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol Biochem. 2018;121:185–192. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Rubio V. (2017) Book review: ggplot2 – elegant graphics for data analysis (2nd edition). J Statist Soft 77 (Book review 2)
- Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Statist Soft. 2007;22:1–19. [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Hae-Min J, Jung-Sook L, Heon-Meen B, Tae-Hoo Y, Se-Young K, Sung-Taik L, Wan-Taek I. Inquilinus ginsengisoli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol. 2011;61:201. doi: 10.1099/ijs.0.018689-0. [DOI] [PubMed] [Google Scholar]
- Hamilton N. An extension to 'ggplot2', for the creation of ternary diagrams [r package ggtern version 2.2.1] Anesthesiology. 2015;64:72–86. [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Sudo H. Economic importance of licorice. Plant Biotechnol. 2009;26:101–104. [Google Scholar]
- Hosseinzadeh H, Nassiri-Asl M. Pharmacological effects of Glycyrrhiza spp. and its bioactive constituents: update and review. Phytother Res. 2016;29:1868–1886. doi: 10.1002/ptr.5487. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Multcomp: simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Huang W, Long C, Lam E. Roles of plant-associated microbiota in traditional herbal medicine. Trends Plant Sci. 2018;23:559–562. doi: 10.1016/j.tplants.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Jan S, Mir JI, Singh DB, Faktoo SZ, Sharma A, Alyemeni MN, Ahmad P. Effect of environmental variables on phytonutrients of Origanum vulgare L. in the sub-humid region of the northwestern Himalayas. Environ Monit Assess. 2018;190:571. doi: 10.1007/s10661-018-6951-5. [DOI] [PubMed] [Google Scholar]
- Jiao S, Xu Y, Zhang J, Hao X, Lu Y. Core microbiota in agricultural soils and their potential associations with nutrient cycling. mSystems. 2019;4:e00313–e00318. doi: 10.1128/mSystems.00313-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S, Zhang Z, Yang F, Lin Y, Chen W, Wei G. Temporal dynamics of microbial communities in microcosms in response to pollutants. Mol Ecol. 2017;26:923–936. doi: 10.1111/mec.13978. [DOI] [PubMed] [Google Scholar]
- Kaewkla O, Franco CMM. Promicromonospora callitridis sp. nov., an endophytic actinobacterium isolated from the surface-sterilized root of an Australian native pine tree. Int J Syst Evol Microbiol. 2017;67:3559–3563. doi: 10.1099/ijsem.0.002165. [DOI] [PubMed] [Google Scholar]
- Koberl M, Schmidt R, Ramadan EM, Bauer R, Berg G. The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front Microb. 2013;4:400. doi: 10.3389/fmicb.2013.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R (2015) Pheatmap: pretty heatmaps. R package version 061
- Kirchhof G, Reis VM, Baldani JI, Eckert B, Döbereiner J, Hartmann A. Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil. 1997;194:45–55. [Google Scholar]
- Kudoyarova G, Arkhipova T, Korshunova T, Bakaeva M, Loginov O, Dodd IC. Phytohormone mediation of interactions between plants and non-symbiotic growth promoting bacteria under edaphic stresses. Front Plant Sci. 2019;10:1368. doi: 10.3389/fpls.2019.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Statist Soft. 2008;25:1–18. [Google Scholar]
- Lauber CL, Ramirez KS, Aanderud Z, Lennon J, Fierer N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013;7:1641–1650. doi: 10.1038/ismej.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GSB, Mackinder B, Lock M. Legume publications of the royal botanic gardens, Kew, London. Curtiss Bot Mag. 2005;14:245–257. [Google Scholar]
- Li WD, Hou JL, Wang WQ, Tang XM, Liu CL, Xing D. Effect of water deficit on biomass production and accumulation of secondary metabolites in roots of Glycyrrhiza uralensis. Russ J Plant Physiol. 2011;58:538–542. doi: 10.1134/S1021443711030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen X, Liu J, Liu T, Cheng J, Wei G, Lin Y. Temporal and spatial succession and dynamics of soil fungal communities in restored grassland on the loess plateau in China. Land Degrad Dev. 2019;30:1273–1287. [Google Scholar]
- Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- Lucas M, Balbín-Suárez A, Smalla K, Vetterlein D, Martinez-Abarca F. Root growth, function and rhizosphere microbiome analyses show local rather than systemic effects in apple plant response to replant disease soil. PLoS One. 2018;13:e0204922. doi: 10.1371/journal.pone.0204922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:125. [Google Scholar]
- Mehrotra M, Duose DY, Singh RR, Barkoh BA, Manekia J, Harmon MA, Patel KP, Routbort MJ, Medeiros LJ, Wistuba II, Luthra R. Versatile ion S5XL sequencer for targeted next generation sequencing of solid tumors in a clinical laboratory. PLoS One. 2017;12:e0181968. doi: 10.1371/journal.pone.0181968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekola JC, White PS. Special paper: the distance decay of similarity in biogeography and ecology. J Biogeogr. 1999;26:867–878. [Google Scholar]
- Nikolayeva O, Robinson MD. EdgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- Nuccio EE, Anderson-Furgeson J, Estera KY, Pett-Ridge J, De VP, Brodie EL, Firestone MK. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology. 2016;97:1307–1318. doi: 10.1890/15-0882.1. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M. Vegan: community ecology package. R Package version. 2007;2:0–4. [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podosokorskaya OA, Kadnikov VV, Gavrilov SN, Mardanov AV, Merkel AY, Karnachuk OV, Ravin NV, Bonch-Osmolovskaya EA, Kublanov IV. Characterization of Melioribacter roseus gen. Nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ Microbiol. 2013;15:1759–1771. doi: 10.1111/1462-2920.12067. [DOI] [PubMed] [Google Scholar]
- Roesch LFW, Camargo FAO, Bento FM, Triplett EW. Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil. 2008;302:91–104. [Google Scholar]
- Schlaeppi K, Dombrowski N, Oter RG, Ver LTE, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A. 2014;111:585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013;7:1493–1506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J, Muchero W, Bonito G, Vilgalys R, Tuskan G, Podar M, Schadt CW. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS One. 2013;8:e76382. doi: 10.1371/journal.pone.0076382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaweenut N, Hachisuka Y, Ando S, Yanagisawa S, Yoneyama T. Two seasons’ study on nifH gene expression and nitrogen fixation by diazotrophic endophytes in sugarcane (Saccharum spp. hybrids): expression of nifH genes similar to those of rhizobia. Plant Soil. 2011;338:435–449. [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Van AL, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun. 2016;7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu WH. Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol. 2017;39:123–128. doi: 10.1016/j.pbi.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V. Corrplot: visualization of a correlation matrix. MMWR. 2013;52:145–151. [Google Scholar]
- Wu L, Chen J, Xiao Z, Zhu X, Wang J, Wu H, Wu Y, Zhang Z, Lin W. Barcoded pyrosequencing reveals a shift in the bacterial community in the rhizosphere and rhizoplane of Rehmannia glutinosa under consecutive monoculture. Int J Mol Sci. 2018;19:850. doi: 10.3390/ijms19030850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen W, Zong L, Yang J, Jiao S, Lin Y, Wang E, Wei G. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol Ecol. 2017;26:1641–1651. doi: 10.1111/mec.14027. [DOI] [PubMed] [Google Scholar]
- Xie W, et al. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza. 2018;28:1–16. doi: 10.1007/s00572-018-0827-y. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang Y, Zhang P, Trivedi P, Riera N, Wang Y, Liu X, Fan G, Tang J, Coletta-Filho HD, Cubero J, Deng X, Ancona V, Lu Z, Zhong B, Roper MC, Capote N, Catara V, Pietersen G, Vernière C, al-Sadi AM, Li L, Yang F, Xu X, Wang J, Yang H, Jin T, Wang N. The structure and function of the global citrus rhizosphere microbiome. Nat Commun. 2018;9:4894. doi: 10.1038/s41467-018-07343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu YX, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J, Cao S, Wang X, Wang C, Wang H, Qu B, Fan G, Yuan L, Garrido-Oter R, Chu C, Bai Y. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol. 2019;37:676–684. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci China Life Sci. 2018;61:3–11. doi: 10.1007/s11427-018-9284-4. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice) J Chromatogr A. 2009;1216:1954–1969. doi: 10.1016/j.chroma.2008.07.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 2201 kb)
(CSV 4 kb)