Abstract
Thromboembolic disease is a common cardio-cerebral vascular disease that threatens human life and health. Thrombin not only affects the exogenous coagulation pathway, but also the endogenous pathway. Thus, it becomes one of the most important targets of anticoagulant drugs. RGD-hirudin is an anticoagulant drug targeting thrombin, but it can only be administered intravenously. We designed a low molecular weight peptide based on RGD-hirudin that could prevent blood clots. We first used NMR to identify the key amino acid residues of RGD-hirudin that interacted with thrombin. Then, we designed a novel direct thrombin inhibitor peptide (DTIP) based on the structure and function of RGD-hirudin using homology modeling. Molecular docking showed that the targeting and binding of DTIP with thrombin were similar to those of RGD-hirudin, suggesting DTIP interacted directly with thrombin. The active amino acids of DTIP were identified by alanine scanning, and mutants were successfully constructed. In blood clotting time tests in vitro, we found that aPTT, PT, and TT in the rat plasma added with DTIP were greatly prolonged than in that added with the mutants. Subcutaneous injection of DTIP in rats also could significantly prolong the clotting time. Thrombelastography analysis revealed that DTIP significantly delayed blood coagulation. Bio-layer interferometry study showed that there were no significant differences between DTIP and the mutants in thrombin affinity constants, suggesting that it might bind to other sites of thrombin rather than to its active center. Our results demonstrate that DTIP with low molecular weight can prevent thrombosis via subcutaneous injection.
Keywords: RGD-hirudin, thrombin inhibitor, direct thrombin inhibitor peptide (DTIP), homology simulation, molecular docking, thrombelastography, bio-layer interferometry, subcutaneous injection
Introduction
Cardiovascular disease (CVD) continues to be the leading cause of death worldwide [1]. Venous and arterial thromboembolic diseases, such as myocardial infarction, stroke, deep vein thrombosis, and pulmonary embolism, are leading causes of cardiovascular-associated death [2]. Available anticoagulants, such as warfarin, heparin, vitamin K antagonists, dabigatran, edoxaban, rivaroxaban, and apixaban, are effective for the treatment and prophylaxis of thrombotic cardiovascular events [3]. Nonetheless, users of warfarin have a serious risk of bleeding [4], especially intracranial hemorrhage [5]. Unfractionated heparin significantly reduces fatality in patients with venous thromboembolism, which contributes to substantial morbidity and mortality worldwide, but its clinical application is limited by its intravenous administration and its short half-life [6]. Rivaroxaban, apixaban, and edoxaban are inhibitors of Factor X, a central enzyme in the coagulation cascade [6].
Thrombin, a serine protease active within the coagulation cascade, mediates the conversion of fibrinogen to fibrin, which is an important factor in the process of coagulation. Thrombin actives blood coagulation Factors V, VIII, XI, and XIII during coagulation [7, 8], activates platelets by cleaving the protease-activated receptors [9], and binds to thrombomodulin and activating protein C [10] to contribute to clot formation. Thrombin has procoagulant and anticoagulant activities and has three functional domains: (1) the active site, which cleaves the appropriate peptide bonds in its substrates (e.g., fibrinogen, protein C) by the catalytic triad (serine, histidine, and aspartic acid); (2) exosite I, which identifies fibrinogen and other substrates, such as thrombomodulin; and (3) exosite II, which recognizes and binds to glycosaminoglycans, such as heparin and heparan sulfate [9, 11, 12]. Further elucidation of the structure and function of thrombin in blood coagulation could be used to develop new prevention and treatment strategies for embolisms.
Many direct thrombin inhibitors (DTIs), which bind directly to thrombin and block its activity, have been reported and include hirudin and recombinant hirudin [13]. One study suggested that there were two binding sites between the bivalent direct thrombin inhibitor hirudin and thrombin: the C-terminal tail of hirudin with exosite I and the N-terminus with a portion of the active site [13]. Our previous work showed that RGD-hirudin binds to thrombin [14] and effectively inhibits the activity of thrombin [15]. However, RGD-hirudin is a protein drug and is administered via intravenous injection, which limits its clinical application [16]. To develop a novel DTI with a low molecular weight, we proposed the following hypothesis. If a number of amino acids, except the C-terminus tail and N-terminus part of RGD-hirudin, are deleted, the remaining molecule will still possess anticoagulative and antithrombotic properties.
Nuclear magnetic resonance (NMR) is a frequently used method for studying protein structures. In addition, homology modeling is a new, timesaving, simple, and accurate method. The protein data bank (PDB) was established at Brookhaven National Laboratory as a repository for three-dimensional (3D) structural data of proteins and nucleic acids in 1971 [17], and the global protein database (wwPDB) (https://www.wwpdb.org/) currently contains ~144,000 experimentally identified 3D protein structures [18]. Homology modeling, a kind of accurate computational structural prediction method [19], is used to determine the 3D structure of a target protein from its amino acid sequence on the basis of one or more templates [20]. Homology modeling can simulate highly accurate 3D structures [21]. Target-template alignment, model building, and model evaluation are performed (step by step, until satisfactory) during this prediction process [22].
To facilitate future clinical applications, we focused on designing a novel direct thrombin inhibitor peptide (DTIP) based on computer simulations. In addition, the validity and rationality of the peptide inhibitor were verified by comparing its functional amino acid residue mutants.
Materials and methods
Three-dimensional structure construction
Discovery studio (DS, version 3.1) was used for molecular simulations (NeoTrident Technology Ltd., China). The amino acid sequence of RGD-hirudin was obtained from our lab. The sequences of RGD-hirudin and DTIP were input into the DS server. Proteins with similar sequences were found and served as templates. Then, sequence alignments between the target and template proteins were completed. Subsequently, the structure of the target protein was simulated using the tertiary structures of the templates. The three-dimensional structures of RGD-hirudin and DTIP were built based on the structures of templates using homology modeling. Finally, a Ramachandran plot was used to evaluate the results of the models.
Protein–protein docking
Docking of RGD-hirudin and thrombin (PDB ID: 4HTC) was performed with DS. The possible docking sites were presented in the results of ZDOCK, which was accomplished by moving the ligand around the receptor. The greatest number of possible poses were listed based on their E-RDOCK. The docking of DTIP and thrombin was performed as described above.
Alanine scanning
Each amino acid residue of DTIP was systematically changed to an alanine, one residue was changed at a time, and the changes in energy are shown. Residues with the highest mutation energies were designated as the functional amino acid residue site.
Exploration of mutants that have the highest energy change when they interact with thrombin
With DS, the energy change was studied using alanine scanning. Hydrogen bonds between DTIP and thrombin were presented in the simulation software. Hydrogen bonds are not constructed when the functional amino acid residues were converted to an alanine.
Preparation of 15N, 13C-labeled RGD-hirudin
15N, 13C-labeled RGD-hirudin was expressed and purified for NMR experiments as previously reported [23]. HPLC-MS (Waters) was used to assess the purity and labeling efficiency of RGD-hirudin. The homogeneity of the purified protein was further confirmed using a Bruker Autoflex II spectrometer.
Chemical shift perturbation (CSP) measurements
Chemical shift perturbations were measured using 15N-Heteronuclear Single Quantum Coherence (HSQC) spectra recorded on a Varian INOVA 600 NMR spectrometer. NMR samples were prepared in 90% PB buffer and 10% D2O at pH 7.4 with 75 mmol/L NaCl, which contained 1 mmol/L 15N, 13C-labeled protein and 5 mmol/L DTT, while the thrombin concentration varied from 0 to 1.2 mmol/L. Two-dimensional (2D) 15N-HSQC spectra were acquired at 37 °C using 64 t1 increments. NMR spectra data were processed using NMR pipe, a multidimensional spectral processing system based on UNIX pipes, and analyzed using NMRView, a computer program for the visualization and analysis of NMR data, and Sparky 3 (University of California, San Francisco, CA, USA).
For a given 15N-HSQC cross-peak, a proton CSP (CSPH) induced by thrombin binding was defined as the difference between the corresponding chemical shifts in the bound (δbound1H) and free (δfree1H) states: CSPH = δbound1H – δfree1H.
Construction of the plasmid, electroporation, and high-expression yeast screening of mutant variants
The mutant genes were amplified using different forward and reverse primers (Supplementary Table S1), which were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Then, the mutant genes were restructured with a pPIC9K vector, which was digested completely with restriction enzymes, XhoI and NotI. pPIC9K was obtained from our lab, and the enzymes were purchased from New England Biolabs (Beijing) Ltd. Recombinant pPIC9K plasmids were transformed into E. coli DH5α cells, and sequencing was accomplished by Sangon Biotech (Shanghai) Co., Ltd. Then, the plasmid was transformed into P. pastoris GS115 cells through electroporation after it was linearized by SalI. Positive and high-expression clones were screened by using different concentrations of G418 (Invitrogen, USA).
Fermentation and purification of DTIP and mutant variants
The single clone with the highest expression level was selected as the seed to enlarge the cultivation and express the protein. Protein expression was induced by methanol during the fermentation period. Ammonium hydroxide was used to maintain the pH. After ~24 h of induction, the supernatant was collected by centrifugation. The protein concentration increased after the supernatant underwent ultrafiltration. Salt ions were removed using gel filtration chromatography. The proteins of interest were harvested after anion exchange chromatography.
Plasma clotting measurements in vitro
Whole blood was drawn from the abdominal aorta of Sprague Dawley (SD) rats after being anesthetized with chloral hydrate (300 mg/kg). SD rats (350 ± 30 g, male) were purchased from the Animal Center of Fudan University. All animal procedures were carried out in accordance with institutional guidelines at Fudan University. Sodium citrate (3.8%) was used at a ratio of 1:9 for anticoagulation. The plasma was collected followed by centrifugation (900 × g, 15 min). DTIP or alanine scanning mutants were added to an equal volume of plasma. Plasma clotting measurements were conducted according to test kits. In the aPTT test, 50 μL ellagic acid was added to 50 μL plasma samples, followed by 50 μL calcium chloride solution. In the PT test, 50 μL Thromborel S was added to 50 μL plasma samples. In the TT test, 50 μL thrombin time test reagent was added to 50 μL plasma samples. These kits were acquired from Shanghai Long lsland Biotech Co., Ltd. The same sample was tested a total of four times. If the coagulation time was equal to or greater than 250 s, it was recorded as 250 s. Bivalirudin was purchased from Meilunbio (Shanghai).
Clotting time tests with the administration of DTIP or mutants in vivo
Thirty-six SD rats were randomly assigned. Rats (350 ± 30 g, male, n = 6) were anesthetized by chloral hydrate (300 mg/kg, intraperitoneal injection). Carotid cannulation was performed, and blood was taken from the carotid canal at 0, 0.5, 1, 2, and 3 h after subcutaneous injection of DTIP or alanine scanning mutants. The blood was anticoagulated with 3.8% sodium citrate (1:9). aPTT, PT and TT were tested with a Sysmex CA-1500 System.
Thromboelastography (TEG)
Fresh blood from healthy volunteers was anticoagulated with sodium citrate (1:9). Normal saline and proteins (at the same concentrations) were added to the same volume of blood. The whole process of blood clotting was detected by a Thromboelastograph Coagulation Analyzer. All participants gave written informed consent. The collection of volunteers' blood conforms to the Declaration of Helsinki and is approved by the Institutional Review of Board of Fudan University.
Bio-layer interferometry (BLI)
An appropriate volume of biotin was added to DTIP (0.01 mg/mL) and mutant variants (0.01 mg/mL), followed by incubating at room temperature (RT) for 1 h. The biotin-labeled samples must be desalinized using a desalting column before use. Thrombin was diluted to different concentrations ranging from 8.64 to 0.27 μmol/L. Binding was assessed with the ForteBio Octet RED 96 software. Enoxaparin sodium was purchased from Meilunbio (Shanghai).
Results
Interaction between RGD-hirudin and thrombin
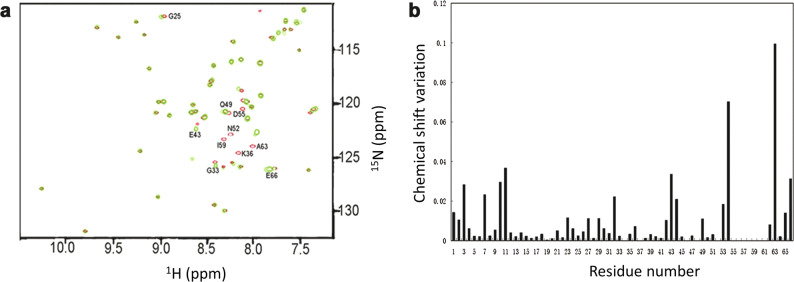
To characterize the interaction between RGD-hirudin and thrombin, we performed 2D 1H-15N HSQC experiments with free and thrombin-bound RGD-hirudin in the presence of hydrogen peroxide (H2O2). Thrombin was titrated to 15N, 13C-labeled RGD-hirudin up to a molar ratio of 1:1.2. Upon binding to thrombin, the amide proton cross-peaks of RGD-hirudin showed a significant change in the chemical shift, indicating that thrombin binds to RGD-hirudin (Fig. 1a).
Fig. 1. The interaction between thrombin and RGD-hirudin.
a Overlay of 1H-15N HSQC spectra of RGDhirudin in the absence (shown in red) and presence of thrombin (shown in green). Thrombin was titrated into RGD-hirudin at a molar ratio of 1.2:1. Both spectra are plotted at the same signal-to-noise level. b Weighted CSPs are plotted versus the amino acid sequence upon titration of RGD-hirudin with thrombin. Most residues with Δδ ≥ 0.01 ppm or missing residues are located on the N-terminus (1–10) and C-terminus (53–66) of RGD-hirudin.
Based on the chemical shift perturbation data of the 2D 1H-15N HSQC spectra, we determined the key amino acids of RGD-hirudin involved in binding to thrombin. The NMR resonance assignment of 1H-15N HSQC cross-peaks of RGD-hirudin showed that the chemical shifts of cross-peaks from the C- and N-termini in RGD-hirudin were largely changed upon binding to thrombin (Fig. 1b). In particular, residues 53–66 from the C-terminus showed a substantial change in their chemical shifts, and cross-peaks from the Asn37, Asn52, Asp55, Phe56, Glu57, Ile59, and Glu61 residues disappeared. In addition, chemical shift perturbations from the N-terminus were also found for Val1, Val2, Tyr3, Thr7, and Gln11 residues. These results indicate that the C- and N-termini of RGD-hirudin are primarily involved in the interaction with thrombin. Therefore, we hypothesized that modification of RGD-hirudin, such that it only contains the C- and N-termini, would still allow RGD-hirudin to interact with thrombin and exert its inhibitory effect.
Three-dimensional structural model of RGD-hirudin
Homology simulations are based on the theory that the conservation of the tertiary structure of a protein is higher than that of the amino acid sequence. Even if the protein primary sequence changes slightly, its tertiary structure changes very little. When there are highly similar first-order amino acid sequences, the three-dimensional structure of an unknown protein (target) can be built based on one or more relevant and known structures (templates). When the similarity of the first-order sequence is higher, the accuracy of the tertiary structure is higher. Homology modeling is considered a valid and strong tool for creating a model [24]. Osmani’s experiments demonstrated that homology modeling offers a high degree of accuracy [25].
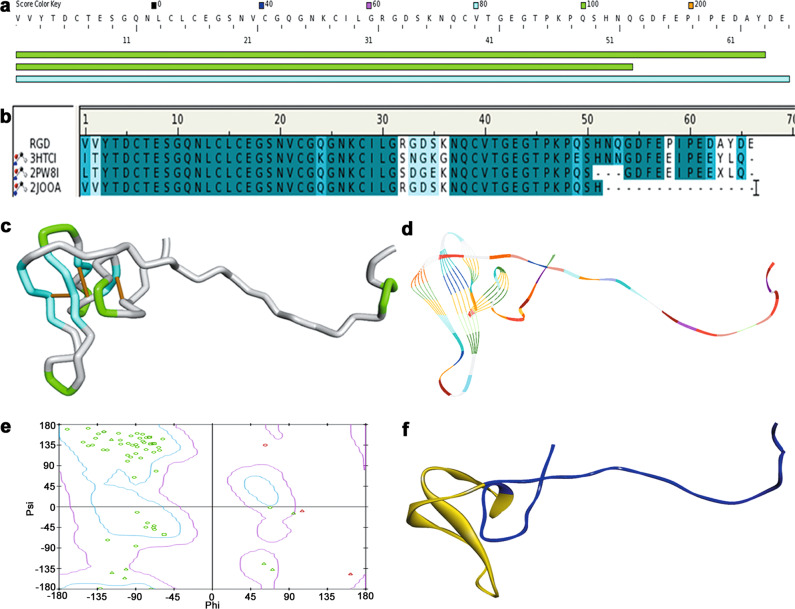
The three-dimensional structure of RGD-hirudin was completed based on homology modeling. Proteins that contain similar amino acid sequences were found when the amino acid sequence of RGD-hirudin was searched in the DS server database; these proteins included 3HTC (PDB ID), recombinant hirudin [26]; 2PW8, sulfo-hirudin [27]; and 2JOO, recombinant RGD-hirudin [28] (Fig. 2a). 3HTCI, 2PW8I, and 2JOOA were designated as templates because of their high consistency. Then, the sequence alignment to the templates was completed successfully (Fig. 2b). The same amino acid residues are shown in dark blue, while similar amino acid residues are shown in light blue (Fig. 2b). Homology models were built based on the templates’ structures. Three disulfide bonds (depicted in yellow) were contained within RGD-hirudin, which are illustrated as a tube (Fig. 2c). In Fig. 2d, the structure of RGD-hirudin is displayed as a line ribbon. Almost all the residues were in the allowed areas, just as the Ramachandran plot manifested (90.2% residues in the allowed region, 7.8% in the marginal region, 2.0% in the disallowed region) (Fig. 2e). The Ramachandran plot is a useful tool for protein structure assessment. Its main application is to detect the quality of the homology modeling. To create a short protein peptide, the fragment from Leu15 to Thr45, which is denoted in yellow, was removed (Fig. 2f), and the remaining peptide was called the direct thrombin inhibitor peptide (DTIP).
Fig. 2. Molecule simulation of RGD-hirudin.
a Similar sequences with RGD-hiudin in the DS server database. b Amino acid sequence alignment of RGD-hirudin with recombinant hirudin (PDB ID: 3HTCI), sulfo-hirudin (PDB ID: 2PW8I), and recombinant RGD-hirudin (PDB ID: 2JOOA). Same amino acid residues are shown in dark blue, while similar amino acid residues are shown in light blue. c Homology modeling of RGD-hiudin in a tube format. d Homology modeling of RGD-hirudin in a line ribbon format. e Ramachandran plot of the RGD-hirudin model. The distribution of the RGD-hirudin residues (green ring) are shown in colors: allowed region (within light blue), marginal region (beyond light blue and within pink) and disallowed region (beyond pink). The percent of residues in allowed the region was 90.2%, in the marginal region 7.8%, in the disallowed region 2.0%. Phi is represented by a peptide alpha carbon left C–N bond rotation angle, and psi is shown as a peptide alpha carbon on the right side of the C–C bond rotation angle. f Homology modeling of RGD-hirudin in a flat ribbon format (blue) and the fragment from Leu15 to Thr45 (yellow).
Three-dimensional structural model of DTIP
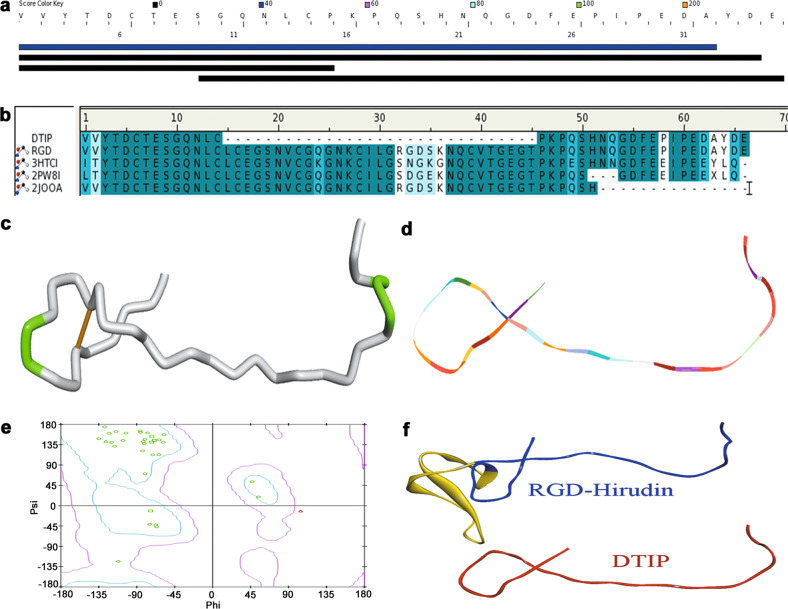
The DTIP structure was simulated after its amino acid sequence had been determined. In the DS server database, similar proteins were found after searching using the DTIP sequence (Fig. 3a). The results of the alignment between DTIP and four templates (RGD-hirudin, recombinant hirudin, sulfo-hirudin, and recombinant RGD-hirudin) are shown in Fig. 3b. In Fig. 3c, a disulfide bond (yellow) was displayed explicitly in a tube format. Homology modeling of DTIP, based on these four templates, can be observed in a line ribbon format (Fig. 3d). The three-dimensional analog structure was evaluated through a Ramachandran plot, which indicated that the model was suitable for structural studies (96.3% residues in the allowed region, 3.7% in the marginal region, and 0.0% in the disallowed region) (Fig. 3e). There was a great similarity of the structures and amino acid sequences between DTIP and RGD-hirudin (Fig. 3f). Therefore, we hypothesized that DTIP might be a direct thrombin inhibitor (DTI).
Fig. 3. Molecule simulation of DTIP.
a Similar sequences with DTIP in DS server database. b Amino acid sequence alignment of DTIP with RGD-hirudin, recombinant hirudin, sulfo-hirudin, and recombinant RGD-hirudin. Same amino acid residues are shown in dark blue, while similar amino acid residues are shown in light blue. c Homology modeling of DTIP in a tube format. d Homology modeling of DTIP in a line ribbon format. e Ramachandran Plot of the DTIP model. The distribution of the DTIP residues (green ring) are shown in colors: allowed region (within light blue), marginal region (beyond light blue and within pink), and disallowed region (beyond pink). The percent of residues in the allowed region was 96.3%, in the marginal region 3.7%, and in the disallowed region 0.0%. f Homology modeling of RGD-hirudin and DTIP in a flat ribbon format (blue and red, respectively) and the fragment from Leu15 to Thr45 (yellow).
Thrombin docked with RGD-hirudin and DTIP
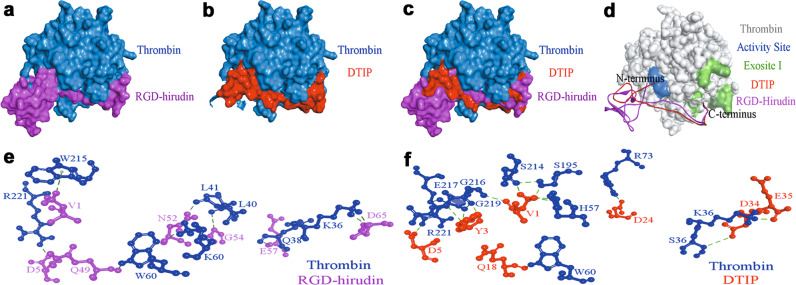
The structure of DTIP is similar to RGD-hirudin. Nevertheless, the interaction of DTIP with thrombin required verification. Molecular docking was performed to explore the binding of DTIP and thrombin. The best docking complex was selected based on the dock score (Supplementary Table S2 and Fig. 4a–c). Docking results (Fig. 4d) revealed that the N-terminus of DTIP or RGD-hirudin bound to the active site of thrombin (in blue) and suggested that DTIP could therefore inhibit thrombin activity. In addition, we identified a binding site between the C-terminus of DTIP or RGD-hirudin and exosite I of thrombin (in atrovirens). To further validate this interaction, intermolecular hydrogen bonds were analyzed. Some hydrogen bonds between RGD-hirudin and thrombin were found; Val1 in the N-terminus of RGD-hirudin with Trp215 of thrombin, Asp5 with Arg221 and Gln49 in the C-terminus of RGD-hirudin with Trp60, Asn52 with Lys60 and Leu41, Gly54 with Leu40, Glu57 with Gln38, and Asp65 with Lys36 (Fig. 4e). Interestingly, many hydrogen bonds were formed between DTIP and thrombin; Val1 in the N-terminus of DTIP with His57 of thrombin, Ser195, Ser214, and Gly216 of thrombin, Tyr3 with Gly216, Gly219, and Glu217, Asp5 with Arg221, Gln18 with Trp60, Asp24 in the N-terminus of DTIP with Arg73 of thrombin, Asp34 with Ser36, and Glu35 with Lys36 (Fig. 4f). The molecular docking results and intermolecular hydrogen-bond interactions led us to hypothesize that DTIP could identify and bind to thrombin.
Fig. 4. Docking of thrombin with RGD-hirudin and DTIP.
a–c Molecular docking between RGD-hirudin (purple) /DTIP (red) and thrombin (blue). d The N-terminus of RGD-hirudin (purple)/DTIP (red) interact with the active site (blue) of thrombin (gray), the C-terminus with exosite I (green). e Intermolecular hydrogen bonds between RGD-hirudin (purple) and thrombin (blue). f Intermolecular hydrogen bonds between DTIP (red) and thrombin (blue).
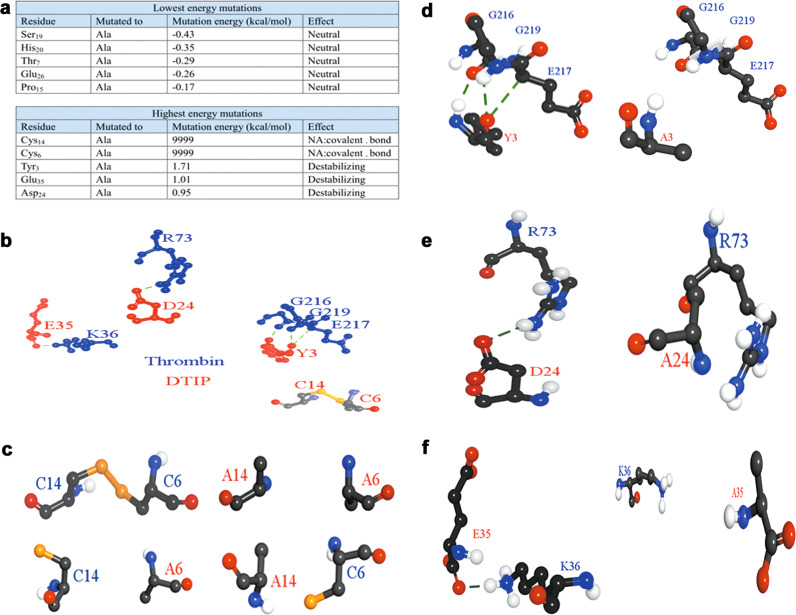
Mutational site screening using alanine scanning
To study the structure and function of DTIP, we selected some amino acid residues to mutate to alanine through alanine scanning in DS (Fig. 5a). Functional amino acid residues were selected according to the change in the energy before and after the mutation. Cys14, Cys6, Tyr3, Glu35, and Asp24 were considered to be functional amino acid residues according their mutation energy. In Fig. 5b, the mutations in which the amino acid residues formed hydrogen bonds with thrombin or disulfide bonds. Once these residues were changed to alanine, hydrogen bonds or disulfide bonds no longer existed (Fig. 5c–f). It is well known that hydrogen and disulfide bonds are important in protein structures. Hence, we suspected DTIP has a different anticoagulant activity from these mutants in which the amino acid residues were mutated to alanine.
Fig. 5. Alanine scanning and the function bonds of five amino acid residues of DTIP.
a The result of alanine scanning. Amino acid residues with lower energy mutations (up); amino acid residues with higher energy mutations (down). b Hydrogen bonds between five amino acid residues of DTIP (red) and thrombin (blue); disulfide bond between Cys6 and Cys14 of DTIP. c–f Comparison of structures before and after mutation.
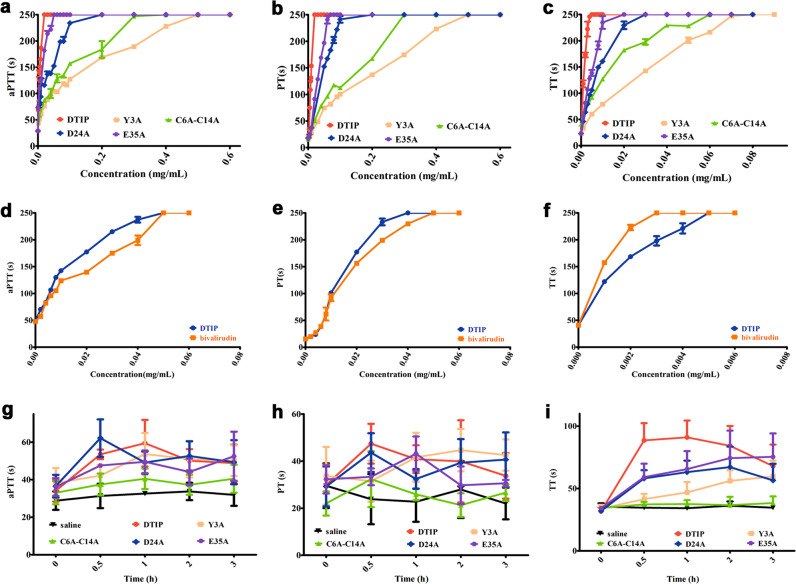
The anticoagulant activity of DTIP was higher than that of mutants in plasma
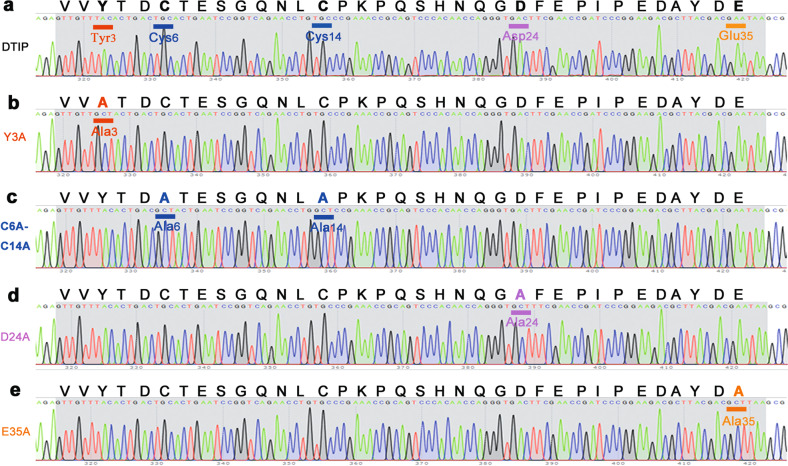
DTIP inhibited thrombin in the molecular simulation results, but experimental verification was still needed. Plasmids carrying DTIP and its mutants were constructed successfully (Fig. 6), and a sufficient protein yield was obtained through the process of fermentation, concentration, and purification. The activated partial thromboplastin time (aPTT), a sensitive and commonly used screening test for endogenous coagulation, was extended to 250 s at a lower concentration of DTIP and a higher concentration of the mutants (Fig. 7a). Prothrombin time (PT) is not only a screening test for the exogenous coagulation system but also an important monitoring index for clinical anticoagulation therapy. The concentration of DTIP at the PT peak (250 s) was also lower than that of the E35A, D24A, C6A-C14A, and Y3A mutants (Fig. 7b). Thrombin time (TT) can be used to reflect the speed of the conversion of fibrinogen to fibrin in the coagulation pathway. Relative to the mutant, the plasma with the lowest concentration of DTIP reached the TT peak (250 s) (Fig. 7c). At the same concentration, the aPTT, PT, and TT of DTIP and those of bivalirudin were on the same order of magnitude (Fig. 7d–f). These experiments showed that at a low concentration, DTIP can delay plasma coagulation through the exogenous, endogenous, or common coagulation system pathway.
Fig. 6. Base sequences of DTIP and the mutants.
a–e Amino acid sequence and base sequence of DTIP and mutants. The mutant bases of Y3A is underlined in red, C6A-C14A in blue, D24A in pink, and E35A in orange.
Fig. 7. DTIP can significantly prolong blood clotting time whether in vitro or vivo.
SD rats were anesthetized with chloral hydrate (300 mg/kg) and blood was taken from the abdominal aorta (3.8% sodium citrate in a ratio of 1 to 9 with blood). Coagulation time was measured with the kits. The time was recorded 250 s if there was no coagulation in the plasma after 250 s. a–c The concentration of DTIP (red) to peak was obviously lower than that of E35A (green), D24A (blue), C6A-C14A (purple), and Y3A (pink) whether testing aPTT, PT, or TT. d–f aPTT, PT, and TT of DTIP (blue) and bivalirudin (orange). g–i After SD rats were anesthetized, carotid cannulation was performed. DTIP and mutant proteins were administered subcutaneously (1 mg/kg). Anticoagulant effects were monitored at 0, 0.5, 1, 2, and 3 h after administration. The result showed that DTIP can prolong blood clotting time, and mutants can inhibit blood coagulation to varying degrees.
DTIP possesses significant anticoagulant activity in vivo
The anticoagulant activity of DTIP was explored in vivo. After the rats were anesthetized, carotid cannulation was performed. Blood was taken from the carotid canal at 0, 0.5, 1, 2, and 3 h after the subcutaneous injection of DTIP and mutants. The results showed that DTIP prolonged the blood clotting time. However, in the other mutant administration groups, the aPTT, PT, and TT were also prolonged, but their inhibition was weaker than that of DTIP (Fig. 7g–i).
TEG was used to study the anticoagulant activity of DTIP in whole blood
We confirmed that DTIP interacted with thrombin in plasma, and we also studied the whole process of blood coagulation. Thromboelastography (TEG) was performed using fresh blood from healthy volunteers. TEG can reflect the dynamic changes of blood coagulation, including fibrin formation rate, dissolved state, robustness, and coagulation elastic dynamics. Experimental results showed that blood with DTIP (0.01 mg/mL) took the longest time to clot (Fig. 8).
Fig. 8. DTIP delayed the clotting in human whole blood.
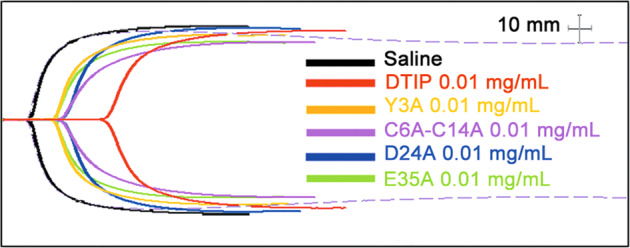
Blood from healthy volunteers was acquired using vacuum blood collection tube. The mutant proteins and DTIP were added to the blood respectively at a final concentration of 0.01 mg/mL. DTIP (red) prolonged the coagulation time of whole blood significantly.
DTIP not only binds to the thrombin activity center but also may bind to exosite I
A study showed that residues Ser195, His57, and Asp189 formed an essential part of the active center of thrombin; residues Lys36, His71, Arg73, Arg75, Tyr76, Arg77a, and Lys109/110 are in exosite I [9]. In the docking results (Table 1), there were some hydrogen bonds between DTIP and thrombin, which included Val1 and His57, Val1 and Ser195, Glu35 and Lys36, and Asp24 and Arg73. Therefore, we determined that DTIP may interact not only with the active center of thrombin but also with exosite I.
Table 1.
Hydrogen bonds between DTIP and thrombin.
| Name | Types | Distance | From | To |
|---|---|---|---|---|
| DTIP: TYR3: HN-H: GLY216: O | Conventional hydrogen bond | 2.01748 | DTIP:TYR3:HN | H: GLY216: O |
| H: GLU217: CA-DTIP: TYR3: O | Carbon hydrogen bond | 3.11836 | H:GLU217:CA | DTIP: TYR3: O |
| H: GLY219: HN-DTIP: TYR3: O | Conventional hydrogen bond | 2.37161 | H:GLY219:HN | DTIP: TYR3: O |
| H: ARG73: HH22-DTIP: ASP24: OD2 | Conventional hydrogen bond | 2.41177 | H:ARG73:HH22 | DTIP: ASP24: OD2 |
| H: LYS36: HZ2-DTIP: GLU35: OCT1 | Conventional hydrogen bond | 2.04514 | H:LYS36:HZ2 | DTIP: GLU35: OCT1 |
| DTIP: VAL1: HT1-H: HIS57: NE2 | Conventional hydrogen bond | 2.73262 | DTIP:VAL1:HT1 | H: HIS57: NE2 |
| DTIP: VAL1: HT2-H: HIS57 | Pi-Donor hydrogen bond | 2.38777 | DTIP:VAL1:HT2 | H: HIS57 |
| DTIP: VAL1: HT1-H: SER195: OG | Conventional hydrogen bond | 2.00852 | DTIP:VAL1:HT1 | H: SER195: OG |
| DTIP: VAL1: HT3-H: SER214: O | Conventional hydrogen bond | 1.72359 | DTIP:VAL1:HT3 | H: SER214: O |
| H: GLY216: HN-DTIP: VAL1: O | Conventional hydrogen bond | 2.55263 | H:GLY216:HN | DTIP: VAL1: O |
| H: ARG221A: HH12-DTIP: ASP5: OD2 | Salt bridge | 2.7241 | H:ARG221A:HH12 | DTIP: ASP5: OD2 |
| H: TRP60D: HE1-DTIP: GLN18: O | Conventional hydrogen bond | 2.77242 | H:TRP60D:HE1 | DTIP: GLN18: O |
| H: SER36A: CB-DTIP: ASP34: OD2 | Carbon hydrogen bond | 3.72795 | H:SER36A:CB | DTIP: ASP34: OD2 |
There are hydrogen bonds between DTIP and the heavy chain of thrombin (H). In this table, we displayed the atoms of amino acid residues in the peptide chain where hydrogen bond was formed. The types, bond length, and orientation of hydrogen bond were also shown
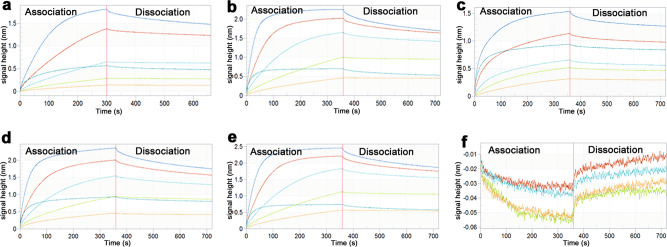
Bio-layer interferometry (BLI), a rapid marker detection technology, is based on the optical interference signal of biological molecular dynamics analysis. This technology was applied in the interaction analysis of biological molecules, such as proteins, nucleic acids, lipids, and sugars. The affinity constant (KD) between biotin-labeled target proteins and thrombin is shown following BLI (Fig. 9 and Table 2). The KD between DTIP and thrombin was 1.64 × 10−7 mol/L (Fig. 9a). Interestingly, there was no distinct diversity among DTIP and the mutant peptides. Enoxaparin sodium, a low molecular-weight heparin (LMWH), prevented the formation of blood clots by binding to antithrombin and potentiating its action [29]. There is no direct interaction between thrombin and enoxaparin sodium (Fig. 9f). The above experiments, such as TEG, demonstrate that the activity of DTIP and the mutants varied. Hence, we speculated that DTIP functions as an anticoagulant on account of the interaction between its N-terminus and the thrombin active site, and the constant KD showed the interaction between the C-terminus and exosite I.
Fig. 9. The affinity constant of DTIP and mutants.
DTIP (0.01 mg/mL) and mutant variants (0.01 mg/mL) were labeled with biotin for about 1 h. Thrombin was diluted to different concentrations from 8.64 μmol/L to 0.27 μmol/L. Binding was proceeded with the software of ForteBio Octet RED 96. Each group was given five concentration gradients. a The binding of thrombin to DTIP, KD = 1.64 × 10−7 M, R2 = 0.9976. b the binding of thrombin to Y3A, KD = 1.62 × 10−7 M, R2 = 0.9936. c The binding of thrombin to C6A-C14A, KD = 1.38 × 10−7 M, R2 = 0.9924. d The binding of thrombin to D24A, KD = 1.94 × 10−7 M, R2 = 0.9957. e The binding of thrombin to E35A, KD = 1.88 × 10−7 M, R2 = 0.9945. f No binding of thrombin with enoxaparin sodium.
Table 2.
Various parameters in the BLI
| Samples |
KD (M) |
KD error |
Kon [1/(M·s)] |
Kon error |
Kdis (1/s) |
Kdis error |
Full X2 |
Full R2 |
|---|---|---|---|---|---|---|---|---|
| DTIP | 1.64 × 10−7 | 2.25 × 10−9 | 2.56 × 103 | 14.3 | 4.18 × 10−4 | 5.25 × 10−6 | 2.5441 | 0.9976 |
| Y3A | 1.62 × 10−7 | 1.86 × 10−9 | 3.76 × 103 | 22.1 | 6.09 × 10−4 | 6.00 × 10−6 | 9.8732 | 0.9936 |
| C6A-C14A | 1.38 × 10−7 | 2.56 × 10−9 | 2.98 × 103 | 20.7 | 4.10 × 10−4 | 7.06 × 10−6 | 4.7585 | 0.9924 |
| D24A | 1.94 × 10−7 | 1.76 × 10−9 | 3.40 × 103 | 16.6 | 6.59 × 10−4 | 5.04 × 10−6 | 6.623 | 0.9957 |
| E35A | 1.88 × 10−7 | 1.90 × 10−9 | 3.38 × 103 | 18.0 | 6.35 × 10−4 | 5.46 × 10−6 | 9.6834 | 0.9945 |
Equilibrium dissociation constant (KD): the degree of affinity
Association rate constant (Kon): rate of product formation per unit time
Dissociation rate constant (Kdis): percentage of product degradation per unit time
Discussion
The number one cause of death globally is cardiovascular disease (CVD) [30]. An estimated 17.3 million people died from CVD in 2008, and almost 23.6 million will die by 2030, mainly due to heart disease and stroke (http://www.who.int/cardiovascular_diseases/about_cvd/en/) [30]. Thrombosis is the primary cause of major cardiovascular diseases, such as ischemic heart disease, ischemic stroke, and venous thromboembolism. There is an urgent need to develop antithrombotic drugs. Thrombin, produced from prothrombin, is a multifunctional serine protease and a key regulator in the hemostatic processes [31]. In the process of coagulation, thrombin contributes to converting soluble fibrinogen into insoluble fibrin and to platelet activation [10]. As a key enzyme in blood coagulation, thrombin has been a target for developing anticoagulant drugs [32]. DTIs can inhibit thrombin directly and do not require antithrombin as a cofactor. Therefore, DTIs have a high-anticoagulant effect. There are many kinds of direct thrombin inhibitors on the market, such as argatroban, dabigatran, dabigatran etexilate, lepirudin, desirudin, and bivalirudin [33]. Argatroban can be applied to coronary artery bypass grafts at an elevated dosage and used for deep vein thrombosis treatment or prophylaxis at a lower dosage [34]. As a direct thrombin inhibitor, dabigatran blocks the conversion of fibrinogen to fibrin [35]. Lepirudin and desirudin are two derivatives of hirudin [33]. There is little doubt they have good anticoagulant effects. However, they have various disadvantages, such as the risk of bleeding. This fact has accelerated the search for new anticoagulant drugs.
Before we defined the sequence of DTIP, we carried out structural studies using NMR spectroscopy, molecular modeling, and site-directed mutagenesis techniques, and demonstrated that the N-terminal (especially for Tyr3) and C-terminal tails of RGD-hirudin bind to exosite I and the active site of thrombin [14, 23]. Next, we preserved the N- (especially for Tyr3) and C-termini of RGD-hirudin and removed the remaining amino acids as much as possible, and then we tested a variety of different truncated peptides.
In the NMR experiments, we found that the C- and N-termini of RGD-hirudin played important roles in the interaction with thrombin. Therefore, a novel peptide (DTIP) was designed with these two termini. To investigate whether DTIP can interact with thrombin, molecular simulations and docking experiments were performed. Homology modeling is a method to simulate the three-dimensional structure of a target protein based on known structures. We simulated the structures of RGD-hirudin and DTIP using known structures from the PDB database; these structures included recombinant hirudin (PDB ID: 3HTC), sulfo-hirudin (PDB ID: 2PW8), and recombinant RGD-hirudin (PDB ID: 2JOO). There were great similarities between their amino acid sequences; theoretically, their high-level structures may be similar to a large extent. Molecular docking results showed that DTIP bound to thrombin in a manner similar to that of RGD-hirudin. In addition, hydrogen bonds formed between DTIP and thrombin, which led us to believe it can inhibit thrombin.
In addition to the homology simulations, a series of experiments were performed to verify the function of DTIP. First, we examined the effects of DTIP on plasma coagulation. Whether examining the aPTT, PT, or TT of plasma, the coagulation was longer with DTIP than with any of the mutants. The DTIP concentration was the lowest when the coagulation time reached its peak (250 s). In addition, DTIP achieved a clotting time similar to that of bivalirudin. These data indicate that DTIP may prevent thrombosis in the coagulation system. Next, the anticoagulant effect on human blood was confirmed. The same effect on whole blood was shown in the TEG experiment. Whole blood with 0.01 mg/mL DTIP had the longest anticoagulant time. In BLI, DTIP had the same affinity constant as the mutants, which reminded us that there was an interaction between the DTIP C-terminus and exosite I of thrombin. RGD-hirudin (0.14 μmol/L) could inhibit ADP-induced platelet aggregation, and this effect on platelet aggregation had a dose-response [23, 36]. However, ADP-induced platelet aggregation was not affected when the final concentration of 5 μmol/L DTIP was added (data not shown).
Certainly, there are several limitations in our study. The pharmacokinetics of DTIP remain to be explored. In future studies, we will focus on solving these problems.
Conclusion
Our results warrant consideration of DTIP as a useful medicine for anticoagulation. We are hopeful that our research methods and results can provide a theoretical basis for the design and development of new anticoagulant drugs.
Supplementary information
Acknowledgements
This study was supported by the National Natural Science Foundation of China (NSFC 81673498) and the Science and Technology Commission of Shanghai Municipality (STCSM 16431904600).
Author contributions
YRL, MFW, and YLZ performed all of the experiments; BZ, YNH, MFW, TYL, and DC participated in the research; YRL and WM designed experiments, analyzed data, and wrote the paper; MY is the supervisor of YRL. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41401-019-0347-0) contains supplementary material, which is available to authorized users.
References
- 1.Timmis A, Townsend N, Gale C, Grobbee R, Maniadakis N, Flather M, et al. European society of cardiology: cardiovascular disease statistics 2017. Eur Heart J. 2018;39:508–79. doi: 10.1093/eurheartj/ehx628. [DOI] [PubMed] [Google Scholar]
- 2.Mackman N. Triggers, targets and treatments for thrombosis. Nature. 2008;451:914–8. doi: 10.1038/nature06797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franchini M, Mengoli C, Cruciani M, Bonfanti C, Mannucci PM. Effects on bleeding complications of pharmacogenetic testing for initial dosing of vitamin K antagonists: a systematic review and meta-analysis. J Thromb Haemost. 2014;12:1480–7. doi: 10.1111/jth.12647. [DOI] [PubMed] [Google Scholar]
- 4.Lane MA, Zeringue A, McDonald JR. Serious bleeding events due to warfarin and antibiotic co-prescription in a cohort of veterans. Am J Med. 2014;127:657–63. doi: 10.1016/j.amjmed.2014.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen AY, Yao JF, Brar SS, Jorgensen MB, Chen W. Racial/ethnic differences in the risk of intracranial hemorrhage among patients with atrial fibrillation. J Am Coll Cardiol. 2007;50:309–15. doi: 10.1016/j.jacc.2007.01.098. [DOI] [PubMed] [Google Scholar]
- 6.Vanassche T, Vandenbriele C, Peerlinck K, Verhamme P. Pharmacotherapy with oral Xa inhibitors for venous thromboembolism. Expert Opin Pharmacother. 2015;16:645–58. doi: 10.1517/14656566.2015.999043. [DOI] [PubMed] [Google Scholar]
- 7.Krenzlin H, Lorenz V, Danckwardt S, Kempski O, Alessandri B. The importance of thrombin in cerebral injury and disease. Int J Mol Sci. 2016;17:84. doi: 10.3390/ijms17010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strukova SM. Role of platelets and serine proteinases in coupling of blood coagulation and inflammation. Biochem (Mosc) 2004;69:1067–81. doi: 10.1023/B:BIRY.0000046880.91848.01. [DOI] [PubMed] [Google Scholar]
- 9.Crawley JT, Zanardelli S, Chion CK, Lane DA. The central role of thrombin in hemostasis. J Thromb Haemost. 2007;5(Suppl 1):95–101. doi: 10.1111/j.1538-7836.2007.02500.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee CJ, Ansell JE. Direct thrombin inhibitors. Br J Clin Pharm. 2011;72:581–92. doi: 10.1111/j.1365-2125.2011.03916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005;3:1861–72. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 12.Lechtenberg BC, Freund SM, Huntington JA. GpIbα interacts exclusively with exosite II of thrombin. J Mol Biol. 2014;426:881–93. doi: 10.1016/j.jmb.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppens M, Eikelboom JW, Gustafsson D, Weitz JI, Hirsh J. Translational success stories: development of direct thrombin inhibitors. Circ Res. 2012;111:920–9. doi: 10.1161/CIRCRESAHA.112.264903. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Zhang Y, Zhao B, Xu Q, Zhou X, Song H, et al. Structural basis of RGD-hirudin binding to thrombin: Tyr3 and five C-terminal residues are crucial for inhibiting thrombin activity. BMC Struct Biol. 2014;14:26. doi: 10.1186/s12900-014-0026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu WF, Mo W, Liu Z, Fu WG, Guo DQ, Wang YQ, et al. The antithrombotic effect of a novel hirudin derivative after reconstruction of carotid artery in rabbits. Thromb Res. 2010;126:e339–43. doi: 10.1016/j.thromres.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Zhang Y, Huang Y, Yu J, Li Y, Wang Q, et al. A novel hirudin derivative inhibiting thrombin without bleeding for subcutaneous injection. Thromb Haemost. 2017;117:44–56. doi: 10.1160/TH16-05-0416. [DOI] [PubMed] [Google Scholar]
- 17.NIGMS. 50 Years of Protein Structure Determination Timeline-HTML Version-National Institute of General Medical Sciences. ed.: https://publications.nigms.nih.gov/psi/timeline_text.html 2011.
- 18.Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 19.Cavasotto CN, Phatak SS. Homology modeling in drug discovery: current trends and applications. Drug Disco Today. 2009;14:676–83. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Muhammed MT, Aki-Yalcin E. Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem Biol Drug Des. 2019;93(Jan):12–20. doi: 10.1111/cbdd.13388. [DOI] [PubMed] [Google Scholar]
- 21.Werner T, Morris MB, Dastmalchi S, Church WB. Structural modelling and dynamics of proteins for insights into drug interactions. Adv Drug Deliv Rev. 2012;64:323–43. doi: 10.1016/j.addr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Zhang Y, Wu Y, Wang J, Liu X, Dai L, et al. Expression, purification, and mass spectrometric analysis of 15N, 13C-labeled RGD-hirudin, expressed in Pichia pastoris, for NMR studies. PLoS ONE. 2012;7:e42207. doi: 10.1371/journal.pone.0042207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuwahara K, Hikosaka M, Kaneko T, Takamatsu A, Nakajima Y, Ogawa R, et al. Analysis of cranial morphology of healthy infants using homologous modeling. J Craniofac Surg. 2019;30:33–8. doi: 10.1097/SCS.0000000000004893. [DOI] [PubMed] [Google Scholar]
- 25.Osmani SA, Bak S, Moller BL. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry. 2009;70:325–47. doi: 10.1016/j.phytochem.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Rydel TJ, Ravichandran KG, Tulinsky A, Bode W, Huber R, Roitsch C, et al. The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 1990;249:277–80. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- 27.Liu CC, Brustad E, Liu W, Schultz PG. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J Am Chem Soc. 2007;129:10648–9. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Mo W, Liu X, Zhu L, Yan X, Song H, et al. The NMR solution structure of recombinant RGD-hirudin. Biochem. Biophys. Res. Commun. 2007;360:103–8. doi: 10.1016/j.bbrc.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Carter NJ, McCormack PL, Plosker GL. Enoxaparin: a review of its use in ST-segment elevation myocardial infarction. Drugs. 2008;68:691–710. doi: 10.2165/00003495-200868050-00012. [DOI] [PubMed] [Google Scholar]
- 30.WHO. About cardiovascular diseases. ed.: http://www.who.int/cardiovascular_diseases/about_cvd/en/.
- 31.Posma JJ, Posthuma JJ, Spronk HM. Coagulation and non-coagulation effects of thrombin. J Thromb Haemost. 2016;14:1908–16. doi: 10.1111/jth.13441. [DOI] [PubMed] [Google Scholar]
- 32.Zavyalova E, Ustinov N, Golovin A, Pavlova G, Kopylov A. G-Quadruplex aptamers to human thrombin versus other direct thrombin inhibitors: the focus on mechanism of action and drug efficiency as anticoagulants. Curr Med Chem. 2016;23:2230–44. doi: 10.2174/0929867323666160517120126. [DOI] [PubMed] [Google Scholar]
- 33.Kong Y, Chen H, Wang YQ, Meng L, Wei JF. Direct thrombin inhibitors: patents 2002-2012 (Review) Mol Med Rep. 2014;9:1506–14. doi: 10.3892/mmr.2014.2025. [DOI] [PubMed] [Google Scholar]
- 34.Fareed J, Jeske WP. Small-molecule direct antithrombins: argatroban. Best Pr. Res. Clin. Haematol. 2004;17:127–38. doi: 10.1016/j.beha.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Fugate JE, Rabinstein AA, McBane RD, Lanzino G. Dabigatran: a primer for neurosurgeons. World Neurosurg. 2013;79:154–8. doi: 10.1016/j.wneu.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Mo W, Zhang YL, Chen HS, Wang LS, Song HY. A novel hirudin derivative characterized with anti-platelet aggregations and thrombin inhibition. J Thromb Thrombolysis. 2009;28:230–7. doi: 10.1007/s11239-008-0251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.